Abstract
Hepatitis C virus (HCV) infection is often associated with hepatic steatosis and yet the molecular mechanisms of HCV-associated steatosis are poorly understood. Because sterol regulatory element-binding proteins (SREBPs) are the major transcriptional factors in lipogenic gene expression including fatty acid synthase (FAS), we examined the effects of HCV nonstructural proteins on the signaling pathways of SREBP. In this study, we demonstrated that HCV nonstructural 4B (NS4B) protein increased the transcriptional activities of SREBPs. We also showed that HCV NS4B enhanced the protein expression levels of SREBPs and FAS. This was further confirmed in the context of viral RNA replication and HCV infection. The up-regulation of both SREBP and FAS by NS4B protein required phosphatidylinositol 3-kinase activity. We also demonstrated that NS4B protein induced a lipid accumulation in hepatoma cells. In addition, NS4B protein synergistically elevated the transcriptional activity of HCV core-mediated SREBP-1. These results strongly suggest that NS4B may play an important role in HCV-associated liver pathogenesis by modulating the SREBP signaling pathway.
Hepatitis C virus (HCV)2 infection has a major impact on public health, affecting more than 170 million people worldwide (1). HCV infection often leads to chronic hepatitis, liver cirrhosis, and ultimately hepatocellular carcinoma (2). Although HCV infection is strongly associated with hepatic steatosis (3), the molecular events that lead to hepatic steatosis during HCV infection are poorly understood. HCV is an enveloped, positive-strand RNA virus classified in the Hepacivirus genus within the Flaviviridae family. HCV is highly heterogeneous and has been classified into six major genotypes and numerous subtypes (4). HCV genome encodes a polyprotein of more than 3,010 amino acids that is cleaved at the endoplasmic reticulum (ER) by host and viral proteases, yielding 3 structural (core, E1, and E2) and 7 nonstructural (p7, NS2 to NS5B) proteins (5). NS4B protein is released from the polyprotein by the NS3/4A serine protease (6). NS4B is a hydrophobic 27-kDa protein located in the ER membrane (7), and has four transmembrane domains with the N and C termini located in the cytoplasm. The N-terminal tail of NS4B has been suggested to be posttranslationally translocated to the ER lumen (8). NS4B protein is known to induce intracellular membrane changes that called a “membranous web” (9). Elazar et al. (10) suggest that a putative amphipathic helix within the N-terminal 26 residues of NS4B mediates membrane association, and these residues are critical for HCV replication in cell culture. Furthermore, NS4B can transform NIH-3T3 cells either in cooperation with Ras (11) or independent of Ras (12).
Hepatic steatosis is defined as an increased fat content in the liver, essentially accounted for by triglycerides (13). The prevalence of steatosis ranges from 40 to 86% in chronic hepatitis C patients (14). Interestingly, steatosis is more frequent in patients infected with HCV genotype 3 than in patients infected with other HCV genotypes, even though not all patients infected with genotype 3 have steatosis (15, 16). It has previously been reported that hepatic steatosis was induced by the HCV core protein through inhibition of the microsomal triglyceride transfer protein activity and very low density lipoprotein secretion (17), impairment of the expression and transcriptional activity of peroxisome proliferators-activated receptor (PPAR) α (18), and activation of the SREBP1 and PPARγ (19). Sterol regulatory element-binding proteins (SREBPs) are ER membrane-bound transcription factors, which activate genes encoding the enzymes that regulate the synthesis of cholesterol and fatty acids, and cellular uptake of lipoproteins (20).
In mammals, there are three SREBP isoforms, designated SREBP-1a, SREBP-1c, and SREBP-2 (20). SREBPs are synthesized as precursors bound to the ER membrane (21). Upon activation by SREBP cleavage-activating protein (SCAP), SREBPs are released from the membrane into the nucleus as mature proteins by sequential cleavage processes (21). The mature SREBP-1a activates target genes involved in both fatty acid and cholesterol biosynthesis, whereas SREBP-1c and SREBP-2 activate target genes involved in fatty acid and cholesterol biosynthesis, respectively (21).
Because both SREBP-1 and SREBP-2 are important transcriptional factors involved in lipid biosynthesis, we investigated the possible involvement of nonstructural proteins of HCV in hepatic lipid accumulation. In this study, we demonstrated that HCV NS4B protein promoted the transcriptional and translational activities of SREBPs and fatty acid synthase (FAS), and this was mediated through the AKT pathway. To our knowledge, this is the first report that NS4B mediates lipogenesis and hence this may provide a novel mechanism of hepatic steatosis associated with HCV infection.
EXPERIMENTAL PROCEDURES
Plasmids—NS4B-Myc of HCV (genotype 1b) was amplified by PCR using the Korean isolate of HCV (22) as a template and subcloned into the pEF6B/His-Myc (Invitrogen) vector. NS3-Myc, NS4B-Myc (genotype 2a, JFH-1), and NS5A-Myc expression plasmids were subcloned into the pEF6B/His-Myc vector. NS5B-Myc expression plasmid was described previously (23). FLAG-core expression plasmid was subcloned into the pFlag-CMV-2 vector. pcDNA3.1-Flag-SREBP-1a (human, amino acids 1–490), pGL2B-FAS-luc (fatty acid synthase promoter), and pSynSRE-luc (hamster HMG-CoA synthase promoter) vectors were kindly provided by Dr. T. R. Osborne (24–26). pcDNA3-HA-SREBP-1c (human, amino acids 1–447) was described previously (27). pBP1c550-Luc, the luciferase gene containing 550-bp fragments of the mouse SREBP-1c promoter, was a gift from Dr. H. Shimano (28).
Cell Culture, Transfection, and HCV Infection—Huh7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin. For transfection, ∼5 × 105 cells plated on 60-mm dishes were transfected with plasmid DNA using either Lipofectamine (Invitrogen) or polyethyleneimine reagent (Sigma) according to instructions from the manufacturer. Huh7 cells containing HCV subgenomic replicons were kindly provided by Dr. C. Seeger (Fox Chase Cancer Center, Philadelphia, PA) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin/streptomycin, 0.1 mm nonessential amino acids (Invitrogen), and 500 μg/ml G418 (Qbiogene, Inc., Irvine, CA). To establish interferon (IFN)-cured cells, HCV replicon cells were treated with 100 units/ml IFN-α (Sigma) for 2 weeks. Elimination of HCV replicon RNA was confirmed by reverse transcription-PCR, Western blotting, and loss of resistance to G418. The infectious HCVs were generated as described previously (29).
Establishment of Stable Cells Expressing NS4B-Myc Protein—To make the cell lines stably expressing NS4B-Myc, Huh7 cells were transfected with the pEF6B-NS4B (genotype 1b) expression plasmid and cultured for 4 weeks in the presence of 10 μg/ml blasticidin. Single positive clones were selected by Western blot analysis using anti-Myc monoclonal antibody. Huh7 cells transfected with empty vector (pEF6B only) were selected as described above and used as a control.
Reporter Assays—Huh7 cells were seeded in a 12-well culture plate and transfected with 0.2 μg of reporter plasmid (FAS-Luc, SRE-Luc, 550-Luc, individually) and 0.1 μg of pCH110 reference plasmid (Amersham Biosciences) containing the Escherichia coli lacZ gene under control of the simian virus 40 promoter. The total DNA amount in each transfection was kept constant by adjustment with empty vector. At 36 h after transfection, cells were harvested and luciferase activities were determined by measuring luminescence activity. Data were normalized by measuring β-galactosidase activity. Luciferase and β-galactosidase assays were performed as described previously (30).
Immunoblot Analysis—Cells were lysed in cell lysis buffer containing 50 mmol/liter Tris-HCl (pH 7.5), 150 mmol/liter NaCl, 1 mmol/liter EDTA, 1% Nonidet P-40, 10% glycerol, and protease inhibitor mixture (Roche) for 20 min on ice. The protein concentration was determined by the Bradford assay (Bio-Rad). Equal amounts of proteins were subjected to 10% SDS-PAGE and electrotransferred to a nitrocellulose membrane. The membrane was blocked in phosphate-buffered saline (PBS) containing 5% nonfat dry milk for 1 h and then incubated 2 h at room temperature with one of following antibodies: anti-FAS antibody (BD Transduction Laboratories), anti-SREBP-1 antibody (BD Transduction Laboratories, Santa Cruz Biotechnology, Inc.), anti-SREBP-2 antibody (BD Transduction Laboratories), anti-AKT and p-AKT antibody (Cell signaling Technology, Beverly, MA), anti-β-actin and anti-FLAG antibody (Sigma), anti-Myc and anti-HA antibody (Santa Cruz Biotechnology, Inc.), anti-NS4B antibody (Virostat), and rabbit anti-NS5A polyclonal antibody in Tris-buffered saline/Tween (20 mmol/liter Tris-HCl (pH 7.5), 500 mmol/liter NaCl, and 0.05% Tween 20). Following two washes in Tris-buffered saline/Tween, the membrane was incubated with either horseradish peroxidase-conjugated goat anti-rabbit antibody or goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in Tris-buffered saline/Tween for 1 h at room temperature. Proteins were detected using an ECL kit (Amersham Biosciences).
Confocal Microscopy—Huh7 cells grown on coverglass (Superior, 18 × 18 mm) were transfected with the NS4B-myc expression plasmid. At 36 h after transfection, cells were washed in PBS and fixed in 4% paraformaldehyde and 0.1% Triton X-100 for 20 min at 37 °C. Cells were incubated in 5% bovine serum albumin for 20 min at 37 °C and then incubated with anti-Myc (Santa Cruz) monoclonal antibody for 2 h at 37 °C. After being washed three times in PBS, cells were further incubated with TRITC-conjugated goat anti-mouse IgG (American Qualex, San Clemente, CA) and BODIPY 493/503 (1 μm, Invitrogen) for 1 h at 37 °C. After two washes with 0.1% Triton X-100 in PBS and three washes in PBS, cells were analyzed using the LSM 510 laser confocal microscopy system and BODIPY intensity was measured by imaging analysis (Carl Zeiss, Inc., Thornwood, NY).
RNA Isolation and Reverse Transcription-PCR—Total RNAs were isolated from vector, NS4B-myc stable, IFN-cured, and HCV subgenomic-replicon cells using TRIzol reagent (Invitrogen). The cDNAs were synthesized by avian myeloblastosis virus (AMV) reverse transcriptase (Promega) from 1 μg of total RNAs using oligo(dT) primer or poly(dA) primers (for NS4B amplification in replicon cells). To estimate transcriptional levels of SREBP-1 and its target genes, a one-tenth aliquot of cDNA was subjected to PCR amplification using SREBP-1, FAS, SCD, ACC, NS4B, and GAPDH primers: SREBP-1-f, 5-ACGGCAGCCCCTGTAACGACCACTGTGA-3 and SREBP-1-r, 5-TGCCAAGATGGTTCCGCCACTCACCAGG-3; FAS-f, 5-GAAACTGCAGGAGCTGTC-3 and FAS-r, 5-CACGGAGTTGAGGCGGAT-3; SCD-f, 5-CCTCTACTTGGAAGACGACATTCGC-3 and SCD-r, 5-GCAGCCGAGCTTTGTAAGAGCGGT-3; ACC-f, 5-GCTGCTCGGATCACTAGTGAA-3 and ACC-r, 5-TTCTGCTATCAGTCTGTCCAG-3; NS4B-f, 5-cgcggatccatgGCCTCACAACTTCCT-3 and NS4B-r, 5-ggcgaattccaTCCGCTGATGAAATT-3; GAPDH-f, 5-GCTCTCCAGAACATCATCCCTGCC-3 and GAPDH-r, 5-CGTTGTCATACCAGGAAATGAGCTT-3. Italic letters represent irrelevant sequences and restriction sites (BamHI and EcoRI) for cloning. Amplified DNA was analyzed by agarose gel electrophoresis.
Statistical Analysis—The data are presented as mean ± S.D. The Student's t test was used for statistical analysis. p < 0.05 was considered statistically significant.
RESULTS
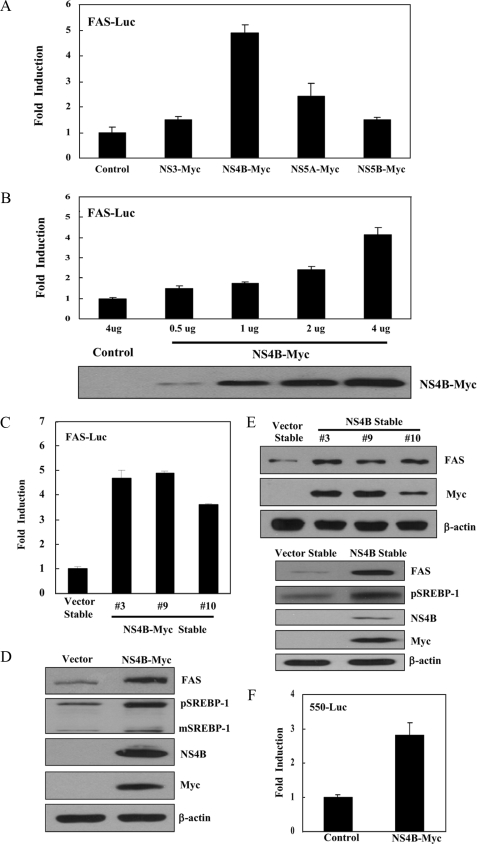
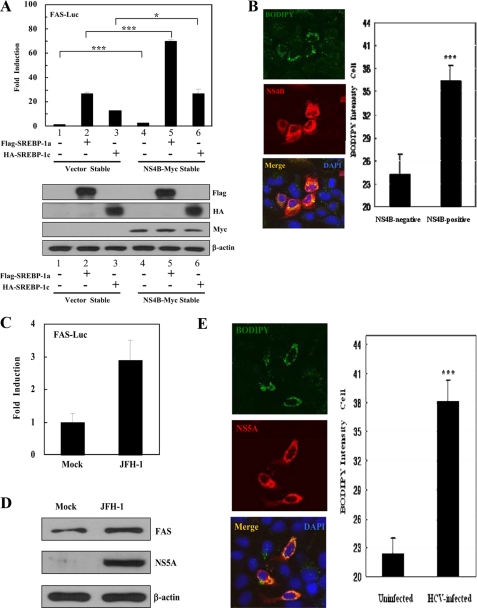
HCV NS4B Protein Increases the Transcriptional Activity of SREBP-1—Because SREBPs are the major transcription factors for lipogenic gene expression, we investigated the effects of HCV nonstructural proteins on transcriptional activity of SREBP-1. Huh7 cells were cotransfected with FAS-Luc reporter plasmid (reporter containing FAS promoter) and NS3-Myc, NS4B-Myc, NS5A-Myc, and NS5B-Myc expression plasmid, individually. We show that the NS4B protein strongly increases the promoter activity of the FAS gene (Fig. 1A). However, both NS3 and NS5B proteins do not increase the promoter activity of the FAS gene although the NS5A protein increases ∼2-fold the promoter activity of the FAS gene as compared with the vector control. Because FAS is one of the downstream targets of SREBP-1, this result indicates that NS4B protein increases the transcriptional activity of SREBP-1. We found that the NS4B protein increased the promoter activity of the FAS gene in a dose-dependent manner (Fig. 1B). To further demonstrate the effect of NS4B on transcriptional activity of SREBP-1, we established NS4B stable cell lines by transfecting Huh7 cells with the NS4B-Myc expression plasmid. We identified 3 clones (3, 9, and 10) by selection with blasticidin. As demonstrated in Fig. 1C, the promoter activity of the FAS gene was also increased in all NS4B stable cells as compared with the vector stable cells. Next, we analyzed the effect of NS4B on gene expression of SREBP-1 using an immunoblot. For this purpose, Huh7 cells were transfected with either vector or NS4B-Myc expression plasmid and total cell lysates were immunoblotted with SREBP-1 antibody. As demonstrated in Fig. 1D, the protein expression level of FAS was increased in Huh7 cells transiently expressing the NS4B protein as compared with the vector control. It is noteworthy that protein expression levels of both precursor and mature forms of SREBP-1 (pSREBP-1 and mSREBP-1) were significantly increased in cells expressing NS4B protein. We further confirmed that protein expression levels of FAS were increased in all NS4B stable cells as compared with the vector stable cells (Fig. 1E, upper panel). In addition, the protein expression level of pSREBP-1 was also increased in the number 3 NS4B stable cell line (Fig. 1E, lower panel) as well as numbers 9 and 10 stable cells (data not shown). We then investigated whether promoter activity of SREBP-1 was increased by the NS4B protein. For this purpose, Huh7 cells were cotransfected with pBP1c550-Luc (28), the luciferase gene containing 550-bp fragments of the mouse SRBBP-1c promoter (550-Luc), and NS4B expression plasmid. As expected, luciferase activity was increased ∼3-fold in cells expressing NS4B as compared with the control vector (Fig. 1F).
FIGURE 1.
HCV NS4B protein increases the transcriptional activity of SREBP-1. A, Huh7 cells were cotransfected with FAS-Luc reporter plasmid together with the indicated expression plasmids. At 36 h after transfection, cells were harvested and then luciferase activities were determined. The amount of DNA in each transfection was kept constant by adding an appropriate amount of pEF6-Myc empty vector. Data represent the mean of two independent experiments. B, HCV NS4B increases the transcriptional activity of SREBP-1 in a dose-dependent manner. Huh7 cells were cotransfected with FAS-Luc reporter plasmid together with increasing amounts of Myc-tagged NS4B expression plasmid. At 36 h after transfection, cells were harvested, and luciferase activities were determined (upper panel). Equal amounts of cell lysates were subjected to immunoblotting with anti-Myc monoclonal antibody (lower panel). C, both vector and NS4B-Myc stable cells were transfected with FAS-Luc reporter plasmid. At 36 h after transfection, cells were harvested and then luciferase activities were determined. D, Huh7 cells were transfected with either vector control or NS4B-Myc expression plasmid. At 36 h after transfection, total cell lysates were immunoblotted with the indicated antibodies. E, both vector stable and NS4B-Myc stable cell (#3) lysates harvested from each cell line were immunoblotted with the indicated antibodies. Protein expression of β-actin was used as a loading control for the same amount of cell lysates. F, Huh7 cells were cotransfected with 550-Luc reporter plasmid together with NS4B-Myc expression plasmid. At 36 h after transfection, cells were harvested and then luciferase activities were determined. Data represent the mean of two independent experiments.
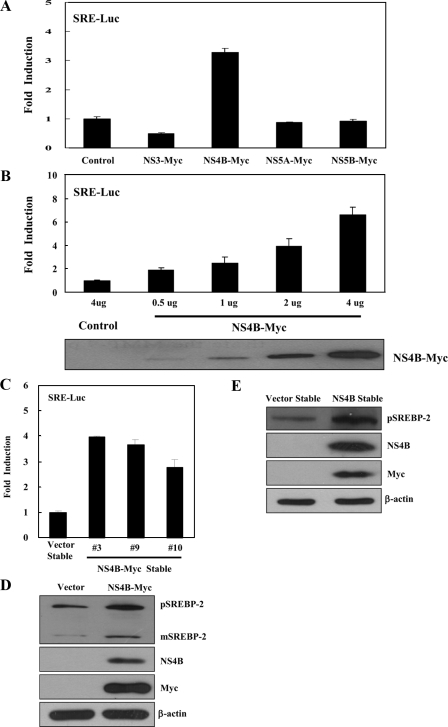
HCV NS4B Protein Increases the Transcriptional Activity and Gene Expression of SREBP-2—Because SREBP-2 is also a transcription factor for lipogenic gene expression, we investigated the effects of HCV nonstructural proteins on transcriptional activity of SREBP-2. Huh7 cells were cotransfected with pSyn-SRE-Luc (hamster HMG-CoA synthase promoter construct, SRE-Luc) and NS3-Myc, NS4B-Myc, NS5A-Myc, and NS5B-Myc expression plasmids, individually. We demonstrated that NS4B increased ∼3-fold the promoter activity of the HMG-CoA synthase gene as compared with the vector control (Fig. 2A). However, other nonstructural proteins did not increase the promoter activity of the HMG-CoA synthase gene. Because HMG-CoA synthase is one of the downstream targets of SREBP-2, this indicates that the NS4B protein also increases the transcriptional activity of SREBP-2. We further showed that the NS4B protein increased promoter activity of the HMG-CoA synthase gene in a dose-dependent manner (Fig. 2B). We then investigated whether transcriptional activity of the SREBP-2 was increased in NS4B stable cells. For this purpose, we transfected either vector stable or NS4B-Myc stable cells with pSynSRE-Luc (SRE-Luc). As shown in Fig. 2C, the promoter activity of the HMG-CoA synthase gene was also increased in all NS4B stable cell lines as compared with the vector stable cells. We then analyzed the effect of NS4B on gene expression of SREBP-2 by immunoblot analysis. We found that protein expression levels of both pSREBP-2 and mSREBP-2 were increased in Huh7 cells transiently expressing the NS4B protein as compared with the vector control (Fig. 2D). We have further shown that the protein expression level of pSREBP-2 was also increased in clone 9 NS4B stable cells (Fig. 2E) as well as clones 3 and 10 stable cells (data not shown).
FIGURE 2.
HCV NS4B protein increases the transcriptional activity of SREBP-2. A, Huh7 cells were cotransfected with SRE-Luc reporter plasmid together with the indicated expression plasmids. At 36 h after transfection, cells were harvested, and luciferase activities were determined. The amount of DNA in each transfection was kept constant by adding an appropriate amount of pEF6-Myc empty vector. Data represent the mean of two independent experiments. B, HCV NS4B increases the transcriptional activity of SREBP-2 in a dose-dependent manner. Huh7 cells were cotransfected with SRE-Luc reporter plasmid together with increasing amounts of Myc-tagged NS4B expression plasmid. At 36 h after transfection, cells were harvested, and then luciferase activities were determined (upper panel). Equal amounts of cell lysates were subjected to immunoblotting with anti-Myc monoclonal antibody (lower panel). C, both vector and NS4B-Myc stable cells were transfected with SRE-Luc reporter plasmid. At 36 h after transfection, cells were harvested and then luciferase activities were determined. D, Huh7 cells were transfected with either vector control or NS4B-Myc expression plasmid. At 36 h after transfection, total cell lysates were immunoblotted with the indicated antibodies. E, total cell lysates harvested from both vector stable and NS4B-Myc stable cells were immunoblotted with the indicated antibodies. Protein expression of β-actin was used as a loading control for the same amount of cell lysates.
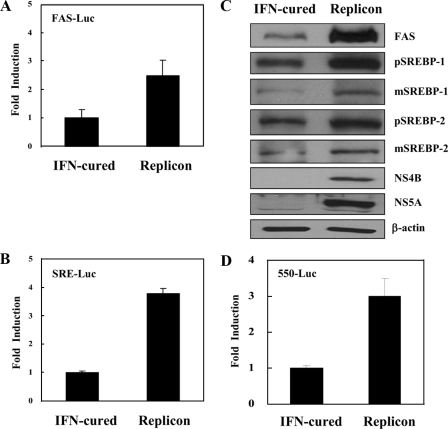
HCV Replicon Cells Increase the Transcriptional Activities and Gene Expressions of SREBP-1 and SREBP-2—To further investigate whether transcriptional and translational activities of SREBP-1 and SREBP-2 were regulated by viral protein in the context of HCV RNA replication, we transfected either FAS-Luc plasmid or SRE-Luc plasmid in IFN-cured and HCV replicon cells, and reporter activities were analyzed. As shown in Fig. 3, A and B, transcriptional activities of both SREBP-1 and SREBP-2 were increased in the replicon cells as compared with the IFN-cured cells. Furthermore, protein expression levels of FAS, pSREBP-1, mSREBP-1, pSREBP-2, and mSREBP-2 were significantly increased in HCV subgenomic replicon cells as compared with the IFN-cured cells (Fig. 3C). We further confirmed that pBP1c550-Luc reporter activity was also increased ∼3-fold in replicon cells as compared with the IFN-cured cells (Fig. 3D).
FIGURE 3.
HCV subgenomic replicon increases both SREBP-1 and SREBP-2 activities. Both IFN-cured and HCV subgenomic replicon cells were transfected with either FAS-Luc (A) or SRE-Luc (B) reporter plasmid. At 36 h after transfection, cells were harvested and then luciferase activities were determined. Data represent the mean of two independent experiments. C, total cell lysates harvested from IFN-cured and HCV subgenomic replicon cells were immunoblotted with the indicated antibodies. Protein expression of β-actin was used as a loading control for the same amount of cell lysates. D, both IFN-cured and HCV subgenomic replicon cells were transfected with 550-Luc reporter plasmid. At 36 h after transfection, cells were harvested, and luciferase activities were determined. Data represent the mean of two independent experiments.
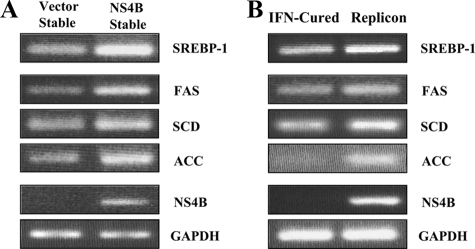
HCV NS4B Protein Increases the mRNA Levels of Lipogenic Genes—Because FAS, stearoyl-CoA desaturase (SCD), and acetyl-CoA carboxylase (ACC) are target genes of SREBP-1 (20), we further examined whether mRNA levels of lipogenic genes were modulated by the HCV NS4B protein. For this purpose, total RNAs isolated from vector stable, NS4B-Myc stable, IFN-cured, and HCV subgenomic replicon cells were compared for transcriptional levels of SREBP-1, FAS, SCD, ACC using cDNAs. As demonstrated in Fig. 4A, both SREBP-1 and its target gene (FAS, SCD, and ACC) expressions were elevated in NS4B stable cells as compared with vector stable cells. This result was further confirmed in the context of HCV RNA replication of replicon cells (Fig. 4B). These data indicated that NS4B-mediated increases of transcriptional and translational activities of SREBP-1 were due to up-regulation of the RNA level of SREBP-1.
FIGURE 4.
HCV NS4B protein up-regulates RNA levels of both SREBP-1 and its target genes. A and B, total RNAs were isolated from vector stable, NS4B-Myc stable, IFN-cured, and HCV subgenomic replicon cells. The cDNAs were synthesized by avian myeloblastosis virus reverse transcriptase from 1 μg of total RNAs. To estimate transcriptional levels of SREBP-1 and its target genes (FAS, SCD, and ACC), cDNAs were subjected to PCR amplification using gene-specific primers, respectively. The amplified DNA was analyzed by agarose gel electrophoresis.
HCV NS4B Protein Increases the Transcriptional Activity of Exogenous Mature SREBP-1 and Induces Lipid Accumulation—To further investigate whether NS4B protein was able to modulate transcriptional activity of SREBP-1, both vector and NS4B-Myc stable cells were cotransfected with FAS-Luc reporter and either mature Flag-SREBP-1a (1–490 amino acids) or mature HA-SREBP-1c (1–447 amino acids) expression plasmids. Because both SREBP-1a and SREBP-1c are mature forms, these can directly translocate to the nucleus and activate transcription of target genes. Indeed, both SREBPs (mature form) activated FAS transcriptional activities in vector stable cells, and SREBP (mature form)-mediated FAS activations were significantly increased by NS4B protein (Fig. 5A). Because NS4B increases endogenous SREBP activation (Fig. 5A, lane 1 versus lane 4 in upper panel), SREBP (mature form)-mediated FAS activations (Fig. 5A, lanes 2 and 3 versus lanes 5 and 6) showing that NS4B increases the transcriptional activity of SREBPs are additional evidence. These data suggest that NS4B stimulates SREBPs transcriptionally. Because we used NS4B protein-activated lipogenic signaling, we then asked whether lipid was accumulated by the NS4B protein. For this purpose, Huh7 cells transfected with either NS4B-Myc or vector plasmid were treated with BODIPY 493/503 for staining of lipid. Indeed, BODIPY signals were greatly increased in cells expressing NS4B as compared with vector control cells (Fig. 5B, left panel). This was further confirmed by quantification analysis (Fig. 5B, right panel). We have confirmed that the total amounts of lipid were increased in NS4B stable cells, and in HCV subgenomic replicon cells as compared with vector stable and IFN-cured cells, respectively (data not shown). To further confirm whether transcriptional activity of SREBP-1 was also increased in HCV-infected cells, we used the infectious JFH-1 strain of HCV (29). At 3 days after infection with HCV, Huh7 cells were transfected with the FAS-Luc reporter plasmid, harvested at 24 h after transfection, and then luciferase activities were determined (Fig. 5C). HCV infection was confirmed by immunostaining cells with NS5A antibody (data not shown). Fig. 5C showed that the promoter activity of the FAS gene was increased ∼3-fold in HCV-infected cells as compared with mock-infected cells. Furthermore, the protein level of FAS was also increased in HCV-infected cells (Fig. 5D). We have further shown that more lipids were accumulated in HCV-infected cells as compared with un-infected cells (Fig. 5E, left panel). This was verified by quantification analysis (Fig. 5E, right panel). These results strongly suggest that the HCV NS4B protein may mediate hepatic lipid accumulation via activation of SREBPs.
FIGURE 5.
HCV NS4B protein induces lipid accumulation through activation of SREBPs. A, both vector and NS4B-Myc stable cells were cotransfected with FAS-Luc reporter and Flag-SREBP-1a (1–490 amino acids) or HA-SREBP-1c (1–447 amino acids) expression plasmids. At 36 h after transfection, cells were harvested and then luciferase activities were determined (top panel). Data represent the mean of two independent experiments. ***, p < 0.001, vector stable versus NS4B stable cells transfected with Flag-SREBP-1a. *, p < 0.05, vector stable versus NS4B stable cells transfected with HA-SREBP-1c. Equal amounts of cell lysates were subjected to immunoblotting with anti-FLAG, anti-HA, anti-Myc, and anti-β-actin monoclonal antibody (bottom panel). B, Huh7 cells were transfected with NS4B-Myc expression plasmid. At 36 h after transfection, cells were fixed and incubated with anti-Myc monoclonal antibody for 2 h. After being washed with PBS, cells were further incubated with TRITC-conjugated goat anti-mouse IgG and BODIPY 493/503 (1 μm, Invitrogen) for 1 h. Samples were analyzed for immunofluorescence staining using the LSM 510 laser confocal microscopy system and BODIPY intensity was quantified. Each bar represents the average intensity of BODIPY staining. ***, p < 0.001, NS4B-negative cells versus NS4B-positive cells. C, Huh7 cells were either mock-infected or infected with HCV JFH-1. At 3 days after infection, cells were transfected with FAS-Luc reporter plasmid. At 24 h after transfection, cells were harvested and then luciferase activities were determined. D, at 3 days after infection, total cell lysates were immunoblotted with either anti-FAS antibody (top panel) or anti-NS5A antibody (middle panel). Protein expression of β-actin was used as a loading control for the same amount of cell lysates (bottom panel). E, at 3 days after infection, cells were fixed and incubated with anti-NS5A polyclonal antibody for 2 h. After being washed with PBS, cells were further incubated with TRITC-conjugated goat anti-mouse IgG and BODIPY 493/503 (1 μm, Invitrogen) for 1 h. Immunofluorescence staining was performed as described in B. ***, p < 0.001, mock-infected cells versus HCV-infected cells. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei.
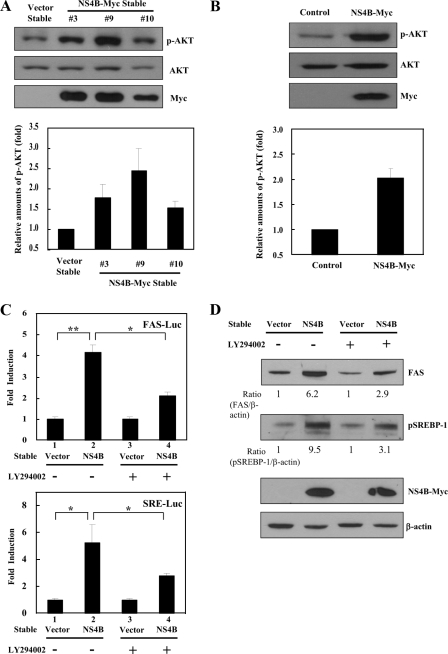
AKT Activation Is Required for NS4B-mediated SREBP Activation—It has previously been reported that activation of AKT kinase-induced gene expression of SREBP-1 (31), and hepatic overexpression of AKT resulted in steatosis (32). To determine whether AKT was involved in NS4B-mediated activation of SREBP-1, we investigated expression levels of both total AKT and phospho-AKT (p-AKT) proteins in vector stable and NS4B stable cells. We found that p-AKT protein levels were significantly higher in all three NS4B stable cells than vector stable cells (Fig. 6A). This was also evident in Huh7 cells transiently expressing the NS4B protein (Fig. 6B). We further confirmed that p-AKT levels were higher in HCV subgenomic replicon cells than IFN-cured cells (data not shown). We then investigated whether SREBP-1 activation in cells expressing NS4B was mediated through the phosphatidylinositol 3-kinase/AKT signaling pathway. For this purpose, both vector and NS4B stable cells transfected with either FAS-Luc or SRE-Luc reporters were either left untreated or treated with LY294002 (phosphatidylinositol 3-kinase inhibitor), and then luciferase activities were determined. As shown in Fig. 6C, NS4B-mediated promoter activations of FAS (upper panel) and HMG-CoA synthase (lower panel) genes were significantly reduced by LY294002. Furthermore, NS4B-mediated increases of FAS and pSREBP-1 protein levels were also significantly decreased by LY294002 (Fig. 6D). These results indicate that NS4B-induced SREBP activation was mediated through the AKT signaling pathway.
FIGURE 6.
AKT is required for NS4B-mediated SREBP activation. A, total cell lysates harvested from both vector stable and NS4B-Myc stable cells were immunoblotted with the indicated antibodies (upper panel). Triplicate experimental data of p-AKT levels were quantified (lower panel). B, Huh7 cells were transiently transfected with either vector or NS4B-Myc expression plasmid. At 24 h after transfection, cells were harvested and immunoblotted with the indicated antibodies (upper panel). Data from triplicate experiments were quantified and each bar represents the average intensity of p-AKT level (lower panel). C, both vector stable and NS4B-Myc stable cells were transfected with either FAS-Luc (upper panel) or SRE-Luc (lower panel) reporter plasmid. At 24 h after transfection, cells were either left untreated (Me2SO) or treated with LY294002 (5 μm) for an additional 12 h. Cells were harvested and then luciferase activities were determined. Data represent the mean of two independent experiments. D, total cellular extracts used in C were immunoblotted with the indicated antibodies. Quantification of the band intensity was determined by using a calibrated GS-800 densitometer.
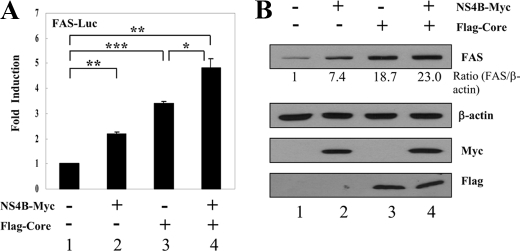
HCV NS4B Protein Synergistically Elevates the Core-mediated SREBP-1 Transcriptional Activation—We have previously reported that HCV core induced hepatic lipid accumulation by activating SREBP1 and PPARγ (19). We therefore asked whether the NS4B protein has synergistic effects on HCV core-mediated transcriptional activation of SREBP-1. For this purpose, Huh7 cells were cotransfected with FAS-Luc reporter plasmid and either NS4B or core, or both NS4B and core. Fig. 7A showed that FAS-Luc reporter activity was increased by either NS4B or core alone. Interestingly, promoter activity of the FAS gene was more elevated by HCV core protein than NS4B protein. As shown in Fig. 7A, FAS luciferase activity was significantly increased by cotransfection of Huh7 cells with NS4B and core expressing plasmids (lane 4). Likewise, the protein level of FAS was increased in cells expressing either NS4B or core protein (Fig. 7B). Furthermore, the protein level of FAS was more elevated in Huh7 cells cotransfected with NS4B and core as compared with either core or NS4B alone (Fig. 7B, lane 4). These data indicate that HCV NS4B protein synergistically activates the transcriptional activity of HCV core-mediated SREBP-1.
FIGURE 7.
HCV core-mediated transcriptional activation of SREBP-1 was synergistically elevated by NS4B protein. A, Huh7 cells were transfected with vector, NS4B-Myc, FLAG-core, individually or cotransfected with NS4B-Myc and FLAG-core in the presence of FAS-Luc reporter plasmid. At 36 h after transfection, cells were harvested, and luciferase activities were determined. Data represent the mean of two independent experiments. *, p < 0.05, core versus HCV NS4B/core. **, p < 0.01, vector versus HCV NS4B. ***, p < 0.001, vector versus HCV core. B, total cellular extracts used in A were immunoblotted with the indicated antibodies. Quantification of the band intensity was determined by using a calibrated GS-800 densitometer.
DISCUSSION
HCV infection is strongly associated with hepatic steatosis (3) and steatosis occurs in 40 to 86% of patients with chronic hepatitis C (14). It has been previously reported that the HCV core protein induced steatosis in in vitro and transgenic mice studies (17, 33, 34). Interestingly, steatosis is more frequent in patients infected with HCV genotype 3 although not all patients with genotype 3 infection have steatosis (15, 16). It has been suggested that the phenylalanine residue positioned at 164 of the core protein can increase the steatosis (35) and polymorphism of HCV core protein is also associated with intracellular accumulation of lipid (36). Furthermore, FAS gene is highly expressed in HCV-infected chimpanzees (37). Recent studies have shown that HCV infection enhances the proteolytic processing of SREBPs in hepatic cells (38) and HCV NS2 protein can up-regulate the transcription of SREBP-1c and FAS (39). Nevertheless, the molecular mechanisms underlying lipogenic signaling by either HCV or cellular factors are not fully understood.
In the present study, we set out to investigate the possible effects of HCV nonstructural proteins on lipogenic gene activities. Our results demonstrated that transcriptional activities of SREBPs were increased by NS4B protein but not by other nonstructural proteins in Huh7 cells. Both mRNA and protein levels of the FAS gene were also up-regulated by NS4B. Furthermore, NS4B in the context of the HCV subgenomic replicon activated the SREBPs and increased both mRNA and protein levels of the FAS gene. We have further shown that HCV NS4B increased the protein levels of both precursor and mature forms of SREBP-1 and SREBP-2. These results indicate that the NS4B protein enhances SREBP-1 and SREBP-2 protein expression at the transcriptional level.
It has been previously reported that HCV RNA replication requires the fatty acid biosynthetic pathway (40–42). In addition, a putative amphipathic helix within the N-terminal 26 residues of NS4B mediates membrane association, and these residues are critical for HCV replication in cell culture (10). In this study, we demonstrate that the HCV NS4B protein increases the FAS and SREBP activities via AKT signaling pathway. Therefore, we tempt to speculate that HCV NS4B-mediated lipogenesis may contribute to efficient HCV replication.
Several studies have shown that activations of PPAR by HCV core protein and hepatitis B virus X protein were required for hepatic accumulation of lipid (19, 27, 43). Because PPARα regulates constitutive transcription of genes encoding fatty acid metabolizing enzymes (44), and PPARγ is a master regulator of genes involved in fatty acid and glucose metabolism (45), further studies are required to determine the role of NS4B in PPAR regulation.
Recently, it has been reported that HCV infection enhanced the proteolytic processing of SREBPs in hepatic cells (38). In the present study, we have demonstrated that HCV infection increased the transcriptional activity of SREBP-1 and the protein level of FAS, and induced lipid accumulation in Huh7 cells. However, HCV infection had only a 3-fold effect on FAS expression. This effect is relatively modest as compared with the effect of HCV core and the NS4B co-expression system (23-fold). In fact, there is a genotype difference in steatosis induction (46). Because we used HCV genotype 2, the only available infectious HCV clone (JFH1) in the cell culture system (29), the fold discrepancy in FAS expression between two systems may be partly due to genotype difference, as we compared NS4B derived from either genotype 1b or genotype 2a (JFH-1) for its effects on transcriptional activation of SREBP-1 and found that NS4B protein derived from genotype 1b increased to a higher level of transcriptional activation of SREBP-1 than genotype 2a (data not shown). Alternatively, fold difference in FAS expression in two systems may be due to viral protein expression levels because only 20–30% of total cells are observed to be infected with the JFH-1 cell culture system. On the other hand, HCV core and NS4B proteins were overexpressed by the cotransfection experiment.
Because HCV core protein is involved in lipogenesis (19, 38), we investigated the co-expression effects of NS4B and core proteins on lipogenic signaling. Indeed, the HCV NS4B protein synergistically elevated the transcriptional activity of HCV core-mediated SREBP-1. We noticed that the effect of NS4B on SREBP activation was relatively modest. However, this effect was significant enough to increase both RNA and protein levels of lipogenic genes. This increase in turn resulted in an induction of lipid accumulation in NS4B expressing cells. Indeed, BODIPY staining data showed the similar level of lipid content between HCV NS4B-transfected cells (Fig. 5B) and HCV-infected cells (Fig. 5E), implying that NS4B alone substantially contributes to the accumulation of lipid. It has been reported that hepatic steatosis is an important factor of the progression of fibrosis (14, 47) and hepatocellular carcinoma (48) in patients with chronic HCV infection. In this regard, we speculate that NS4B-mediated lipid accumulation may contribute to the development of hepatocellular carcinoma.
In conclusion, we have demonstrated that HCV NS4B stimulated the expression of SREBPs and FAS protein and this was accomplished by activation of the AKT signaling pathway. However, how AKT activation affects SREBP gene expression is still not fully understood. Currently, both AKT (49) and liver X receptor (50) are known to be involved in SREBP activation. Nevertheless, how these proteins regulate SREBP is not understood. Therefore, further research is required to understand how NS4B may mediate its effect on SREBP. Collectively, our data imply that NS4B-mediated up-regulation of SREBPs may be associated with HCV-induced steatosis. These results provide a novel mechanism of liver pathogenesis associated with HCV-mediated lipogenesis.
This work was supported by the National R&D Program for Cancer Control, Ministry of Health and Welfare Grant 0620100, Korea, the Research Program for New Drug Target Discovery (M10601000044-06N0100-04410) and Biotechnology Development (2008-04100), and National Research Laboratory Grant ROA-2007-000-20051-0 from the Ministry of Education, Science and Technology, Korea.
Footnotes
The abbreviations used are: HCV, hepatitis C virus; NS4B, nonstructural 4B; HA, hemagglutinin; IFN, interferon; PBS, phosphate-buffered saline; TRITC, tetramethylrhodamine isothiocyanate; AKT, protein kinase B; FAS, fatty acid synthase; SREBP, sterol regulatory element-binding protein; ER, endoplasmic reticulum; PPAR, peroxisome proliferators-activated receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SCD, stearoyl-CoA desaturase; ACC, acetyl-CoA carboxylase.
References
- 1.Lauer, G. M., and Walker, B. D. (2001) N. Engl. J. Med. 345 41-52 [DOI] [PubMed] [Google Scholar]
- 2.Saito, I., Miyamura, T., Ohbayashi, A., Harada, H., Katayama, T., Kikuchi, S., Watanabe, Y., Koi, S., Onji, M., Ohta, Y., Choo, Q.-L., Houghton, M., and Kuo, G. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6547-6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramalho, F. (2003) Antiviral Res. 60 125-127 [DOI] [PubMed] [Google Scholar]
- 4.Simmonds, P. (2004) J. Gen. Virol. 85 3173-3188 [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach, B. D., and Rice, C. M. (2005) Nature 436 933-938 [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., Lohmann, V., Wilkinson, T., and Koch, J. O. (1995) J. Virol. 69 7519-7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hügle, T., Fehrmann, F., Bieck, E., Kohara, M., Kräusslich, H.-G., Rice, C. M., Blum, H. E., and Moradpour, D. (2001) Virology 284 70-81 [DOI] [PubMed] [Google Scholar]
- 8.Lundin, M., Lindstrom, H., Grönwall, C., and Persson, M. A. (2006) J. Gen. Virol. 87 3263-3272 [DOI] [PubMed] [Google Scholar]
- 9.Egger, D., Wölk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D., and Bienz, K. (2002) J. Virol. 76 5974-5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elazar, M., Liu, P., Rice, C. M., and Glenn, J. S. (2004) J. Virol. 78 11393-11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park, J.-S., Yang, J. M., and Min, M.-K. (2000) Biochem. Biophys. Res. Commun. 267 581-587 [DOI] [PubMed] [Google Scholar]
- 12.Einav, S., Sklan, E. H., Moon, H. M., Gehrig, E., Liu, P., Hao, Y., Lowe, A. W., and Glenn, J. S. (2008) Hepatology 47 827-835 [DOI] [PubMed] [Google Scholar]
- 13.Negro, F. (2006) World J. Gastroenterol. 12 6756-6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asselah, T., Rubbia-Brandt, L., Marcellin, P., and Negro, F. (2006) Gut 55 123-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihm, S., Fayyazi, A., Hartmann, H., and Ramadori, G. (1997) Hepatology 25 735-739 [DOI] [PubMed] [Google Scholar]
- 16.Rubbia-Brandt, L., Fabris, P., Paganin, S., Leandro, G., Male, P.-J., Giostra, E., Carlotto, A., Bozzola, L., Smedile, A., and Negro, F. (2004) Gut 53 406-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlemuter, G., Sabile, A., Letteron, P., Vona, G., Topilco, A., Chrétien, Y., Koike, K., Pessayre, D., Chapman, J., Barba, G., and Bréchot, C. (2002) FASEB J. 16 185-194 [DOI] [PubMed] [Google Scholar]
- 18.Dharancy, S., Malapel, M., Perlemuter, G., Roskams, T., Cheng, Y., Dubuquoy, L., Podevin, P., Conti, F., Canva, V., Philippe, D., Gambiez, L., Mathurin, P., Paris, J. C., Schoonjans, K., Calmus, Y., Pol, S., Auwerx, J., and Desreumaux, P. (2005) Gastroenterology 128 334-342 [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. H., Hong, S. P., Kim, K., Park, M. J., Kim, K. J., and Cheong, J. (2007) Biochem. Biophys. Res. Commun. 355 883-888 [DOI] [PubMed] [Google Scholar]
- 20.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, M. S., and Goldstein, J. L. (1997) Cell 89 331-340 [DOI] [PubMed] [Google Scholar]
- 22.Cho, Y. G., Yoon, J. W., Jang, K. L., Kim, C. M., and Sung, Y. C. (1993) Mol. Cells 3 195-202 [Google Scholar]
- 23.Choi, S.-H., Park, K.-J., Ahn, B.-Y., Jung, G., Lai, M. M., and Hwang, S. B. (2006) Mol. Cell. Biol. 26 3048-3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toth, J. I., Datta, S., Athanikar, J. N., Freedman, L. P., and Osborne, T. F. (2004) Mol. Cell. Biol. 24 8288-8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett, M. K., Lopez, J. M., Sanchez, H. B., and Osborne, T. F. (1995) J. Biol. Chem. 270 25578-25583 [DOI] [PubMed] [Google Scholar]
- 26.Dooley, K. A., Millinder, S., and Osborne, T. F. (1998) J. Biol. Chem. 273 1349-1356 [DOI] [PubMed] [Google Scholar]
- 27.Kim, K. H., Shin, H.-J., Kim, K., Choi, H. M., Rhee, S. H., Moon, H.-B., Kim, H. H., Yang, U. S., Yu, D.-Y., and Cheong, J. (2007) Gastroenterology 132 1955-1967 [DOI] [PubMed] [Google Scholar]
- 28.Yoshikawa, T., Shimano, H., Amemiya-Kudo, M., Yahagi, N., Hasty, A. H., Matsuzaka, T., Okazaki, H., Tamura, Y., Iizuka, Y., Ohashi, K., Osuga, J., Harada, K., Gotoda, T., Kimura, S., Ishibashi, S., and Yamada, N. (2001) Mol. Cell. Biol. 21 2991-3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakita, T., Pietschmann, T., Kato, T., Date, T., Miyamoto, M., Zhao, Z., Murthy, K., Habermann, A., Kräusslich, H.-G., Mizokami, M., Bartenschlager, R., and Liang, T. J. (2005) Nat. Med. 11 791-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi, S.-H., Jeong, S.-H., and Hwang, S. B. (2007) Gastroenterology 132 343-357 [DOI] [PubMed] [Google Scholar]
- 31.Fleischmann, M., and Iynedjian, P. B. (2000) Biochem. J. 349 13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, H., Shimano, H., Katagiri, H., Yahagi, N., Sakoda, H., Onishi, Y., Anai, M., Ogihara, T., Fujishiro, M., Viana, A. Y., Fukushima, Y., Abe, M., Shojima, N., Kikuchi, M., Yamada, N., Oka, Y., and Asano, T. (2003) Diabetes 52 2905-2913 [DOI] [PubMed] [Google Scholar]
- 33.Barba, G., Harper, F., Harada, T., Kohara, M., Goulinet, S., Matsuura, Y., Eder, G., Schaff, Z., Chapman, M. J., Miyamura, T., and Bréchot, C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 1200-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriya, K., Yotsuyanagi, H., Shintani, Y., Fujie, H., Ishibashi, K., Matsuura, Y., Miyamura, T., and Koike, K. (1997) J. Gen. Virol. 78 1527-1531 [DOI] [PubMed] [Google Scholar]
- 35.Jhaveri, R., McHutchison, J., Patel, K., Qiang, G., and Diehl, A. M. (2008) J. Infect. Dis. 197 283-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hourioux, C., Patient, R., Morin, A., Blanchard, E., Moreau, A., Trassard, S., Giraudeau, B., and Roingeard, P. (2007) Gut 56 1302-1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su, A. I., Pezacki, J. P., Wodicka, L., Brideau, A. D., Supekova, L., Thimme, R., Wieland, S., Bukh, J., Purcell, R. H., Schultz, P. G., and Chisari, F. V. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15669-15674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waris, G., Felmlee, D. J., Negro, F., and Siddiqui, A. (2007) J. Virol. 81 8122-8130 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Oem, J.-K., Jackel-Cram, C., Li, Y.-P., Zhou, Y., Zhong, J., Shimano, H., Babiuk, L. A., and Liu, Q. (2008) J. Gen. Virol. 89 1225-1230 [DOI] [PubMed] [Google Scholar]
- 40.Ye, J., Wang, C., Sumpter, R., Jr., Brown, M. S., Goldstein, J. L., and Gale, M., Jr. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 15865-15870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapadia, S. B., and Chisari, F. V. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2561-2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, S. S., Peng, L. F., Lin, W., Choe, W.-H., Sakamoto, N., Kato, N., Ikeda, M., Schreiber, S. L., and Chung, R. T. (2007) Gastroenterology 132 311-320 [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, N., Moriya, K., Kiyosawa, K., Koike, K., Gonzalez, F. J., and Aoyama, T. (2008) J. Clin. Investig. 118 683-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyama, T., Peters, J. M., Iritani, N., Nakajima, T., Furihata, K., Hashimoto, T., and Gonzalez, F. J. (1998) J. Biol. Chem. 273 5678-5684 [DOI] [PubMed] [Google Scholar]
- 45.Tontonoz, P., Hu, E., and Spiegelman, B. M. (1994) Cell 79 1147-1156 [DOI] [PubMed] [Google Scholar]
- 46.Abid, K., Pazienza, V., de Gottardi, A., Rubbia-Brandt, L., Conne, B., Pugnale, P., Rossi, C., Mangia, A., and Negro, F. (2005) Hepatology 42 744-751 [DOI] [PubMed] [Google Scholar]
- 47.Fartoux, L., Chazouillères, O., Wendum, D., Poupon, R., and Serfaty, L. (2005) Hepatology 41 82-87 [DOI] [PubMed] [Google Scholar]
- 48.Ohata, K., Hamasaki, K., Toriyama, K., Matsumoto, K., Saeki, A., Yanagi, K., Abiru, S., Nakagawa, Y., Shigeno, M., Miyazoe, S., Ichikawa, T., Ishikawa, H., Nakao, K., and Eguchi, K. (2003) Cancer 97 3036-3043 [DOI] [PubMed] [Google Scholar]
- 49.Furuta, E., Pai, S. K., Zhan, R., Bandyopadhyay, S., Watabe, M., Mo, Y. Y., Hirota, S., Hosobe, S., Tsukada, T., Miura, K., Kamada, S., Saito, K., Iiizumi, M., Liu, W., Ericsson, J., and Watabe, K. (2008) Cancer Res. 68 1003-1011 [DOI] [PubMed] [Google Scholar]
- 50.Higuchi, N., Kato, M., Shundo, Y., Tajiri, H., Tanaka, M., Yamashita, N., Kohjima, M., Kotoh, K., Nakamuta, M., Takayanagi, R., and Enjoji, M. (2008) Hepatol. Res. 38 1122-1129 [DOI] [PubMed] [Google Scholar]