Abstract
Increased conformational flexibility is the prevailing explanation for the high catalytic efficiency of cold-adapted enzymes at low temperatures. However, less is known about the structural determinants of flexibility. We reported two novel cold-adapted zinc metalloproteases in the thermolysin family, vibriolysin MCP-02 from a deep sea bacterium and vibriolysin E495 from an Arctic sea ice bacterium, and compared them with their mesophilic homolog, pseudolysin from a terrestrial bacterium. Their catalytic efficiencies, kcat/Km (10–40 °C), followed the order pseudolysin < MCP-02 < E495 with a ratio of ∼1:2:4. MCP-02 and E495 have the same optimal temperature (Topt, 57 °C, 5 °C lower than pseudolysin) and apparent melting temperature (Tm = 64 °C, ∼10 °C lower than pseudolysin). Structural analysis showed that the slightly lower stabilities resulted from a decrease in the number of salt bridges. Fluorescence quenching experiments and molecular dynamics simulations showed that the flexibilities of the proteins were pseudolysin < MCP-02 < E495, suggesting that optimization of flexibility is a strategy for cold adaptation. Molecular dynamics results showed that the ordinal increase in flexibility from pseudolysin to MCP-02 and E495, especially the increase from MCP-02 to E495, mainly resulted from the decrease of hydrogen-bond stability in the dynamic structure, which was due to the increase in asparagine, serine, and threonine residues. Finally, a model for the cold adaptation of MCP-02 and E495 was proposed. This is the first report of the optimization of hydrogen-bonding dynamics as a strategy for cold adaptation and provides new insights into the structural basis underlying conformational flexibility.
Cold-adapted enzymes, produced by organisms thriving in permanently cold habitats (deep sea, polar regions, and alpine regions), are characterized by high catalytic efficiencies at low temperatures and low stabilities at high temperatures (1–4). Different enzymes may adopt different strategies for cold adaptation, e.g. optimization of flexibility (2, 5) and optimization of electrostatic potential (6–8). The prevailing hypothesis is that cold-adapted enzymes obtain high catalytic efficiencies by increasing conformational flexibility at the expense of stability (4, 9). The comparison of crystal structures has revealed some structural features that might be related to the cold adaptation of enzymes, including more non-branching or small residues, more asparagines, less arginines, more negative net charges, larger hydrophobic accessible surfaces, less salt bridges, and longer loops (3, 10). However, less is currently known about how the conformational flexibility is determined by these features, because no structural feature is present in all cold-adapted enzymes, and no structural features consistently correlate with cold adaptation (3). Moreover, there are also reports about cold-adapted enzymes that possess both high activities and high stabilities (11–13). Laboratory evolution studies also show that high catalytic efficiency and high stability could be obtained simultaneously in a single molecule (14, 15). Therefore, further studies are needed to gain new insights into the structural basis underlying flexibility.
The thermolysin family (M4) is a family of zinc metalloproteases in the MA(E) subclan of the MA clan (16). All peptidases in this family contain a single catalytic zinc ion. This zinc ion is bonded by three ligands, the two histidines in the HEXXH motif and one glutamic acid beyond this motif. The glutamic acid in the HEXXH motif is the catalytic residue. The peptidases in this family are produced by bacteria from various habitats (16). For example, thermolysin, the typical example of this family, is from the thermophilic bacterium Bacillus thermoproteolyticus, pseudolysin is from the mesophilic bacterium Pseudomonas aeruginosa PAO1, and a putative vibriolysin in this family is from the Antarctic bacterium strain 643 (vibriolysin E643). Vibriolysin E643 might be the only known enzyme in this family from a permanently cold habitat (17). Although its sequence has been deposited in a public database (GenBank™ accession number AF272770, NCBI accession number AAF78076), no biochemical data of this vibriolysin has been reported.
In this report, two enzymes of the thermolysin family from permanently cold habitats are reported. The first enzyme, vibriolysin MCP-02, is an extracellular protease secreted by the deep sea bacterium Pseudoalteromonas sp. SM9913. MCP-02 was reported to be a mesophilic protease because of its high optimal temperature (Topt)3 (18), and further analysis in this report showed that it possesses some characteristics of cold-adapted enzymes. The second enzyme, vibriolysin E495, is an extracellular protease secreted by the Arctic sea ice bacterium Pseudoalteromonas sp. SM495. Analysis suggested that E495 has been optimized for higher catalytic efficiency at low temperatures than MCP-02. These two enzymes were compared with their mesophilic homolog, pseudolysin from P. aeruginosa PAO1. As shown in the results, these three enzymes made up a good system to study the structural basis underlying cold adaptation, because (i) the three enzymes are from typical habitats (terrestrial, deep sea, and Arctic) and the comparison of two enzymes at different cold-adaptation levels may provide more information than just comparing a cold-adapted enzyme with a mesophilic enzyme, and (ii) these three enzymes are highly homologous (identity between MCP-02 and E495 was ∼90%, and between MCP-02/E495 and pseudolysin was ∼60%), thus allowing an easy interpretation of the contributions of different structural features to cold adaptation.
In the first part of this study, the sequences, catalytic properties and structural properties of MCP-02 and E495 are presented and compared with that of pseudolysin. In the second part, homology modeling and long molecular dynamics (MD) simulations and systematic structural analysis of the three enzymes are reported to uncover the structural basis underlying the cold adaptation of deep sea MCP-02 and Arctic E495.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions—Pseudoalteromonas sp. SM9913 was cultured at 12 °C. Pseudoalteromonas sp. SM495 was isolated from a sea ice core sample 122–124 cm deep from the ice surface, which was taken from the Canadian Basin, Arctic Ocean (sampling site: 75°28′52″N 152°51′18″W) during the 2nd Chinese National Arctic Research Expedition, summer of 2003. Pseudoalteromonas sp. SM495 was cultured at 12 °C. The P. aeruginosa PAO1 strain was a generous gift from Yu Li-Yan (National Laboratory for Screening New Microbial Drugs, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences (CAMS) and Peking Union Medical College (PUMC), China) and was grown at 30 °C. Escherichia coli DH5α and E. coli BL21(DE3) (Novagen) were grown at 37 °C in Luria-Bertani (LB) medium supplemented with ampicillin for the selection of transformants.
Gene Cloning—Vibriolysin MCP-02 was purified from Pseudoalteromonas sp. SM9913 as described previously (18). Its N-terminal amino acid sequence was determined using automated Edman degradation with PROCISE491 (Applied Biosystems). A 5′-terminal degenerate primer based on the N-terminal sequence and a 3′-terminal degenerate primer based on the conserved fragment of vibriolysin E643 (AF272770) were designed to clone the conserved sequence segment of the mcp-02 gene. The flanking sequences were amplified by thermal asymmetric interlaced PCR as described previously (19). The full-length gene sequence was obtained by overlapping the amplified DNA fragments together. The mcp-02 gene was cloned from Pseudoalteromonas sp. SM9913 based on this full-length sequence and was sequenced again to ensure that a correct sequence was obtained. The nucleic acid sequence for the vibriolysin mcp-02 gene has been deposited in the GenBank™ data base under accession number EF029091. The amino acid sequence of this protein can be accessed through the NCBI Protein Database under NCBI accession number ABL06977.
Pseudoalteromonas sp. SM495 was cultured at 12 °C in a seawater liquid medium as described before (18). Vibriolysin E495 was obtained from the culture supernatant of Pseudoalteromonas sp. SM495 by ammonium sulfate precipitation and ion-exchange chromatography using a DEAE-Sepharose Fast Flow column (Amersham Biosciences). Its N-terminal amino acid sequence was determined using automated Edman degradation with PROCISE491 (Applied Biosystems). The gene sequence of vibriolysin E495 was obtained with the same method used for MCP-02. The nucleic acid sequence for the vibriolysin E495 gene has been deposited in the GenBank™ data base under accession number FJ211191. The amino acid sequence of this protein can be accessed through NCBI Protein Database under accession number ACI28452.
Expression and Purification—The full-length genes of MCP-02, E495, and pseudolysin precursors were amplified by PCR with 5′ primers containing NdeI cleavage sites and 3′ primers, including XhoI cleavage sites. The amplification product was digested with the NdeI and XhoI restriction enzymes and then cloned into the expression vector pET-22b (Novagen). The sequenced plasmid was introduced into the E. coli BL21(DE3) strain. The induction conditions were 1 mm isopropyl 1-thio-β-d-galactopyranoside at 28 °C for 40 h for MCP-02, 0.02 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C for 20 h for E495, and 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C overnight for pseudolysin.
Crude recombinant MCP-02 was obtained by direct addition of solid ammonium sulfate to the culture supernatant. Crude recombinant pseudolysin and recombinant E495 were obtained by disruption of the cells using sonication and then precipitation with solid ammonium sulfate. The crude proteins were then dialyzed and purified using a DEAE-Sepharose Fast Flow column (Amersham Biosciences) with 50 mm Tris-HCl buffer. The buffer pHs were 8.0 for MCP-02, 8.5 for E495, and 9.5 for pseudolysin. The proteins were eluted using a linear NaCl gradient from 0 to 0.5 m. Fractions with high protease activities were collected, and purity was determined by 12.5% SDS-PAGE (supplemental Fig. S1). The fractions with purified enzyme were pooled together and dialyzed against 50 mm Tris-HCl buffer to remove NaCl.
The N-terminal sequences of the three purified recombinant proteases were confirmed by automated Edman degradation with PROCISE491 (Applied Biosystems). The molecular weights of the mature enzymes of MCP-02 and E495 were determined using mass spectrometry (supplemental Figs. S2 and S3). Protein concentrations were determined by the BCA (bicinchoninic acid) method using bovine serum albumin (Sigma) as a standard.
Enzymatic Assays—Casein was used as the substrate for enzymatic activity assays as described (18). All of the buffers used in the enzymatic assays, and spectral measurements were 50 mm Tris-HCl unless otherwise specified. Topt was determined in the range of 0 °C to 80 °C. The optimal pH was determined in the working range of Tris-HCl buffer, 7.0–9.0. For the determination of heat inactivation at 55 °C and 60 °C, the enzymes were incubated at the corresponding temperature for 15–120 min and then immediately placed on ice. The residual activities were determined at 55 °C.
The dipeptide substrate 3-(2-furylacryloyl)glycyl-l-leucine amide (FAGLA) was purchased from Bachem A (Bubendorf, Switzerland). The concentrations of FAGLA were determined spectrophotometrically using ε345 nm = 766 m-1cm-1 (20). The enzymatic activity toward FAGLA was determined by measuring the decrease of absorbance at 345 nm (Δε345 = -317 m-1cm-1) with Feder's method (20). The absorbance was monitored every 1 s for 10 min using a Jasco V-550 spectrophotometer. The temperature was kept constant using a Julabo thermostat. The specific constant kcat/Km was determined using the following relationship: kcat/Km = v0/S0/E0, which is valid at S0 << Km.
Fluorescence Measurements—The thermal unfolding curves were recorded with intrinsic fluorescence as a probe using a Jasco FP-6500 spectrofluorometer equipped with a Peltier computer-controlled thermostat. The excitation wavelength was 280 nm, and the emission wavelength was 334 nm for pseudolysin and E495 and 337 nm for MCP-02. Both excitation and emission bandwidths were 3 nm. The temperature was scanned at a rate of 1 °C per min and was monitored using an external sensor. Cuvettes with a 1-cm path length were used. The protein concentrations were ∼5 μg/ml for MCP-02 and E495 and ∼10 μg/ml for pseudolysin.
CD Measurements—The thermal unfolding curves were also recorded using a Jasco J-810 spectropolarimeter equipped with a Julabo computer-controlled thermostat. The signal was recorded at 222 nm with a bandwidth of 2 nm. The temperature was monitored using an internal sensor, and the heating rate was 0.5 °C per min. A 0.1-cm path length cell was used. The protein concentration was ∼0.2 mg/ml.
Dynamic Quenching of Fluorescence—The conformational flexibility was detected using dynamic fluorescence quenching experiments with acrylamide as the quencher. The samples were in 50 mm Tris buffer (pH 8.0), and the protein concentrations were ∼5 μg/ml for MCP-02 and E495 and ∼10 μg/ml for pseudolysin. Aliquots of the 1.2 m acrylamide stock solution were added to the 2.4-ml sample solution so that the acrylamide concentration increased by steps of ∼5 mm each. The sample was stirred for 30 s before the fluorescence was recorded. The fluorescence was monitored using a Jasco FP-6500 spectrofluorometer equipped with a Peltier computer-controlled thermostat. The excitation wavelength was 280 nm, and the emission wavelength was 334 nm for pseudolysin and E495 and 337 nm for MCP-02. Both the excitation and emission bandwidths were 3 nm. The sensitivity was high, and the response time was 1 s. The experiments were carried out at 15 °C and 40 °C. The data were corrected because of the absorbance of the acrylamide (ε280 nm = 4.3 m-1 cm-1), according to Fcorr = Fobs10ε[Q]/2, where Fcorr is the corrected fluorescence intensity, Fobs the intensity observed in the presence of the quencher and [Q] the quencher concentration. The obtained data were fit to the Stern-Volmer equation F0/Fcorr = 1 + KSV[Q], where F0 is the fluorescence intensity obtained in the absence of the quencher, Fcorr the corrected fluorescence intensity, KSV the Stern-Volmer quenching constant, and [Q] the quencher concentration.
Bioinformatics—Searches for sequences similar to MCP-02 and E495 were done with the BLAST tool available at National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov). The conserved domains in the enzymes were analyzed using the Conserved Domain Database (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (21). The MEROPS peptidase database (Release 8.2, merops.sanger.ac.uk) was used to aid the classification of the enzymes (22). The signal peptides of the enzymes were predicted using the SignalP 3.0 server (www.cbs.dtu.dk/services/SignalP/) (23). Sequences were aligned using ClustalX 2 (24) and were edited using BioEdit (25). The coordinates file of pseudolysin (1EZM.pdb (26)) was obtained from the RCSB Protein Data Bank (PDB, www.rcsb.org/pdb/home/home.do (27)).
Homology Modeling and Quality Validation of Models—The homology models of MCP-02 and E495 were modeled using Modeler 9v2 (28) with the crystal structure of pseudolysin (1EZM.pdb) as the template (26). For each protein, 1000 models of the catalytic domain without zinc and calcium were created and then were selected according to the objective function scores and the qualities of the models. The qualities of the models were checked using the SAVES server (nihserver.mbi.ucla.edu/SAVES/) and PSQS server (www1.jcsg.org/psqs/). The models with low objective function scores and high quality scores were selected to model the ligands zinc and calcium. Then the model qualities were checked again using the SAVES server and PSQS server. Finally, one model for each protein, with both low objective function scores and high quality scores, was selected to be used as the starting structure for the following MD simulations. The quality scores of the selected structural models (Table 1) suggested that the models of MCP-02 and E495 were reliable.
TABLE 1.
Quality scores of protein models calculated using the SAVES server (nihserver.mbi.ucla.edu/SAVES/) and PSQS server (www1.jcsg.org/psqs/)
| Model | Pseudolysin | Virbriolysin MCP-02 | Vibriolysin E495 |
|---|---|---|---|
| ProChecka | 89.4% | 88.9% | 89.8% |
| Verify 3Db | 100% | 100% | 100% |
| Erratc | 91.034% | 91.304% | 90.476% |
| Proved | 98.6% | 97.4% | 96.7% |
| PSQSe | –0.2207 | –0.1985 | –0.2185 |
Ratio of residues in the core region of the Ramachandran plot
Ratio of residues with an average three- to one-dimensional score >0.2
Ratio of residues with error value below the 95% rejection limit. Good high resolution structures generally produced values around 95% or higher. For lower resolution structures (2.5–3 Å), the average overall quality factor was around 91%
Ratio of residues not classified as outliers
Average PSQS for PDB structures was –0.3
MD Simulations—All of the MD simulations were performed using the program NAMD 2.6 (29) and the CHARMM22 force field (30) on a Linux cluster of four nodes each equipped with one Intel® core™ 2 Quad 2.4-GHz processor. The missing atoms in the coordinate file (1EZM.pdb) of pseudolysin were added using Swiss-PdbViewer 3.7 (31). The psfgen package of the program VMD (32) was used to add hydrogen atoms to each protein. The models were then solvated in TIP3P water boxes with the solvate package in VMD. Next, the systems were neutralized by adding counter ions with the autoionize package in VMD. A cutoff distance of 9 Å for van der Waals interactions was assumed with a switching function starting at 7 Å. The non-bonded pair list distance was 11 Å. The SHAKE program was used to restrain the lengths of bonds involving hydrogen atoms to allow a time step of 2 fs for covalent bonds. Periodic boundary conditions were used with a flexible cell. The particle mesh Ewald method was used to compute long range electrostatic interactions, and the density of grid points was ≤1Å-3. The non-bonded pair list was updated every 20 steps. See Table 2 for details of the MD simulations.
TABLE 2.
Setup of the MD simulations of pseudolysin, vibriolysin MCP-02, and vibriolysin E495
| Protein | Total atoms | Protein residues | Protein atoms | Water atoms | Na+ ions | Temp. | Box size | Ensemble | Time |
|---|---|---|---|---|---|---|---|---|---|
| K | Å3 | ns | |||||||
| Pseudolysin | 22,963 | 298 | 4,464 | 18,498 | 1 | 280 | ∼55 × 54 × 74 | NPTa | 30 |
| 310 | ∼56 × 54 × 75 | NPTa | 30 | ||||||
| Vibriolysin MCP-02 | 25,295 | 307 | 4,456 | 20,829 | 10 | 280 | ∼56 × 56 × 78 | NPTa | 30 |
| 310 | ∼58 × 57 × 76 | NPTa | 30 | ||||||
| Vibriolysin E495 | 25,240 | 307 | 4,448 | 20,784 | 8 | 280 | ∼53 × 78 × 59 | NPTa | 30 |
| 310 | ∼53 × 80 × 59 | NPTa | 30 |
The isothermal-isobaric ensemble
The ensemble was minimized first for 2,000 steps with the positions of the Cα atoms fixed, and then for five 500-step cycles with decreasing harmonic constraints (100, 50, 10, 1, and 0 kcal mol-1 Å-2) on the Cα atoms in each cycle. Next, the temperature was increased gradually from 0 K to the final temperature (280 K or 310 K) in 5,000 steps with temperature assigned every 100 steps and with the Cα atoms under a harmonic constraint of 100 kcal mol-1 Å-2. Thereafter, the ensemble was equilibrated for four 5000-step cycles with decreasing harmonic constraints (100, 50, 10, and 1 kcal mol-1 Å-2) on the Cα atoms in each cycle. The final equilibration was carried out for 75,000 steps (i.e. 125 ps) without harmonic constraints. The final equilibrated conformations at each temperature were used as the starting point of the 30-ns Langevin dynamics simulations. A Langevin damping coefficient of 5 ps-1 was used during the equilibration and the final simulation. The ensemble was kept at 1 atm using a modified Nosé-Hoover method. In the simulation, the coordinates were saved every 5,000 time steps (i.e. 10 ps), and those of the last 15 ns were used in the hydrogen bond (HB) and salt bridge analyses.
Analysis of Simulations—The analyses were conducted with the help of VMD. The backbone atoms of residues 2–298 for MCP-02 and E495 and 2–297 for pseudolysin were used in the following fitting procedures. For each trajectory, the root mean square deviation (r.m.s.d.) of the atomic positions of the backbone atoms was calculated. The average structure of the last 15-ns simulation was also calculated after the structure of each frame was fitted to the starting frame of the last 15-ns simulation. Then, each frame of the last 15-ns simulation was fitted to the average structure, and the root mean square fluctuation (r.m.s.f.) of the backbone atoms was calculated as an index of the flexibility for each residue. The average r.m.s.f. of all residues in the protein was used as an index of protein global flexibility.
Structure Analysis—The HBs and salt bridges were detected for each frame of the last 15-ns simulation using self-written c codes. The HBs were detected following the HBPLUS (33) manual, except that the hydrogen positions were directly obtained from the simulation trajectories. The default geometric criteria of HBPLUS were used: maximum H-A distance of 2.5 Å, minimum angles of 90° for D-H-A, D-A-AA, and H-A-AA, with H for hydrogen, D for hydrogen donor, A for acceptor, and AA for acceptor antecedent. The salt bridges were detected with a maximum distance 4 Å between positively and negatively charged atoms of the residue pairs. The bond persistency was defined as the percentage of the frames in which the bond exists and was used as an index of bond stability.
RESULTS
Sequence and Homology Modeling—The mcp-02 gene encodes a precursor of 727 residues (Table 3). The BLAST search of the NCBI databases using the MCP-02 precursor sequence as a query revealed several sequences with high identities. The metalloprotease BmpI precursor (BAG70540) from Pseudoalteromonas sp. SB-B1 from fish farm sediments, which was recently deposited in the public databases, showed the highest identity (96%, full sequence identity for the BLAST results in this study unless otherwise specified) to the MCP-02 precursor. The E495 precursor in this study showed the second highest identity (80%) to the MCP-02 precursor. The metalloprotease precursor (BAB85124) from the marine bacterium Pseudoalteromonas sp. A28 (34) showed the third highest identity (78%). The SignalP 3.0 server predicted that the MCP-02 precursor contains a signal peptide (1–24). The BLAST search showed that the MCP-02 precursor sequence contains six domains: one FTP domain (63–112), one PepSY domain (142–199), one Peptidase_M4 domain (211–354), one Peptidase_M4_C domain (356–500), and two PPC domains (537–607 and 647–713). The molecular mass of mature MCP-02 was ∼33,929 Da as determined using mass spectrometry (supplemental Fig. S2). Mature MCP-02 contains 315 residues (Ala-205 to Asp-519), which was deduced from its N-terminal sequence and molecular weight.
TABLE 3.
Sequence information, catalytic and structural properties, and MD simulations of pseudolysin, vibriolysin MCP-02, and vibriolysin E495
| Enzyme | Pseudolysin | Vibriolysin MCP-02 | Vibriolysin E495 |
|---|---|---|---|
| Sequence | |||
| Precursor/mature sequence lengtha | 498/301 (301) | 727/315 (302) | 730/310 (302) |
| Pairwise identity MCP-02 | 59.5% | ||
| Pairwise identity E495 | 58.9% | 90.7% | |
| Leu number (ratio) | 14 (4.65%) | 20 (6.62%) | 18 (5.96%) |
| Ile number (ratio) | 9 (2.99%) | 7 (2.32%) | 7 (2.32%) |
| Val number (ratio) | 19 (6.31%) | 16 (5.30%) | 16 (5.30%) |
| Asn number (ratio) | 20 (6.64%) | 34 (11.3%) | 32 (10.6%) |
| Arg number (ratio) | 15 (4.98%) | 5 (1.66%) | 4 (1.32%) |
| Ser number (ratio) | 25 (8.31%) | 28 (9.27%) | 33 (10.9%) |
| Thr number (ratio) | 18 (5.98%) | 16 (5.30%) | 21 (6.95%) |
| Tyr number (ratio) | 22 (7.31%) | 18 (5.96%) | 17 (5.63%) |
| Trp number (ratio) | 4 (1.33%) | 6 (1.99%) | 6 (1.99%) |
| His number (ratio) | 7 (2.33%) | 5 (1.66%) | 5 (1.66%) |
| Net charges (Arg+Lys-Asp-Glu)b | –5 | –13 | –12 |
| Catalytic and structural properties | |||
| Optimal pH (casein, 60 °C)c | 7.5 | 8.0 | 7.5 |
| Topt (casein, optimal pH)d | 62 °C | 57 °C | 57 °C |
| kcat/Km (FAGLA, 25 °C) (s–1 mm–1)e | 0.64 ± 0.01 | 1.32 ± 0.01 | 2.55 ± 0.05 |
| Half-life t½ (55 °C) (min)f | 530 | 90 | 82 |
| Half-life t½ (60 °C) (min)f | 160 | 23 | 20 |
| Inactivation rate kinact (55 °C) (s–1)f | 2.2 × 10–5 | 1.3 × 10–4 | 1.4 × 10–4 |
| Inactivation rate kinact (60 °C) (s–1)f | 7.3 × 10–5 | 5.1 × 10–4 | 5.8 × 10–4 |
| Apparent Tm (fluorescence)g | 75 °C | 64 °C | 63 °C |
| Apparent Tm (CD)g | 72 °C | 65 °C | 65 °C |
| ΔKSV (40–15 °C, acrylamide) (m–1)h | 2.6 ± 0.1 | 5.3 ± 0.1 | 5.7 ± 0.1 |
| MD simulations (280 K/310 K)i | |||
| Mean r.m.s.f. (backbone) (Å) | 0.65/0.84 | 0.67/0.88 | 0.78/0.93 |
| Total HBs | 750/925 | 778/1023 | 887/1117 |
| Unstable HBs (persistency < 5%) | 367/500 | 429/621 | 484/680 |
| Average HBs in each frame | 219 ± 6/206 ± 7 | 221 ± 7/210 ± 7 | 219 ± 7/209 ± 7 |
| Average HB persistency | 29.2%/22.3% | 28.4%/20.5% | 24.7%/18.7% |
| Total salt bridges | 23/26 | 14/15 | 16/18 |
| Average salt bridges in each frame | 13.4 ± 0.8/14.4 ± 1.1 | 8.1 ± 1.0/8.1 ± 1.0 | 5.8 ± 0.9/8.0 ± 1.0 |
| Average salt bridge persistency | 58%/55% | 58%/54% | 36%/44% |
The numbers in parentheses are the residues used in the calculation of sequence identity and amino acid composition
Because each enzyme contains one zinc ion and one calcium ion, the total number of charges for each enzyme will be –1 for pseudolysin, –9 for vibriolysin MCP-02, and –8 for vibriolysin E495
See supplemental Fig. S4
The experiments were carried out at the optimal pH for each enzyme with 1% (w/w) casein as the substrate. See Fig. 4A
The pH was 8.0. See Fig. 3
The experiments were carried out at the optimal pH for each enzyme with 1% (w/w) casein as the substrate
The experiments were carried out at pH 8.0. See Fig. 4 (B and C)
The increase in the Stern-Volmer quenching constant KSV from 15 °C to 40 °C. The slopes of lines are presented in Fig. 5
The r.m.s.f., HB, and salt bridge data were calculated from the last 15-ns data of 30-ns simulations and are presented in the form of 280 K/310 K
The E495 gene encodes a precursor of 730 residues. The BLAST search showed that it has the highest identity (90%) to vibriolysin E643 precursor (AAF78076). Both metalloprotease BmpI precursor (BAG70540) from Pseudoalteromonas sp. SB-B1 and metalloprotease precursor (BAB85124) from Pseudoalteromonas sp. A28 showed the second highest identity (81%). The signal peptide is 1–24 as predicted by the SignalP server. The E495 precursor contains five domains: one FTP domain (63–113), one Peptidease_M4 domain (213–356), one Peptidease_M4_C domain (358–496) and two PPC domains (539–609 and 650–716) as identified by BLAST. The molecular mass of mature E495 was ∼33,376 Da as determined using mass spectrometry (supplemental Fig. S3). The deduced length of mature E495 is 310 residues (Ala-207 to Thr-516), which was deduced from the N-terminal sequence and molecular weight.
The alignment of mature sequences of pseudolysin, MCP-02, and E495 is shown in Fig. 1. Of the first 304 positions in the alignment, the identity between MCP-02 and pseudolysin is 59.5%, that between E495 and pseudolysin is 58.9%, and that between MCP-02 and E495 is 90.7% (Table 3 and Fig. 1).
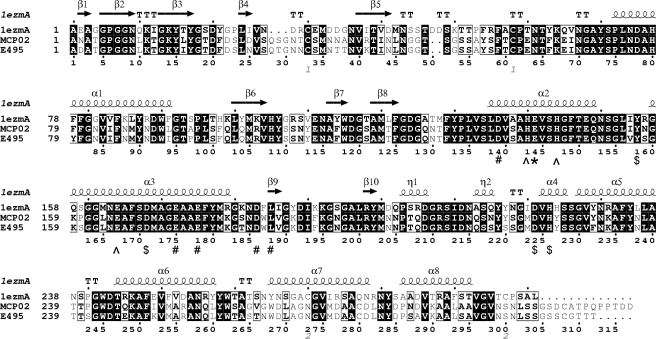
FIGURE 1.
Alignment of mature sequences of pseudolysin (1EZM), vibriolysin MCP-02 and vibriolysin E495. The N-terminal sequences of the three mature enzymes were determined using the Edman degradation method. The C termini of MCP-02 and E495 were deduced from the molecular weights (determined using mass spectrometry). The C terminus of pseudolysin shown is the C terminus of the pseudolysin precursor. The catalytic residue is indicated by “*,” the zinc ligands are indicated by “^,” the calcium ligands are indicated by “#,” and other important residues that might be involved in catalysis are indicated by “$.” The secondary structure elements of pseudolysin (top) were shown. The figure was made with ESPript 2.2 (44).
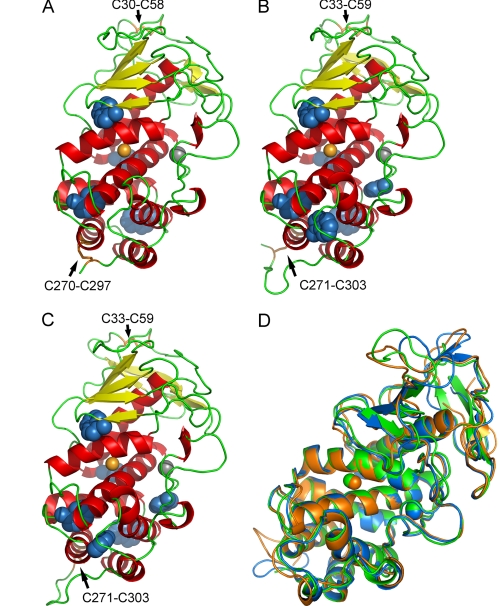
In the thermolysin family, only the structures of thermolysin (35), pseudolysin (26), and aureolysin (36) have been resolved. Among them, pseudolysin (1EZM.pdb) has the highest identity to MCP-02 and E495. The high sequence identities between MCP-02/E495 and pseudolysin allowed the homology modeling of the MCP-02 and E495 structures with pseudolysin (1EZM.pdb) as a template. The molecular structures of pseudolysin, MCP-02, and E495 are shown in Fig. 2 (A–C). Generally, the template pseudolysin and the models of MCP-02 and E495 have very similar structures. They contain an N-terminal domain composed of six (or eight) strands of anti-parallel β-sheets and two α-helices and a C-terminal domain composed mainly of eight α-helices. The zinc ion is linked by the three ligands at the cleft formed by the two domains. One calcium ion was also modeled in MCP-02 and E495. The models have two disulfide bonds, C33–C59 and C271–C303, corresponding to C30–C58 and C270–C297 in pseudolysin (Fig. 1).
FIGURE 2.
Comparison of three-dimensional structures of pseudolysin, vibriolysin MCP-02, and vibriolysin E495. A–C, ribbon representation of the template pseudolysin (A) and homology models of MCP-02 (B) and E495 (C). The zinc ion (orange ball), calcium ion (gray ball), the tryptophan residues (blue balls), and the disulfide bonds (orange sticks) are shown. D, the superposition of the average structures of the 280 K MD simulation trajectories (15–30 ns) of pseudolysin (green), MCP-02 (blue), and E495 (orange).
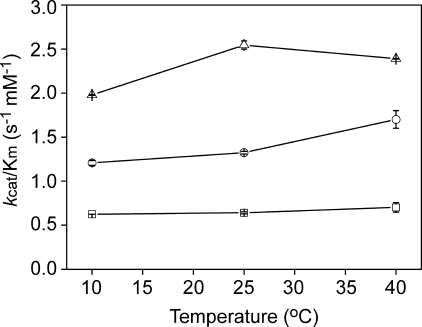
Activity—Using 1% (w/w) casein as the substrate, the optimal pH of MCP-02 was 8.0, whereas that of E495 and pseudolysin was 7.5 (supplemental Fig. S4). The activity toward synthetic FAGLA was compared at pH 8.0 (Fig. 3). In the temperature range 10–40 °C, the order of catalytic efficiency kcat/Km was terrestrial pseudolysin < deep sea MCP-02 < Arctic E495, with a ratio of ∼1:2:4. This result suggested that, although deep sea MCP-02 showed high thermal stability as described in the previous study (18), its activity has been optimized for efficient catalysis at the low temperatures of deep sea. Arctic E495 had higher activity than MCP-02 at low and medium temperatures (10–40 °C), suggesting a more efficient optimization for high catalytic efficiency in E495.
FIGURE 3.
Comparison of catalytic efficiencies kcat/Km toward the synthetic substrate FAGLA of pseudolysin (squares), vibriolysin MCP-02 (circles), and vibriolysin E495 (triangles) at different temperatures. The experiments were carried out using 50 mm Tris-HCl buffer (pH 8.0).
Thermal Stability—The parameters for the thermal stability of the three enzymes are shown in Table 3, and the thermo-dependence of activity and thermal unfolding curves are shown in Fig. 4. Both deep sea MCP-02 and Arctic E495 had a high Topt of 57 °C, although these were 5 °C lower than that of pseudolysin (62 °C) (Fig. 4A). MCP-02 and E495 had very similar parameters of thermal inactivation: the half-life (t½) was 90 min for MCP-02 and 82 min for E495 at 55 °C and 23 min for MCP-02 and 20 min for E495 at 60 °C, both of which were lower than that of pseudolysin (530 min at 55 °C and 160 min at 60 °C).
FIGURE 4.
Comparison of thermo-dependence of activity (A) and thermal unfolding measured by fluorescence (B) and CD (C) of pseudolysin (squares), vibriolysin MCP-02 (circles), and vibriolysin E495 (triangles). A, the thermo-dependence of activity was measured in 50 mm Tris-HCl buffer at pH 7.5 (for pseudolysin and vibriolysin E495) and pH 8.0 (for vibriolysin MCP-02), using 1% (w/w) casein as the substrate. B, the tryptophan fluorescence was monitored at 334 nm (for pseudolysin and vibriolysin E495) and at 337 nm (for vibriolysin MCP-02). The temperature was monitored using an external sensor with a gradient of 1 °C per min. C, the CD was monitored at 222 nm. The temperature was monitored using an internal sensor with a gradient of 0.5 °C per min.
The thermal unfolding of the three enzymes was monitored with the fluorescence of tryptophan as a probe (Fig. 4B). The results showed that the apparent melting temperatures (Tm) were 64 °C for MCP-02 and 63 °C for E495, both of which were ∼11 °C lower than that of pseudolysin (75 °C). Thermal unfolding was also monitored with CD (Fig. 4C). The results showed that the apparent Tm of MCP-02 and E495 was 7 °C lower than pseudolysin (65 °C for both MCP-02 and E495 and 72 °C for pseudolysin).
All of the above results came to the same order for thermal stability: terrestrial pseudolysin > deep sea MCP-02 ≈ Arctic E495. Compared with the catalytic efficiency order (pseudolysin < MCP-02 < Arctic E495) at low and medium temperatures, it could be concluded that different structural features have evolved in cold adaptation from pseudolysin to MCP-02 and from MCP-02 to E495, because the stability was compromised from pseudolysin to MCP-02, but not from MCP-02 to E495.
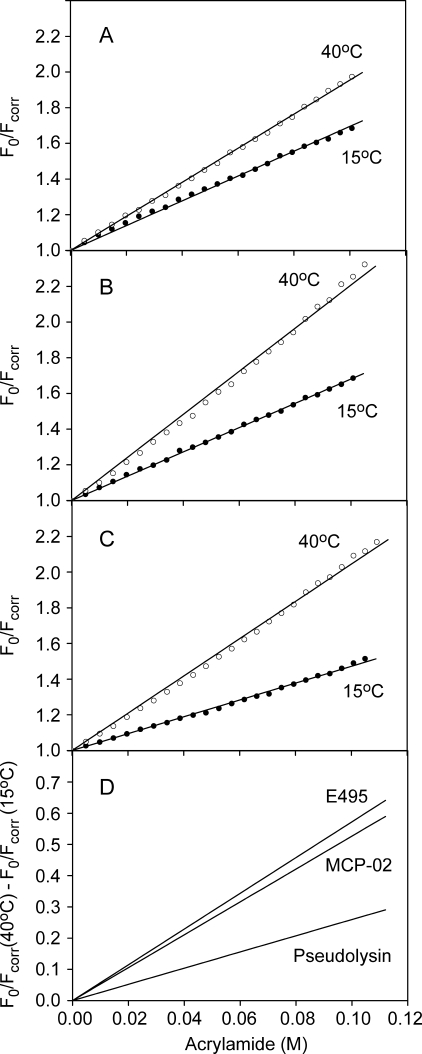
Fluorescence Quenching—The fluorescence quenching experiment was used to compare the conformational flexibility of the enzymes. The Stern-Volmer plots at 15 °C and 40 °C for pseudolysin, MCP-02, and E495 are shown in Fig. 5. Because the numbers and local environments of tryptophans in these three enzymes are different (Fig. 2, A–C), the absolute values of the Stern-Volmer quenching constant KSV cannot be compared directly. Instead, the difference in KSV at different temperatures (40 °C and 15 °C in this study, i.e. ΔKSV (40–15 °C)) was used as a parameter to represent the conformational flexibility of the enzyme. As shown in Fig. 5D, the ΔKSV (40–15 °C) value was 2.6 ± 0.1 m-1 for pseudolysin, 5.3 ± 0.1 m-1 for MCP-02, and 5.7 ± 0.1 m-1 for E495, suggesting that pseudolysin had the most rigid structure, deep sea MCP-02 had a more flexible structure, and Arctic E495 had the most flexible structure. The different flexibilities of E495 and MCP-02 from cold habitats suggested that the Arctic E495 underwent further optimization of flexibility compared with deep sea MCP-02.
FIGURE 5.
Stern-Volmer plots of fluorescence quenching by acrylamide. The fluorescence quenching was carried out at 15 °C (filled circles) and 40 °C (open circles) for pseudolysin (A), vibriolysin MCP-02 (B), and vibriolysin E495 (C), and the difference in fluorescence quenching between 15 °C and 40 °C was obtained by subtracting the regression lines of the Stern-Volmer plots at the individual temperatures (D). The quenching constants KSV corresponding to the plot slope are 6.95 (±0.04) m-1 at 15 °C and 9.54 (±0.03) m-1 at 40 °C for pseudolysin, 6.80 (±0.02) m-1 at 15 °C and 12.07 (±0.09) m-1 at 40 °C for vibriolysin MCP-02, and 4.72 (±0.03) m-1 at 15 °C and 10.44 (±0.07) m-1 at 40 °C for vibriolysin E495.
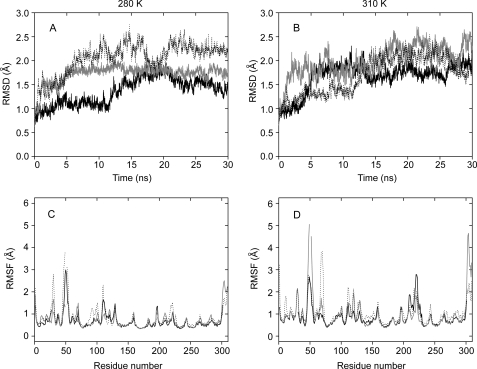
MD Simulations—Long (30 ns) MD simulations of these three enzymes were carried out at 280 K and 310 K. During the simulations, the three enzymes showed very similar structures, and the α-helices fitted well with each other (Fig. 2D). Both zinc and calcium ions were stably bonded to their ligands in the proteins during the simulations. Generally, the α-helices showed the lowest fluctuations in the enzyme, and the loops showed the highest fluctuations (Fig. 6, C and D). The average r.m.s.f. value of all residues (backbone) in the protein was used as an index of the global flexibility. The results showed that both deep sea MCP-02 and Arctic E495 had higher r.m.s.f. values than mesophilic pseudolysin (0.67 Å for MCP-02 and 0.78 Å for E495, versus 0.65 Å for pseudolysin at 280 K; 0.88 Å for MCP-02 and 0.93 Å for E495, versus 0.84 Å for pseudolysin at 310 K), indicating that the global structures of deep sea MCP-02 and Arctic E495 were more flexible than that of mesophilic pseudolysin (Table 3). It was also noted that the average r.m.s.f. of Arctic E495 was higher than that of deep sea MCP-02, suggesting a more flexible structure for Arctic E495 than deep sea MCP-02. The flexibility order of pseudolysin < deep sea MCP-02 < Arctic E495 agreed with that obtained from the fluorescence quenching experiment.
FIGURE 6.
The backbone r.m.s.d. (A, 280 K and B, 310 K) and residue r.m.s.f. (C, 280 K and D, 310 K) values of the MD simulations for pseudolysin (thick), vibriolysin MCP-02 (thin), and vibriolysin E495 (dot). The residue r.m.s.f. values were calculated from the 15- to 30-ns trajectories. The residue numbers in C and D are the same as the numbers in the alignment in Fig. 1.
Salt Bridges—The numbers of salt bridges in each frame in deep sea MCP-02 (8.1 ± 1.0 at 280 K and 8.1 ± 1.0 at 310 K) and Arctic E495 (5.8 ± 0.9 at 280 K and 8.0 ± 1.0 at 310 K) were smaller than that in mesophilic pseudolysin (13.4 ± 0.8 at 280 K and 14.4 ± 1.1 at 310 K) (Table 3). The salt bridges of the three proteins are listed in Table 4. It was noted that pseudolysin contained a salt bridge network consisting of five residues. In pseudolysin, Arg-279 simultaneously interacted with Glu-249 and Asp-253, while Glu-249 interacted with Arg-245 and Asp-253 interacted with Arg-256. The size of this five-member network could be further enlarged by another salt bridge, Asp-243 to Lys-246, which was located close to Glu-249 to Arg-245. The size of this network was remarkably smaller in both MCP-02 and E495. At the corresponding positions, MCP-02 contained only one salt bridge, Asp-279 to Arg-254, whereas E495 contained one salt bridge, Asp-279 to Arg-254, plus an additional weak salt bridge, Glu-246 to Lys-250. The result that MCP-02 and E495 contained less salt bridges than pseudolysin correlated well with the lower thermal stabilities of MCP-02 and E495 (Table 3), suggesting that decreased number of salt bridges in deep sea MCP-02 and Arctic E495 might be the origin of their decreased thermal stabilities. Comparison of amino acid compositions suggested that the decreased number of salt bridges in MCP-02/E495 was a result of the decreased number of arginines (Table 3).
TABLE 4.
Salt bridges (persistency > 40%) in the 15- to 30-ns MD simulation trajectories of pseudolysin, vibriolysin MCP-02, and vibriolysin E495
|
Pseudolysin
|
Vibriolysin MCP-02
|
Vibriolysin E495
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Persistency
|
Persistency
|
Persistency
|
|||||||||
| 280 K | 310 K | 280 K | 310 K | 280 K | 310 K | ||||||
| % | % | % | |||||||||
| E31-R29 | 100 | 100 | |||||||||
| E31-R55 | 100 | ||||||||||
| D41-K50 | 45 | 45 | |||||||||
| E66-K65 | 90 | ||||||||||
| D89-K85 | 51 | ||||||||||
| D89-R88 | 92 | 96 | |||||||||
| D117-R104 | 51 | 51 | |||||||||
| E165-R199 | 100 | 100 | |||||||||
| D168-R198 | 100 | 100 | D169-R199 | 100 | 100 | D169-R199 | 100 | 100 | |||
| E175-R179 | 98 | 90 | E176-K180 | 91 | 68 | E176-K180 | 92 | 79 | |||
| D184-K189 | 91 | 94 | D184-K189 | 84 | 78 | ||||||
| D189-R179 | 98 | 82 | |||||||||
| D190-K189 | 69 | ||||||||||
| D201-R205 | 100 | 75 | |||||||||
| D206-R208 | 100 | D207-R209 | 100 | 100 | |||||||
| D212-K232 | 100 | 100 | D212-K232 | 100 | |||||||
| D243-K246 | 96 | 99 | D244-K247 | 86 | 98 | D244-K247 | 66 | ||||
| E246-K250 | 54 | ||||||||||
| E249-R245 | 97 | 94 | |||||||||
| E249-R279 | 95 | 94 | |||||||||
| D253-R256 | 100 | 100 | |||||||||
| D253-R279a | 100 | 100 | D279-R254a | 98 | D279-R254a | 100 | 100 | ||||
| D285-R288 | 100 | 96 | |||||||||
| D305-K288 | 87 | 59 | |||||||||
Note the different positive-negative order in vibriolysins MCP-02 and E495 compared to pseudolysin
Intramolecular Hydrogen Bonding—HBs are the most important non-covalent forces that maintain protein three-dimensional structure and thus tend to be optimized during cold adaptation. However, comparison of HB numbers between different proteins may be hampered by several factors. The number of HBs in a crystal structure may be affected by factors like crystal packing. Thus, a direct comparison of crystal structures may result in an ambiguous result. The direct comparison of homology models may also be unreliable, because HBs are highly directional (3). To address the above issues, comparison of structures from the MD simulations rather than direct comparison of homology models was employed to study the structural features of homologous proteins. The advantages of comparing structures from the MD simulations are (i) that it provides detailed information about the dynamics of protein conformations and interactions and (ii) that it could minimize the uncertainty caused by crystal packing and homology modeling.
The results showed that the number of HBs in each frame of the 15- to 30-ns simulations was 219 ± 6 for pseudolysin, 221 ± 7 for deep sea MCP-02, and 219 ± 7 for Arctic E495 at 280 K and 206 ± 7 for pseudolysin, 210 ± 7 for deep sea MCP-02 and 209 ± 7 for Arctic E495 at 310 K (Table 3). This result showed that the three enzymes had nearly the same number of HBs in the static structures.
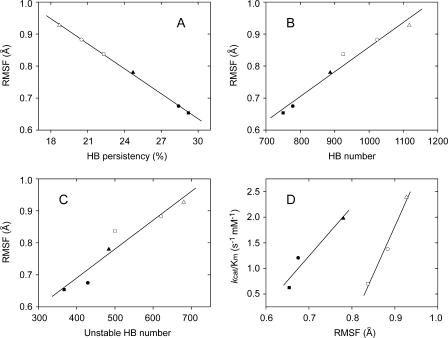
Further analysis showed that, in the 15- to 30-ns simulations, a total of 750 HBs at 280 K and 925 HBs at 310 K was detected in pseudolysin, whereas these values were larger in MCP-02 (778 at 280 K and 1023 at 310K) and the largest in E495 (887 at 280 K and 1117 at 310 K) (Table 3). This result suggested that, although the three enzymes had nearly the same number of HBs in the static structure, the average HB persistency (i.e. average HB stability) was different for the different enzymes: pseudolysin > deep sea MCP-02 > Arctic E495 (Table 3). As shown in Fig. 7A, this average HB persistency inversely correlated with the average r.m.s.f. To the authors' best knowledge, this is the first report showing strong correlations between hydrogen bonding in the dynamic structure and conformational flexibility. This result suggested that the average HB persistency has been optimized to improve the conformational flexibility during cold adaptation of deep sea MCP-02 and Arctic E495.
FIGURE 7.
The relationships between global flexibility and HB persistency (A), between global flexibility and number of HBs in the dynamic structures (B), between global flexibility and number of unstable HBs in the dynamic structures (C) and between catalytic efficiency and global flexibility (D). A, the average r.m.s.f. values (backbone) are inversely related to the average persistency of HBs in the structure. R =-0.9995; S.D. = 0.0039. B, the average r.m.s.f. values (backbone) are related to the number of HBs in the dynamic structure. R = 0.9820; S.D. = 0.0234. C, the average r.m.s.f. values (backbone) are related to the number of unstable HBs (persistency < 5%) in the dynamic structure. R = 0.9547; S.D. = 0.0369. A–C, the values were obtained from the 280 K (filled symbols) and 310 K (open symbols) MD simulation trajectories (15–30 ns) of pseudolysin (squares), vibriolysin MCP-02 (circles), and vibriolysin E495 (triangles). D, the catalytic efficiencies are related to the average r.m.s.f. values (backbone) of the protein. The kcat/Km values obtained at 10 °C are plotted against r.m.s.f. values obtained at 280 K (filled symbols), and the kcat/Km values obtained at 40 °C are plotted against r.m.s.f. values obtained at 310 K (open symbols), for pseudolysin (squares), vibriolysin MCP-02 (circles), and vibriolysin E495 (triangles).
The number of HBs in the static structure remained unchanged during cold adaptation, while the average HB persistency in the dynamic structure decreased from pseudolysin to MCP-02 and then to E495, suggesting that the increase in HB number in the dynamic structure played a key role in the decrease of the HB persistency (see Table 3 for HB number and Fig. 7B for the correlation between HB number and r.m.s.f.). Further analysis showed that the increase of HB number depended mainly on the increase in unstable HBs (persistency < 5%) in the dynamic structure (see Fig. 7C for the correlation between unstable HB number and r.m.s.f. and Fig. 8 for distribution of HB persistency).
FIGURE 8.
The distribution of HB persistency of the 280 K (A) and 310 K (B) MD simulations. The data were obtained from the 15- to 30-ns simulation trajectories of pseudolysin (black), vibriolysin MCP-02 (gray), and vibriolysin E495 (white).
The high sequence identity between E495 and MCP-02 allowed for a direct comparison of the HBs in the dynamic structures at sites where substitutions occurred (a total of 30 substituted sites in the mature sequence). As shown in Table 5, at 280 K, 17 of the total 30 sites in E495 formed more HBs than in MCP-02. At 310 K, 19 of the total 30 sites in E495 formed more HBs than in MCP-02. As a result, the 30 substituted sites could form 43 (280 K) and 57 (310 K) more HBs in E495 than in MCP-02. Therefore, the contribution of the substituted sites, which accounted for only 9.8% of the total 307 residues and participated in ∼20% of all HBs in the protein dynamic structure, constituted up to 39.4% (280 K) and 60.6% (310 K) of the total increase in the HB number (109 for 280 K and 94 for 310 K) (Table 5). The above observations strongly suggested that these substituted sites have been selected to improve the number of HBs in the dynamic structure of the protein.
TABLE 5.
Number of HBs formed at substituted sites of MCP-02 and E495 in the dynamic structures
|
Residues, MCP-02, E495
|
280 K
|
310 K
|
||||
|---|---|---|---|---|---|---|
| MCP-02 | E495 | Δ | MCP-02 | E495 | Δ | |
| N2D | 7 | 6 | –1 | 10 | 5 | –5 |
| L16I | 5 | 6 | 1 | 5 | 6 | 1 |
| D25N | 7 | 9 | 2 | 4 | 16 | 12 |
| S29T | 5 | 3 | –2 | 5 | 13 | 8 |
| T32N | 6 | 9 | 3 | 10 | 13 | 3 |
| N37T | 11 | 11 | 0 | 17 | 13 | –4 |
| A38T | 2 | 4 | 2 | 4 | 5 | 1 |
| R41K | 11 | 9 | –2 | 17 | 10 | –7 |
| F79Y | 4 | 8 | 4 | 5 | 9 | 4 |
| L92I | 4 | 3 | –1 | 3 | 4 | 1 |
| A95P | 2 | 3 | 1 | 5 | 3 | –2 |
| I155V | 1 | 7 | 6 | 3 | 7 | 4 |
| N157S | 2 | 6 | 4 | 5 | 10 | 5 |
| P160S | 2 | 11 | 9 | 2 | 15 | 13 |
| S182T | 3 | 2 | –1 | 5 | 4 | –1 |
| N216S | 8 | 9 | 1 | 11 | 10 | –1 |
| Y218S | 5 | 4 | –1 | 5 | 5 | 0 |
| Y230F | 5 | 3 | –2 | 6 | 3 | –3 |
| P241S | 0 | 0 | 0 | 7 | 2 | –5 |
| Q246E | 6 | 6 | 0 | 4 | 6 | 2 |
| I250K | 4 | 5 | 1 | 4 | 6 | 2 |
| S261T | 3 | 9 | 6 | 2 | 7 | 5 |
| G263S | 1 | 4 | 3 | 1 | 5 | 4 |
| V264T | 4 | 12 | 8 | 3 | 13 | 10 |
| G265N | 2 | 9 | 7 | 2 | 11 | 9 |
| N285S | 10 | 3 | –7 | 12 | 13 | 1 |
| D286A | 7 | 4 | –3 | 7 | 6 | –1 |
| A292S | 3 | 5 | 2 | 4 | 8 | 4 |
| D305S | 5 | 8 | 3 | 18 | 21 | 3 |
| A307G | 5 | 5 | 0 | 18 | 12 | –6 |
| Sum of substituteda | 140 | 183 | 43 | 204 | 261 | 57 |
| Sum of Proteinb | 778 | 887 | 109 | 1023 | 1117 | 94 |
| Ratioc | 18.0% | 20.6% | 39.4% | 19.9% | 23.4% | 60.6% |
The sum of HBs of the 30 substituted sites
The sum of HBs of all 307 sites in the protein
The percentage of HBs of the substituted sites in all the HBs in protein
Comparison of amino acid compositions suggested that the major strategy for increasing the number of HBs was to increase the number of residues with HB-forming side chains in the protein. As shown in Table 3, the numbers of asparagines, serines, and threonines increased from pseudolysin to MCP-02, and the numbers of serines and threonines continued to increase from deep sea MCP-02 to Arctic E495. This result suggested that the increased numbers (and decreased persistency) of HBs in the MCP-02/E495 dynamic structures were not the result of increased flexibility of the protein structures, but instead, was the major structural origin of the increased flexibilities of MCP-02/E495.
DISCUSSION
Catalytic and Structural Properties—The catalytic efficiency order (pseudolysin < deep sea MCP-02 < Arctic E495) at low and medium temperatures (10–40 °C) indicated that these three enzymes constituted a good system to study cold adaptation. Both deep sea MCP-02 and Arctic E495 had higher catalytic efficiencies at 10-40 °C than the terrestrial pseudolysin. This phenomenon agrees with the general idea that enzymes from permanently cold habitats have higher catalytic efficiencies at low temperatures than their mesophilic homologs. Compared with deep sea MCP-02, Arctic E495 showed an elevated catalytic efficiency, suggesting that Arctic E495 was much more cold adapted than deep sea MCP-02. This could be related to two factors. The first is that the Arctic sea ice bacterium survives in a much colder environment than the deep sea bacterium. The second is that the two enzymes account for different portions of the extracellular enzymes. MCP-02 makes up only a minor portion of the total extracellular enzymes of deep sea Pseudoalteromonas sp. SM9913 (18), whereas in contrast, E495 is the major extracellular enzyme of Pseudoalteromonas sp. SM495.4 This suggests that E495 is subject to more stringent evolutionary pressure than MCP-02.
Comparisons between cold-adapted and mesophilic enzymes show that cold-adapted enzymes often have lower stabilities than their mesophilic homologs. In some cases, the difference in stabilities (represented by Topt or Tm, etc.) may be as large as 20–30 °C (37–40), and in other cases, the difference is only ∼10 °C (13, 41, 42). However, there are also examples showing that cold-adapted enzymes have higher stabilities than their mesophilic homologs (12). This suggests that the low stability is not an inherent property of cold-adapted enzymes. Deep sea MCP-02 and Arctic E495 in this study also showed high stabilities. Topt of deep sea MCP-02 was as high as 57 °C, which was only 5 °C lower than the mesophilic pseudolysin. Moreover, Arctic E495 had nearly the same stability as deep sea MCP-02, although the catalytic efficiency of E495 has been further optimized to surpass that of MCP-02.
The psychrophilic alkaline metalloprotease (PAP) from P. TAC II 18 is a Zn2+-Ca2+ protease (42, 43). Compared with the t½ of ∼20 min for PAP at 45 °C (42), the t½ values of deep sea MCP-02 and Arctic E495 at 60 °C were ∼20 min, suggesting much higher stabilities (15 °C higher) of MCP-02 and E495 than PAP. Moreover, deep sea MCP-02 and Arctic E495 are even 5 °C more stable than the mesophilic homolog of PAP, alkaline metalloprotease (AP) from P. aeruginosa, because the t½ of AP at 55 °C is ∼20 min (42). This might be caused by different tertiary structures. The thermolysin family belongs to the MA(E) subclan (16), and proteins in the thermolysin family are composed mainly of rigid α-helices, with the catalytic center located on two α-helices. On the contrary, AP and PAP belong to the MA(M) subclan (16), and their structures are rich in β-sheets rather than α-helices, with the catalytic center located on one α-helix plus one loop. The high stabilities of MCP-02 and E495 seem to benefit from the large proportion of α-helices in the tertiary structures.
Dynamic fluorescence quenching experiments and MD simulations were used to compare the flexibility of the enzymes. Both methods came to the same flexibility order, pseudolysin < deep sea MCP-02 < Arctic E495, suggesting a continuous optimization of flexibility during the course of cold adaptation from deep sea MCP-02 to Arctic E495. However, there were some disagreements between the results from the two methods. The MD simulations showed that the difference in flexibility between E495 and MCP-02 was bigger than that between MCP-02 and pseudolysin, but the dynamic fluorescence quenching experiments suggested that the difference in flexibility between E495 and MCP-02 was smaller. One possible explanation was that the fluorescence quenching data represented the local flexibility where the tryptophan residues were located, while the average r.m.s.f. values represent the global flexibility. An alternative explanation is that the ΔKSV (40–15 °C) values obtained from the fluorescence quenching experiment were an index of protein permeability, and thus just rough estimates of protein flexibility.
Structural Basis Underlying Cold Adaptation—The high activity, high flexibility, and low stability of cold-adapted enzymes have been noticed for a long time. A general explanation for this high activity is that it originates from the high flexibility, whereas the high flexibility is mainly due to the decreased stability (4, 9). However, the presence of enzymes with both high activity and high stability (11–13) suggests that care must be taken when interpreting the activity/flexibility/stability relationships.
To explain the stability/flexibility relationship, we tried to distinguish the dynamic structure from the static structure. The number of HBs in the static structure was almost the same for pseudolysin, MCP-02 and E495, which is in accordance with the previous observations that the difference in HB number of crystal structures is often small (3, 10). However, the analysis of hydrogen bonding dynamics showed that the number of HBs in the dynamic structure increased during the adaptation from pseudolysin to MCP-02 and then from MCP-02 to E495, which means that the average persistency of HBs in the dynamic structure decreased from pseudolysin to MCP-02 and then from MCP-02 to E495. This persistency order inversely correlated with the global flexibility (represented by the average r.m.s.f.), strongly suggesting that hydrogen bonding has been optimized to improve flexibility.
The stability order pseudolysin > MCP-02 ≈ E495 could be explained by the HB numbers and salt bridge numbers in the static structures. The high stabilities of the three enzymes (although those of MCP-02/E495 were slightly lower than pseudolysin) could be related to the observation that the three enzymes had nearly the same numbers of HBs in the static structure. In addition, the slightly lower stabilities of MCP-02/E495 may come from the decreased number of salt bridges in the static structure. Thus, during cold adaptation from pseudolysin to MCP-02 and then to E495, protein stability was determined by the number and strength of interactions (HBs and salt bridges) in the static structure, whereas the protein flexibility was determined by the HB persistency in the dynamic structure. Therefore, the emerging picture is that protein stability and flexibility are only loosely coupled, because they are governed by related but different structural factors. The result that Arctic E495 has higher flexibility than deep sea MCP-02, but nearly the same stability (Table 1) clearly demonstrated this point.
An argument may be made that the decreased HB persistency was the origin of elevated flexibility rather than the result of elevated flexibility. There are two points of explanation. First, flexibility, which is used to describe the conversions between different conformations, originates from molecular thermal motions and is restricted by the forces that maintain the three-dimensional structure. Weaker bonds will allow easier conversions between conformations and thus result in higher flexibility. Second, the number of amino acid residues with HB-forming side chains (arginines, serines, and threonines) increased from pseudolysin to MCP-02 to E495, suggesting that the strategy, employed by MCP-02/E495 to decrease HB persistency, was to introduce more HB-forming conformations by increasing residues with HB-forming side chains.
In addition, it should be noted that, although optimization of hydrogen bonding in the dynamic structure seems to be the major strategy for optimization of flexibility, other strategies cannot be ruled out. For example, during the adaptation from pseudolysin to MCP-02, the decreased number of salt bridges in MCP-02 may also have contributed to the increased flexibility. The longer sequence of MCP-02 than pseudolysin might be another origin of the increased flexibility. The increased flexibility of MCP-02 might also benefit from the increased number of leucines (long and non-branching amino acids) in the molecule. Moreover, the protein flexibility might also be affected by the slightly different positions of disulfide bonds in the protein (Figs. 1 and 2).
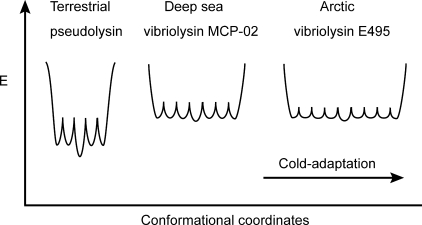
An Energy Landscape Model—Feller et al. (37) suggested a model of folding funnels for psychrophilic and thermophilic enzymes. Here, this model was modified and used to show how deep sea MCP-02 and Arctic E495 are adapted to cold environments by optimizing flexibility (Fig. 9). In the model, the bottom of the folding funnel for each enzyme is shown. The deep sea MCP-02 was slightly less stable than pseudolysin, and thus the average energy of the conformations at the bottom of the funnels should be higher than that of pseudolysin. This may be caused by the decreased number of salt bridges in MCP-02. The greater flexibility of MCP-02 than pseudolysin could be related to the longer sequence, more long and non-branching residues, and the increased number of HBs in the dynamic structure and was represented in the model by lowered energy barriers between local energy minima. The increased number of HBs in the dynamic structure also suggested that MCP-02 has more local energy minima and thus a wider funnel. E495 and MCP-02 had very similar stabilities and were represented in the model by the same energy level of their funnel bottoms. The adaptation from deep sea MCP-02 to Arctic E495 was closely related to the direct optimization of hydrogen bonding. The average HB persistency of Arctic E495 was smaller than deep sea MCP-02, suggesting smaller energy barriers between different local energy minima in Arctic E495. Arctic E495 contained more serines and threonines, and therefore had more HB-forming conformations. This was represented in the model by more local energy minima in E495.
FIGURE 9.
An energy landscape model for the cold adaptation of metalloproteases in the thermolysin family.
Supplementary Material
The amino acid sequences reported in this work can be accessed through NCBI Protein Database under NCBI accession numbers ABL06977 and ACI28452.
The nucleotide sequences reported in this paper have been submitted to the GenBank™/EBI Data Bank with accession number(s) EF029091 and FJ211191.
The work was supported by the Hi-Tech Research and Development Program of China (Grants 2007AA091903 and 2007AA021306), National Natural Science Foundation of China (Grants 30770040 and 40706001), National Basic Research Program of China (Grant 2004CB719601), Program for New Century Excellent Talents in University (Grant NCET-06-0578), the China Ocean Mineral Resources R&D Association (COMRA) Program (Grant DYXM-115-02-2-6), and the Chinese Polar Science Stratagem Fund (Grant 200601). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: Topt, optimal temperature of activity; FAGLA, 3-(2-furylacryloyl)glycyl-l-leucine amide; HB, hydrogen bond; MD, molecular dynamics; r.m.s.d., root mean square deviation; r.m.s.f., root mean square fluctuation; Tm, melting temperature; AP, alkaline metalloprotease; PAP, psychrophilic alkaline metalloprotease.
B.-B. Xie, F. Bian, X.-L. Chen, H.-L. He, J. Guo, X. Gao, Y.-X. Zeng, B. Chen, B.-C. Zhou, and Y.-Z. Zhang, unpublished data.
References
- 1.Feller, G., and Gerday, C. (2003) Nat. Rev. Microbiol. 1 200-208 [DOI] [PubMed] [Google Scholar]
- 2.Lonhienne, T., Gerday, C., and Feller, G. (2000) Biochim. Biophys. Acta 1543 1-10 [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui, K. S., and Cavicchioli, R. (2006) Annu. Rev. Biochem. 75 403-433 [DOI] [PubMed] [Google Scholar]
- 4.Georlette, D., Blaise, V., Collins, T., D'Amico, S., Gratia, E., Hoyoux, A., Marx, J. C., Sonan, G., Feller, G., and Gerday, C. (2004) FEMS Microbiol. Rev. 28 25-42 [DOI] [PubMed] [Google Scholar]
- 5.Olufsen, M., Smalas, A. O., Moe, E., and Brandsdal, B. O. (2005) J. Biol. Chem. 280 18042-18048 [DOI] [PubMed] [Google Scholar]
- 6.Russell, R. J., Gerike, U., Danson, M. J., Hough, D. W., and Taylor, G. L. (1998) Structure 6 351-361 [DOI] [PubMed] [Google Scholar]
- 7.Brandsdal, B. O., Smalas, A. O., and Aqvist, J. (2001) FEBS Lett. 499 171-175 [DOI] [PubMed] [Google Scholar]
- 8.Moe, E., Leiros, I., Riise, E. K., Olufsen, M., Lanes, O., Smalas, A., and Willassen, N. P. (2004) J. Mol. Biol. 343 1221-1230 [DOI] [PubMed] [Google Scholar]
- 9.D'Amico, S., Claverie, P., Collins, T., Georlette, D., Gratia, E., Hoyoux, A., Meuwis, M. A., Feller, G., and Gerday, C. (2002) Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 917-925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gianese, G., Bossa, F., and Pascarella, S. (2002) Proteins 47 236-249 [DOI] [PubMed] [Google Scholar]
- 11.Leiros, H. K., Pey, A. L., Innselset, M., Moe, E., Leiros, I., Steen, I. H., and Martinez, A. (2007) J. Biol. Chem. 282 21973-21986 [DOI] [PubMed] [Google Scholar]
- 12.Fedoy, A. E., Yang, N., Martinez, A., Leiros, H. K., and Steen, I. H. (2007) J. Mol. Biol. 372 130-149 [DOI] [PubMed] [Google Scholar]
- 13.Svingor, A., Kardos, J., Hajdu, I., Nemeth, A., and Zavodszky, P. (2001) J. Biol. Chem. 276 28121-28125 [DOI] [PubMed] [Google Scholar]
- 14.Wintrode, P. L., Miyazaki, K., and Arnold, F. H. (2000) J. Biol. Chem. 275 31635-31640 [DOI] [PubMed] [Google Scholar]
- 15.Miyazaki, K., Wintrode, P. L., Grayling, R. A., Rubingh, D. N., and Arnold, F. H. (2000) J. Mol. Biol. 297 1015-1026 [DOI] [PubMed] [Google Scholar]
- 16.Barrett, A. J., Rawlings, N. D., and Woessner, J. F. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., pp. 374-412, Elsevier Academic Press, London
- 17.Adekoya, O. A., Helland, R., Willassen, N. P., and Sylte, I. (2006) Proteins 62 435-449 [DOI] [PubMed] [Google Scholar]
- 18.Chen, X. L., Zhang, Y. Z., Gao, P. J., and Luan, X. W. (2003) Mar. Biol. 143 989-993 [Google Scholar]
- 19.Liu, Y.-G., and Whitter, R. F. (1995) Genomics 25 674-681 [DOI] [PubMed] [Google Scholar]
- 20.Feder, J. (1968) Biochem. Biophys. Res. Commun. 32 326-332 [DOI] [PubMed] [Google Scholar]
- 21.Marchler-Bauer, A., Anderson, J. B., Derbyshire, M. K., Weese-Scott, C., Gonzales, N. R., Gwadz, M., Hao, L., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z., Krylov, D., Lanczycki, C. J., Liebert, C. A., Liu, C., Lu, F., Lu, S., Marchler, G. H., Mullokandov, M., Song, J. S., Thanki, N., Yamashita, R. A., Yin, J. J., Zhang, D., and Bryant, S. H. (2007) Nucleic Acids Res. 35 D237-D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, N. D., Morton, F. R., Kok, C. Y., Kong, J., and Barrett, A. J. (2008) Nucleic Acids Res. 36 D320-D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emanuelsson, O., Brunak, S., von, H. G., and Nielsen, H. (2007) Nat. Protoc. 2 953-971 [DOI] [PubMed] [Google Scholar]
- 24.Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007) Bioinformatics 23 2947-2948 [DOI] [PubMed] [Google Scholar]
- 25.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41 95-98 [Google Scholar]
- 26.Thayer, M. M., Flaherty, K. M., and McKay, D. B. (1991) J. Biol. Chem. 266 2864-2871 [DOI] [PubMed] [Google Scholar]
- 27.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000) Nucleic Acids Res. 28 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
- 29.Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kale, L., and Schulten, K. (2005) J. Comput. Chem. 26 1781-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph-McCarthy, D., Kuchnir, L., Kuczera, K., Lau, F. T. K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D. T., Prodhom, B., Reiher, W. E., Roux, B., Schlenkrich, M., Smith, J. C., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 31.Guex, N., and Peitsch, M. C. (1997) Electrophoresis 18 2714-2723 [DOI] [PubMed] [Google Scholar]
- 32.Humphrey, W., Dalke, A., and Schulten, K. (1996) J. Mol. Graph. 14 33-38 [DOI] [PubMed] [Google Scholar]
- 33.McDonald, I. K., and Thornton, J. M. (1994) J. Mol. Biol. 238 777-793 [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. O., Kato, J., Nakashima, K., Kuroda, A., Ikeda, T., Takiguchi, N., and Ohtake, H. (2002) Biosci. Biotechnol. Biochem. 66 1366-1369 [DOI] [PubMed] [Google Scholar]
- 35.Tronrud, D. E., Monzingo, A. F., and Matthews, B. W. (1986) Eur. J. Biochem. 157 261-268 [DOI] [PubMed] [Google Scholar]
- 36.Banbula, A., Potempa, J., Travis, J., Fernandez-Catalan, C., Mann, K., Huber, R., Bode, W., and Medrano, F. (1998) Structure. 6 1185-1193 [DOI] [PubMed] [Google Scholar]
- 37.D'Amico, S., Marx, J. C., Gerday, C., and Feller, G. (2003) J. Biol. Chem. 278 7891-7896 [DOI] [PubMed] [Google Scholar]
- 38.Xu, Y., Feller, G., Gerday, C., and Glansdorff, N. (2003) J. Bacteriol. 185 2161-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins, T., Meuwis, M. A., Gerday, C., and Feller, G. (2003) J. Mol. Biol. 328 419-428 [DOI] [PubMed] [Google Scholar]
- 40.Georlette, D., Damien, B., Blaise, V., Depiereux, E., Uversky, V. N., Gerday, C., and Feller, G. (2003) J. Biol. Chem. 278 37015-37023 [DOI] [PubMed] [Google Scholar]
- 41.Bae, E., and Phillips, G. N., Jr. (2004) J. Biol. Chem. 279 28202-28208 [DOI] [PubMed] [Google Scholar]
- 42.Chessa, J. P., Petrescu, I., Bentahir, M., Van, B. J., and Gerday, C. (2000) Biochim. Biophys. Acta 1479 265-274 [DOI] [PubMed] [Google Scholar]
- 43.Aghajari, N., Van, P. F., Villeret, V., Chessa, J. P., Gerday, C., Haser, R., and Van, B. J. (2003) Proteins 50 636-647 [DOI] [PubMed] [Google Scholar]
- 44.Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999) Bioinformatics 15 305-308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.