Abstract
Glutaredoxins (Grxs) are efficient catalysts for the reduction of mixed disulfides in glutathionylated proteins, using glutathione or thioredoxin reductases for their regeneration. Using GFP fusion, we have shown that poplar GrxS12, which possesses a monothiol 28WCSYS32 active site, is localized in chloroplasts. In the presence of reduced glutathione, the recombinant protein is able to reduce in vitro substrates, such as hydroxyethyldisulfide and dehydroascorbate, and to regenerate the glutathionylated glyceraldehyde-3-phosphate dehydrogenase. Although the protein possesses two conserved cysteines, it is functioning through a monothiol mechanism, the conserved C terminus cysteine (Cys87) being dispensable, since the C87S variant is fully active in all activity assays. Biochemical and crystallographic studies revealed that Cys87 exhibits a certain reactivity, since its pKa is around 5.6. Coupled with thiol titration, fluorescence, and mass spectrometry analyses, the resolution of poplar GrxS12 x-ray crystal structure shows that the only oxidation state is a glutathionylated derivative of the active site cysteine (Cys29) and that the enzyme does not form inter- or intramolecular disulfides. Contrary to some plant Grxs, GrxS12 does not incorporate an iron-sulfur cluster in its wild-type form, but when the active site is mutated into YCSYS, it binds a [2Fe-2S] cluster, indicating that the single Trp residue prevents this incorporation.
Glutaredoxins (Grxs)5 are GSH- or thioredoxin reductase-dependent oxidoreductases involved in the maintenance of cellular redox homeostasis. When Grxs are recycled by GSH, the GSSG formed is in turn reduced by NADPH and glutathione reductase (GR), forming the GSH/Grx reducing system. The first Grxs characterized usually contained the active site motif Cys-Pro-Tyr-Cys, the second active site cysteine being generally not essential for Grx activity (1, 2). It has been shown, however, to be required for a few reactions, such as the reduction of some low molecular weight disulfides or of disulfide bonds in E. coli ribonucleotide reductase and phosphoadenylyl-sulfate reductase (1, 3, 4). In most cases, Grxs rather reduce specifically protein-glutathione adducts via two distinct mechanisms. The monothiol mechanism requires only the more N terminus active site cysteine together with two glutathione molecules, and the dithiol mechanism requires either the two active site cysteines or the N terminus active site cysteine and a conserved extra active site C terminus cysteine. Both types of disulfides formed on Grx are reduced in vitro by GSH or thioredoxin reductases (1, 4–7). In comparison, thioredoxins (Trxs) efficiently reduce protein disulfides but have low or no activity with mixed disulfides (7, 8). Sharing high structural and functional homologies with Trx, Grxs display a common Trx fold (9). The CXXC motif in Trx and Grx is located in a partially exposed surface loop downstream of a β-strand and at the N terminus of an α-helix (9). Interestingly, some Trxs and Grxs only contain the N terminus cysteine of the CXXC motif, the second cysteine being very often replaced by a serine. In plants, this variation of active site sequence exists in approximately half of the 30 Grxs existing (10). Grxs were initially categorized, based on the active site sequence, into two groups, a dithiol (CPY/FC motif) and a monothiol (CGFS motif) subgroup (11). Nevertheless, the increasing number of Grxs with primary structures that deviate from these standard motifs has led to the proposal of a more appropriate and thorough classification of plant Grxs (10, 12, 13). Three subclasses have been defined based on their active site structures. Subclass I includes proteins with CXX(C/S) active sites other than CGFS, subclass II contains exclusively Grxs with a CGFS motif, and subclass III, which is specific to land plants, corresponds to Grxs with a peculiar CCXX active site.
Many structures of dithiol Grxs from human (Protein Data Bank codes 1JHB, 2CQ9, 2FLS, 2HT9, 1B4Q), pig (Protein Data Bank code 1KTE), mouse (PDB codes 1T1V, 1WJK), plant (Protein Data Bank codes 1Z7P, 1Z7R, 2E7P), yeast (Protein Data Bank code 2JAC), bacteria (Protein Data Bank codes 1EGR, 1EGO, 1FOV, 1GRX, 1G7O, 1H75, 2AYT, 1J08, 1R7H, 2YWM), virus (Protein Data Bank codes 2HZE, 2HZF), or T4 bacteriophage (Protein Data Bank codes 1DE1, 1DE2, 1AAZ, 1ABA, 1QFN, 3GRX) have been solved by NMR spectroscopy or x-ray crystallography both in the oxidized and reduced forms. Some of these Grx structures contain either a low molecular weight substrate or an interacting peptide partner. Only one NMR structure (Escherichia coli Grx4, PDB code 1YKA) of monothiol Grx is available despite the abundance of genes reported.
In this study, we have investigated the structure-function relationship of a monothiol Grx isoform (GrxS12) found in Populus tremula × tremuloides, which possesses an unusual 28WCSYS32 active site sequence, unique to plants. Phylogenetic analyses indicate that GrxS12 belongs to Grx subclass I, along with classical dithiol Grxs. In addition to the active site cysteine, there is an additional C terminus cysteine in position 87, which is present in many dithiol or monothiol Grxs. Its role remains obscure, although it has been demonstrated that it can serve as a resolving cysteine of the glutathionylated catalytic cysteine in a few organisms (5–7). Biochemical and enzymatic studies of mutated proteins together with the resolution of liganded GrxS12 structures allowed us (i) to identify the GSH binding site of this enzyme, (ii) to investigate the role of the C terminus cysteine in the catalytic mechanism of GrxS12, and (iii) to understand why an iron-sulfur cluster is not present in GrxS12 despite its apparently favorable CSYS motif.
EXPERIMENTAL PROCEDURES
Materials—NAP-5 columns were purchased from GE Healthcare. Hydroxyethyldisulfide (HED) and 5,5′-dithiobis-2-nitrobenzoic acid were from Aldrich and Pierce, respectively. All other reagents were from Sigma.
Cloning and Construction of GrxS12 Mutants by Site-directed Mutagenesis—The open reading frame sequence encoding poplar GrxS12 was amplified from a P. tremula × tremuloides leaf cDNA library using GrxS12 forward and reverse primers (supplemental Table 1) and cloned into the NcoI and BamHI restriction sites (underlined in the primers) of pET3d (Novagen). The sequence amplified encodes a protein deprived of the first 74 amino acids corresponding to the putative targeting sequence and in which a methionine and an alanine have been added during cloning. The protein starts thus with the N terminus sequence 1MASFGSRL8 and ends with 98AKKSQG113 at the C terminus (see supplemental Fig. 1). Using two complementary mutagenic primers, the two cysteines of GrxS12 were individually substituted into serines, the Trp in position 28 was mutated into Tyr, and the active site was also entirely modified from WCSYS into YCGYC. The primers are listed in supplemental Table 1. The mutated proteins are called GrxS12 W28Y, C29S, and C87S and GrxS12 YCGYC.
Expression and Purification of the Recombinant Proteins—For protein production, the E. coli BL21(DE3) strain, containing the pSBET plasmid, was co-transformed with the different recombinant plasmids (14). Cultures were successively amplified up to 2.4 l in LB medium supplemented with ampicillin and kanamycin at 37 °C. Protein expression was induced at exponential phase by adding 100 μm isopropyl β-d-thiogalactopyranoside for 4 h at 37 °C.
The cultures were then centrifuged for 15 min at 4400 × g. The pellets were resuspended in 30 ml of TE NaCl (30 mm Tris-HCl, pH 8.0, 1 mm EDTA, 200 mm NaCl) buffer, and the suspension was conserved at -20 °C.
Cell lysis was performed by sonication (3 × 1 min with intervals of 1 min), and the soluble and insoluble fractions were separated by centrifugation for 30 min at 27,000 × g. The soluble part was then fractionated with ammonium sulfate in two steps, and the protein fraction precipitating between 40 and 80% of the saturation contained the recombinant protein, as estimated by 15% SDS-PAGE. The protein was purified by size exclusion chromatography after loading on an ACA44 (5 × 75-cm) column equilibrated in TE NaCl buffer. The fractions containing the protein were pooled, dialyzed by ultrafiltration to remove NaCl, and loaded onto a DEAE-cellulose column (Sigma) in TE (30 mm Tris-HCl, pH 8.0, 1 mm EDTA) buffer. All of the proteins (GrxS12 WT, W28Y, C29S, C87S, and YCGYC) passed through the DEAE column and were subsequently loaded onto a carboxymethylcellulose column (Sigma) in TE buffer. The proteins were eluted using a 0–0.4 m NaCl gradient. Finally, the fractions of interest were pooled, dialyzed, concentrated by ultrafiltration under nitrogen pressure (YM10 membrane; Amicon), and stored in TE buffer at -20 °C. Purity was checked by SDS-PAGE. Protein concentrations were determined spectrophotometrically using a molar extinction coefficient at 280 nm of 9970 m-1 cm-1 for the GrxS12 WT, C29S, and C87S and 5960 m-1 cm-1 for GrxS12 W28Y and GrxS12 YCGYC.
In Vivo Subcellular Localization—A fragment of 285 nucleotides coding for the 95 first amino acids of GrxS12 was cloned in the 5′-part of the GFP coding sequence under the control of a double 35S promoter into the plasmid pCK-GFP3 using GrxS12 pCK forward and reverse primers (supplemental Table 1). Nicotiana benthamiana epidermal leaf cells were transfected by bombardment of the abaxial side of young leaves with tungsten particles coated with plasmid DNA. Images were obtained 18 h later with a Zeiss LSM510 confocal microscope. Stomata cells were preferentially imaged, because of their small size and because typically only one of the two guard cells is transfected and expresses the GFP construction, whereas the untransfected cell serves the role of internal negative control. Chloroplasts were visualized by the natural fluorescence of chlorophyll.
Reduction and Oxidation of Wild-type and Mutated GrxS12—The proteins (50–100 μm) were reduced by 10 mm DTT for 1 h at 25 °C, followed by desalting on NAP-5 column pre-equilibrated with 30 mm Tris-HCl, pH 7.9. Oxidized Grxs were prepared by incubation of prereduced with 10 mm oxidized DTT or 1–5 mm GSSG for 1–2 h at 25 °C. GSSG-oxidized GrxS12 (WT or mutants) were desalted as above and treated by 10 mm reduced DTT or by 2 mm GSH in the presence of 6 μg/ml yeast glutathione reductase and 0.5 mm NADPH.
Mass Spectrometry Analysis—Reduced and oxidized GrxS12 WT and C87S and trypsin-cleaved proteins were analyzed by MALDI-TOF MS as described in Ref. 15.
Fluorescence Properties of Wild-type and Mutated GrxS12—The fluorescence characteristics of GrxS12 and GrxS12 C87S in the reduced and oxidized forms were recorded with a spectrofluorometer (Cary Eclipse; VARIAN) in TE buffer with a 10 μm concentration of each protein.
Determination of Free Thiol Groups—The number of free thiol groups in untreated, reduced, or oxidized proteins was determined spectrophotometrically with 5,5′-dithiobis-2-nitrobenzoic acid, as described in Ref. 7.
pKa Determination of GrxS12 Sulfhydryls with (2-pyridyl)dithiobimane (PDT-bimane)—The reaction of PDT-bimane with cysteine forms pyridine-2-thione, which has a maximum absorption wavelength of 343 nm (16). The stock solution of PDT-bimane was made in DMSO, and the concentration was determined using the absorbance extinction coefficient at 380 nm, ε380 = 5000 m-1 cm-1 in ethanol. Reactions were started by the addition of PDT-bimane to a final concentration of 25 μm into a cuvette containing 10 μm reduced proteins in 500 μl of sodium citrate or phosphate buffer ranging from pH 3.0 to 8.0 and rapidly mixed, and the absorbance at 343 nm was recorded over 120 min with a Varian Cary 50 spectrophotometer. Absorbance data were fitted directly to the Michaelis-Menten equation with the GraphPad Prism 5 program and the t½ (the time to reach half-maximal reactivity as monitored by half-maximal release of pyridyl-2-thione) at each pH was determined. Those values were plotted against pH using sigmoidal curve fit and GraphPad Prism 5 (GraphPad software).
Activity Measurements—The activity measurements of WT or mutant GrxS12 in the HED assay or for reduction of DHA or glutathionylated GAPDH were performed as described in Zaffagnini et al. (7).
Crystallization, Data Collection, Structure Determination, and Crystallographic Refinement—The complex of poplar GrxS12 with glutathione (GrxS12·GSH) was directly obtained using the purified recombinant protein. The complex of GrxS12 with both GSH and β-mercaptoethanol (βMSH) (GrxS12·GSH·βMSH) was prepared after purification steps by the addition of 2 mm GSH and 10 mm HED to the purified recombinant protein for 2 h at room temperature prior to extensive dialyses against TE buffer to remove unbound GSH, HED, or βMSH.
Crystals were grown at 20 °C by the microbatch under oil (paraffin) method. Optimal crystallization conditions were screened based on the sparse matrix crystallization approach. The protein at an initial concentration of 10–15 mg ml-1 in TE buffer was mixed with similar precipitant solutions (for each protein) in a 1:1 ratio. The GrxS12·GSH crystals were obtained by using 0.1 m Na-HEPES (pH 7.5) and 20% polyethylene glycol 8000 solution (JBS 5-B4), whereas GrxS12·GSH·βMSH crystallized in 0.1 m Na-HEPES (pH 7.5) and 25% polyethylene glycol 1000 solution (JBS 1-C3). Grx crystals grew rapidly within 2 days.
X-ray diffraction data were collected from single crystals flash-cooled in a nitrogen stream at 100 K. Data of both complexes, GrxS12·GSH and GrxS12·GSH·βMSH, were collected on a MAR165 CCD detector at beamlines X11 and X13 (DESY/EMBL, Hamburg, Germany), respectively. All crystallographic data were indexed, processed, and scaled with HKL-2000 (17). Data collection and refinement statistics are summarized in Table 1. The crystal structure of GrxS12·GSH was determined by molecular replacement with MOLREP (18, 19), using human Grx2 (Protein Data Bank code 2FLS) as a model (20). The resulting solution coordinates were used for automatic model building using the program ARP/wARP (21). After 75 cycles of autobuilding, 99% of the model (786 atoms were refined) was built automatically. Hence, the initial model of GrxS12·GSH comprises 100 residues with a connectivity index of 0.96 and R factor of 19.5%. This structure was then refined using REFMAC version 5.4 (18, 22) interspersed with manual inspection using COOT (23). A GSH molecule was added almost toward the end of the refinement of the GrxS12·GSH model. The coordinates of GrxS12·GSH were then used to solve the structure of GrxS12·GSH·βMSH, also by molecular replacement. The structure refinement of the latter was done as for the template. Positions of water molecules were identified with ARP/wARP and were checked manually. The validation of both crystal structures was performed with PROCHECK (24). All figures were prepared with PyMOL (25). Structure superimpositions were performed using the LSQMAN program from the DEJAVU package (26) or Lsqkab (superpose) program of the CCP4 package.
TABLE 1.
Data collection and refinement statistics for GrxS12·GSH and GrxS12·GSH·βMSH crystals
| Data set | GrxS12·GSH | GrxS12·GSH·βMSH |
|---|---|---|
| Data collection and processing statistics | ||
| Data collection site | X11 DESY/EMBL-Hamburg | X13 DESY/EMBL-Hamburg |
| Wavelength (Å) | 0.8150 | 0.8063 |
| Space group | P212121 | P212121 |
| Unit cell dimensions (Å) (a, b, c) | 39.03, 47.27, 55.62 | 38.83, 46.82, 55.36 |
| Asymmetric unit | 1 subunit | 1 subunit |
| Resolution range (Å)a | 50.00–1.70 (1.73–1.70) | 50.00–1.80 (1.86–1.80) |
| Redundancya | 4.49 (4.16) | 6.69 (6.85) |
| Completeness (%)a | 99.7 (95.0) | 99.6 (100.0) |
| I/σIa | 13.37 (2.26) | 23.26 (16.95) |
| Rmergea,b | 0.045 (0.239) | 0.048 (0.107) |
| Refinement statistics | ||
| Resolution range (Å) | 36.0–1.70 | 30.0–1.80 |
| Reflections used | 11,234 | 9301 |
| Rcrystc (Rfree)d | 19.28 (23.95) | 16.78 (21.45) |
| Protein/waters/GSH/HED | 106 residues/186/1/0 | 106 residues/186/1/0 |
| Mean B factor (Å2) | ||
| Main chain | 16.53 | 12.04 |
| Side chain | 23.33 | 14.79 |
| Water | 32.57 | 27.88 |
| Ligand | 15.31 | 17.00 |
| All | 20.52 | 16.24 |
| r.m.s. deviation from ideal geometry | ||
| Bond lengths (Å) | 0.012 | 0.011 |
| Bond angles (degrees) | 1.4 | 1.4 |
| Dihedral angles (degrees) | 23.5 | 23.7 |
| Improper angles (degrees) | 1.61 | 1.75 |
| Ramachandran plot | ||
| Residues in most favored regions (%) | 96.7 | 95.6 |
| Residues in additionally allowed regions (%) | 3.3 | 4.4 |
| Residues in generously allowed regions (%) | 0.0 | 0.0 |
Rcryst = Σ|Fo – Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively
The Rfree value was calculated from 5% of all data that were not used in the refinement
Rcryst = Σ|Fo – Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively
Rfree is as for Rcryst but calculated for a test set comprising reflections not used in the refinement (5%)
RESULTS
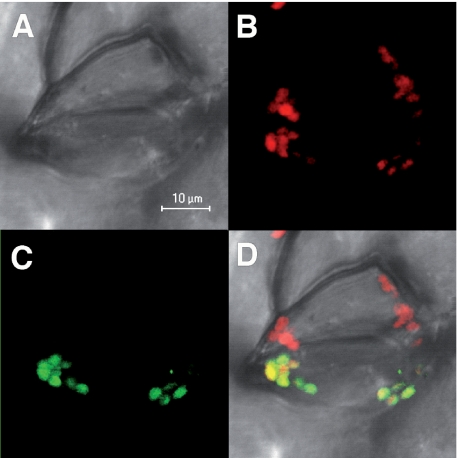
Subcellular Localization of GrxS12—The genome analysis of Populus trichocarpa suggests that at least four Grxs should be located in the chloroplast. Two Grxs of subgroup II, possessing a CGFS active site and called GrxS14 and GrxS16, have been experimentally shown to be located in plastids and would be involved in iron-sulfur biogenesis, since they incorporate an iron-sulfur cluster that can be transferred very quickly to apoferredoxin (27). The GrxS12 from P. tremula × tremuloides is a protein of 185 amino acids with a predicted chloroplastic N terminus-targeting sequence. In order to experimentally confirm its localization, the sequence coding the first 95 amino acids, including thus the putative targeting sequence and the first α-helix and β-strand, was fused to the GFP coding sequence and used to bombard tobacco leaf cells. As shown in Fig. 1, the fluorescence associated with this construction, transfected into one of the two guard cells of a stomate, strictly co-localizes with the autofluorescence of chlorophyll, indicating that the protein is indeed chloroplastic.
FIGURE 1.
GFP localization of GrxS12 in plant guard cells. A, cells under visible light; B, autofluorescence of chlorophyll (red); C, fluorescence of the GFP construction; D, merged images.
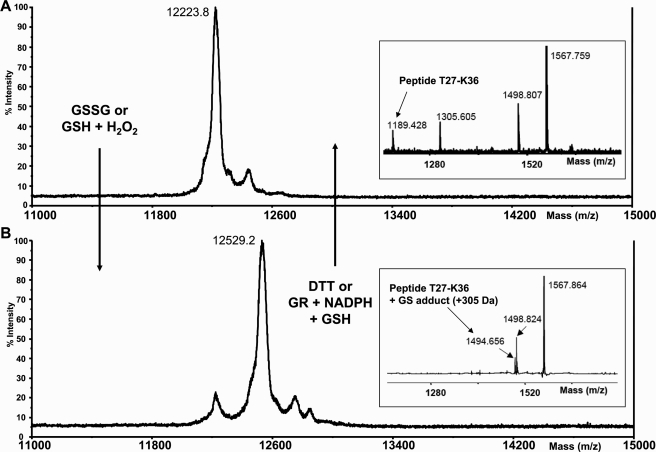
Determination of GrxS12 Redox States by Fluorescence, Thiol Titration, and Mass Spectrometry—The recombinant protein produced in E. coli is a protein containing 113 residues devoid of the first 74 N terminus residues (which represent the transit peptide) and in which a methionine and an alanine have been added in the new N terminus end. The predicted molecular mass and pI are 12,360 Da and 8.56, respectively. A MALDI-TOF analysis of the purified GrxS12 WT revealed two protein peaks with molecular masses of 12,229.1 and 12,535.7 Da (data not shown). The first one is consistent with a protein where the methionine is cleaved, which is not surprising, since the second residue is an alanine, and the second peak is consistent with a glutathionylated protein also deprived of the methionine (306.6 Da mass increase compared with peak 1). There are two conserved cysteine residues in GrxS12, the active site cysteine in position 29 and a C terminus cysteine in position 87. In some CGFS Grxs, the latter cysteine can form an intramolecular disulfide with the catalytic cysteine (5–7). In order to check the possibility that Cys87 acts as a recycling cysteine in GrxS12, we measured the number of free thiol groups under reducing and oxidizing conditions in GrxS12 WT and C87S (Table 2). After purification and in the absence of any reducing or oxidizing treatment (native form), GrxS12 WT possesses about one free thiol group, whereas there is no free thiol in GrxS12 C87S. A reduction by DTTred led to the expected number of free thiols, two for the WT and one for the C87S mutant. After a prereduction step of WT GrxS12, a subsequent oxidation by DTTox over a long period (2 h) did not change these values, indicating that no intra- or intermolecular disulfide bridges can be formed. Such disulfides have not been observed on nonreducing SDS-polyacrylamide gels either (data not shown). On the contrary, subsequent oxidation using 1 mm GSSG gave the same values as those found originally in the untreated proteins, one free thiol group for GrxS12 WT but no free thiol in GrxS12 C87S. Together, these results suggest the presence of a glutathione adduct on the catalytic cysteine (Cys29), whereas the second cysteine (Cys87) is not modified. In order to confirm these results, we analyzed by MALDI-TOF mass spectrometry prereduced GrxS12 before or after glutathionylation treatments in the presence of GSSG or GSH and H2O2 (Fig. 2). After treatment, the mass of the protein increased by ∼305 Da, a feature consistent with the formation of one glutathione adduct. This increase was reversed by treatment with either DTTred or GSH/GR/NADPH. A similar behavior was observed for WT or C87S GrxS12, indicating that Cys29 is indeed the residue modified by glutathionylation. This was further confirmed by tryptic digestion and peptide mass fingerprinting of reduced or glutathionylated GrxS12. Indeed, the peptide (Thr27–Lys36) containing Cys29 is shifted by 305 Da in the glutathionylated protein. All of these results indicate that Cys29 is undergoing glutathionylation, whereas Cys87 is not glutathionylated in any of the conditions tested.
TABLE 2.
Number of free thiols in WT and C87S GrxS12 under various redox conditions The untreated column is indicative of the protein thiol content measured after purification. GrxS12 was reduced by DTTred and subsequently oxidized by DTTox or GSSG. After each treatment (DTTred, DTTox, and GSSG), samples were desalted. The thiol content per protein was quantified by 5,5′-dithiobis(nitrobenzoic acid). Data are represented as mean ± S.D. (n = 4). ND, not determined.
| Untreated | DTTred treatment | DTTox treatment | GSSG treatment | |
|---|---|---|---|---|
| GrxS12 | 0.821 ± 0.078 | 1.98 ± 0.269 | 1.9 ± 0.1 | 0.808 ± 0.029 |
| GrxS12 C87S | 0.115 ± 0.017 | 1.056 ± 0.125 | ND | 0.132 ± 0.053 |
FIGURE 2.
MALDI-TOF mass spectrometry analysis of reduced or glutathionylated GrxS12. MALDI-TOF spectra of whole protein or after tryptic digestion (insets) were determined for reduced GrxS12 before (A) or after (B) glutathionylation treatments (5 mm GSSG or 0.1 mm H2O2 plus 0.5 mm GSH for 1 h). The shifted peptide (Thr27–Lys36) is indicated. The 305 Da shift after glutathionylation treatment could be reversed by treatments with 10 mm DTT or with 2 mm GSH in the presence of 6 μg/ml yeast glutathione reductase and 0.5 mm NADPH. Similar results were obtained with the WT and C87S GrxS12.
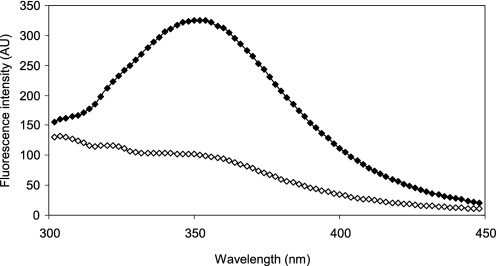
GrxS12 contains a single tryptophan adjacent to the active site. Given that proximity, we have investigated whether the intrinsic fluorescence of GrxS12 could change under reducing (GSH or DTTred) or oxidizing (GSSG or DTTox) conditions. Reduced GrxS12 displays an emission spectrum with a maximum at 350 nm characteristic of a fluorescence signal strongly dominated by Trp (Fig. 3). Adding 1 mm GSSG to reduced GrxS12 immediately led to the disappearance of the fluorescence signal. The addition of DTTred restored the initial spectrum (data not shown). Similar results were obtained with GrxS12 C87S, indicating that the tryptophan environment strongly changed after glutathionylation of the first active site cysteine. In accordance with thiol titrations, the addition of DTTox did not change the GrxS12 fluorescence spectrum.
FIGURE 3.
Fluorescence spectra of poplar GrxS12 under different redox states. Emission spectra of reduced (♦) and oxidized (⋄) GrxS12 (excitation at 290 nm) were recorded with 10 μm protein at 25 °C in TE, pH 8.0, buffer. The reduced GrxS12 was obtained by a pretreatment of the protein with 10 mm DTT. GrxS12 was oxidized using 1 mm GSSG after DTT prereduction. In each case, residual compounds were removed by gel filtration.
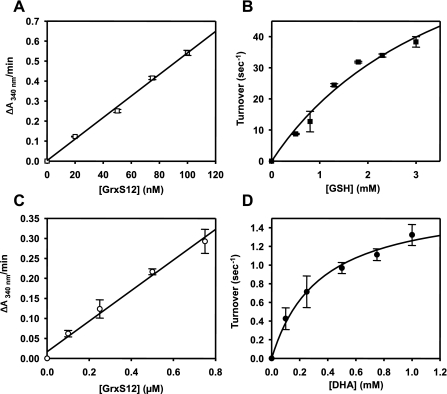
Dehydroascorbate Reductase Activity, HED Assay, and Reduction of Glutathionylated A4-GAPDH—GrxS12 was found to be active in the two classical GRX assays: the reduction of DHA and HED assays. The activity of GrxS12 in these two assays displayed a linear relationship with increasing protein concentrations in the 0–0.75 μm and 0–100 nm ranges, respectively (Fig. 4, A and C). The kinetic analyses revealed catalytic efficiency values (kcat/Km) comparable to those reported for other GrxS to subclass I. (Fig. 4, B and D, and Table 3).
FIGURE 4.
HED and DHA reductase activity of poplar GrxS12. A and C, linear dependence of HED (A) and DHA reductase (C) activity on GrxS12 concentration expressed as ΔA340/min. The data are represented as mean ± S.D. B and D, variations of the apparent turnover number during an HED assay catalyzed by 50 nm GrxS12 in the presence of GSH concentrations ranging from 0.5 to 3 mm (B) and during DHA reduction catalyzed by 0.25 μm GrxS12 in the presence of varying DHA concentrations ranging from 0.1 to 1 mm (D). Turnover represents mol of NADPH oxidized/s by 1 mol of GrxS12. Activity was calculated after subtracting the spontaneous reduction rate observed in the absence of GrxS12. Three separate experiments were performed, and the data are represented as mean ± S.D. The best fit was obtained using the Michaelis-Menten equation.
TABLE 3.
Kinetic parameters of GrxS12 in HED and DHA activity assays The apparent Km value for GSH in the HED assay was determined using a GSH concentration range of 0.5–3.0 mm in the presence of 0.7 mm HED. The apparent Km value for DHA was determined using a concentration range of 0.1–1 mm in the presence of 2 mm GSH. The apparent Km and apparent turnover values (kcat) were calculated by nonlinear regression using the Michaelis-Menten equation. Data are represented as mean ± S.D. (n = 3).
| Km | kcat | kcat/Km | |
|---|---|---|---|
| mm | s–1 | m–1 s–1 | |
| DHA | 0.38 ± 0.19 | 1.73 ± 0.19 | 4.6 × 103 |
| GSH | 3.99 ± 0.48 | 92.78 ± 5.32 | 2.33 × 104 |
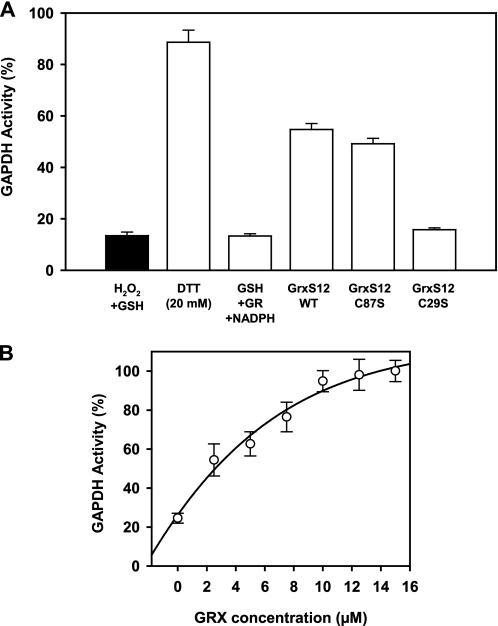
We have recently demonstrated that A4-GAPDH activity is reversibly inhibited by glutathionylation (28). This chloroplastic protein, which participates in the Calvin cycle, can be used as a more physiological substrate to test deglutathionylation activity. In order to obtain glutathionylated A4-GAPDH, the protein was incubated in the presence of 0.1 mm H2O2 plus 0.5 mm GSH. Subsequently, the ability of WT and monocysteinic variants of GrxS12 to recover GAPDH activity was determined (Fig. 5A). Using a GSH/GR/NADPH reduction system, WT and C87S GrxS12 allowed recovery of ∼50% of GAPDH initial activity, whereas no reactivation was observed with the C29S mutant. The recovery of GAPDH activity obtained in the presence of GrxS12 did not reach the levels obtained in the presence of DTT (∼90%). This can be explained by the fact that H2O2 plus GSH leads to glutathionylation but also to primary oxidation of the catalytic cysteine of GAPDH to sulfenic acid, the latter being more efficiently reduced by 20 mm DTT than by the Grx system (28). The kinetic parameters of GAPDH deglutathionylation by GrxS12 were determined (Fig. 5B). With an S0.5 of 3.7 ± 0.7 μm and a t0.5 around 2–3 min, GrxS12 exhibits a catalytic efficiency comparable with other Grxs from Chlamydomonas reinhardtii (7).
FIGURE 5.
Reactivation of glutathionylated A4-GAPDH. A4-GAPDH was inactivated by incubation with 0.1 mm H2O2 in the presence of 0.5 mm GSH for 15 min at 25 °C and subsequently treated with 35 μm 1,3-bisphosphoglycerate. A, the reactivation assays were performed under the following conditions: (i) 20 mm DTT, (ii) 2 mm GSH in the presence of 6 μg/ml yeast glutathione reductase and 0.2 mm NADPH alone or (iii) in the presence of 10 μm GrxS12 WT, C29S, or C87S. The NADPH-dependent activity was determined before (black bar) and after the different reactivation treatments (white bars). B, reactivation of glutathionylated A4-GAPDH with 2 mm GSH, 6 μg/ml yeast glutathione reductase, 0.2 mm NADPH in the presence of varying concentrations of GrxS12 ranging from 2.5 to 15 μm. Activities are represented as a percentage of the initial activity measured before the inactivation treatment. The data are shown as mean ± S.D.
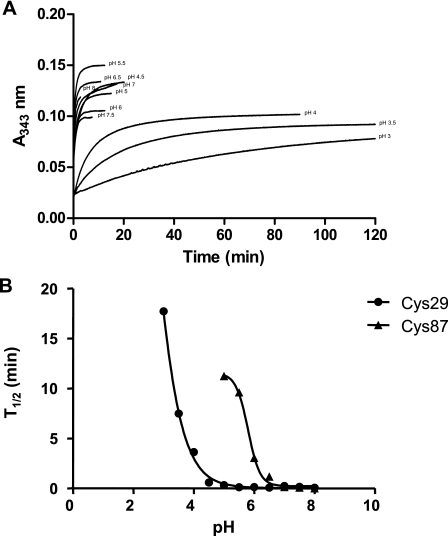
In all of these assays (DHA, HED, and GAPDH assays), GrxS12 C87S appeared as efficient as the WT protein. These results indicate that in these assays, GrxS12 activity relies on a monothiol mechanism involving only Cys29. Nevertheless, at higher concentrations (10 times more than the WT), the GrxS12 C29S mutant exhibited an activity slightly above the background activity in the HED assay (data not shown). In order to know whether Cys87 is reactive in this mutant, the pKa of both cysteines was measured using the thiol-cleavable fluorophore PDT-bimane (Fig. 6). From the plot of the t½ of half-maximal release of pyridyl-2-thiolate against pH, the pKa of Cys29 is ∼2.8, a value in the range of those determined for mammalian Grxs, and the pKa of Cys87 is ∼5.6 (29). Thus, the C terminus Cys87 is in the thiolate form at the pH of the activity assays and thus at physiological pH. In addition, since this cysteine is surface exposed (see below), it should thus be able to perform a nucleophilic attack on an accessible disulfide.
FIGURE 6.
pKa determination of GrxS12 sulfhydryl groups. A, reaction of GrxS12 C87S with PDT-bimane was monitored at 343 nm at pH values ranging from 3.0 to 8.0. The increase at 343 nm results from the release of pyridyl-2-thione from PDT-bimane. Each curve was fit to the Michaelis-Menten equation. B, t½ for the reactions of PDT-bimane with GrxS12 C29S and C87S were plotted as a function of pH, and the results were fitted to a sigmoidal curve. From this plot, sulfhydryl pKa values of 2.84 ± 0.08 and 5.63 ± 0.17 were determined for Cys29 and Cys87, respectively.
Structure of GrxS12 and Quality of the Models—The two liganded forms of the enzyme (GrxS12·GSH and GrxS12·GSH·βMSH) crystallized in the orthorhombic system with similar unit cell parameters and with a monomer in the asymmetric unit. Both structures have been determined at 1.70 and 1.80 Å resolutions for GrxS12·GSH and GrxS12·GSH·βMSH, respectively.
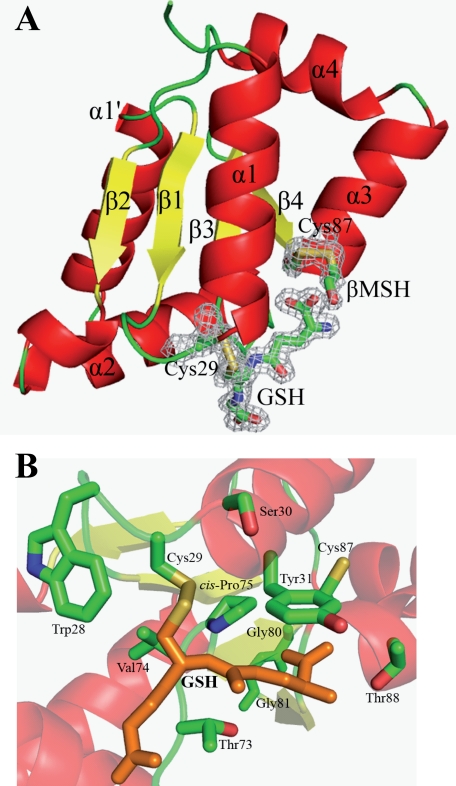
Overall structures are well defined in the 2Fo - Fc electron density map except for a few N terminus residues and some lateral chains that are solvent-exposed. The final models contain 106 amino acid residues, corresponding to residues from Gly5 to Lys110. A GSH molecule could be easily located in both models, at the active site covalently bound to Cys29. In the GrxS12·GSH·βMSH structure, a βMSH molecule can also be modeled covalently bound to the extra cysteine (Cys87) at the C terminus of the glutathionylated enzyme.
Superimposition between both liganded structures gives the r.m.s. deviation value of 0.162 Å (based on alignments of 106 Cα positions). These low r.m.s. deviation values suggest that binding of a βMSH molecule did not induce significant variations within these structures. Therefore, unless indicated otherwise, our discussion by default will be based on the structure of GrxS12·GSH because of its better structure resolution.
The overall subunit structure of GrxS12 shows no significant differences from other known Grxs. They all display a Trx fold with/without some secondary structure extensions. Poplar GrxS12 has an N terminus α helix (namely α1′ in this study; see Fig. 7 and supplemental Table 2) in addition to the Trx fold. This additional secondary structure is also present in most Grxs, including human Grx2 (Protein Data Bank code 2FLS), poplar GrxC1 (PDB code 2E7P), yeast Grx1P (Protein Data Bank code 2JAC), and E. coli monothiol Grx4 (Protein Data Bank code 1YKA) but not in T4 bacteriophage Grx (Protein Data Bank code 1ABA) nor in E. coli Grx1 (Protein Data Bank code 1GRX) and Grx3 (Protein Data Bank code 3GRX). A structural evaluation of the conserved residues between GrxS12 and its orthologs reveals that residues that constitute β1, β3, and β4 are highly conserved (supplemental Fig. 1).
FIGURE 7.
Schematic representations of GrxS12·GSH·βMSH structure (A), highlighting the active site of the protein in B. All α-helices are shown in red, whereas β-strands are in yellow and connecting loops are in green. Corresponding secondary structures are labeled. Both Cys29 and Cys87, GSH, and βMSH molecules are highlighted in sticks, with final 2Fo - Fc electron densities (1.2σ level) covering chosen residues and ligands for clarity. The catalytic Cys29 and the bound GSH molecule are highlighted in orange in B for clarity.
Active Site—The active site of the enzyme is hosting a GSH molecule that is covalently bound to Cys29 and noncovalently stabilized by three neighboring loops between β1-α1′, α2-β3, and β4-α3 (composed of 28WCSY31, 73TVP75, and 86GCT88; referred to as the GSH binding site in this paper; see Fig. 7). The boundary loops that support the glutathione binding are in their usual conformations as observed for other Grxs. We describe here, for the first time, the crystal structure of glutaredoxin of subclass I with the active site motif 29CSYS32 (in comparison with the classical CPY/FC motif for dithiol Grxs and CGFS for monothiol Grxs). Despite the replacement of a Pro present in the Grx counterparts possessing a CPY/FC motif (with φ and Ψ values of approximately -53° and -40°, respectively) by a Ser (Ser30) in the CSYS motif of GrxS12 (with φ and Ψ values of -60.47° and -40.18°, respectively), they adopt the same backbone conformation. GrxS12 also possesses a second Ser residue at the active site in which the Ser32 replaces the common C terminus active site Cys of dithiol Grxs. Ser32 is hydrogen-bonded (2.50 Å) to another nearby conserved serine residue (Ser25). The latter residue could play an important role in stabilizing the active site, since it is conserved in all Grxs with a CXXS active site motif.
It is interesting to note that residue Trp28 at the active site of GrxS12 (WCSYS) is well conserved in all plant GrxS12 orthologs and also in Arabidopsis GrxC5 (WCSYC) orthologs but not in other known Grxs. In the poplar GrxS12 structure, the position of Trp28 (χ1 value =+52.71°) is similar to most other classical thioredoxins (χ1 values between +40° and +55°), and its side chain covers an important part of the active site surface in the glutathionylated form, but fluorescence experiments indicate that it would adopt another conformation in the reduced form. The χ1 value of Trp28 is similar to those described for other residues at the corresponding position in other Grxs (+57.57° for Tyr29 of poplar GrxC1 and +67.64° for Ser36 of human Grx2).
In GrxS12, the GSH binding site constitutes a groove, mainly populated by polar residues, along which the backbone atoms of GSH are positioned (this is where GrxS12 accommodates the cysteinyl and glutamyl moieties of GSH; see Table 4). Situated at the loop between β4 and α3, the backbone amino groups of Cys87 and Thr88 stabilize the glutamyl group of GSH through hydrogen bonds. These interactions are strengthened by another hydrogen bond between the side chain of Thr88 and the N terminus of GSH (Table 4). The two residues immediately preceding Cys87, 85GG86 (GG kink), are strictly conserved in all GrxS12 orthologs and in the majority of the subclass I Grx members. This GG kink is in proximity of the cis-Pro75 (Fig. 7). cis-Pro75 plays an important structural role (30, 31), and the GG kink may be important in determining the backbone geometry of the following residues that belong to the α-helix 3. On the other hand, no interaction was observed between the GlyGSH and residues of GrxS12, thus suggesting that the GSH molecule is rather flexible on its C terminus side.
TABLE 4.
Hydrogen bonding interactions at the GrxS12-GSH interface
|
Interacted atoms
|
Distance
|
|||
|---|---|---|---|---|
| GrxS12 | GSH | GrxS12·GSH | GrxS12·GSH·βMSH | |
| Cysteine residue | Å | |||
| Val74 N | O2 | 2.89 | 2.80 | |
| Val74 O | N2 | 2.83 | 2.81 | |
| Glutamate residue | ||||
| Cys87 N | O11 | 2.83 | 2.83 | |
| Cys87 N | O12 | 3.21 | 3.21 | |
| Thr88 N | O12 | 2.80 | 2.87 | |
| Thr88 Oγ1 | N1 | 3.09 | 3.44 | |
In the model of GrxS12·GSH·βMSH, a βMSH molecule is covalently bound to the extra C terminus cysteine (Cys87) and stabilized by conserved Tyr31 and Glu34 of the glutathionylated enzyme. This structural observation suggests that Cys87 potentially possesses some reactivity. This is probably true, since in the classical Grx HED assay (see results above), residual activity is detected for the C29S variant. The distance between the Sγ atoms of both cysteine residues of the enzyme is ∼8 Å, both cysteines being separated by the active site Tyr31.
Comparison of the GSH Binding Site with Other Glutaredoxins—Of the available dithiol Grx structures, only those with a GSH present have been retrieved (see supplemental Fig. 2). In three cases, the second active site cysteine is mutated, and the GSH is covalently bound (human Grx1 (32), E. coli Grx1 (33), and E. coli Grx3 (34)); in one case, the GSH is not covalently bound to the wild type enzyme (human Grx2) (20). In these Grx structures, including the poplar GrxS12 structure, GSH is in an antiparallel orientation with respect to the main chains of the well preserved -TVP-loop segment for hydrogen bond formation (see supplemental Fig. 2). Favorable contacts with GSH are enhanced by the presence of a cis-Pro near the substrate binding site, the geometry of the latter residue increasing the stringency of the fold and ensuring the correct conformation of the preceding valine. The -GCT-segment is conserved with only some variations (Ser, Ala, or Asp) at the position of Thr. A polar residue present at the edge of a GSH binding groove coming into contact with GluGSH might be important to harbor the tripeptide molecule at the groove, by making hydrogen bonds with the amino group. This is indeed the case for all liganded Grx structures except for human Grx1. The position and orientation of the GSH diverge slightly at the glycine residue of GSH in all of the above mentioned liganded structures. The carboxylate group of GlyGSH is interacting with the side chain of Arg67 in human Grx1, whereas no similar interaction is found in other Grx complex structures.
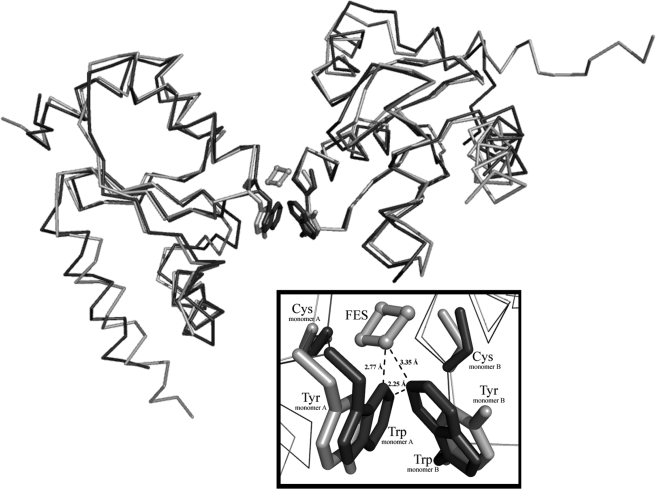
Iron-Sulfur Cluster Assembly in GrxS12—The natural occurrence of an iron-sulfur cluster in poplar GrxC1 (YCGYC active site) and the possibility to incorporate such a prosthetic group in other Grxs from subgroup I when their active site is mutated from YC(P/F)YC into YCGYC but the absence of cluster in a WCGYS-mutated variant of GrxS12 raised the question of the role of the Trp residue and possibly of the cysteine to serine substitution in the second position of GrxS12 active site (35). Although the latter residue is not involved in the cluster ligation, it is probably involved in its stabilization (35). To test this, two mutations have been introduced in GrxS12, leading to YCSYS and YCGYC active sites. The UV-visible spectra obtained at the end of an aerobic purification of the two proteins indicate that they possess the typical [2Fe-2S] cluster observed previously in GrxC1 (supplemental Fig. 3) (data not shown). This supports the proposal that the active site Trp structurally prevents the incorporation of an iron-sulfur cluster in GrxS12. Moreover, when the monomer of the GrxS12 structure is superimposed onto the poplar GrxC1 structure, which contains an iron-sulfur cluster bridged between two monomers, it is obvious that the side chain of Trp28 acts as the major deterrent that prevents the incorporation of the cluster in GrxS12 (Fig. 8).
FIGURE 8.
Superimposition of the hypothetical dimer of GrxS12 (dark gray) to the poplar GrxC1 dimer (light gray). A close-up view detailing the ISC at the active site of poplar GrxC1 is presented in the inset. Upon the superimposition of the hypothetical dimer of GrxS12, Trp28 of GrxS12 is the major hindrance of dimerization and the incorporation of the ISC. The dimer of poplar GrxC1 is bridged by a [2Fe-2S] cluster depicted as spheres with sticks (light gray) showing the coordination. The close distances between the ISC and both Trp residues of GrxS12 are labeled.
DISCUSSION
With about 30 genes coding for Grxs, distributed in three major subclasses, higher plants have an expanded Grx family compared with other organisms that do not contain more than seven Grxs, as in Saccharomyces cerevisiae (36). When starting with such multigene families, one objective is to understand whether those genes and proteins have specific or redundant functions. The specificity is generally thought to be related to differences in activity, to differential gene and protein expression or protein localization (37). Some other explanations are linked to novel and unexpected functions. In poplar chloroplast, GrxS14 and GrxS16 are able to incorporate an iron-sulfur cluster and to transfer it to acceptor proteins (27). In addition, the apoproteins are not reduced by glutathione but rather by ferredoxin thioredoxin reductases (FTR).6 Although belonging to subclass I Grxs, which contain dithiol Grxs, GrxS12 lacks the C terminus active site cysteine and displays a monothiol 28WCSYS32 motif. Nevertheless, there is an additional C terminus cysteine in position 87 (poplar GrxS12 numbering), present in all GrxS12 orthologs and conserved in many other dithiol or monothiol Grxs, including CGFS Grxs (subclass II). From a physiological point of view, it is worth mentioning that, in the chloroplast, GrxS12 is very different from the CGFS Grxs, since it efficiently reduces chloroplastic thiol-dependent antioxidant enzymes using glutathione as a substrate, indicating that plant GrxS12 may have several specific functions related to oxidative stress and signaling by regulating the thiol status of key proteins through deglutathionylation reactions.
Overall, the poplar GrxS12 structure shows no significant difference as compared with other known Grx structures. A glutathione is covalently bound to the active site cysteine of GrxS12. Structural analyses of numerous glutathione-liganded Grxs (20, 31, 32, 38, 39), including poplar GrxS12, reveal that their glutathione binding sites share the following characteristics: (i) the presence of a CXX(C/S) active site motif localized at the N terminus of helix α1′; (ii) the presence of a Tyr or a Phe at close vicinity of the catalytic Cys; (iii) the presence of 73TVP75 (numbering in GrxS12) loop motif with the Pro always in the cis conformation; (iv) the presence of a GG kink (in the loop between β3 and β4) at the proximity of the active site; and (v) conservation of charged residues at both edges of the substrate binding groove (GSH binding pocket).
Role of GrxS12 Active Site Organization for Regeneration Mechanism—The WCSYS active site sequence of GrxS12 is unique to plants and well conserved in all GrxS12 orthologs. The closest active sites are in mammalian Grx2 (SCSYC), in S. cerevisiae Grx6 (TCSYS), and in Azorhizobium caulinodans (WCSYC, accession number YP_001527212).
The position corresponding to Trp28 in GrxS12 is usually occupied by Phe, Tyr, Ser, Thr, or even Gly in other known Grxs. In GrxS12, Trp28 cooperates with Tyr31 at the active site for the stabilization of the mixed disulfide between Cys29 and CysGSH. This enables a favorable interaction between the hydrophobic and highly polarizable sulfurs and the hydrophobic and polarizable phenol ring of Tyr31 and the indole ring of Trp28, providing an additional determinant of peptide specificity toward GSH (particularly toward the γ-GluGSH moiety) or glutathionylated substrates. Interestingly, we were not able to obtain a GrxS12 crystal structure without a bound GSH molecule, and moreover a reduced GrxS12 is strongly precipitating over time, suggesting that the glutathionylated state might be more stable. By contrast, the W28Y mutant of GrxS12 is rather stable. It is also interesting to note that Trp (corresponding to the Trp28 in GrxS12) has been attributed as a key residue in thioredoxins for the recognition of its substrate (40, 41). Therefore, one can envisage that Trp28 could play a similar additional role in GrxS12.
The Ser30 in GrxS12 replaces the classical Pro found in most traditional Grxs. Mutagenesis on human mitochondrial Grx2 demonstrated that this Pro to Ser replacement in the active site dipeptide is the major determinant for the affinity toward glutathionylated substrates (4). This difference, however, is rather small to trigger drastic modification in the three-dimensional structure of the GrxS12 active site, as shown for human Grx2 (20). Nevertheless, the serine residue in the active site motif could provide more flexibility to the main chain. All plant Grxs tested so far, including GrxS12, exhibit similar deglutathionylation activities with the classical in vitro substrate of Grx (HED), regardless of their active site motifs (CPYC, CSYS) (4, 7, 38).7 For deglutathionylation of GAPDH, GrxS12 appeared slightly less efficient than classical Grxs harboring a CPYC active site, but its deglutathionylation activity is comparable with that reported for a chloroplastic CGFS containing Grx (7).
In fact, the regeneration mechanism proposed in previous studies for two identified partners of GrxS12, methionine sulfoxide reductase MsrB1 and peroxiredoxin Prx IIE, most likely occurs via a glutathionylated protein intermediate (42, 43). This is supported by the capacity of GrxS12 but also of most other Grxs to catalyze efficiently deglutathionylation reactions.
Iron-Sulfur Cluster Assembly—Another important point is that human Grx2 (SCSYC active site), poplar GrxC1 (YCGYC), S. cerevisiae Grx6 (TCSYS), and Trypanosoma brucei Grx1 (MCAYS) are able to bind an iron-sulfur cluster, most likely a [2Fe-2S] cluster (20, 35, 44, 45). The crystal structures of Grx2 and GrxC1 in the holoform are similar and showed that the incorporation of the cluster requires the dimerization of the Grx and the presence of two external glutathione molecules (20, 35, 46). It has been demonstrated that all other poplar Grxs from subgroup I (GrxC2, -C3, and -C4; YCP(Y/F)C active sites), except GrxS12 (WCSYS), were also able to incorporate an iron-sulfur cluster when the active site was changed into CGYC, leading to the conclusion that the presence of a glycine or another small residue such as a serine but not a proline is critical for iron-sulfur cluster incorporation (35). It was also suggested in the same study that the presence of the tryptophan could prevent this incorporation. The mutation of the GrxS12 WSCYS active site into YCSYS or YCGYC, which allowed the assembly of an iron-sulfur center in these variants, confirmed this hypothesis. In addition, the resolution of the GrxS12 structure allowed us to model a hypothetic dimer in which the bulky tryptophan residue (Trp28) has been clearly identified as the major deterrent for the incorporation of the ISC in the active site of GrxS12 (Fig. 8). Moreover, bad contacts between the two indole side chains of Trp28 of the two subunits also suggest that dimerization of GrxS12 is almost impossible without a conformational change of the Trp side chain (Fig. 8). Apart from the above-mentioned Trp, the presence of a cis-Pro in the C terminus of proteins of the Trx superfamily was proposed recently to prevent metal binding in these proteins, whereas almost all Grxs that incorporate an iron-sulfur cluster do possess it (47).
Is There a Role for the Additional Conserved Cysteine?—Many Trxs and Grxs have at least one additional conserved Cys residue outside the active site whose exact role has not always been elucidated. Some CGFS Grxs have been reported to possess a disulfide bridge involving an extra active site cysteine at the same position as in GrxS12 (5–7). For example, in S. cerevisiae Grx5, the importance of this disulfide is not clear, since it is required in vitro for deglutathionylation activity but not in vivo for the iron-sulfur biogenesis (48). For C. reinhardtii Grx3, it is also crucial in vitro, acting as a resolving cysteine for deglutathionylation reactions and then for its ferredoxin thioredoxin reductase-dependent regeneration (7). Many other Grxs from several organisms having additional cysteines have been found in vitro to undergo different types of oxidations and can be glutathionylated or nitrosylated or form intramolecular or intermolecular disulfide bridges (20, 30, 49–51).
The additional cysteine (Cys87) in GrxS12 (corresponding to the position of Cys83 in E. coli Grx1) is solvent-exposed and is situated ∼8.4 Å away from the catalytic Cys29, separated by the aromatic side chain of Tyr31. The corresponding conserved active site Tyr residue has been reported to undergo conformational rearrangement in E. coli Grx3 from the free to the substrate-bound form or vice versa (52). Therefore, based on this Tyr flexibility, one could envisage that GrxS12 could undergo similar in situ redox-driven conformational changes. Nevertheless, the hypothesis that Cys87 is able to reduce the Cys29-SSG mixed disulfide spontaneously is unlikely for the following reasons: (i) the C87S GrxS12 variant appeared as efficient as the WT protein in all activity assays; (ii) the recombinant protein is partially glutathionylated on the first active site cysteine, and once the protein is fully glutathionylated on Cys29, the mixed disulfide is stable; (iii) we have been unable to form a fully oxidized form of GrxS12 (i.e. an intramolecular disulfide Cys29–Cys87), neither using a treatment with DTTox nor during crystallization. Nevertheless, because of its low pKa (∼5.6), Cys87 of GrxS12 is fairly reactive. This is supported by the residual activity observed for GrxS12 C29S and by the presence of a βMSH covalently bound to Cys87. Therefore, the possibility cannot be excluded that this cysteine residue may have another role(s) apart from the GSH-specific oxidoreductase activity of GrxS12, serving as a target of post-translational modification (e.g. nitrosylation), as observed for human Grx1 and hence regulating Grx activity (50).
Conclusions—GrxS12 structures described in this study are the first structures of a plant monothiol Grx belonging to subclass I and the first crystallographic structure of a GSH-protein mixed disulfide in a natural CXXS active site motif. This study enabled the identification of a highly conserved GSH binding site in GrxS12 orthologs (28WCSY31, 73TVP75, and 86GCT88), similar to that described before. Concerning the active site residues, Trp28 prevents the incorporation of an iron-sulfur cluster, and Tyr31, although having a certain degree of flexibility, prevents the formation of an intramolecular disulfide between Cys29 and Cys87. Nevertheless, we cannot completely rule out the possibility of Cys87 playing a role as a recycling cysteine only under certain circumstances (e.g. substrate-dependent conditions (nonglutathionylated substrates)). Thus, contrary to another chloroplastic Grxs (C. reinhardtii Grx3 with a CGFS active site), which probably uses a dithiol mechanism for deglutathionylation reactions (an intramolecular disulfide bridge is reduced by ferredoxin thioredoxin reductase), it appears from the structure and the activity of the cysteinic variants that the GrxS12-dependent deglutathionylation occurs through a monothiol mechanism involving only the first active site cysteine. It would first attack the target protein-glutathione mixed disulfide, the Grx becoming itself glutathionylated before a glutathione molecule solves this mixed disulfide to regenerate the reduced Grx.
Supplementary Material
Acknowledgments
We are very grateful to the DESY team at the EMBL-Hamburg Outstation for providing access to beamlines X11 and X13.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) ACJ60637.
The atomic coordinates and structure factors (codes 3FZ9 and 3FZA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Agence Nationale de la Recherche Grants GNP05010G and JC07_204825 (to N. R. and J. C.) and Grant JC-45751 (to M. Z. and S. D. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–3.
Footnotes
The abbreviations used are: Grx, glutaredoxin; βMSH, β-mercaptoethanol; DHA, dehydroascorbate; DTTred, reduced dithiothreitol; DTTox, oxidized dithiothreitol; GR, glutathione reductase; HED, hydroxyethyldisulfide; Trx, thioredoxin; WT, wild type; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; MS, mass spectrometry; PDT-bimane, (2-pyridyl)dithiobimane; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; r.m.s., root mean square; ISC, iron-sulfur cluster.
N. Rouhier, M. Zaffagnini, and S. D. Lemaire unpublished results.
N. Rouhier, unpublished data.
References
- 1.Bushweller, J. H., Aslund, F., Wuthrich, K., and Holmgren, A. (1992) Biochemistry 31 9288-9293 [DOI] [PubMed] [Google Scholar]
- 2.Rouhier, N., Gelhaye, E., and Jacquot, J. P. (2002) FEBS Lett. 511 145-149 [DOI] [PubMed] [Google Scholar]
- 3.Lillig, C. H., Prior, A., Schwenn, J. D., Aslund, F., Ritz, D., Vlamis-Gardikas, A., and Holmgren, A. (1999) J. Biol. Chem. 74 7695-7698 [DOI] [PubMed] [Google Scholar]
- 4.Johansson, C., Lillig, C. H., and Holmgren, A. (2004) J. Biol. Chem. 279 7537-7543 [DOI] [PubMed] [Google Scholar]
- 5.Tamarit, J., Bellí, G., Cabiscol, E., Herrero, E., and Ros, J. (2003) J. Biol. Chem. 278 25745-25751 [DOI] [PubMed] [Google Scholar]
- 6.Fernandes, A. P., Fladvad, M., Berndt, C., Andrésen, C., Lillig, C. H., Neubauer, P., Sunnerhagen, M., Holmgren, A., and Vlamis-Gardikas, A. (2005) J. Biol. Chem. 280 24544-24552 [DOI] [PubMed] [Google Scholar]
- 7.Zaffagnini, M., Michelet, L., Massot, V., Trost, P., and Lemaire, S. D. (2008) J. Biol. Chem. 283 8868-8876 [DOI] [PubMed] [Google Scholar]
- 8.Jung, C. H., and Thomas, J. A. (1996) Arch. Biochem. Biophys. 335 61-72 [DOI] [PubMed] [Google Scholar]
- 9.Martin, J. L. (1995) Structure 3 245-250 [DOI] [PubMed] [Google Scholar]
- 10.Rouhier, N., Gelhaye, E., and Jacquot, J.-P. (2004) Cell. Mol. Life Sci. 61 1266-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Manzaneque, M. T., Ros, J., Cabiscol, E., Sorribas, A., and Herrero, E. (1999) Mol. Cell. Biol. 19 8180-8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaire, S. D. (2004) Photosynth. Res. 79 305-318 [DOI] [PubMed] [Google Scholar]
- 13.Rouhier, N., Couturier, J., and Jacquot, J.-P. (2006) J. Exp. Bot. 57 1685-1696 [DOI] [PubMed] [Google Scholar]
- 14.Schenk, P. M., Baumann, S., Mattes, R., and Steinbiss, H. H. (1995) BioTechniques 19 196-200 [PubMed] [Google Scholar]
- 15.Augusto, L., Decottignies, P., Synguelakis, M., Nicaise, M., Le Maréchal, P., and Chaby, R. (2003) Biochemistry 42 3929-3938 [DOI] [PubMed] [Google Scholar]
- 16.Mansoor, S. E., and Farrens, D. L. (2004) Biochemistry 43 9426-9438 [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project 4 (1994) Acta Crystallogr. Sect. D 50 760-76315299374 [Google Scholar]
- 19.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 30 1022-1025 [Google Scholar]
- 20.Johansson, C., Kavanagh, K. L., Gileadi, O., and Opperman, U. (2007) J. Biol. Chem. 282 3077-3082 [DOI] [PubMed] [Google Scholar]
- 21.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 22.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 23.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 25.Delano, W. L. (2002) PyMol, DeLano Scientific, Palo Alto, CA
- 26.Kleywegt, G. J., and Jones, T. A. (1997) Acta Crystallogr. Sect. D 53 179-185 [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay, S., Gama, F., Molina-Navarro, M. M., Gualberto, J. M., Claxton, R., Naik, S. G., Huynh, B. H., Herrero, E., Jacquot, J. P., Johnson, M. K., and Rouhier, N. (2008) EMBO J. 27 1122-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaffagnini, M., Michelet, L., Marchand, C., Sparla, F., Decottignies, P., Le Marechal, P., Miginiac-Maslow, M., Noctor, G., Trost, P., and Lemaire, S. D. (2007) FEBS J. 274 212-226 [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan, U., Mieyal, P. A., and Mieyal, J. J. (1997) Biochemistry 36 3199-3206 [DOI] [PubMed] [Google Scholar]
- 30.Bushweller, J. H., Billeter, M., Holmgren, A., and Wüthrich, K. (1994) J. Mol. Biol. 235 1585-1597 [DOI] [PubMed] [Google Scholar]
- 31.Nordstrand, K., Aslund, F., Holmgren, A., Otting, G., and Berndt, K. D. (1999) J. Mol. Biol. 286 541-552 [DOI] [PubMed] [Google Scholar]
- 32.Yang, Y., Jao, S., Nanduri, S., Starke, D. W., Mieyal, J. J., and Qin, J. (1998) Biochemistry 37 17145-17156 [DOI] [PubMed] [Google Scholar]
- 33.Sodano, P., Xia, T. H., Bushweller, J. H., Bjornberg, O., Holmgren, A., Billeter, M., and Wuthrich, K. (1991) J. Mol. Biol. 221 1311-1324 [DOI] [PubMed] [Google Scholar]
- 34.Aslund, F., Nordstrand, K., Berndt, K. D., Nikkola, M., Bergman, T., Ponstingl, H., Jornvall, H., Otting, G., and Holmgren, A. (1996) J. Biol. Chem. 271 6736-6745 [DOI] [PubMed] [Google Scholar]
- 35.Rouhier, N., Unno, H., Bandyopadhay, S., Masip, L., Kim, S. K., Hirasawa, M., Gualberto, J., Lattard, V., Kusunoki, M., Knaff, D. B., Georgiou, G., Hase, T., Johnson, M. K., and Jacquot, J. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7379-7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morel, M., Kohler, A., Martin, F., Gelhaye, E., and Rouhier, N. (2008) New Phytol. 180 391-407 [DOI] [PubMed] [Google Scholar]
- 37.Rouhier, N., Lemaire, S. D., and Jacquot, J. P. (2008) Annu. Rev. Plant. Biol. 59 143-166 [DOI] [PubMed] [Google Scholar]
- 38.Nikkola, M., Gleason, F. K., Saarinen, M., Joelson, T., Bjornberg, O., and Eklund, H. (1991) J. Biol. Chem. 266 16105-16112 [PubMed] [Google Scholar]
- 39.Xia, T. H., Bushweller, J. H., Sodano, P., Billeter, M., Bjornberg, O., Holmgren, A., and Wuthrich, K. (1992) Protein Sci. 1 310-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause, G., and Holmgren, A. (1991) J. Biol. Chem. 266 4056-4066 [PubMed] [Google Scholar]
- 41.Menchise, V., Corbier, C., Didierjean, C., Jacquot, J. P., Benedetti, E., Saviano, M., and Aubry, A. (2000) Biopolymers 56 1-7 [DOI] [PubMed] [Google Scholar]
- 42.Vieira Dos Santos, C., Laugier, E., Tarrago, L., Massot, V., Issakidis-Bourguet, E., Rouhier, N., and Rey, P. (2007) FEBS Lett. 581 4371-4376 [DOI] [PubMed] [Google Scholar]
- 43.Gama, F., Bréhélin, C., Gelhaye, E., Meyer, Y., Jacquot, J. P., Rey, P., and Rouhier, N. (2008) Physiol. Plant. 133 599-610 [DOI] [PubMed] [Google Scholar]
- 44.Mesecke, N., Mittler, S., Eckers, E., Herrmann, J. M., and Deponte, M. (2008) Biochemistry 47 1452-1463 [DOI] [PubMed] [Google Scholar]
- 45.Comini, M. A., Rettig, J., Dirdjaja, N., Hanschmann, E. M., Berndt, C., and Krauth-Siegel, R. L. (2008) J. Biol. Chem. 283 27785-27798 [DOI] [PubMed] [Google Scholar]
- 46.Feng, Y., Shong, N., Rouhier, N., Hase, T., Kusunoki, M., Jacquot, J. P., Jin, C., and Xia, B. (2006) Biochemistry 45 7998-8008 [DOI] [PubMed] [Google Scholar]
- 47.Su, D., Berndt, C., Fomenko, D. E., Holmgren, A., and Gladyshev, V. N. (2007) Biochemistry 46 6903-6910 [DOI] [PubMed] [Google Scholar]
- 48.Bellí, G., Polaina, J., Tamarit, J., De La Torre, M. A., Rodríguez-Manzaneque, M. T., Ros, J., and Herrero, E. (2002) J. Biol. Chem. 277 37590-37596 [DOI] [PubMed] [Google Scholar]
- 49.Sun, C., Berardi, M. J., and Bushweller, J. H. (1998) J. Mol. Biol. 280 687-687 [DOI] [PubMed] [Google Scholar]
- 50.Hashemy, S. I., Johansson, C., Berndt, C., Lillig, C. H., and Holmgren, A. (2007) J. Biol. Chem. 282 14428-14436 [DOI] [PubMed] [Google Scholar]
- 51.Melchers, J., Dirdjaja, N., Ruppert, T., and Krauth-Siegel, R. L. (2007) J. Biol. Chem. 282 8678-8694 [DOI] [PubMed] [Google Scholar]
- 52.Nordstrand, K., Sandström, A., Åslund, F., Holmgren, A., Otting, G., and Berndt, K. D. (2000) J. Mol. Biol. 303 423-432 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.