Abstract
Leptin activates multiple signaling pathways in cells, including the phosphatidylinositol 3-kinase pathway, indicating a degree of cross-talk with insulin signaling. The exact mechanisms by which leptin alters this signaling pathway and how it relates to functional outputs are unclear at present. A previous study has established that leptin inhibits the activity of the phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome 10), an important tumor suppressor and modifier of phosphoinositide signaling. In this study we demonstrate that leptin phosphorylates multiple sites on the C-terminal tail of PTEN in hypothalamic and pancreatic β-cells, an action not replicated by insulin. Inhibitors of the protein kinases CK2 and glycogen synthase kinase 3 (GSK3) block leptin-mediated PTEN phosphorylation. PTEN phosphorylation mutants reveal the critical role these sites play in transmission of the leptin signal to F-actin depolymerization. CK2 and GSK3 inhibitors also prevent leptin-mediated F-actin depolymerization and consequent ATP-sensitive K+ channel opening. GSK3 kinase activity is inhibited by insulin but not leptin in hypothalamic cells. Both hormones increase N-terminal GSK3 serine phosphorylation, but in hypothalamic cells this action of leptin is transient. Leptin, not insulin, increases GSK3 tyrosine phosphorylation in both cell types. These results demonstrate a significant role for PTEN in leptin signal transmission and identify GSK3 as a potential important signaling node contributing to divergent outputs for these hormones.
Efficient signaling by leptin and insulin is essential for the maintenance of body energy homeostasis, with disruptions in these processes strongly associated with diabetes and obesity (1, 2) and, at least for insulin, neurodegenerative disorders such as Alzheimer disease (3, 4). In recent years there has been a significant increase in understanding the intracellular signaling processes associated with the actions of insulin on a wide variety of cell types (5). However, our knowledge of leptin signaling is less advanced, with most studies indicating that leptin and insulin share many signaling intermediates in common, often leading to similar cellular outcomes (6, 7). In particular, signaling through the STAT (signal transducers and activators of transcription), mitogen-activated protein kinase, and PI3K3 pathways have been reported extensively in numerous cell types for both leptin and insulin (5, 8).
Nevertheless, leptin and insulin can cause differing and sometimes opposing cellular outputs, even on the same cell type. This is demonstrated in hypothalamic neurons, where electrophysiological or imaging studies show differential outcomes for leptin and insulin action (9–11). Thus, although superficially leptin may utilize the same signaling pathways as insulin, the exact nature of the leptin-induced signaling intermediates and their interplay with one another and with individual effectors is still relatively unknown. Recently, it was demonstrated that although leptin, like insulin, raises PtdIns(3,4,5)P3 levels in hypothalamic cells, the mechanism underlying the increase by leptin differs from that of insulin. Whereas insulin increased PI3K activity, leptin had little effect on PI3K activity. Instead, leptin inhibited the lipid and protein phosphatase, PTEN, which resulted in increased PtdIns(3,4,5)P3 levels in the presence of active PI3K (12). Previously, PI3K-dependent leptin signaling had been shown to open ATP-sensitive (KATP) channels in rat hypothalamic neurons (13) and in rat and mouse insulin-secreting cells (12, 14, 15), resulting in cell hyperpolarization and inhibition of firing. KATP activation by leptin is dependent on actin depolymerization in both cell types (12–15). The connection between leptin-driven PI3K activity, actin re-modeling, and KATP opening appears not to be due simply to increased PtdIns(3,4,5)P3 but may also require coincident inhibition of PTEN protein and lipid phosphatase activity through increased PTEN phosphorylation (12).
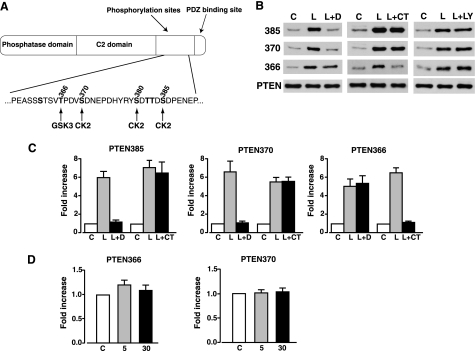
In this study we have focused on the mechanisms by which leptin alters PTEN phosphorylation in hypothalamic and insulin-secreting cells. The biological activity of PTEN toward anionic lipid substrates is considered to be dependent on its phosphorylation status at various residues between Ser-362 and Ser-385, located in the PTEN C-terminal tail (Fig. 1A), with phosphorylation maintaining PTEN in an inactive state (16). The major sites of phosphorylation are Ser-370 and Ser-385 (17, 18), with significant phosphorylation at a cluster of sites (Ser-380, Thr-382, and Thr-383) close to Ser-385. Mutations of the cluster site residues have significant effects on PTEN stability and membrane and protein binding (17, 19, 20). It is thought that PTEN is constitutively phosphorylated and not linked to physiological signaling events such as stimulation of cells with insulin or growth factors (21, 22). However, we previously demonstrated that leptin stimulation of hypothalamic cells reduced PTEN phosphatase activity and increased PTEN phosphorylation at the cluster site (12). This increased phosphorylation was sensitive to inhibition of protein kinase CK2, previously implicated as the main kinase phosphorylating PTEN at these residues (17, 18). CK2 has also been reported to phosphorylate PTEN at Ser-370 and Ser-385 (Fig. 1A) using in vitro kinase assays (17, 23), resulting in a 30% depression of PTEN phosphatase activity (18). In addition, GSK3β has been reported to phosphorylate PTEN at Ser-362 and Thr-366 in human embryonic kidney 293T cells, with Thr-366 phosphorylation reducing the biological activity of PTEN.
FIGURE 1.
Inhibitors of CK2 and GSK3 reduce leptin-mediated phosphorylation of PTEN in N29/4 cells. A, schematic representation of PTEN. The sequence of amino acids for part of the C-terminal tail domain is shown, with the main phosphorylation sites denoted in bold, and the putative phosphorylating kinase shown below. B, phosphorylated PTEN levels in N29/4 cells using the phospho-specific PTEN antibodies indicated under non-stimulated conditions (C), in response to 10 nm leptin alone (L), and in the presence of 10 μm DMAT (L+D), 2 μm CT99021 (L+CT), and 10 μm LY294002 (L+LY). C, the bar graphs show mean normalized levels of phosphorylated PTEN at Ser-385, Ser-370, and Thr-366 under non-stimulated conditions (C) and for leptin in the absence (L) and presence of DMAT (L+D) and CT99021 (L+CT; n = 6 for each). D, bar graphs showing mean normalized levels of phosphorylated PTEN at Thr-366 and Ser-370 under non-stimulated conditions (C) and after stimulation of N29/4 cells for 5 or 30 min with 10 nm insulin.
Consequently, we have assessed whether leptin mediates phosphorylation of PTEN at Ser-370, Ser-385, and Thr-366 in a CK2- and GSK3-dependent manner and further examined the contribution of Ser-370 and Thr-366 to downstream events such as actin re-organization and KATP channel opening using hypothalamic and insulin-secreting cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Islet Isolation—The mouse hypothalamic cell line N29/4 was cultured as previously described (24). MIN6 cells (obtained with permission from Jun-ichi Miyazaki, Osaka University) were used between passages 24 and 30 and grown in Dulbecco's modified Eagle's medium containing 25 mm glucose and supplemented with 12.5% heat-inactivated fetal calf serum, 4 mm l-glutamine, 100 μm 2-mercaptoethanol, and 1% penicillin and streptomycin in a humidified atmosphere of 95% air and 5% CO2. Wild-type C57Bl/6 mice (20–25 g) were used for islet isolation and killed by cervical dislocation immediately before the islet isolation procedure. All animal procedures conformed to the UK Animals (Scientific Procedures) Act 1986 and were approved by our institutional ethical review committee. Pancreatic islets were isolated from mice under sterile conditions, and isolated cells were cultured as described previously (12).
Cell Transfection, Staining, and Actin Analysis—N29/4 cells were plated in 6-well plates at 40% confluence 24 h before transfection. 5 μl of Lipofectamine (Invitrogen) and 2 μg of PTEN DNA were mixed with 100 μl of serum-free Opti medium, incubated at room temperature for 20 min to form the transfection complexes, and diluted with 500 μl of culture medium. The mixture was applied to the cells, left overnight at 37 °C, replaced with culture medium, and incubated for 48 h before use. In experiments where wild-type and mutant PTEN constructs were overexpressed, F-actin or PtdIns(3,4,5)P3 levels were determined from 60 cells for control and each treatment from a total of 4 experiments (comparing transfected cells, as determined by PTEN antibody staining, with control cells). Similarly, 72 cells were examined from 4 experiments to measure F-actin in control and leptin with or without CT99021 and/or 2-dimethyl-amino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT). Cells were treated with DMAT (Calbiochem) and/or CT99021 (25) in normal saline (135 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, and 0.2% BSA, pH 7.4) for 20 min before challenge with leptin or saline in the continuous presence of inhibitor for 60 min before fixing as described previously (13). For staining, cells were washed in phosphate-buffered saline (PBS), permeabilized in PBS, 0.5% Triton X-100 for 10 min, rinsed, blocked with 20% goat serum (Sigma), and incubated with rhodamine-conjugated phalloidin or 2 units ml-1 Alexa 488-phalloidin (Molecular Probes) for 90–120 min. Immunocytochemical staining was performed as described previously (13) using a monoclonal antibody against PTEN (Chemicon) or PtdIns(3,4,5)P3 (Molecular Probes). The secondary antibodies were Cy3 or fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson ImmunoResearch). Fluorescence-labeled proteins or lipids in cells were observed with a 63× oil objective, and images were acquired by confocal microscopy (Zeiss LSM 510). For quantitative analysis of F-actin and PtdIns(3,4,5)P3 levels, Aida/two-dimensional densitometry was used to measure pixel intensity of defined areas of transfected, treated, and control cells. After subtraction of background, intensity values for treated cells were expressed as a ratio of control cell intensity.
Western Blotting—N29/4 and MIN6 cells, grown in 6-well plates, were exposed to normal saline for 30 min before treatment with leptin, insulin, or saline in the presence and absence of LY294002 (Sigma), DMAT, or CT99021 for the required time (inhibitors added 20–30 min before challenge). Protein isolation, content, immunoblotting, and analysis procedures were as previously described (13). PTEN and phospho-PTEN antibodies, Ser-380/Thr-382/Thr-383 were from Cell Signaling (1:5000), Ser-385 was from BIOSOURCE (1:500), and Ser-370 and Thr-366 were from Nick Leslie (1:500). phosphorylated (p)-PKB (Ser-473) and p-GSK3 (Ser-9/Ser-21) were obtained from Cell Signaling and used at 1:1000. p-GSK3 (Tyr-216/Tyr-279), PKB, and GSK3 antibodies (all polyclonal) were obtained from BD Transduction Laboratories and used at 1:2500 (p-GSK3) and 1:1000 (PKB and GSK3). Monoclonal anti β-actin antibody (used at 1:10000) was from MP Biochemicals. Protein bands on gels were quantified by densitometry, where total density was determined with respect to constant area, background was subtracted, and average relative band density was calculated.
Immunoprecipitation and Assay of GSK3—Cell extracts (0.5 mg) were incubated with protein G-Sepharose conjugated to 2.5 mg of anti-GSK3α/β antibody, Upstate Biotechnology, Inc. (Lake Placid, NY) for 1 h at 4 °C on a shaking platform. The immunocomplexes were pelleted and washed with 1 ml of lysis buffer containing 0.5 m NaCl and twice with 1 ml of assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.03% (v/v) Brij-35, and 0.1% (v/v) β-mercaptoethanol). The immunoprecipitated GSK3 was incubated at 30 °C for 20 min in a total volume of 50 ml containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.03% (v/v) Brij-35, 0.1% (v/v) β-mercaptoethanol, 10 mm MgCl2, 0.1 mm [γ-32P]ATP (0.5 × 106 cpm/nmol), and 30 mm phosphoglycogen synthase 2 peptide (available from Upstate). Radiolabeled peptide produced was separated from unreacted ATP by binding to P81 paper and washing in 75 mm H3PO4 before counting in 2 ml of scintillant. One unit of kinase activity is defined as the amount that catalyzes the phosphorylation of 1 nmol of substrate in 1 h.
Electrophysiology—Mouse cultured β-cells were superfused at room temperature (22–25 °C) with normal saline. Wholecell current and voltage clamp recordings were used to monitor membrane potential and macroscopic current, respectively, using an Axopatch 200B amplifier. Whole-cell experiments were maintained in current-clamp mode to monitor the cell resting membrane potential, with short excursions into voltage clamp mode to examine macroscopic current-voltage relations. In voltage-clamp recordings the membrane potential was held at -70- and 20-mV steps of 2 s duration, with 20 ms between pulses applied (range of voltages, -160 to -40 mV). Whole cell voltage clamp data were analyzed using pCLAMP 8.2 software (Molecular Devices). Current clamp data were recorded and stored onto digital audio tapes and replayed for analysis and illustration on a Gould TA240 chart recorder. Recording electrodes were pulled from borosilicate glass and had resistances of 5–10 megaohms when filled with pipette solution, which comprised 140 mm KCl, 5 mm MgCl2, 3.8 mm CaCl2, 3 mm MgATP, 10 mm EGTA, 10 mm HEPES, pH 7.2 (free [Ca2+] of 100 nm). Application of drugs was made via the bath perfusion system except for DMAT and the GSK3 inhibitors, which were present in the pipette solution.
Statistical Analysis—Data are presented as the means ± S.E. unless otherwise stated. All statistical analyses were performed using analysis of variance, one sample t test, Student's paired or unpaired t tests. A probability level of 0.05 was considered significant.
RESULTS
We used the leptin- and insulin-sensitive hypothalamic cell line N29/4 (12, 24) to examine PTEN phosphorylation at Thr-366, Ser-370, and Ser-385 (Fig. 1A) on stimulation with leptin (10 nm) or insulin (10 nm). Leptin rapidly (<5 min) increased the level of p-PTEN at all three sites (Fig. 1B), an action sustained for at least 60 min for each site (data not shown). This leptin-mediated increase in p-PTEN was unaffected by the presence of the PI3K inhibitor, LY294002 (10 μm; n = 6 for each; Fig. 1B). Previous studies indicated that the cluster site, Ser-385 and Ser-370, may be phosphorylated by protein kinase CK2 (17, 18, 23), and Thr-366 may be phosphorylated by GSK3β (23). Consequently, we examined leptin phosphorylation of Thr-366, Ser-370, and Ser-385 PTEN residues in the absence and presence of DMAT (a CK2 inhibitor (26, 27) and CT99021 (a specific GSK3 inhibitor (28, 29)). DMAT (10 μm) prevented leptin from phosphorylating Ser-370 and Ser-385 but not Thr-366 in N29/4 cells (Fig. 1, B and C). Conversely, leptin phosphorylation of Thr-366 was prevented by CT99021 (2 μm), whereas phosphorylation of Ser-370 and Ser-385 was unaffected by this inhibitor (Fig. 1, B and C). In contrast, 10 nm (Fig. 1D) or 50 nm (data not shown) insulin had no effect on p-PTEN levels in N29/4 cells at Thr-366 or Ser-370 residues, with the Ser-385 site not tested.
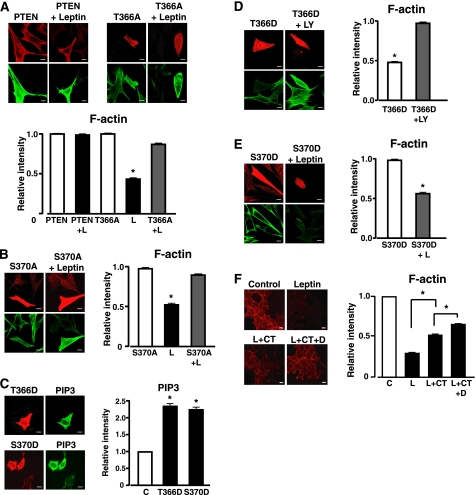
Leptin re-organization of the actin cytoskeleton is dependent on PI3K activity and inhibition of the lipid and protein phosphatase activity of PTEN. Overexpression of cluster site p-PTEN mutants demonstrated that phosphorylation at this site was required to limit PTEN phosphatase activity and prevent leptin-induced F-actin depolymerization (12). Accordingly, we have examined whether phosphorylation of PTEN at Ser-370 and/or Thr-366 is also required for transmission of the leptin signal to F-actin. First, we sought to determine whether phosphorylation of Thr-366 or Ser-370 altered the activity of PTEN in cells by overexpression of PTEN mutants with alanine or aspartic acid substitutions. PtdIns(3,4,5)P3 and F-actin levels were assessed by staining with a monoclonal PtdIns(3,4,5)P3 antibody and Alexa 488-conjugated phalloidin respectively, after transfection of PTEN mutants into N29/4 cells. In control experiments overexpression of wild-type PTEN had no effect per se but prevented leptin from decreasing F-actin (Fig. 2A). Overexpression of T366A PTEN or S370A PTEN reduced PtdIns(3,4,5)P3 levels (data not shown), indicative of enhanced PtdIns(3,4,5)P3 phosphatase activity, did not affect F-actin in the absence of leptin and inhibited leptin-induced F-actin reduction (Fig. 2, A and B). In contrast, overexpression of T366D PTEN or S370D PTEN increased PtdIns(3,4,5)P3 levels (Fig. 2C). The presence of the 366D PTEN mutant in N29/4 cells decreased F-actin levels in the absence of leptin in a PI3K-dependent manner (Fig. 2D). Surprisingly, overexpression of S370D PTEN, although increasing PtdIns(3,4,5)P3, did not per se alter F-actin staining in the absence of leptin nor occlude the effect of leptin on F-actin (Fig. 2E). This action of S370D PTEN indicates, as observed previously (12), that increasing cellular PtdIns(3,4,5)P3 levels alone is not a predictor or indeed sufficient to initiate PTEN-dependent downstream signaling events such as F-actin depolymerization.
FIGURE 2.
Effect of PTEN phosphorylation mutants on PtdIns(3,4,5)P3 and F-actin levels in N29/4 cells. Wild-type PTEN and T366A PTEN (A) and S370A PTEN (B) were overexpressed in N29/4 cells (upper panels, Cy3 secondary antibody on cells treated with monoclonal PTEN antibody) and F-actin visualized with Alexa 488-phalloidin (lower panels) with and without stimulation by 10 nm leptin. Note that untransfected cells in the leptin (L)-treated group are not easily visible in the right lower panels due to leptin-mediated F-actin depolymerization. Bar graphs showing mean levels of F-actin after treatment of N29/4 cells ± overexpression of PTEN or T366A PTEN (A) or S370A PTEN (B) with 10 nm leptin. Data are shown as a ratio of control, untreated cells. C, PtdIns(3,4,5)P3 levels in N29/4 cells transfected with T366D PTEN or S370D PTEN were investigated using a PtdIns(3,4,5)P3 antibody. The bar graph shows mean levels of PtdIns(3,4,5)P3 (as a ratio of control) in N29/4 cells after treatment with these PTEN phosphorylation mutants. D and E, F-actin levels were visualized (lower panels) in N29/4 cells transfected (upper panels) with T366D PTEN (D) or S370D PTEN (E) in the absence (left) and presence (right) of 10 μm LY294002 (LY, D) or 10 nm leptin (E). The bar graphs show mean levels of F-actin after treatment of N29/4 cells overexpressing T366D PTEN (D) or S370D PTEN (E). Data are shown as a ratio of control, untreated cells. F, cellular F-actin levels in the absence or presence of 10 nm leptin and in cells pretreated with 2 μm CT99021 alone (CT) or in combination with 10 μm DMAT (D) before leptin challenge. Note that F-actin is visualized in this series of experiments with rhodamine-conjugated phalloidin. The bar graph denotes mean levels of F-actin-treated cells. Data are shown as the ratio of control, untreated cells. Values represent the mean ± S.E. for 3–4 experiments per group. *, p < 0.05 compared with the appropriate labeled control for each treatment. Scale bars, 10 μm. PIP3, PtdIns(3,4,5)P3.
The dependence of F-actin depolymerization by leptin on the phosphorylation status of the cluster (12) and Thr-366 sites led us to explore whether this action of leptin was sensitive to CK2 and/or GSK3 inhibition. The presence of the GSK3 inhibitor, CT99021 (2 μm) or the CK2 inhibitor DMAT (10 μm) alone had no effect on F-actin (data not shown). Leptin decreased the mean F-actin level, and in the presence of CT99021 this was significantly reduced (Fig. 2F). The combined presence of CT99021 and DMAT inhibited F-actin depolymerization by leptin significantly more than CT99021 alone but did not completely occlude leptin from affecting F-actin levels (Fig. 2F). Nevertheless, the presence of the GSK3 and CK2 inhibitors at concentrations that significantly reduced PTEN phosphorylation also significantly reduced the ability of leptin to depolymerize F-actin.
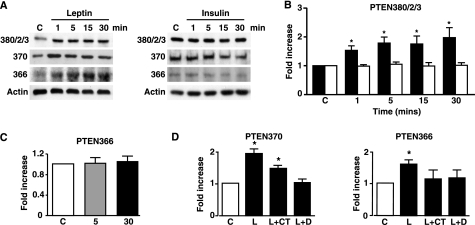
Phosphorylation of PTEN in β-Cells and KATP Activation—It was previously demonstrated that KATP channel opening and hyperpolarization of mouse pancreatic β-cells by leptin could be inhibited by the lipid phosphatase dead G129E PTEN mutant and mimicked by the dual lipid and protein phosphatase dead C124S PTEN mutant (12). This is consistent with inhibition of PTEN lipid and protein phosphatase activity for the successful transmission of the leptin signal to KATP channels in β-cells. As leptin has not been shown to alter p-PTEN status in β-cells, we used MIN6 β-cells to explore this action of leptin. Although the effects of leptin on p-PTEN were not as robust as observed for N29/4 cells, leptin (10 nm) rapidly (≤1 min) increased phosphorylation of the cluster site, Ser-370 and Thr-366, actions sustained for at least 30 min in each case (Fig. 3, A and B). Insulin (10 nm) had no effect on PTEN phosphorylation levels at any of these sites over this time period (Fig. 3, A–C). The CK2 inhibitor DMAT (2 μm) inhibited leptin-mediated PTEN phosphorylation at Ser-370 and Thr-366, whereas the GSK3 inhibitor CT99021 (10 μm) inhibited phosphorylation at Thr-366 only (Fig. 3D).
FIGURE 3.
Leptin increases PTEN phosphorylation in MIN6 pancreatic β-cells. A, phosphorylated PTEN levels in MIN6 cells using the phospho-specific PTEN antibodies indicated (cluster site, Ser-370 and Thr-366), in response to 10 nm leptin or 10 nm insulin for the indicated times (minutes). B, the bar graph shows mean normalized levels of phosphorylated PTEN at the cluster phosphorylation site (380/2/3) for untreated cells and for cells stimulated with leptin (filled bars; n = 6) and insulin (open bars; n = 8) up to 30 min (*, p < 0.05, versus control (C) or insulin). C, bar graph shows the mean normalized levels of phosphorylated Thr-366 PTEN after stimulation with 10 nm insulin for 5 and 30 min. D, bar graphs showing mean normalized levels of phosphorylated Thr-366 PTEN and Ser-370 PTEN under non-stimulated conditions (C) and after stimulation with 10 nm leptin in the absence (L) and presence of DMAT (L+D) and CT99021 (L+CT; n = 7 for each). *, p < 0.05.
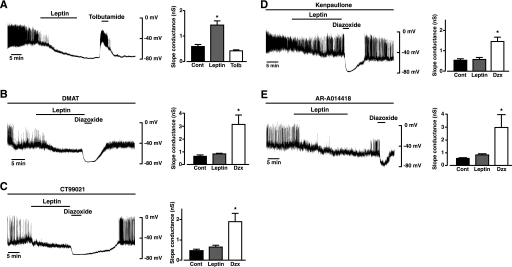
Consequently, we predicted that the F-actin reduction and opening of KATP channels by leptin (12, 13) would be susceptible to inhibition of CK2 and GSK3. Therefore, we examined the ability of leptin to hyperpolarize the membrane potential and activate KATP currents in whole-cell current and voltage clamp recordings, respectively, from mouse primary cultured pancreatic β-cells in the absence and presence of DMAT and various GSK3 inhibitors. In control experiments β-cells were dialyzed with an electrode solution containing 3 mm ATP, which resulted in a mean membrane potential of -41.8 ± 1.5 mV (n = 22) under current clamp conditions. Application of leptin (10 nm) resulted in a slowly developing hyperpolarization (Fig. 4A) to a mean value of -69.4 ± 1.6 mV (n = 22, p < 0.05), which was reversibly inhibited (to -34.5 ± 1.9 mV; n = 17, p < 0.05) by the sulfonylurea tolbutamide (100 μm). Voltage clamp analysis (Fig. 4A) showed that leptin increased the mean cell slope conductance from 0.73 ± 0.11 to 1.62 ± 0.22 nanosiemens (n = 19; p < 0.05), an action reduced by tolbutamide to 0.42 ± 0.04 nanosiemens (n = 14; p < 0.05), indicating that the increase in current responsible for the augmented slope conductance and β-cell hyperpolarization by leptin is due to KATP channel opening (14, 15, 30). The presence of DMAT (10 μm) in the electrode solution (and, therefore, cell interior) resulted in a mean resting membrane potential of -44.8 ± 3.4 mV (p > 0.05 compared with control). Subsequent addition of leptin caused a small, insignificant (p > 0.05) hyperpolarization to -52.4 ± 8.4 mV (n = 5; Fig. 4B), which was significantly different (p < 0.05) from the control leptin response. Voltage clamp analysis showed that DMAT also prevented leptin from increasing the β-cell mean slope conductance (Fig. 4B). The KATP channels were operational in the presence of DMAT as β-cells were readily responsive (Fig. 4B) to the direct-acting KATP activator, diazoxide (200 μm), which caused hyperpolarization to -75.2 ± 1.7 mV (p < 0.05) and increased slope conductance from 0.82 ± 0.07 to 3.1 ± 0.8 nanosiemens (n = 5; p < 0.05). CT99021 (2 μm), in the electrode solution, resulted in a mean β-cell membrane potential of -41.5 ± 3.3 mV. Application of leptin (Fig. 4C) caused hyperpolarization to -51.5 ± 4.1 mV, which although less than the control leptin response, was significantly different (n = 5, p < 0.05) from the mean resting potential before leptin challenge. However, voltage clamp analysis showed that in the presence of CT99021 leptin did not significantly increase the β-cell slope conductance (Fig. 4C). As a small, but significant hyperpolarization of β-cells was elicited by leptin in the presence of CT99021, we decided to confirm that GSK3 inhibition does occlude leptin-mediated β-cell hyperpolarization and increase in KATP current. Consequently, we examined the effects of the GSK3 inhibitors, AR-A0144-18 (28, 31) and kenpaullone (29) on leptin-induced opening of KATP. The presence of AR-A0144-18 (2 μm) or kenpaullone (10 μm) in the electrode solution did not alter (p < 0.05) β-cell membrane potential (-40.4 ± 1.6 mV and -35.2 ± 2.1 mV, respectively) compared with control recordings. Leptin (10 nm) caused a small hyperpolarization of the β-cells for both AR-A014418 (to -51.8 ± 6.4; n = 5) and kenpaullone (to -45.5 ± 4.6 mV; n = 6), which was not significant (p > 0.05) for either inhibitor (Fig. 4, D and E). Voltage clamp analysis confirmed that the presence of either GSK3 inhibitor prevented an increase in slope conductance by leptin (Fig. 4, D and E). Diazoxide (200 μm), in contrast, reversibly hyperpolarized β-cells in the presence of each GSK3 inhibitor (Fig. 4, C–E, CT99021, to -73.0 ± 2.2 mV (n = 5; p < 0.05), AR-A014418 to -72.4 ± 4.1 mV (n = 5, p < 0.05), and kenpaullone, to -72.3 ± 2.4 mV (n = 6, p < 0.05) and significantly increased the mean slope conductance to values comparable for control β-cells (p < 0.05).
FIGURE 4.
Activation of KATP channels in mouse pancreatic β-cells by leptin is occluded by the presence of CK2 or GSK3 inhibitors. Representative whole-cell current clamp recordings from a mouse cultured β-cell dialyzed with an electrode solution containing 3 mm ATP. In this and subsequent panels the trace begins ∼2 min after obtaining the recording configuration. Leptin (10 nm), tolbutamide (Tolb, 100 μm), or diazoxide (Dzx, 200 μm) were applied for the time indicated on each trace (A–E). Cont, control. The action of leptin on β-cell membrane potential was determined in the absence (A) and presence of 10 μm DMAT (B), 2 μm CT99021 (C), 10 μm kenpaullone (D), and 2 μm AR-A0144–18 (E) added to the electrode solution and, hence, cell interior. The bar graphs to the right of each trace denote the mean slope conductance, determined from voltage clamp current-voltage relations in the absence and presence of leptin. In control cells (A) the hyperpolarization and increased slope conductance induced by leptin are inhibited by the presence of tolbutamide. For drug-treated cells leptin has no effect on mean cell conductance, although in each case application of 200 μm diazoxide hyperpolarizes the cells, which is associated with a significant increase in slope conductance. *, p < 0.05, compared with control.
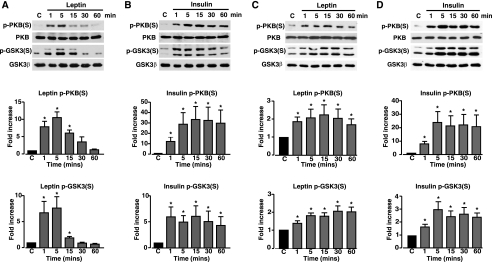
Phosphorylation and Kinase Activity of GSK3—These results taken together with those of Ning et al. (12) indicate that leptin inhibits PTEN phosphatase activity via phosphorylation at multiple sites on the C terminus of PTEN, which results in increased PtdIns(3,4,5)P3 levels, actin restructuring, and KATP activation. These events appear to be mediated through CK2- and GSK3-dependent mechanisms. However, the dependence on PTEN Thr-366 phosphorylation on GSK3 activity and consequent influence on downstream signaling events such as KATP channel regulation gives rise to a quandary. Leptin stimulation of rat arcuate increases PKB phosphorylation with subsequent GSK3 phosphorylation at Ser-9/Ser-21 (13), actions also reported for leptin on mouse skeletal muscle cells (32, 33), mouse cortical neurons (34), and MCF-7 breast cancer cells (35). These latter actions of leptin replicate those of insulin, which increases PI3K activity and raises PtdIns(3,4,5)P3 levels with subsequent increased PKB and GSK3 phosphorylation (5). Importantly, the phosphorylation of GSK3 at these serine residues by PKB is associated with reduced kinase activity (36, 37).
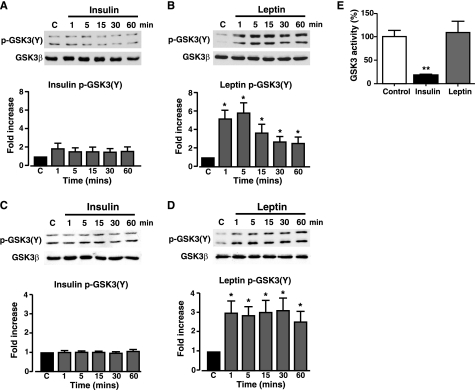
This outcome is inconsistent with the requirement for active GSK3 causing phosphorylation of PTEN Thr-366 allowing subsequent downstream signaling. Indeed, leptin (10 nm) increases the phosphorylation of PKB (Ser-473) and GSK3 (Ser-9/Ser-21) in N29/4 hypothalamic cells (Fig. 5A), although this is not well sustained in the hypothalamic cells, as previously reported for rat arcuate (13) and mouse muscle (32, 33). Insulin (10 nm) induced a sustained rise in PKB (Ser-473) and GSK3 (Ser-9/Ser-21) phosphorylation in N29/4 cells (Fig. 5B). Similar results were obtained for MIN6 β-cells, although in this case both leptin (10 nm) and insulin (10 nm) produced sustained increases (Fig. 5, C and D) in the levels of phosphorylated PKB (Ser-473) and GSK3 (Ser-9/Ser-21). These results are ostensibly inconsistent with the view that leptin signaling to actin and KATP channels requires GSK3-dependent phosphorylation and inhibition of PTEN activity. However, GSK3 also has a tyrosine residue in the activation loop of the enzyme (Tyr-216 in GSK3β and Tyr-279 in GSK3α), the phosphorylation of which is important for GSK3 activity (38, 39). Most studies conclude that this site is not physiologically regulated by growth factors or cytokines (40). Insulin (10 nm) did not affect GSK3 Tyr-216/Tyr-279 phosphorylation levels in either N29/4 cells (Fig. 6A) or MIN6 β-cells (Fig. 6C). In contrast, leptin (10 nm) rapidly increased GSK3 phosphorylation at Tyr-216/Tyr-279 in N29/4 (Fig. 6B), and MIN6 (Fig. 6D) cells, which was sustained for at least 1 h. Thus, in contrast to insulin, which increases phosphorylation of GSK3 serine sites only (consistent with kinase inhibition), leptin increases the phosphorylation level of both serine and tyrosine sites on GSK3. To define the outcome of this dual phosphorylation of GSK3, we examined the effects of leptin (10 nm) and insulin (50 nm) on GSK3 activity in N29/4 cells. Stimulation of N29/4 cells by insulin for 30 min results in a substantial inhibition of GSK3 activity, as expected from increased PKB activity causing GSK3 Ser-9/Ser-21 phosphorylation. In contrast, stimulation of N29/4 cells with leptin (10 nm) for 30 min, a time when PTEN phosphorylation is increased, PtdIns(3,4,5)P3 levels are high, F-actin is depolymerized, and β-cells are hyperpolarized, results in maintained GSK3 activity (Fig. 6E). This outcome is consistent with the finding that, although both hormones increase GSK3 Ser-9/Ser-21 phosphorylation levels, leptin, but not insulin, also increases GSK3 Tyr-216/Tyr-279 phosphorylation.
FIGURE 5.
Leptin and insulin increase PKB and GSK3 serine phosphorylation in N29/4 and MIN6 cells. PKB (Ser-473 (S)) and GSK3 (Ser-9/Ser-21) phosphorylation was determined in N29/4 (A and B) and MIN6 (C and D) cells, stimulated with 10 nm leptin (A and C), or 10 nm insulin (B and D) for the indicated times (minutes). The bar graphs below each set of blots show mean normalized p-PKB and p-GSK3 levels for untreated cells and cells stimulated with leptin (N29/4 (A, n = 9), MIN6 (C, n = 6)) or with insulin (N29/4 (B, n = 9), MIN6 (D, n = 6)). *, p < 0.05, compared with control level for each treatment. Cont, control.
FIGURE 6.
Leptin, but not insulin, increases GSK3 tyrosine phosphorylation in N29/4 and MIN6 cells. GSK3 (Tyr-216/Tyr-279 (Y)) phosphorylation was determined in N29/4 (A and B) and MIN6 cells (C and D) stimulated with 10 nm insulin (A and C) or 10 nm leptin (B and D) for the indicated times (minutes). The bar graphs below each set of blots show mean normalized p-GSK3 for untreated cells and cells stimulated with insulin (N29/4 (A, n = 9), MIN6 (C, n = 6)) or with leptin (N29/4 (B, n = 9), MIN6 (D, n = 6). *, p < 0.05, compared with control level for each treatment. E, N29/4 cells were stimulated with either 10 nm leptin or 50 nm insulin for 30 min. The bar graph denotes the mean normalized GSK3 activity and shows that insulin significantly inhibited GSK3 activity, whereas leptin had no effect on GSK3 activity (n = 3 for each). Cont, control.
DISCUSSION
Leptin phosphorylation of PTEN at the cluster site is CK2-dependent and leads to inhibition of PTEN phosphatase activity with a consequent increase in PtdIns(3,4,5)P3 levels in N29/4 cells (12). Here, we show that leptin increases PTEN phosphorylation at three further sites, Thr-366, Ser-370, and Ser-385 in N29/4 cells. We also show that leptin increases p-PTEN at the cluster site, Thr-366 and Ser-370, in MIN6 pancreatic β-cells. As demonstrated for the cluster site (12), phosphorylation of PTEN in N29/4 cells at Thr-366 and Ser-370 is important for transfer of the leptin signal downstream to cause F-actin depolymerization. Leptin-mediated reduction in F-actin levels was previously shown to be dependent on basal PI3K activity and inhibition of both lipid and protein phosphatase activity of PTEN (12). Correspondingly, overexpression of the PTEN phosphorylation site mutants, S370A PTEN and T366A PTEN, in N29/4 cells occluded leptin-driven reduction in F-actin. The Thr-366 and Ser-370 alanine and aspartic acid PTEN mutants, when overexpressed in N29/4 cells, alter PtdIns(3,4,5)P3 levels exactly as reported for the cluster site mutants. This outcome does not agree with in vitro assays, which show no alteration in enzyme activity associated with these mutants (41), unlike the corresponding mutations of the cluster site residues (12). Thus, S370D and T366D both raised PtdIns(3,4,5)P3 levels, indicative of reduced lipid phosphatase activity per se and dominant negative behavior with respect to native PTEN activity. Interestingly, these mutants produced different outcomes on F-actin levels and leptin signal transduction. Overexpression of T366D PTEN per se mimicked the effects of leptin by increasing PtdIns(3,4,5)P3 and inducing F-actin depolymerization in a PI3K-dependent manner. This replicates the effect of overexpression of C124S PTEN and D3PTEN (cluster site mutant) in these cells (12) and is consistent with phosphorylation of the Thr-366 site reducing PTEN lipid and protein phosphatase activity. In contrast, overexpression of S370D PTEN was unable to elicit F-actin depolymerization per se even in the face of increased cellular PtdIns(3,4,5)P3 levels and did not occlude leptin-mediated F-actin depolymerization. Consequently, mimicking phosphorylation by aspartate substitution at Ser-370 does not exactly replicate the effect of aspartate substitution at Thr-366 or the cluster phosphorylation site (12), with all three mutant forms of PTEN increasing PtdIns(3,4,5)P3 levels but only the latter two reducing F-actin levels. The S370D PTEN mutant also does not duplicate the action of the lipid phosphatase dead, protein phosphatase active, mutant G129E PTEN. Overexpression of G129E PTEN in N29/4 cells increases PtdIns(3,4,5)P3 levels with no reduction in F-actin and prevents leptin-mediated F-actin depolymerization (12), whereas the presence of S370D PTEN allows leptin depolymerization of actin. Our conclusion from these data is that S370D is lipid phosphatase dead but protein phosphatase active and that leptin inhibits the protein phosphatase activity by phosphorylation at the Thr-366 and cluster sites, whereas the protein phosphatase activity of G129E PTEN cannot be overcome by phosphorylation at these sites. Thus, phosphorylation of PTEN at Ser-370 is adequate to inhibit PTEN lipid phosphatase activity and increase PtdIns(3,4,5)P3 in N29/4 cells but is not sufficient per se for efficacious transmission of the leptin signal to F-actin.
A number of phosphorylation sites have been identified within the PTEN C-terminal tail, with the protein kinase, CK2, purported to phosphorylate multiple residues, including the cluster site, Ser-385 and Ser-370, but not Thr-366 (17–19, 23, 41). PTEN protein stability and activity are considered to be dependent on phosphorylation status of the C-terminal tail (17, 18), with phosphorylation argued to reduce association of PTEN with the plasma membrane (20, 42, 43). It is presently unclear what effect leptin-induced phosphorylation will have on PTEN protein stability, as it has been reported that T366A and S370A are more stable than wild-type PTEN and GSK3 inhibition increases the stability of wild-type PTEN (41), whereas alanine mutation of the cluster site results in reduced protein stability (17, 19). The inhibition of leptin-induced phosphorylation of the cluster site (12), Ser-370 and Ser-385, in N29/4 cells and of the cluster site and Ser-370 in MIN6 cells by DMAT supports the view that CK2 is the kinase responsible for phosphorylation at these sites by this hormone. Inhibition of PI3K had no effect on leptin-mediated phosphorylation at Ser-370, Ser-385, or Thr-366 in N29/4 cells, indicating no evidence for PI3K-dependent feedback phosphorylation (44) consistent with leptin-induced phosphorylation of the cluster site. For GSK3 to phosphorylate its substrate, the substrate has to be primed by phosphorylation of a site four or five residues C-terminal of the target Ser/Thr of GSK3 (45, 46). Thus, phosphorylation of Ser-370 by CK2 is considered a priming event for subsequent GSK3 phosphorylation of Thr-366 (23), and inhibition of GSK3 blocks phosphorylation of PTEN on Thr-366 (41). Occlusion of leptin driven-increased p-PTEN on Thr-366 by the GSK3 inhibitor CT99021 in N29/4 and MIN6 cells agrees with these findings. Consequently, we expected DMAT to reduce or inhibit leptin-mediated phosphorylation of Thr-366 in these cells. This was observed for MIN6 pancreatic β-cells but not for the hypothalamic cells, where DMAT prevented leptin-induced phosphorylation of Ser-370 but not Thr-366. Thus, either an additional unidentified priming kinase is responsible or there is sufficient phosphorylation of Ser-370 remaining in DMAT-treated N29/4 cells to allow GSK3 priming. Interestingly, an in vitro study of PTEN phosphorylation has shown that phosphorylation of Ser-370 by CK2 is less efficient than that of the cluster site and that GSK3 is capable of phosphorylating PTEN at Thr-366 without priming, although at low efficiency (41). Thus, we predicted that inhibition of CK2 and GSK3 would prevent or reduce transmission of the leptin signal via PTEN to actin filament restructuring and KATP channel opening. Our results show that the GSK3 inhibitor CT99021 per se or in combination with DMAT significantly reduces leptin-mediated F-actin depolymerization. Furthermore, leptin-driven hyperpolarization of pancreatic β-cells by KATP activation was prevented by the presence of DMAT or any one of three structurally dissimilar GSK3 inhibitors. This is consistent with the view that leptin-mediated phosphorylation of PTEN and subsequent inhibition of its phosphatase activity is a necessary step for signal transmission to KATP channels.
In contrast, insulin does not cause F-actin depolymerization in N29/4 cells nor does it hyperpolarize pancreatic β-cells by opening KATP channels, and this differential output may be mediated via PTEN (12). The results presented here indicate a key role for PTEN Thr-366 phosphorylation via GSK3 activity for successful transmission of the leptin signal. For this to occur, GSK3 activity has to be maintained or increased by leptin. However, leptin, like insulin, increases PKB phosphorylation with the resultant phosphorylation of GSK3 at Ser-9/Ser-21 consistent with inhibition of GSK3 activity. Indeed, both insulin and leptin increased the phosphorylation of GSK3 at Ser-9/Ser-21 in N29/4 and MIN6 cells. In contrast, leptin, but not insulin, caused a rapid and sustained increase in GSK3 phosphorylation at Tyr-216/Tyr-279 in both cell types. Phosphorylation of GSK3 at this site is considered to be activating and necessary for functional activity (38). Indeed, direct GSK3 kinase assay showed that insulin stimulation of N29/4 cells substantially reduced GSK3 activity, whereas leptin had no effect. Taken together, the semiquantitative Western blot data show that insulin significantly alters N-terminal serine phosphorylation of GSK3, which would reduce its activity toward primed substrates. However, phosphorylation of PTEN at Thr-366 in response to leptin does not appear to require priming (phosphorylation of this site is unaffected by DMAT), and this may explain why insulin does not reduce Thr-366 phosphorylation. In contrast, GSK3 in leptin-treated cells increased phosphate in Tyr-216/Tyr-279 compared with control or insulin-treated cells. This is a possible explanation why leptin and not insulin induces Thr-366 phosphorylation. However, there are other ways to regulate GSK3, including protein-protein interaction (e.g. Wnt signaling) and Thr-380 phosphorylation (e.g. cytokine signaling) that also require further investigation. Identification of the kinase (or phosphatase) responsible for the increase in tyrosine phosphorylation of GSK3 and development of a quantitative multiple reaction monitoring liquid chromatographymass spectrometric approach for each of the phosphorylation sites on GSK3 would be required to elucidate the exact mechanism of control by leptin. These subtle signaling differences downstream of insulin and leptin receptors are likely to contribute to the ability of each to perform their distinct physiological actions through modulation of the same signaling pathways/molecules.
In conclusion, this study has demonstrated that leptin increases the phosphorylation of PTEN at multiple residues in a CK2- and GSK3-dependent manner. This action of leptin is required for successful transmission of the leptin signal to cause cytoskeletal remodeling, the opening of KATP channels, and subsequent hyperpolarization of membrane potential. Interestingly, complete loss of PTEN activity in mouse hypothalamic proopiomelanocortin neurons permanently activates KATP channels and hyperpolarizes the neurons resulting in hyperphagia, leptin resistance, and diet-induced obesity (10). The role of GSK3 in PTEN regulation is intriguing. Insulin signaling inhibits GSK3 activity, and this may act via an alteration in PTEN Thr-366 phosphorylation as a negative feedback loop for leptin signaling in cells. In contrast, leptin, regardless of whether it stimulates PI3K activity in all cells or not, by maintaining GSK3 activity may result in a positive feedback to PI3K signaling in cells. Such interplay between leptin and insulin signaling in cells may have important implications for downstream signaling events. For example, inhibition of PTEN in peripheral tissues such as liver, muscle, and fat demonstrate improved insulin sensitivity and systemic glucose tolerance under normal and high fat diets (48). Dysregulation of PTEN (lipid and protein phosphatase) activity has also been implicated in certain cancers (49) and neurodegenerative disease (e.g. Parkinson disease (47)). Thus, modulation of PTEN function plays multiple and important roles in numerous physiological and pathological phenomena.
Acknowledgments
We thank Dr. Nick Leslie for providing the PTEN mutants and PTEN Ser-370 and Thr-366 phospho-antibodies and for helpful comments.
This work was supported by Wellcome Trust Grant 068692 (to M. L. J. A.), Tenovus Scotland, and a Biotechnology and Biological Sciences Research Council Studentship (to L. C. M.).

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: PI3K, phosphatidylinositol 3-kinase; DMAT, 2-dimethyl-amino-4,5,6,7-tetrabromo-1H-benzimidazole; p-, phosphorylated; PtdIns(3,4,5)P3, phosphatidylinositol-3,4,5-trisphosphate; PKB, protein kinase B; GSK3, glycogen synthase kinase 3.
References
- 1.Schwartz, M. W., and Porte, D., Jr. (2005) Science 307 375-379 [DOI] [PubMed] [Google Scholar]
- 2.O'Rahilly, S. (2007) Biochem. Soc. Trans. 35 3-36 [DOI] [PubMed] [Google Scholar]
- 3.Messier, C., and Teutenberg, K. (2005) Neural Plast. 12 311-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plum, L., Schubert, M., and Brüning, J. C. (2005) Trends Endocrinol. Metab. 16 59-65 [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi, C.M., Emanuelli, B., and Khan, C.R. (2006) Nat. Rev. Mol. Cell Biol. 7 85-96 [DOI] [PubMed] [Google Scholar]
- 6.Niswender, K. D., and Schwartz, M. W. (2003) Front. Neuroendocrinol. 24 1-10 [DOI] [PubMed] [Google Scholar]
- 7.Niswender, K. D., Baskin, D. G., and Schwartz, M. W. (2004) Trends Endocrinol. Metab. 15 362-369 [DOI] [PubMed] [Google Scholar]
- 8.Frübeck, G. (2006) Biochem. J. 393 7-2016336196 [Google Scholar]
- 9.Choudhury, A. I., Heffron, H., Smith, M. A., Al-Qassab, H., Xu, A. W., Selman, C., Simmgen, M., Clements, M., Claret, M., MacColl, G., Bedford, D. C., Hisadome, K., Diakonov, I., Moosajee, V., Bell, J. D., Speakman, J. R., Batterham, R. L., Barsh, G. S., Ashford, M. L. J., and Withers, D. J. (2005) J. Clin. Investig. 115 940-950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plum, L., Ma, X., Hampel, B., Balthasar, N., Coppari, R., Münzberg, H., Shanabrough, M., Burdakov, D., Rother, E., Janoschek, R., Alber, J., Belgardt, B. F., Koch, L., Seibler, J., Schwenk, F., Fekete, C., Suzuki, A., Mak, T.W., Krone, W., Horvath, T. L., Ashcroft, F. M., and Brüning, J. C. (2006) J. Clin. Investig. 116 1886-1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, A. W., Kaelin, C. B., Takeda, K., Akira, S., Schwartz, M. W., and Barsh, G. S. (2005) J. Clin. Investig. 115 951-958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning, K., Miller, L. C., Laidlaw, H. A., Burgess, L. A., Perera, N. M., Downes, C. P., Leslie, N. R., and Ashford, M. L. J. (2006) EMBO J. 25 2377-2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirshamsi, S., Laidlaw, H. A., Ning, K., Anderson, E., Burgess, L. A., Gray, A., Sutherland, C., and Ashford, M. L. J. (2004) BMC Neurosci 5 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey, J., McKay, N. G., Walker, K. S., Van der Kaay, J., Downes, C. P., and Ashford, M. L. J. (2000) J. Biol. Chem. 275 4660-4669 [DOI] [PubMed] [Google Scholar]
- 15.Harvey, J., Hardy, S. C., Irving, A. I., and Ashford, M. L. J. (2000) J. Physiol. (Lond.) 527 95-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downes, C. P., Ross, S., Maccario, H., Perera, N., Davidson, L., and Leslie, N. R. (2007) Adv. Enzyme Regul. 47 184-194 [DOI] [PubMed] [Google Scholar]
- 17.Torres, J., and Pulido, R. (2001) J. Biol. Chem. 276 993-998 [DOI] [PubMed] [Google Scholar]
- 18.Miller, S. J., Lou, D. Y., Seldin, D. C., Lane, W. S., and Neel, B. G. (2002) FEBS Lett. 528 145-153 [DOI] [PubMed] [Google Scholar]
- 19.Vasquez, F., Ramaswamy, S., Nakamura, N., and Sellers, W. R. (2000) Mol. Cell. Biol. 20 5010-5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasquez, F., Grossman, S. R., Takahashi, Y., Rokas, M. V., Nakamura, N., and Sellars, W. R. (2001) J. Biol. Chem. 276 48627-48630 [DOI] [PubMed] [Google Scholar]
- 21.Leslie, N. R., and Downes, C. P. (2002) Cell. Signal. 14 285-295 [DOI] [PubMed] [Google Scholar]
- 22.Leslie, N. R., and Downes, C. P. (2004) Biochem. J. 382 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Khouri, A. M., Ma, Y., Togo, S. H., Williams, S., and Mustelin, T. (2005) J. Biol. Chem. 280 35195-35202 [DOI] [PubMed] [Google Scholar]
- 24.Belsham, D. D., Cai, F., Cui, H., Smukler, S. R., Salapatek, A. F., and Shkreta, L. (2004) Endocrinology 145 393-400 [DOI] [PubMed] [Google Scholar]
- 25.Finlay, D., Patel, S., Dickson, L. M., Shpiro, N., Marquez, R., Rhodes, C. J., and Sutherland, C. (2004) BMC Mol. Biol. 5 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagano, M.A., Meggio, F., Ruzzene, M., Andrzejewska, M., Kazimierczuk, Z., and Pinna, L.A. (2004) Biochem. Biophys. Res. Commun. 321 1040-1044 [DOI] [PubMed] [Google Scholar]
- 27.Sarno, S., Ruzzene, M., Frascella, P., Pagano, M. A., Meggio, F., Zambon, A., Mazzorana, M., Di Maira, G., Lucchini, V., and Pinna, L. A. (2005) Mol. Cell. Biochem. 274 69-76 [DOI] [PubMed] [Google Scholar]
- 28.Murray, J. T., Campbell, D. G., Morrice, N., Auld, G., Shpiro, N., Marquez, R., Peggie, M., Bain, J., Bloomberg, G. B., Grahammer, F., Lanf, F., Wulff, P., Kuhl, D., and Cohen, P. (2004) Biochem. J. 384 477-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bain, J., Plater, L., Elliot, M., Shpiro, N., Hastie, C. J., McLauchlan, H., Klevernic, I., Arthur, J. S. C., Alessi, D. R., and Cohen, P. (2007) Biochem. J. 408 297-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey, J., McKenna, F., Herson, P. S., Spanswick, D., and Ashford, M. L. J. (1997) J. Physiol. (Lond.) 504 527-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat, R., Xue, Y., Berg, S., Hellberg, S., Ormö, M., Nilsson, Y., Radesäter, A., Jerning, E., Markgren, P.-O., Borgegård, T., Nylöf, M., Giménez-Cassina, A., Hernández, F., Lucas, J. J., Díaz-Nido, J., and Avila, J. (2003) J. Biol. Chem. 278 45937-45945 [DOI] [PubMed] [Google Scholar]
- 32.Maroni, P., Bendinelli, P., and Piccoletti, R. (2003) Mol. Cell. Endocrinol. 201 109-121 [DOI] [PubMed] [Google Scholar]
- 33.Maroni, P., Bendinelli, P., and Piccoletti, R. (2005) Cell Biol. Int. 29 542-550 [DOI] [PubMed] [Google Scholar]
- 34.Valerio, A., Ghisi, V., Dossena, M., Tonello, C., Giordana, A., Frontini, A., Ferrario, M., Pizzi, M., Spano, P., Carruba, M. O., and Nisoli, E. (2006) J. Biol. Chem. 281 12950-12958 [DOI] [PubMed] [Google Scholar]
- 35.Garofalo, C., Sisci, D., and Surmacz, E. (2004) Clin. Cancer Res. 10 6466-6475 [DOI] [PubMed] [Google Scholar]
- 36.Sutherland, C., Leighton, I. A., and Cohen, P. (1993) Biochem. J. 296 15-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785-789 [DOI] [PubMed] [Google Scholar]
- 38.Hughes, K., Nikolakaki, E., Plyte, S. E., Totty, N. F., and Woodgett, J. R. (1993) EMBO J. 12 803-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Q. M., Fiol, C. J., DePaoli-Roach, A. A., and Roach, P. J. (1994) J. Biol. Chem. 269 14566-14574 [PubMed] [Google Scholar]
- 40.Cole, A., Frame, S., and Cohen, P. (2004) Biochem. J. 377 249-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maccario, H., Perera, N. M., Davidson, L., Downes, C. P., and Leslie, N. R. (2007) Biochem. J. 405 439-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasquez, F., Matsuoka, S., Sellers, W. R., Yanagida, T., Ueda, M., and Devreotes, P. N. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3633-3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das, S., Dixon, J. E., and Cho, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7491-7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birle, D., Bottini, N., Williams, S., Huynh, H., deBelle, I., Adamson, E., and Mustelin, T. (2002) J. Immunol. 169 286-291 [DOI] [PubMed] [Google Scholar]
- 45.Frame, S., and Cohen, P. (2001) Biochem. J. 359 1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cole, A. R., Knebel, A., Morrice, N. A., Robertson, L. A., Irving, A. J., Connolly, C. N., and Sutherland, C. (2004) J. Biol. Chem. 279 50176-50180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, R. H., and Mak, T. W. (2006) Br. J. Cancer 94 620-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasaoka, T., Wada, T., and Tsuneki, H. (2006) Pharmacol. Ther. 112 799-809 [DOI] [PubMed] [Google Scholar]
- 49.Knobbe, C. B., Lapin, V., Suzuki, A., and Mak, T. W. (2008) Oncogene 27 5398-5415 [DOI] [PubMed] [Google Scholar]