Abstract
Homologous pairing, an essential process for homologous recombination, is the formation of a heteroduplex joint by an invading single-stranded DNA tail and a complementary sequence within double-stranded DNA (dsDNA). The base rotation of the parental dsDNA, to switch from parental base pairs to heteroduplex ones with the invading single-stranded DNA, sterically requires vertical extension between adjacent base pairs, which inevitably induces untwisting of the dsDNA. RecA is a prototype of the RecA/Rad51/Dmc1 family proteins, which promote ATP-dependent homologous pairing in homologous DNA recombination in vivo, except in mitochondria. As predicted by the requirement for the untwisting, dsDNA bound to RecA is extended and untwisted, and homologous pairing by RecA in vitro is extensively stimulated by the negative supercoils of dsDNA substrates. D-loop formation in negatively supercoiled dsDNA, which serves as an assay for homologous pairing, is also catalyzed in an ATP-independent manner by proteins structurally unrelated to RecA, such as Mhr1. Mhr1 is required for yeast mitochondrial DNA recombination instead of RecA family proteins. Inconsistent with the topological requirements, tests for the effects of negative supercoils revealed that Mhr1 catalyzes homologous pairing with relaxed closed circular dsDNA much more efficiently than with negatively supercoiled dsDNA. Topological analyses indicated that neither the process nor the products of homologous pairing by Mhr1 involve a net topological change of closed circular dsDNA. This would be favorable for homologous recombination in mitochondria, where dsDNA is unlikely to be under topological stress toward unwinding. We propose a novel topological mechanism wherein Mhr1 induces untwisting without net topological change.
Homologous DNA recombination plays critical roles in the repair of double-stranded DNA (dsDNA)3 breaks and meiotic disjunctions of homologous chromosomes in the nuclear genome (for reviews, see Refs. 1 and 2) and in DNA replication as well as DNA repair in yeast mitochondria (3) (see Ref. 4 for review). A key intermediate step in homologous recombination is homologous pairing, in which a single-stranded DNA (ssDNA) tail derived from a double-stranded break invades a homologous sequence within intact dsDNA, resulting in heteroduplex joint formation with the complementary sequence of the dsDNA by replacing a parental strand. Heteroduplex joints are formed by switching base pairs of the parental dsDNA to heteroduplex base pairs between the invading ssDNA and the complementary sequence of the dsDNA, which requires extension between adjacent base pairs of the dsDNA to provide a space for the rotation of bases for the base pair switch (5). The extension of dsDNA is necessarily associated with its untwisting (6, 7), which generates positive supercoiling in the remaining region in closed circular dsDNA (cc-dsDNA; Fig. 1, from DNA 1 to DNA 2). A negatively supercoiled cc-dsDNA substrate neutralizes the positive supercoils, but a relaxed cc-dsDNA substrate accumulates the positive supercoils, which will resist the formation of heteroduplex joints.
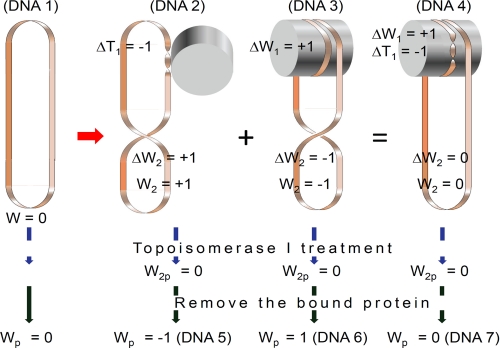
FIGURE 1.
Topological consequences of cc-dsDNA binding to a protein and a model for the binding of cc-dsDNA to Mhr1. The extension and associated untwisting of cc-dsDNA that are required for homologous pairing are induced by RecA/Rad51 binding (29) and generate positive (or left-handed) supercoils in the remaining part of the cc-dsDNA (DNA 2). This type of untwisting is observed as the formation of negatively supercoiled cc-dsDNA by the topoisomerase I treatment of cc-dsDNA bound to RecA/Rad51, followed by the removal of proteins (DNA 5). When the substrate cc-dsDNA for homologous pairing is negatively supercoiled, the positive supercoils that are generated during the reaction are neutralized, and thus, the negative supercoils stimulate homologous pairing by RecA (11). On the other hand, the right-handed wrapping around a protein generates negative (or right-handed) supercoils (DNA 3). This is observed as the generation of positively supercoiled cc-dsDNA, when the cc-dsDNA bound to human Rad52 is treated with topoisomerase I and the proteins are removed (DNA 6) (41). The positive supercoils generated by untwisting can be canceled by the negative supercoils generated by the right-handed wrapping (DNA 4). If the substrate cc-dsDNA is negatively supercoiled, then the negative supercoils prevent right-handed wrapping. These phenomena explain our finding that the negative supercoils of cc-dsDNA prevent homologous pairing by Mhr1 (Figs. 3, 4, 6, and 7) and that the treatment of the cc-dsDNA-Mhr1 complex with topoisomerase generates neither positive nor negative supercoils, as shown in Fig. 8B. The cancellation of the positive supercoils and the negative supercoils is not necessarily complete. In the case of the Rad52-cc-dsDNA complex, the negative supercoils generated by the wrapping are larger than the positive supercoils generated by the untwisting. This figure represents the change by a single unit of supercoil or twist, caused by the binding of originally relaxed cc-dsDNA (DNA 1) to a protein for simplicity. W, writhing number of the entire cc-dsDNA; Wn, local writhing number; ΔWn, local changes in writhe (positive values, left-handed supercoiling or right-handed wrapping); ΔTn, local change in twist (negative values, untwisting) in region n. Subscripts 1 and 2 indicate the protein-bound region of the DNA molecule and the remaining region of the DNA (where ΔT2 = 0), respectively. The subscript p indicates the values after topoisomerase treatment. Before topoisomerase treatment, the sum of the local changes in writhe and twist is zero, because there is no change in the linking number. If the substrate dsDNA is relaxed (W = 0), then W2 is equal to ΔW2. The treatment of the protein-DNA complex by topoisomerase I results in the relaxing of W2 (to become 0). After the removal of proteins, Wp, which is the sum of W1 and W2 (equal to 0), is equal to the sum of ΔT1 and ΔW1.
Homologous pairing activities are typically assayed in vitro by examining the formation of D-loops, using negatively supercoiled cc-dsDNA (natural cc-dsDNA, called “form I”) and homologous ssDNA oligonucleotides as substrates (8, 9). A D-loop consists of a heteroduplex joint and a loop of a displaced parental strand of the dsDNA, with the associated untwisting of the parental dsDNA (9, 10). Thus, the formation of a D-loop in cc-dsDNA by itself also generates positive supercoils. Consistent with these topological requirements in homologous pairing and D-loop formation, negative supercoils were shown to be essential for uncatalyzed D-loop formation (10) and to stimulate RecA- and Rad51-catalyzed D-loop formation extensively, as compared with the initial velocity with dsDNA without supercoils (11, 12). RecA is the prototype of the RecA/Rad51/Dmc1 family proteins, which are essential to catalyze homologous pairing through an ATP-dependent reaction in homologous DNA recombination in prokaryotic genomes and eukaryotic nuclei (8, 13–17). Cellular DNA is likely to be relaxed by the potent activities of DNA topoisomerases, but the factors required to fulfill the topological requirement for homologous pairing in vivo remain unidentified, especially in mitochondria.
Homologous recombination in bacterial viruses and the mitochondria of yeasts and mammals is independent of the RecA/Rad51/Dmc1 family proteins. Instead, we found that in yeast, mtDNA recombination depends on Mhr1, a protein structurally unrelated to RecA/Rad51 (3, 18) and that Mhr1-catalyzes in vitro D-loop formation in an ATP-independent manner (19). Mitochondrial DNA (mtDNA) encodes essential components of the machinery that drives oxidative respiration-dependent energy production. mtDNA undergoes extensive mitotic homologous recombination in the yeast Saccharomyces cerevisiae (20) and humans (21, 22). In addition, Mhr1 is a critical player in the vegetative segregation of heteroalleles that leads to a genetic state in which all of the copies of mtDNA in each cell or in each individual (in the case of healthy human babies) share an identical sequence (called “homoplasmy”) (20). The function of Mhr1 in homoplasmy is in sharp contrast to the generally accepted roles of homologous recombination in nuclear genomes; i.e. homologous recombination, and in particular meiotic recombination, contributes to genetic diversification. Mhr1 functions in homoplasmy to initiate the rolling circle mode of mtDNA replication to form concatemers, which are selectively transmitted to daughter cells, leading to homoplasmy (19, 23). RecA does not initiate this mode of DNA replication in wild type Escherichia coli cells (24). Thus, it is important to identify the differences and similarities between the homologous pairing catalyzed by the RecA/Rad51/Dmc1 family proteins and that catalyzed by Mhr1 to provide insights into the differences between the two genetic outcomes.
The ATP-independent D-loop formation mediated by Mhr1 and other proteins structurally unrelated to RecA may be a type of complementary strand annealing (i.e. a two-strand reaction) through the mechanism proposed for uncatalyzed D-loop formation. This idea is based on the following observations. Negatively supercoiled dsDNA tends to melt and create ssDNA regions (9), and some non-RecA proteins (λ phage β protein, RecT, Rad52, and RecO) have potent activity to anneal complementary ssDNA molecules (25–28). If this model is correct, then Mhr1 would absolutely require negative supercoiling for the apparently homologous pairing. Alternatively, the D-loop formation catalyzed by non-RecA proteins is a true homologous pairing reaction, including ssDNA invading an internal, homologous sequence in dsDNA (i.e. a three-strand reaction). This model was proposed based on structural analysis of an ssDNA oligonucleotide bound to either RecA or Rad51 in the presence of a nonhydrolyzable ATP analogue, which suggested that homologous pairing is an intrinsic function of DNA molecules rather than a specific function of proteins (5, 29).
In this study, topological analyses originally aimed at testing the above alternative models revealed an unexpected feature of the Mhr1-catalyzed homologous pairing, which is prevented by negative supercoils and generates no net topological changes of dsDNA substrates during and after the reactions. This finding suggests a novel mechanism wherein Mhr1 extends and untwists dsDNA to promote homologous pairing.
EXPERIMENTAL PROCEDURES
Purification of Mhr1—The purified Mhr1 used in this study was obtained as previously described (19).
Standard Reaction Buffer—The standard buffer consisted of 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, and 1 mm dithiothreitol.
DNA Substrates—The methods for the preparation of supercoiled pGsat4 and fX174 RF I (replicative form I) cc-dsDNA and ssDNA oligonucleotide (50-mer oligonucleotides) and for the 5′-end labeling of the ssDNA oligonucleotide with 32P were described previously (19). Negatively supercoiled pUC18 plasmid DNA was prepared from pUC18 plasmid DNA-harboring E. coli XL2-blue cells (endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F′ [::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK- mK+)) (Stratagene), using an illustra plasmidPrep Mini Spin Kit (GE Healthcare). The amounts of DNA are expressed as the amounts of nucleotides, unless otherwise stated.
Preparation of Relaxed cc-dsDNA—Partially or fully relaxed cc-dsDNA was prepared by treating negatively supercoiled pUC18 plasmid DNA (209 μm in nucleotides) with 0.025 units/μl or 0.1 units/μl calf thymus topoisomerase I (Takara Shuzo Co., Kyoto, Japan) in a 20-μl reaction mixture at 37 °C for 30 min. The topoisomerase I was then completely removed from the reaction solution by a phenol/chloroform extraction, and the DNA was purified with an illustra plasmidPrep Mini Spin Kit. When we analyzed the Mhr1-catalyzed formation of three-stranded structures in the presence of active topoisomerase I, which should release any topological stress during the reaction, the topoisomerase removal and DNA purification procedures were not performed. Two-dimensional gel electrophoresis was performed as described previously to analyze the relaxed cc-dsDNA (30, 31).
Standard D-loop Formation Assay—The standard reaction mixture (20.5 μl) for the assay consisted of 15.6 mm Tris-HCl (pH 7.5), 1.8 mm dithiothreitol, 88 μg/ml bovine serum albumin, and 1 mm MgCl2. After the 32P-labeled ssDNA oligonucleotide (1.0 μm final concentration after dsDNA was added, unless otherwise stated) was incubated with 1.95 μm (final concentration) Mhr1 in the reaction mixture (19.5 μl) for 5 min at 37 °C, 1 μl of dsDNA was added to a concentration of 15 μm, and the mixture was incubated at 37 °C for the indicated period of time. For the reaction with E. coli RecA, 1.3 mm ATP and MgCl2 (final concentration, 13 mm) were added together with the dsDNA. After the proteins were removed, the products (D-loops) were assayed by agarose gel electrophoresis. After electrophoresis, the gel was dried and exposed to an imaging plate, which was analyzed with a Fuji BAS2000 image analyzer. Details of the assay have been described previously (19).
A Restriction Endonuclease Protection Assay for Homologous Pairing—The homologous three-stranded structures, formed as a result of homologous pairing, are protected from restriction enzymes. A restriction endonuclease protection assay based on this principle was performed according to a previously described method (32), with a slight modification. In four sets of the reaction mixture (7.0 μl), Mhr1 at various concentrations was incubated with an ssDNA oligonucleotide (1.3 μm final concentration after dsDNA was added) bearing the sequence encompassing either the NdeI site from pUC18 (NdeI ssDNA oligonucleotide, 5′-CGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGAT-3′; in two sets) or the SspI site from pUC18 (SspI ssDNA oligonucleotide, 5′-AATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATT-3′; in two sets) in the standard reaction buffer at 37 °C for 30 min. After the incubation, to each sample containing the Mhr1-ssDNA oligonucleotide complexes (7.0 μl), 3.0 μl of supercoiled or relaxed pUC18 dsDNA or ScaI-linearized pUC18 dsDNA (to a final concentration of 41.8 μm) were added, and the mixture was incubated at 37 °C for 30 min. To each one from the two sets of reaction mixtures containing the same ssDNA oligonucleotide, the NdeI or SspI restriction endonuclease (1.0 unit/μl or 0.5 units/μl, respectively) was then added, and the samples were incubated at 37 °C for 30 min to allow DNA cleavage.
When the formation of homologous three-stranded structures by Mhr1 was analyzed in the absence of topological stress (i.e. in the presence of eukaryotic topoisomerase I, which relaxes both negative and positive supercoils), we first relaxed the samples of negatively supercoiled pUC18 cc-dsDNA (209 μm) with 0.025 units/μl or 1.0 unit/μl calf thymus topoisomerase I at 37 °C for 30 min in a 20-μl reaction mixture. After a 1.5-fold dilution of the reaction solution containing the relaxed pUC18 cc-dsDNA (final concentration, 41.8 μm) and active topoisomerase I (final concentration, 0.005 or 0.2 units/μl) by the standard reaction buffer, 3 μl of the diluted dsDNA containing the topoisomerase were added to the Mhr1-ssDNA oligonucleotide complexes (7.0 μl) and mixed, and the mixture was incubated at 37 °C for 30 min. The NdeI was then added, and the samples were incubated at 37 °C for 30 min.
After the DNA substrates were incubated with Mhr1 and then treated with a restriction enzyme, all of the proteins were removed by the addition of 1 μl of 10% SDS and 1 μl of 10 mg/ml proteinase K, followed by an incubation at 37 °C for 15 min. The DNA was then analyzed by electrophoresis on 1% agarose gels in the presence of 0.3 μg/ml ethidium bromide. The signals from the various DNA species in the gel were quantified by a Southern blot analysis, using [32P] pUC18 DNA as a probe, after the DNA was transferred to N+ nylon membranes. The signals were analyzed with a Fuji BAS2000 image analyzer (19).
Electrophoretic Assay for the Protein-free Products of Homologous Pairing by Mhr1 in the Presence of Topoisomerase I Using cc-dsDNA Substrates—In each reaction (20 μl), negatively supercoiled cc-dsDNA (pUC18 plasmid DNA (245 μm) or φX174 RF I cc-dsDNA (172 μm)) was treated in the standard reaction buffer with 1.0 unit/μl calf thymus topoisomerase I at 37 °C for 30 min. Then 3.0 μl of relaxed cc-dsDNA (final concentration in the reaction mixture, 49.0 μm for pUC18 plasmid DNA and 34.5 μm for φX174 RF I cc-dsDNA) in standard buffer containing active topoisomerase were added to 7.0 μl of a solution containing 32P-labeled ssDNA oligonucleotide (1.3 μm calculated final concentration, assuming that all DNA was recovered during the labeling and purifying processes) and 0 or 3.9 μm (final concentration) Mhr1 in the standard reaction buffer, and the reaction mixture was incubated at 37 °C for 30 min in the presence of 0.2 units/μl topoisomerase I. After the proteins were removed by adding 1 μl of 10% SDS and 1 μl of 10 mg/ml proteinase K at 37 °C for 20 min, the DNA products were analyzed by electrophoresis on 1% agarose gels (2 V/cm) for 11 h in a cold room (4 °C). The products were also assayed by two-dimensional electrophoresis on 1% agarose gels in the cold room, as described (30, 31). After electrophoresis, the gel was dried and exposed to an imaging plate, which was analyzed with a Fuji BAS2000 image analyzer, as described previously (19).
Assay for the Topological Status of the Products of Homologous Pairing by Mhr1—Relaxed cc-dsDNA was prepared by incubating negatively supercoiled pUC18 plasmid DNA (209 μm) with 1.0 unit/μl calf thymus topoisomerase I at 37 °C for 30 min. To initiate Mhr1-catalyzed homologous pairing, the relaxed pUC18 cc-dsDNA and active calf thymus topoisomerase I were added to the reaction mixture (the final dsDNA concentration and the amount of the topoisomerase I were ∼41.8 μm and 0.2 units/μl, respectively) containing Mhr1-NdeI ssDNA oligonucleotide complexes, formed by the incubation of 1.3 μm NdeI-ssDNA oligonucleotides with 3.9 μm Mhr1. The reaction mixture was incubated at 37 °C for 30 min. As a control, Mhr1 was omitted from the reaction. After a phenol/chloroform extraction, the DNA products were alkaline-treated (pH 13.0) to dissociate the ssDNA oligonucleotide from the cc-dsDNA, neutralized to reform the double helix, and then analyzed by two-dimensional electrophoresis on 1% agarose gels. The first and second dimensions of gel electrophoresis were both performed at 1.3 V/cm for 11 h at room temperature, but the second one was performed in the presence of 0.02 μg/ml ethidium bromide at room temperature, as described (30, 31).
Assay for cc-dsDNA Binding by Mhr1—In each reaction (10 μl), negatively supercoiled cc-dsDNA (pUC18 plasmid DNA, 41.8 μm in nucleotides) was mixed with Mhr1 at various concentrations (0, 0.93, and 1.9 μm) in the standard reaction buffer and incubated at 37 °C for 30 min. The Mhr1-cc-dsDNA complexes were then subjected to a gel shift assay, as previously described (19).
Assay for Topological Changes of cc-dsDNA Due to Mhr1 Binding—In each reaction (20 μl), negatively supercoiled cc-dsDNA (pUC18 plasmid DNA; 209 μm) was treated with 1.0 unit/μl calf thymus topoisomerase I at 37 °C for 30 min. Then the reaction mixture was diluted 1.5-fold by the standard reaction buffer, 3 μl of the diluted solution (containing relaxed cc-dsDNA, final concentration 41.8 μm) were added to 7.0 μl of a solution containing Mhr1 at various concentrations (0, 0.93, and 1.9 μm), and the solution was incubated at 37 °C for 30 min in the presence of 0.2 units/μl topoisomerase I. The DNA products were analyzed by electrophoresis on 1% agarose gels (2 V/cm) for 11 h at room temperature, after the topoisomerase I was removed by a phenol/chloroform extraction.
RESULTS
Mhr1 Promotes D-loop Formation in Vitro from Negatively Supercoiled cc-dsDNA and Homologous ssDNA Oligonucleotides—When negatively supercoiled cc-dsDNA and homologous ssDNA oligonucleotide fragments (or ssDNA oligonucleotides) are incubated in the presence of an excess amount of an ATP-dependent homologous pairing protein (RecA or Dmc1), D-loops are quickly formed and then are actively dissociated in an ATP-hydrolysis-dependent manner. This process occurs as a part of “the D-loop cycle” (34–36). We found that purified Mhr1 promotes D-loop formation from these substrates via a reaction that is independent of ATP (19). Unlike the RecA/Dmc1-catalyzed reaction, in the Mhr1-catalyzed reaction, the D-loops were formed as a result of a simple time-dependent reaction (Fig. 2). D-loop formation depended on the presence of Mhr1 and the homology between the dsDNA and ssDNA oligonucleotide (Fig. 2). After the proteins were removed, the products of the Mhr1-promoted reaction were determined to be authentic D-loops, because they migrated on gels with the same velocity as that observed for D-loops formed by RecA, and they dissociated following the cleavage of the dsDNA outside the homologous region (19). This is critical proof for D-loop formation by Mhr1, since it is well established that in the absence of a topological constraint toward untwisting (generated by negative supercoils of dsDNA substrate), D-loops spontaneously and quickly dissociate (37).
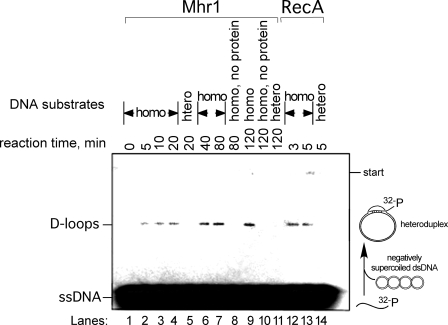
FIGURE 2.
D-loop formation promoted by Mhr1. [32P]ssDNA oligonucleotides (1.0 μm final concentration after dsDNA was added; SAT-1 from pGsat4) were incubated with 1.95 μm Mhr1 for 5 min, and then negatively supercoiled homologous (pGsat4; lanes 1–4, 6, 7, 9, and 19) or heterologous (φX174) cc-dsDNA (15 μm) was added, and the samples were incubated at 37 °C for the indicated periods of time. After the proteins were removed, the products were analyzed by agarose gel electrophoresis. Control samples were as follows. Lanes 5, 11, and 14, heterologous combination; lanes 8 and 10, no protein; lanes 12–14, E. coli RecA (5 μm) instead of Mhr1 with 13 mm Mg2+ and ATP.
We next studied the effects of negative supercoils and topological constraints of cc-dsDNA substrates on Mhr1-catalyzed homologous pairing. Due to the characteristics described above, the D-loop formation assay is not suitable to detect the products of homologous pairing using dsDNA substrates with variable topological conditions. Therefore, we quantified the Mhr1-catalyzed homologous pairing activity using the restriction endonuclease protection assay described by Ferrin and Camerini-Otero (38) as a reliable assay for homologous pairing, to test the effects of supercoils and topological constraints. This assay is based on the principle that the three-stranded structures formed through homologous pairing of dsDNA and a ssDNA oligonucleotide by homologous DNA-pairing proteins, such as RecA, are resistant to restriction endonucleases (38). To detect the formation of homologous three-stranded structures, we first used negatively supercoiled plasmid cc-dsDNA (pUC18) bearing a single NdeI cleavage site and a single SspI site as the dsDNA substrate and ssDNA oligonucleotides containing a sequence homologous to the region around the NdeI (NdeI ssDNA oligonucleotide) or SspI (SspI ssDNA oligonucleotide) cleavage site. If three-stranded structures formed as a result of homologous pairing after the incubation of the dsDNA with the NdeI or SspI ssDNA oligonucleotide and a homologous pairing protein, then the dsDNA would become resistant to NdeI or SspI, respectively, whereas it would remain sensitive to SspI or NdeI, respectively (Fig. 3A, a). The resistance was dependent on the incubation with Mhr1 and the combination of the oligonucleotide and the restriction endonuclease that recognized the restriction site within the oligonucleotide (Fig. 3B, a). For optimal protection of the dsDNA by Mhr1, the ssDNA oligonucleotide and Mhr1 had to be incubated together before the addition of dsDNA; the simultaneous addition of the ssDNA oligonucleotide and dsDNA decreased the degree of protection, whereas the incubation of the dsDNA with Mhr1 before the addition of the ssDNA oligonucleotide did not protect the dsDNA from the endonuclease (data not shown).
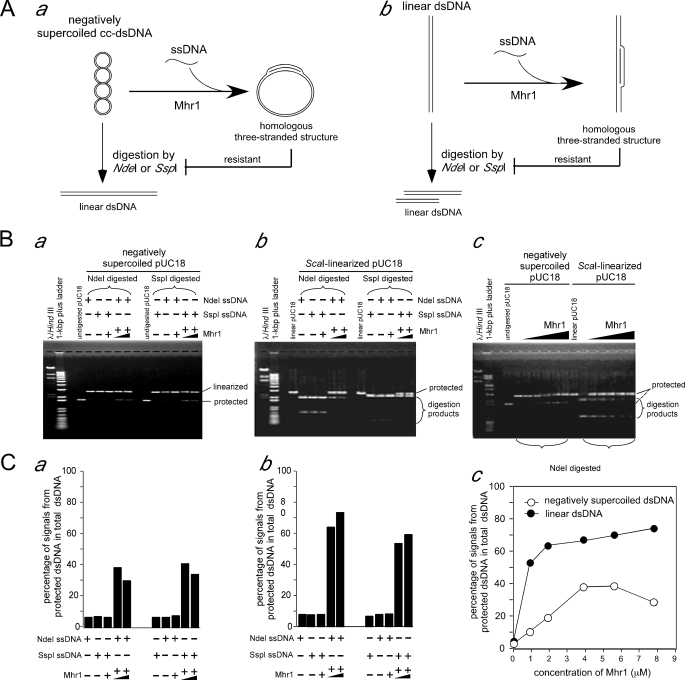
FIGURE 3.
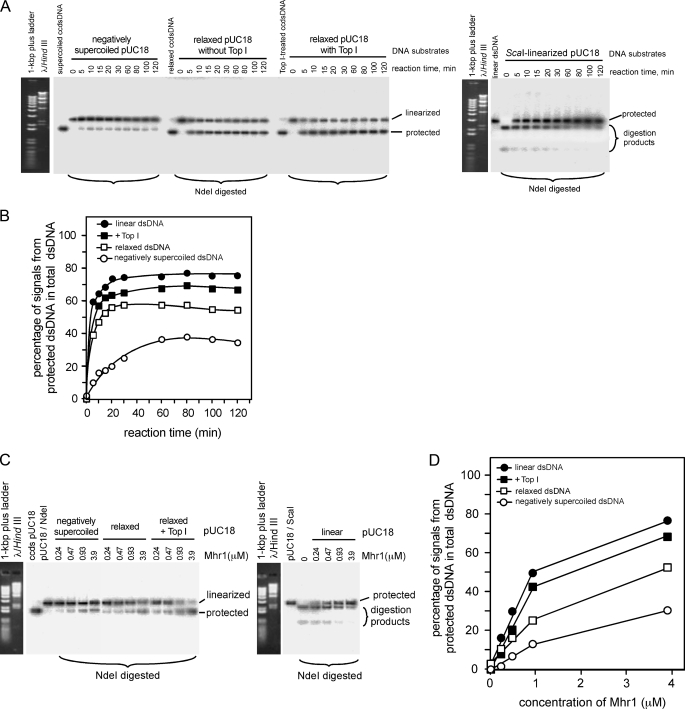
Mhr1-catalyzed homologous three-stranded structure formation with negatively supercoiled cc-dsDNA or linear dsDNA measured by a restriction endonuclease protection assay. A, schematic diagrams of the restriction endonuclease protection assay for the homologous pairing of a ssDNA oligonucleotide with negatively supercoiled cc-dsDNA (a) or linear dsDNA (b). B, homologous three-stranded structures formed using negatively supercoiled cc-dsDNA (a and the left half of c) or linear dsDNA (b and the right half of c). After the Mhr1-ssDNA oligonucleotide complexes were formed by incubating Mhr1 with the NdeI ssDNA oligonucleotide or the SspI ssDNA oligonucleotide at 37 °C for 30 min, the dsDNA solution was then added to initiate the Mhr1-catalyzed reaction at 37 °C for 30 min. All of the DNA and protein concentrations are the final concentrations after the formation of the complete reaction mixture, unless otherwise stated. In this experiment, two sets (in a and b) or a single set of 41.8 μm negatively supercoiled pUC18 cc-dsDNA (in a and c) or linear dsDNA (in b and c) were incubated with 1.3 μm NdeI ssDNA oligonucleotide (in a–c) or SspI ssDNA oligonucleotide (in a and b) and Mhr1 (3.9 or 7.8 μm in a and b; 0, 0.93, 1.9, 3.9, 5.6, or 7.8 μm in c) at 37 °C for 30 min. In these experiments, ssDNA oligonucleotide and Mhr1 were incubated at 37 °C for 30 min before the addition of dsDNA. After digestion with NdeI or SspI, the proteins were removed by treatments with SDS and proteinase K, and the DNA products were subjected to electrophoresis on 1% agarose gels in the presence of 0.3 μg/ml ethidium bromide. NdeI and SspI each cleave pUC18-dsDNA at a single site. The homologous three-stranded structures formed with the NdeI or SspI ssDNA oligonucleotide, at the NdeI or SspI site, protected the dsDNA from cleavage by NdeI or SspI and are thus resistant to the treatment with the restriction endonucleases, respectively, but are still sensitive to SspI or Nde1, respectively. C, quantitative analysis of the DNA bands in the gels shown in B. The DNA species were detected by a Southern blot analysis, using 32P-labeled pUC18 DNA as a probe. The percentages of signals derived from NdeI- or SspI-resistant supercoiled pUC18 DNA or linear DNA among those from the total dsDNA are plotted.
The signals representing the protected dsDNA with a three-stranded structure became smaller when an excess amount of Mhr1 was added (Fig. 3, B (a and c) and C (a and c)). This and previous results (19) showed that excessive levels of the enzyme prevent Mhr1-catalyzed homologous pairing of dsDNA and the ssDNA oligonucleotide, as also observed with two other ATP-independent D-loop-forming proteins: Rad52 (33) and RecT (39).
Mhr1 Efficiently Promotes Homologous Pairing of Linear dsDNA with an ssDNA Oligonucleotide—To study whether supercoils are required for the homologous pairing of dsDNA and an ssDNA oligonucleotide mediated by Mhr1, we tested linear dsDNA as a DNA substrate. The linear dsDNA contained internal regions that were homologous to the partner ssDNA oligonucleotides. This substrate allowed us to avoid the effects of the dsDNA termini. Joint molecule formation mediated by homologous pairing, followed by strand exchange between circular homologous ssDNA and linear dsDNA that has the terminus (or termini) homologous to the ssDNA, has been used to assess proteins for their ability to promote homologous recombination. However, the termini of linear dsDNA tend to be converted to short ssDNA regions by the action of a contaminating exonuclease in the protein preparations, and even the blunt termini of dsDNA consisting of G-C pairs show some characteristics of ssDNA (40). These potential ssDNA regions might act as substrates for annealing rather than homologous pairing.
To detect homologous three-stranded structures resistant to NdeI or SspI, pUC18 cc-dsDNA was linearized with ScaI and was used in the assay for the activity of Mhr1 as a DNA substrate bearing NdeI and SspI cleavage sites far from the strand termini (Fig. 3A, b). We confirmed the absence of internal single-stranded breaks or gaps in the linearized dsDNA that might cause false positive signals by showing that the dsDNA was completely resistant to S1 nuclease (data not shown), which converts internal single-stranded breaks or gaps into double-stranded breaks. Considering the topological requirements described in the Introduction, an unexpected result was obtained. The signals for the formation of the three-stranded structures of linearized pUC18 dsDNA and a homologous ssDNA oligonucleotide by Mhr1 were even stronger than those obtained with negatively supercoiled cc-dsDNA, particularly when Mhr1 was added in excess (Fig. 3, B (b and c) and C (b and c)).
The Effects of Removing Topological Constraints on D-loop Formation Mediated by Mhr1—The preferential Mhr1-mediated formation of homologous three-stranded structures with linear dsDNA may result from an interaction between Mhr1 and the termini of the dsDNA. To eliminate this possibility, we first produced relaxed circular pUC18 dsDNA using calf thymus topoisomerase I, which relaxes both negative and positive supercoils. Then, together with active topoisomerase I, the relaxed circular dsDNA was added to a reaction mixture that included preincubated ssDNA oligonucleotides and the same concentrations of Mhr1 that were used for the formation of homologous three-stranded structures with negatively supercoiled dsDNA. The products were then subjected to the restriction endonuclease protection assay (Fig. 4A). Treatment with a larger amount of topoisomerase I markedly increased the formation of homologous three-stranded structures to levels that were significantly higher than those obtained with negatively supercoiled cc-dsDNA (Fig. 4, B and C) and were similar to those obtained with linear dsDNA (compare Fig. 4 with Fig. 3; see Fig. 7). This result clearly indicates that topological stress is a limiting factor for Mhr1-catalyzed homologous pairing.
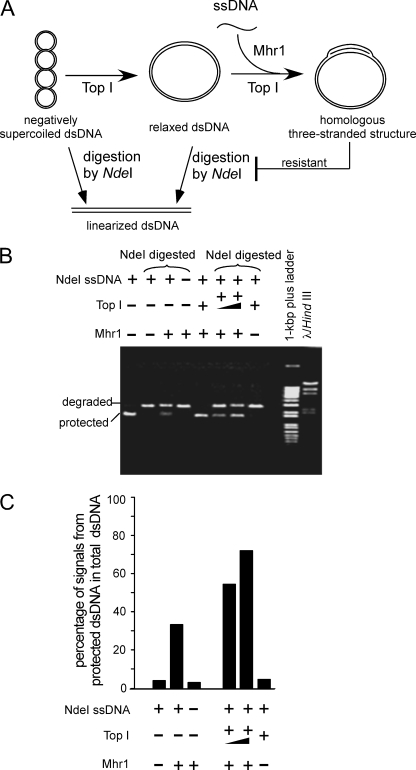
FIGURE 4.
Preferential Mhr1-catalyzed homologous three-stranded structure formation in the presence of eukaryotic topoisomerase I. A, schematic diagrams of the formation of homologous three-stranded structures by Mhr1 with relaxed cc-dsDNA and the ssDNA oligonucleotide in the presence of calf thymus topoisomerase I. B, Mhr1-catalyzed homologous three-stranded structure formation. Negatively supercoiled pUC18 cc-dsDNA was relaxed by the treatment with calf thymus topoisomerase I (Top I). To initiate Mhr1-catalyzed homologous pairing, the relaxed pUC18 cc-dsDNA and active calf thymus topoisomerase I were added to the reaction mixture containing the Mhr1-NdeI ssDNA oligonucleotide complexes. Relaxed pUC18 cc-dsDNA (41.8 μm) was incubated with 1.3 μm NdeI ssDNA oligonucleotide, Mhr1 (0 and 3.9 μm) and calf thymus topoisomerase I at 37 °C for 30 min. As controls, untreated negatively supercoiled pUC18 cc-dsDNA (41.8 μm) was incubated with 1.3 μm NdeI ssDNA oligonucleotide and Mhr1 (0 or 3.9 μm) (without calf thymus topoisomerase I) at 37 °C for 30 min. After the incubation, the DNA products were digested with NdeI, the proteins were removed by SDS and proteinase K treatments, and the products were analyzed by gel electrophoresis, as described in the legend to Fig. 3B. Note that for the topoisomerase-minus control (Top1-), no calf thymus topoisomerase I was added during the entire experimental process. C, a quantitative representation of the results shown in B. The DNA species were detected by a Southern blot analysis, and the percentages of the signals derived from the NdeI-resistant dsDNA among those from the total dsDNA are plotted, as described in the legend to Fig. 3C.
FIGURE 7.
Comparison of negatively supercoiled cc-dsDNA and relaxed cc-dsDNA as the dsDNA substrate for Mhr1-catalyzed formation of homologous three-stranded structures in the presence or absence of eukaryotic topoisomerase I. A, time dependence of Mhr1-catalyzed homologous three-stranded structure formation. Negatively supercoiled pUC18 cc-dsDNA, relaxed pUC18 cc-dsDNA, relaxed pUC18 cc-dsDNA containing active calf thymus topoisomerase I, or ScaI-linearized pUC18 dsDNA (∼41.8 μm) was incubated with the reaction mixture containing 1.3 μm NdeI ssDNA oligonucleotide and Mhr1 (0 or 3.9 μm) at 37 °C for the indicated time periods in the order described in the legends to Figs. 3B and 4B. After the incubation, the DNA products were digested with NdeI, and the proteins were removed by SDS and proteinase K treatments. The DNA products were then subjected to electrophoresis on 1% agarose gels containing ethidium bromide, and the DNA products in the gel were transferred to nylon plus membranes. The signals representing the DNA species were detected by a Southern blot analysis, using 32P-labeled pUC18 as a probe. B, quantitative representations of the results shown in A. The percentages of the signals representing the DNA species detected by a Southern blot analysis among those from the total dsDNA are plotted, as describedin the legend to Fig. 3C. •, linear dsDNA; ▪, relaxed cc-dsDNA with the topoisomerase I; □, relaxed cc-dsDNA (in the absence of topoisomerase); ○, negatively supercoiled cc-dsDNA. C, dependence of the three-stranded structure formation on the Mhr1 concentration. Negatively supercoiled pUC18 cc-dsDNA, relaxed pUC18 cc-dsDNA, relaxed pUC18 cc-dsDNA containing active calf thymus topoisomerase I, or ScaI-linearized pUC18 dsDNA (∼41.8 μm) was incubated with the reaction mixture containing 1.3 μm NdeI ssDNA oligonucleotide and the indicated amounts of Mhr1 at 37 °C for 30 min, as in A. After the incubation, the DNA products were digested with NdeI, and the proteins were removed by SDS and proteinase K treatments. The DNA products were then analyzed as described in A. D, quantitative representations of the results shown in C. Symbols are the same as those in B.
From these results, we concluded that Mhr1 does not require negative supercoils in dsDNA to form three-stranded structures, and thus, it catalyzes true homologous pairing. In addition, Mhr1 promotes homologous pairing of dsDNA and an ssDNA oligonucleotide in the absence of topological constraints more efficiently than in the presence of negative supercoils.
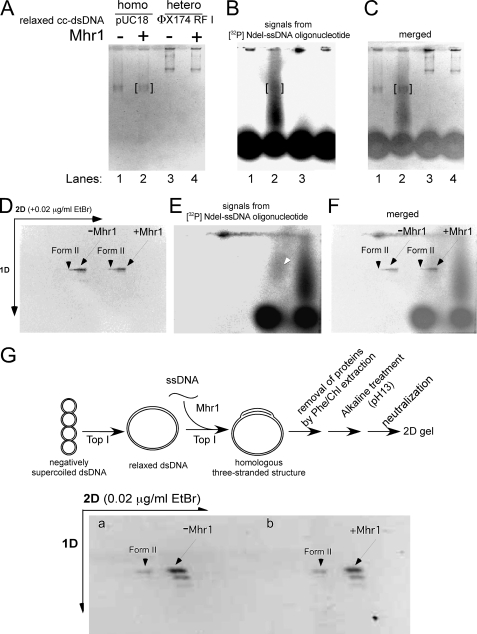
The Entity Detected by the Restriction Enzyme Protection Assay Is Truly a Homologous Three-stranded Structure—Since the restriction endonuclease protection assay used for the detection of homologous three-stranded structures formed on relaxed or linear dsDNA by Mhr1 was performed in the presence of Mhr1, this assay might have an ambiguity; i.e. it is possible that this assay might not directly detect a homologous three-stranded structure but might detect a remaining event (e.g. a specific modification of Mhr1 binding) initiated by the formation of the homologous three-stranded structure. It is known that in the absence of topological stress toward untwisting, D-loops quickly dissociate (37). Thus, we further examined the products of Mhr1-catalyzed homologous pairing by agarose gel electrophoresis to determine whether the entity detected by the restriction enzyme protection assay contained an associated homologous ssDNA oligonucleotide and whether the homologous three-stranded structures formed on cc-dsDNA with homologous [32P]ssDNA (60 nucleotides long) by Mhr1 in the presence of topoisomerase I survive, even after all of the proteins are removed by SDS and proteinase K treatments at 37 °C. As shown in Fig. 5B, substantial amounts of the [32P]ssDNA oligonucleotide migrated at a slower rate than that of the free [32P]ssDNA oligonucleotide and formed a distinct signal from that of the free ssDNA oligonucleotide. The fraction of 32P signals within the slowly migrating ssDNA was 35 ± 4% of the total 32P signals, which corresponds to 83 ± 10% of dsDNA by a calculation, considering that the number of ssDNA oligonucleotide molecules is in excess over that of cc-dsDNA molecule (22 and 9.1 nm in DNA molecules, respectively). Since the true yield of the ssDNA oligonucleotide during the 32P-labeling process could not be calculated (because of a limitation of our Radio Isotope facility), we assumed 100% yield in the above calculation, and thus, the signal strength of the slowly moving 32P correlated well with the amounts of protected dsDNA in the restriction enzyme assay (60–70%; Figs. 4 and 7). The signal for the dsDNA derived from the homologous three-stranded structures overlapped with the tail of the 32P signal (Fig. 5C). When the [32P]ssDNA oligonucleotide was incubated with heterologous dsDNA and Mhr1, there was no difference in the migration of the [32P]ssDNA oligonucleotide and the free ssDNA oligonucleotide (Fig. 5B). Although most of the [32P]ssDNA oligonucleotide migrated as the free ssDNA oligonucleotide in the second dimension of the two-dimensional gel electrophoresis (Fig. 5E), a detectable amount of the [32P]ssDNA oligonucleotide still migrated with the homologous dsDNA (Fig. 5F). These results indicate that the homologous three-stranded structure formed on cc-dsDNA with the homologous [32P]ssDNA oligonucleotide by Mhr1, in the presence of the topoisomerase, survived the protein removal process by the SDS and proteinase K treatments and that the [32P]ssDNA oligonucleotide gradually and continuously dissociated from the homologous cc-dsDNA during the first and second dimensions of the electrophoresis, with some remaining on the cc-dsDNA.
FIGURE 5.
Gel analysis of the protein-free products of homologous three-stranded structure formation by Mhr1 in the presence of calf thymus topoisomerase I. Negatively supercoiled cc-dsDNA (pUC18 and φX174 RF I) was treated with calf thymus topoisomerase I at 37 °C for 30 min. To initiate Mhr1-catalyzed homologous pairing, the relaxed cc-dsDNA and active calf thymus topoisomerase I were added to the reaction mixture containing Mhr1-[32P]NdeI ssDNA oligonucleotide complexes. The solution of homologous (pUC18; 49.0 μm) or heterologous (φX174 RF I; 34.5 μm) relaxed cc-dsDNA was incubated with the reaction mixture of [32P]NdeI ssDNA oligonucleotide (1.3 μm, calculated by assuming that the yield of each 32P labeling step was 100%), Mhr1 (0 and 3.9 μm), and calf thymus topoisomerase I at 37 °C for 30 min. After the proteins were removed by treatments with SDS and proteinase K, the DNA products were analyzed by electrophoresis on 1% agarose gels (2 V/cm) for 11 h in a cold room (4 °C). The second dimension of gel electrophoresis was performed in the presence of 0.02 μg/ml ethidium bromide. After electrophoresis, the gel was dried and exposed to an imaging plate, which was analyzed with a Fuji BAS2000 image analyzer. Due to the limitations of our Radio Isotope facility, it was not possible to obtain images in A and D with the same quality as those in G and Fig. 6. A, the negative print of the first dimensional gel image after ethidium bromide staining. Lanes 1 and 2, homologous combinations in the absence and presence of Mhr1, respectively. Lanes 3 and 4, heterologous combinations in the absence and presence of Mhr1, respectively. B, homologous pairing products represented by the 32P-labeled NdeI-ssDNA oligonucleotide in the same gel shown in A, as detected by exposure to an imaging plate. The positions of pUC18 cc-dsDNA are framed in red. The [32P]NdeI-ssDNA oligonucleotide gradually and continuously dissociated from the homologous pairing products during electrophoresis but formed a distinct signal from the unreacted or free 32P-labeled NdeI-ssDNA oligonucleotide. C, a merged image of A and B. D, the negative print of the second dimensional gel image after ethidium bromide staining. Nicked circular dsDNA (Form II) and cc-dsDNA are indicated by arrowheads and arrows, respectively. -Mhr1, control without Mhr1; +Mhr1, the products from the complete system for the formation of a homologous three-stranded structure by Mhr1 in the presence of the topoisomerase. E, homologous pairing products represented by the 32P-labeled NdeI-ssDNA oligonucleotide in the same gel shown in D, as detected by exposure to an imaging plate. Most of the 32P-labeled NdeI-ssDNA oligonucleotide dissociated from the homologous pairing products but formed a distinct set of signals from a dotlike signal of unreacted 32P-labeled NdeI-ssDNA oligonucleotide. Only a small fraction of the 32P-labeled NdeI-ssDNA oligonucleotide formed a signal at the position of cc-dsDNA, indicated by an arrow. F, a merged image of D and E. G, the negative print showing the topological state of the products of homologous pairing by Mhr1. Homologous (pUC18) relaxed cc-dsDNA (41.8 μm) was incubated with 1.3 μm unlabeled NdeI ssDNA oligonucleotide, Mhr1 (0 and 3.9 μm), and calf thymus topoisomerase I at 37 °C for 30 min. After a phenol/chloroform extraction, the DNA products were subjected to alkaline treatment (pH, 13.0) to dissociate all of the ssDNA oligonucleotide from the cc-dsDNA. After the DNA solution was neutralized to recover the double helix, the cc-dsDNA molecules were analyzed by two-dimensional electrophoresis. The second dimension of gel electrophoresis was performed in the presence of 0.02 μg/ml ethidium bromide. The outline of the experiment is illustrated above the gel image. Nicked circular dsDNA and cc-dsDNA are indicated with arrowheads and arrows, respectively. 1D, first dimension; 2D, second dimension.
Topological Status of dsDNA in the Products of Homologous Pairing by Mhr1 in the Presence of the Topoisomerase—We noticed that the signal of the dsDNA derived from the homologous three-stranded structures formed by Mhr1 in the presence of the topoisomerase I migrated at exactly the same velocity as the free dsDNA of a control without Mhr1 (Fig. 5A) and that the cc-dsDNA that had borne the homologous three-stranded structure seemed to be as completely relaxed as the control dsDNA (Fig. 5D).
Therefore, we examined the topological status of the parental dsDNA in the products of homologous pairing by Mhr1 in the presence of topoisomerase I, after removing the associated ssDNA oligonucleotides. We formed the homologous three-stranded structure as described above. After all of the proteins were removed to fix the topological status of the dsDNA, the DNA products were treated at an alkaline pH to dissociate all of the ssDNA oligonucleotide from the homologous three-stranded structure and then neutralized to restore the double helix of the cc-dsDNA. The topological status of the dsDNA was analyzed by two-dimensional gel electrophoresis. It should be noted that the topological status of the cc-dsDNA observed by this assay reflects that of the cc-dsDNA containing a homologous three-stranded structure with the 60-nucleotide-long ssDNA oligonucleotide and that D-loop formation unavoidably untwists the double helix by 6 turns (9). The untwisting in the presence of topoisomerase I results in a decrease in the linking number of the dsDNA, and a decrease in the linking number by 6 can be easily detected by two-dimensional gel electrophoresis as a shift of the bands by 6 steps toward negatively supercoiled dsDNA (see Fig. 6A). A comparison of the control from which Mhr1 was omitted (Fig. 5G, a) and the cc-dsDNA derived from the products of homologous pairing (Fig. 5G, b; note that 60–70% of them once had a homologous three-stranded structure, as described) does not reveal even a single-step change in the linking number by the formation of the homologous three-stranded structure by Mhr1. In addition, although most of the homologous three-stranded structures were dissociated during electrophoresis, the [32P]ssDNA oligonucleotide that remained associated with the cc-dsDNA migrated with the completely relaxed cc-dsDNA in the two-dimensional gel (Fig. 5F). These results indicate that the dsDNA in the homologous three-stranded structure has the same extent of twisting as the dsDNA free of such a structure and thus exclude D-loops as the products of Mhr1-catalyzed homologous pairing in the presence of the topoisomerase.
FIGURE 6.
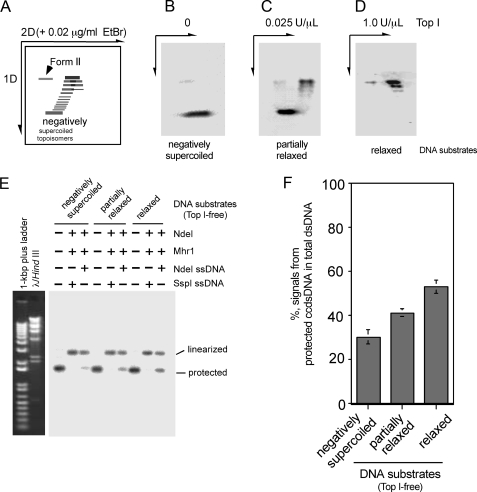
Comparison of negatively supercoiled cc-dsDNA and relaxed cc-dsDNA as the dsDNA substrate for Mhr1-catalyzed homologous three-stranded structure formation in the absence of topoisomerase. A–D, two-dimensional (2D) gel electrophoretic profiles of negatively supercoiled and relaxed pUC18 DNA species. Full or partially relaxed pUC18 cc-dsDNA was prepared by treating supercoiled pUC18 cc-dsDNA with calf thymus topoisomerase I (Top 1), and purifying it from the proteins. The DNA products were analyzed by electrophoresis on 1% agarose gels, and then electrophoresis was performed in the second dimension in the presence of 0.02 μg/ml ethidium bromide. A, schematic diagram. Relaxed cc-dsDNA moves together with nicked circular dsDNA (Form II) in the first dimension (1D) and moves faster than form II DNA in the second dimension. B, the negative print of untreated negatively supercoiled pUC18 after ethidium bromide staining. C, the negative print of partially relaxed pUC18 after ethidium bromide staining. D, the negative print of fully relaxed pUC18 after ethidium bromide staining. E, Mhr1-catalyzed homologous three-stranded structure formation with various dsDNA substrates and ssDNA oligonucleotides. Fully or partially relaxed or negatively supercoiled pUC18 cc-dsDNA (41.8 μm) was incubated with 1.3 μm NdeI ssDNA oligonucleotides or SspI ssDNA oligonucleotides and Mhr1 (0 or 3.9 μm) at 37 °C for 30 min (in the absence of topoisomerase) in the order described in the legend to Fig. 3B. After the incubation, the DNA products were digested with NdeI, and the proteins were removed by SDS and proteinase K treatments. The DNA products were then subjected to electrophoresis on 1% agarose gels containing ethidium bromide. The signals representing the DNA species in the gel were detected by a Southern blot analysis, using 32P-labeled pUC18 as a probe. F, a quantitative representation of the results shown in E. The percentages of the signals representing the DNA species detected by a Southern blot analysis among those from the total dsDNA are plotted, as described in the legend to Fig. 3C.
A Comparison of Relaxed cc-dsDNA and Negatively Supercoiled cc-dsDNA as the dsDNA Substrate for Mhr1-catalyzed Formation of Homologous Three-stranded Structures—Since dsDNA free of topological constraints forms more homologous pairing products by Mhr1 than negatively supercoiled cc-dsDNA, we examined whether positive or negative supercoils are generated as a topological barrier during the Mhr1-catalyzed reaction. In these experiments, we compared relaxed cc-dsDNA and negatively supercoiled cc-dsDNA as the dsDNA substrate for Mhr1-mediated homologous three-stranded structure formation, in the absence of topoisomerase I. We first produced completely and partially relaxed pUC18 cc-dsDNA, using calf thymus topoisomerase I (Fig. 6, A–D). The topoisomerase was then removed, and each relaxed cc-dsDNA sample was added to a reaction mixture in which ssDNA oligonucleotides had been previously incubated with Mhr1. After an incubation at 37 °C, the products were subjected to the restriction endonuclease protection assay. At the heterologous restriction in the dsDNA, no protection was observed. Inconsistent with the topological requirements for homologous pairing (see the Introduction), at the homologous restriction site, the relaxed cc-dsDNA was protected better than the negatively supercoiled DNA, indicating that Mhr1 promotes homologous pairing more efficiently with relaxed cc-dsDNA than with negatively supercoiled cc-dsDNA, and the fully relaxed cc-dsDNA yielded a larger amount of the products, as compared with the results obtained with partially relaxed cc-dsDNA (Fig. 6, E and F). To confirm these results, we compared fully relaxed cc-dsDNA and negatively supercoiled cc-dsDNA after an incubation with different amounts of Mhr1 for various time periods and found that under all of the tested conditions, the relaxed cc-dsDNA formed homologous three-stranded structures more quickly and with larger yields than the negatively supercoiled cc-dsDNA did (Fig. 7). These results argue against a topological barrier in relaxed cc-dsDNA during the process of homologous pairing by Mhr1, but in contrast to RecA-catalyzed homologous pairing (11), the negative supercoils of dsDNA substrates act as a topological barrier in the Mhr1-catalyzed reaction.
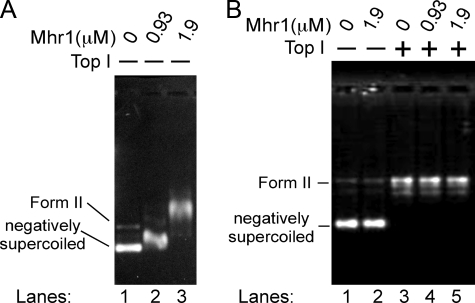
Topological Status of cc-dsDNA Bound to Mhr1—A possible mechanism that would account for both the untwisting required for pairing between homologous sequences and the observed prevention of Mhr1-catalyzed homologous pairing by negative supercoils would be that the cc-dsDNA wraps around Mhr1 in a right-handed manner, as discussed in the case of human Rad52 (41), associated with untwisting of the double helix (Fig. 1, DNA 4). Negative supercoils of cc-dsDNA would prevent the initiation of the right-handed wrapping. In this model, the positive supercoils generated by untwisting of the double helix (Fig. 1, DNA 2) compensate for the negative supercoils generated by the right-handed wrapping (Fig. 1, DNA 3). To confirm this possibility, we formed complexes of Mhr1 and cc-dsDNA in the absence of ssDNA so that all of the cc-dsDNA was integrated in the complex (Fig. 8A) (see Ref. 19) and then treated the samples with calf thymus topoisomerase I. We examined the topological status of the treated DNA after removing all of the proteins by gel electrophoresis. As shown in Fig. 8B, the cc-dsDNA bound to Mhr1 (at 1.9 μm) was relaxed to exactly the same extent as the free cc-dsDNA (control without Mhr1), as expected from the above model (Fig. 1, DNA 4 to DNA 7). This result is not simply the resistance of Mhr1-bound relaxed cc-dsDNA to topoisomerase I, since the yield of homologous three-stranded structures on relaxed cc-dsDNA by Mhr1 was always slightly but significantly higher in the presence of topoisomerase I, as compared with its absence (Fig. 7), indicating that the cc-dsDNA interacting with Mhr1 is sensitive to the action of topoisomerase I. Therefore, this result supports the model described above (Fig. 1, DNA 4).
FIGURE 8.
The binding of cc-dsDNA to Mhr1 and the topological status of Mhr1-bound cc-dsDNA. A, the binding of cc-dsDNA to Mhr1. Negatively supercoiled pUC18 plasmid DNA (41.8 μm) was mixed with the indicated concentrations of Mhr1 in standard buffer and incubated at 37 °C for 30 min. The reaction mixtures containing the Mhr1-cc-dsDNA complexes were then subjected to a gel shift assay, as previously described (19). B, effects of increasing concentrations of Mhr1 on relaxed cc-dsDNA in the presence of calf thymus topoisomerase I (Top I). Relaxed cc-dsDNA was prepared by incubating negatively supercoiled pUC18 plasmid DNA (41.8 μm) with 1.0 unit/μl calf thymus topoisomerase I at 37 °C for 30 min. The relaxed cc-dsDNA (41.8 μm) was mixed with the indicated concentrations of Mhr1 in standard buffer and incubated in the presence of 0.2 units/μl topoisomerase I at 37 °C for 30 min. As a control, negatively supercoiled cc-dsDNA was incubated with the indicated concentrations of Mhr1 under the same conditions, without topoisomerase. After a phenol/chloroform extraction, the DNA products were analyzed by electrophoresis on 1% agarose gels for 11 h at room temperature.
DISCUSSION
The restriction enzyme protection assay might currently be the only reliable means available for studying the effects of supercoils or topological constraints on Mhr1-catalyzed homologous pairing. The restriction enzyme protection assay has very rigorous inner controls; it includes two restriction enzymes recognizing different sequences, two sets of homologous ssDNA oligonucleotides, and dsDNA regions on the same dsDNA molecule, with each set bearing a cleavage site for each of the restriction enzymes, a negative control to monitor the sensitivity of the dsDNA to a restriction enzyme, and essential tests for the homology requirements between the ssDNA oligonucleotide and dsDNA substrates to generate positive signals (i.e. protection against cleavage by the restriction enzymes). The requirement for homology is critical, to exclude various artifacts and to conclude that the products are indeed formed by homologous pairing. Thus, this assay has sufficient rigor for testing the effects of the presence or absence of negative supercoils in the substrate cc-dsDNA. This test detects any homologous three-stranded structure (including the D-loop and triplex structures) formed by a homologous pairing reaction.
Testing the effects of negative supercoils and topological constraints on Mhr1-catalyzed homologous pairing revealed that a markedly higher yield of homologous three-stranded structures was obtained with dsDNA without topological constraints (linear dsDNA and cc-dsDNA in the presence of calf thymus topoisomerase I, which relaxes both positive and negative supercoils; Figs. 3, 4, and 7). This result clearly showed that the topological constraint of negatively supercoiled cc-dsDNA inhibits Mhr1-catalyzed homologous pairing and excludes a simple complementary strand annealing model for Mhr1-catalyzed D-loop formation and clearly showed that the topological constraint of negatively supercoiled cc-dsDNA inhibits Mhr1-catalyzed homologous pairing. We confirmed that the homologous three-stranded structure on cc-dsDNA formed by Mhr1 in the presence of topoisomerase I contained physically associated homologous ssDNA oligonucleotides, even after the removal of Mhr1 by SDS and proteinase K treatments (Fig. 5B). The amounts of [32P]ssDNA oligonucleotides associated with the three-stranded structure correlated with those detected by the restriction endonuclease protection assay. During this test, we noticed that the cc-dsDNA that bears the homologous three-stranded structure formed by Mhr1 in the presence of topoisomerase I and the cc-dsDNA that once bore the homologous three-stranded structure, which was dissociated, have exactly the same linking number as cc-dsDNA relaxed by topoisomerase I in the absence of Mhr1 under the same reaction conditions (Fig. 5, D, F, and G). Thus, the homologous three-stranded structure formed by Mhr1 does not include untwisting of the parental double helix. This finding clearly excludes the possibility that the products of homologous pairing by Mhr1 are D-loops, since D-loops are necessarily associated with the separation of the parental double helix and thus a decrease in the twist.
The best fitting structure satisfying a homologous three-stranded structure without any associated untwisting of the double helix is a parallel triplex, in which the invading ssDNA oligonucleotide forms heteroduplex joints with a complementary strand of parental dsDNA by Watson-Crick type base pairing, and the other strand of the parental DNA sits in the major groove in the heteroduplex joint, with base pair-specific hydrogen bonds (42). The three-stranded structure survived the treatments for removing proteins but gradually dissociated during gel electrophoresis (Fig. 5B).
When we used negatively supercoiled cc-dsDNA, the products detected by gel electrophoresis were authentic D-loops (Fig. 2) (19). These D-loops are probably derived from the triplex or homologous three-stranded structure by releasing the topological stress generated by the negative supercoils of the parental cc-dsDNA after the removal of all proteins (9).
A comparison of partially or fully relaxed cc-dsDNA with negatively supercoiled cc-dsDNA as dsDNA substrates for Mhr1-catalyzed homologous pairing in the absence of topoisomerase I, to analyze the topological barrier derived from negatively supercoiled cc-dsDNA, further revealed that Mhr1 promotes the pairing of relaxed cc-dsDNA with a homologous ssDNA oligonucleotide at a much higher velocity and yield than those observed with negatively supercoiled DNA (Figs. 6 and 7). This clearly indicated that negative supercoils prevent Mhr1-catalyzed homologous pairing and that no net supercoils were generated during the Mhr1-catalyzed reaction. These conclusions were quite surprising, since this appears to be incompatible with the requirements for the extension of dsDNA associated with the untwisting for homologous pairing (see Introduction). In fact, consistent with the requirements for a homologous pairing reaction, the DNA bound to Mhr1 is in an extended conformation, in the same manner as DNA bound to RecA/Rad51, as shown by chemical probing and NMR analyses.4
A solution to this apparent discrepancy first came to light in a recent finding from a study on human Rad52 by Kagawa et al. (41). Like Mhr1, Rad52 is a homologous pairing protein that does not require ATP. It was found that when relaxed cc-dsDNA forms complexes with human Rad52 or its conserved N-terminal domain in the presence of eukaryotic topoisomerase I, the cc-dsDNA purified from the reaction mixture was slightly positively supercoiled, indicating that the interaction between the cc-dsDNA and Rad52 generates negative supercoils. The crystal structure of the conserved Rad52 N-terminal domain revealed a ring containing 11 subunits (43, 44), and the dsDNA binding site is located along the side of the ring (41). These findings suggest that dsDNA wraps around the Rad52 ring in a right-handed manner (Fig. 1, DNA 3). Under the conditions used for homologous three-stranded structure formation, but without the ssDNA oligonucleotide, the treatment of cc-dsDNA complexed with Mhr1 (Fig. 8A) with eukaryotic topoisomerase I relaxed the cc-dsDNA with no detectable supercoiling (Fig. 8B). The extent of positive supercoiling in the presence of an excess of Rad52 was much smaller than that expected according to the assumption that the cc-dsDNA was saturated with bound Rad52, which probably binds at a ratio of 4 base pairs per Rad52 monomer (see Ref. 41). Thus, we must consider the slight positive supercoiling of cc-dsDNA bound to Rad52 following the topoisomerase I treatment (41); the complete relaxation of cc-dsDNA bound to Mhr1 following the same treatment (Fig. 8B); the accomplishment of the necessary untwisting during the homologous pairing process (see the Introduction) while generating few, if any, positive supercoils (Figs. 6 and 7); and the antagonizing effect of negative supercoils of the dsDNA substrates in the Mhr1-catalyzed formation of homologous three-stranded structures (Figs. 6 and 7). A plausible explanation is that Rad52 and Mhr1 form a complex with dsDNA (and ssDNA oligonucleotide), in which the dsDNA wraps around the Rad52 ring or Mhr1 in a right-handed manner, with concomitant untwisting and extending of the double helix to compensate for the negative supercoils generated by the right-handed wrapping (Fig. 1, DNA 4). The negative supercoiling of cc-dsDNA prevents the right-handed wrapping (see Fig. 1, DNA 3). This is a novel mechanism for homologous pairing that may be shared by Mhr1 and Rad52, and it differs from that attributed to the RecA/Rad51 family proteins. A common feature between the Mhr1-catalyzed homologous pairing and the RecA/Rad51-catalyzed reaction is local untwisting of the substrate double helix, which is associated with the extension of the dsDNA for homologous pairing (29).
Since mitochondria contain potent topoisomerase activities, mtDNA is likely to be free of topological constraints. Mhr1 is probably optimized for homologous pairing in mitochondria. The finding that Mhr1 promotes homologous pairing with cc-dsDNA without topological constraints at much higher efficiency than with negative supercoiled cc-dsDNA supports the expected topological conditions of mtDNA within mitochondrial nucleotides, unlike the general notion based on the negative supercoils of isolated closed circular double-stranded mtDNA.
Acknowledgments
We are grateful to Dr. Wataru Kagawa (Systems and Structural Biology Center in RIKEN Yokohama Institute), Dr. Shigeyuki Yokoyama (Systems and Structural Biology Center in RIKEN Yokohama Institute), and Dr. Hitoshi Kurumizaka (Waseda University) for communicating observations before publication. We also thank Dr. Tsutomu Mikawa (Yokohama City University; RIKEN SPring-8) and Tokiha Masuda (Yokohama City University; CMBL in RIKEN ASI) for sharing unpublished observations.
This work was supported by a grant from the Bioarchitect Research Program of RIKEN (to F. L. and M. Y.), a grant from the Life Science Foundation of Japan (to F. L.), Grants-in-Aid for Scientific Research (C) 18570168 and 20570171 from the Japan Society for the Promotion of Science (to F. L.), Grant-in-Aid (B) 19370004 from the Japan Society for the Promotion of Science (to T. S.), and a grant from the RIKEN Strategic Research Program (to F. L.).
Footnotes
The abbreviations used are: dsDNA, double-stranded DNA; mtDNA, mitochondrial DNA; ssDNA, single-stranded DNA; cc-dsDNA, closed circular dsDNA.
T. Masuda and T. Mikawa, personal communication.
References
- 1.O'Driscoll, M., and Jeggo, P. A. (2006) Nat. Rev. Genet. 7 45-54 [DOI] [PubMed] [Google Scholar]
- 2.Neale, M. J., and Keeney, S. (2006) Nature 442 153-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling, F., Morioka, H., Ohtsuka, E., and Shibata, T. (2000) Nucleic Acids Res. 28 4956-4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata, T., and Ling, F. (2007) Mitochondrion 7 17-23 [DOI] [PubMed] [Google Scholar]
- 5.Nishinaka, T., Shinohara, A., Ito, Y., Yokoyama, S., and Shibata, T. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11071-11076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stasiak, A., DiCapua, E., and Koller, T. (1981) J. Mol. Biol. 151 557-564 [DOI] [PubMed] [Google Scholar]
- 7.Stasiak, A., and DiCapua, E. (1982) Nature 299 185-186 [DOI] [PubMed] [Google Scholar]
- 8.Shibata, T., DasGupta, C., Cunningham, R. P., and Radding, C. M. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 1638-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beattie, K. L., Wiegand, R. C., and Radding, C. M. (1977) J. Mol. Biol. 116 783-803 [DOI] [PubMed] [Google Scholar]
- 10.Holloman, W. K., Wiegand, R., Hoessli, C., and Radding, C. M. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 2394-2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata, T., DasGupta, C., Cunningham, R. P., Williams, J. G. K., Osber, L., and Radding, C. M. (1981) J. Biol. Chem. 256 7565-7572 [PubMed] [Google Scholar]
- 12.Van Komen, S., Petukhova, G., Sigurdsson, S., Stratton, S., and Sung, P. (2000) Mol. Cell 6 563-572 [DOI] [PubMed] [Google Scholar]
- 13.McEntee, K., Weinstock, G. M., and Lehman, I. R. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 2615-2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara, A., Ogawa, H., Matsuda, Y., Ushio, N., Ikeo, K., and Ogawa, T. (1993) Nat. Genet. 4 239-243 [DOI] [PubMed] [Google Scholar]
- 15.Sung, P. (1994) Science 265 1241-1243 [DOI] [PubMed] [Google Scholar]
- 16.Bishop, D. K., Park, D., Xu, L. Z., and Kleckner, N. (1992) Cell 69 439-456 [DOI] [PubMed] [Google Scholar]
- 17.Li, Z., Golub, E. I., Gupta, R., and Radding, C. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11221-11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling, F., Makishima, F., Morishima, N., and Shibata, T. (1995) EMBO J. 14 4090-4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling, F., and Shibata, T. (2002) EMBO J. 21 4730-4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dujon, B. (1981) in The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance (Strathern, J. N., Jones, E. W., and Broach, J. R., eds) pp. 505-635, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 21.Kraytsberg, Y., Schwartz, M., Brown, T. A., Ebralidse, K., Kunz, W. S., Clayton, D. A., Vissing, J., and Khrapko, K. (2004) Science 304 981. [DOI] [PubMed] [Google Scholar]
- 22.Zsurka, G., Hampel, K. G., Kudina, T., Kornblum, C., Kraytsberg, Y., Elger, C. E., Khrapko, K., and Kunz, W. S. (2007) Am. J. Hum. Genet. 80 298-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling, F., and Shibata, T. (2004) Mol. Biol. Cell 15 310-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen, A., and Clark, A. J. (1986) J. Bacteriol. 167 327-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muniyappa, K., and Radding, M. C. (1986) J. Biol. Chem. 261 7472-7478 [PubMed] [Google Scholar]
- 26.Hall, S. D., Kane, M. F., and Kolodner, R. D. (1993) J. Bacteriol. 175 277-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortensen, U. H., Bendixen, C., Sunjevaric, I., and Rothstein, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10729-10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiyama, T., New, J. H., and Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6049-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata, T., Nishinaka, T., Mikawa, T., Aihara, H., Kurumizaka, H., Yokoyama, S., and Ito, Y. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8425-8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi, A., and Asai, K. (1984) Nature 309 677-681 [DOI] [PubMed] [Google Scholar]
- 31.Nakasu, S., and Kikuchi, A. (1985) EMBO J. 4 2705-2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezza, R. J., Voloshin, O. N., Vanevski, F., and Camerini-Otero, R. D. (2007) Genes Dev. 21 1758-1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagawa, W., Kurumizaka, H., Ikawa, S., Yokoyama, S., and Shibata, T. (2001) J. Biol. Chem. 276 35201-35208 [DOI] [PubMed] [Google Scholar]
- 34.Shibata, T., Ohtani, T., Iwabuchi, M., and Ando, T. (1982) J. Biol. Chem. 257 13981-13986 [PubMed] [Google Scholar]
- 35.Wu, A. M., Kahn, R., DasGupta, C., and Radding, C. M. (1982) Cell 30 37-44 [DOI] [PubMed] [Google Scholar]
- 36.Enomoto, R., Kinebuchi, T., Sato, M., Yagi, H., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2004) J. Biol. Chem. 279 35263-35272 [DOI] [PubMed] [Google Scholar]
- 37.Radding, C. M., Beattie, K. L., Holloman, W. K., and Wiegand, R. C. (1977) J. Mol. Biol. 116 825-839 [DOI] [PubMed] [Google Scholar]
- 38.Ferrin, L. J., and Camerini-Otero, R. D. (1991) Science 254 1494-1497 [DOI] [PubMed] [Google Scholar]
- 39.Noirot, P., and Kolodner, R. D. (1998) J. Biol. Chem. 273 12274-12280 [DOI] [PubMed] [Google Scholar]
- 40.Kochoyan, M., Leroy, J. L., and Gueron, M. (1987) J. Mol. Biol. 196 599-609 [DOI] [PubMed] [Google Scholar]
- 41.Kagawa, W., Kagawa, A., Saito, K., Ikawa, S., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2008) J. Biol. Chem. 283 24264-24273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlieghe, D., Vanmeervelt, L., Dautant, A., Gallois, B., Precigoux, G., and Kennard, O. (1996) Science 273 1702-1705 [DOI] [PubMed] [Google Scholar]
- 43.Kagawa, W., Kurumizaka, H., Ishitani, R., Fukai, S., Nureki, O., Shibata, T., and Yokoyama, S. (2002) Mol. Cell 10 359-371 [DOI] [PubMed] [Google Scholar]
- 44.Singleton, M. R., Wentzell, L. M., Liu, Y., West, S. C., and Wigley, D. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13492-13497 [DOI] [PMC free article] [PubMed] [Google Scholar]