Abstract
ATP7B is a human P1B-type ATPase that has a crucial role in maintaining copper(I) homeostasis. Mutations in the corresponding gene are the cause of Wilson disease. Among its various distinguishing features is a long (∼630 amino acids) N-terminal cytosolic tail containing six domains that are individually folded and capable of binding one copper(I) ion each. We expressed the entire tail as a single construct in Escherichia coli and investigated its interaction with its copper chaperone (i.e. HAH1) by solution NMR spectroscopy. We observed that all six of the metal-binding domains were metallated by Cu(I)-HAH1, with the first, the second, and the fourth domains forming an adduct with it. This behavior is different from that of the highly similar human ATPase ATP7A, in which only two domains form such an adduct. The distinct behaviors of the different domains were analyzed in terms of the energetics of Cu(I) transfer, hinting at a specific role of the interaction with copper(I)-HAH1 in the overall functional process.
Human ATP7B is a P1B-type ATPase that, like the other human copper(I)-transporting ATPase, ATP7A, can translocate copper(I) to the trans-Golgi network, where the metal is incorporated into copper-dependent enzymes. Copper stimulation results in the redistribution of both ATP7A and ATP7B to the plasma membrane through intracellular vesicles in order to export excess copper out of the cytosol (1–4). Mutations in the ATP7B autosomal gene produce the copper accumulation in tissues that characterizes Wilson disease (5–7). Mutations in ATP7A are instead responsible for Menkes disease (8, 9). ATP7A and ATP7B are often referred to as the MNK and WLN proteins, respectively.
P1B-type ATPases contain four major regions/domains (the N-terminal copper-binding tail, the transmembrane domain, the ATP-binding domain, and the phosphatase domain) as well as a short C-terminal tail (10). The N-terminal copper-binding cytosolic tails of WLN and MNK are both ∼630 amino acids long. They contain six 70-amino acid, independently folded metal-binding domains (MBDs),2 which are relatively similar in sequence and structure (11–15). Each MBD harbors the conserved sequence motif GMXCXXC, through which it can bind one equivalent of copper(I) (16). The N-terminal tail has an important role in tuning the activity of the enzyme (17–19) and in modulating the intracellular trafficking rates of ATP7A and ATP7B (20–22), an aspect of the cellular function of these ATPases that is lacking in less complex organisms, including yeast. The presence of either the intact fifth or intact sixth metal-binding domain in the human proteins is sufficient to support both WLN/MNK activity and intracellular trafficking at essentially normal levels (21, 23–26). This has stimulated researchers to investigate the cellular role of the additional four domains that are present in the two human ATPases as well as in several other mammalian homologues. The KCu binding affinity of the six MBDs of both the MNK (13, 27, 28) and WLN (29, 30) proteins range between 1- and 5-fold the binding affinity of the copper(I) metallochaperone HAH1. Instead, the two MBDs of the yeast Ccc2 ATPase have essentially the same affinity as the Atx1 metallochaperone (31). Copper(I) is transferred from the chaperone to the MBDs of the ATPases via a metal-bridged intermediate (32–35), which may or may not be present at detectable levels in solution. All the six MBDs of the N-terminal tail of MNK are metallated by the metallochaperone. Additionally, two of them form, at relatively high levels, an intermolecular copper(I)-bridged adduct with HAH1 (28).
In this work, we investigated the interaction of copper(I)-HAH1 with the whole N-terminal tail of WLN to characterize the different behavior of its six MBDs metal-binding domains and compare it to the properties of the N-terminal tail of MNK. The variations between these two systems may be relevant for the details of their different functioning in tissues where both proteins are expressed (such as kidney and placenta). The features of the interactions of the MBDs of P1B-type ATPases with their partners are rationalized on the basis of a specific energetic profile of the copper transfer reaction.
EXPERIMENTAL PROCEDURES
Preparation of Protein Samples—A DNA segment corresponding to residues 1–633 of WLN (WLN1-6 hereafter) was amplified by PCR and cloned in the Gateway Entry vector pENTR/TEV/D-TOPO (Invitrogen) to include the tobacco etch virus protease cleavage site at the N-terminal end. This segment was then subcloned into the pETG-20A EMBL (36) through the Gateway LR reaction, yielding a plasmid expressing the protein fused with thioredoxin A and a His tag at the N terminus. Mutants were generated using the QuikChange mutagenesis kit (Stratagene).
Wild type and mutant WLN1-6 were expressed in Escherichia coli Rosetta(DE3) cells (Novagen) in minimal medium cultures and purified using HisTrap chelating FF columns (GE Healthcare). For isotope enrichment, (15NH4)2SO4 and 13C-labeled glucose were used. The His tag and the fusion protein were cleaved with AcTEV (Invitrogen). Protein purity was checked by SDS-PAGE and matrix-assisted laser desorption ionization time-of-flight mass spectra. Copper(I) was added to samples in an inert atmosphere chamber (Coy Lab) by adding an acetonitrile solution of tetrakis(acetonitrile)copper(I) hexafluorophosphate as copper source in the presence of 1 mm dithiothreitol. All samples contained around 0.3 mm protein in 50 mm phosphate, 50 mm arginine, 50 mm glutamate at pH 7.0. The addition of arginine and glutamate to the buffer extended the lifetime of the samples (37), which, however, remained in the range of 2–3 days.
HAH1 samples were prepared as already reported, always without a poly-His tag (38). In titration experiments, we added copper(I)-HAH1 and apo-WLN1-6 directly in the NMR tube under N2 atmosphere, using the same procedure previously followed for MNK1-6 (28).
NMR Spectroscopy—NMR experiments were acquired using Bruker Avance spectrometers operating at proton frequencies of 500, 700, and 900 MHz, all equipped with cryogenically cooled probes. Resonance assignments of WLN1-6 were performed through conventional multidimensional NMR techniques based on triple resonance experiments (39). Titrations of apo-WLN1-6 with copper(I)-HAH1 were followed through 1H-15N SOFAST-heteronuclear multiple quantum coherence experiments (40). The backbone dynamics of copper(I)-HAH1 in the presence of apo-WLN1-6 was investigated through the analysis of 15N R1, R2 relaxation rates (41). More than one sample was needed to complete NMR data collection. A table summarizing the NMR experiments performed is given in the supplemental material (Table S1). All of the figures reporting NMR spectra were generated with the program CARA (42).
RESULTS
The first 633 amino acids of WLN (WLN1-6, with the wild-type sequence), comprising the entire cytosolic tail and in particular the six MBDs, were expressed in E. coli as a single construct; the protein could be enriched in the 15N and 13C stable isotopes, permitting its characterization by NMR spectroscopy. Protein solubility was only about 300 μm at pH 7.0; in addition, the samples were poorly stable, displaying significant degradation after 2–3 days. Fig. S1 reports a two-dimensional 1H-15N HSQC spectrum of apo-WLN1-6. The particularly favorable line widths of the spectra are due to the presence of long linker segments between the various domains, which allow them to tumble regardless of one another. We assigned signals from all six MBDs using triple resonance experiments; the backbone resonance assignments both for the apo and copper(I) forms are reported in Tables S2 and S3. In the other regions of the polypeptide chain, we could observe and assign some signals from the linker connecting domains 1 and 2, domains 3 and 4, and domains 5 and 6. The present assignment was more extensive than that achieved for MNK1-6 (28). Using a labile inorganic copper(I) complex, we could fully metallate the protein. The HSQC spectra of metallated WLN1-6 were used to readily identify metal transfer from copper(I)-HAH1 to the various MBDs during titrations of apo-WLN1-6 with HAH1 (see below).
As mentioned, the major focus of this work was on the analysis of the interaction between apo-WLN1-6 and copper(I)-HAH1. This was investigated by adding the latter to WLN1-6, with only one of the two proteins enriched in 15N so that only its signals appeared in 1H-15N HSQC spectra. The samples contained 1 mm dithiothreitol. The interaction with copper(I)-HAH1 caused a variation of the chemical shifts of the signals from the backbone amide moieties of domains 1, 2, and 4 (Fig. 1), whose magnitude increased upon increasing concentration of the titrant. Instead, for domains 3, 5, and 6, we observed slow or intermediate exchange with the copper(I)-bound species (Fig. 2), whose signals increased in intensity upon increasing the partner concentration. These features progressed to the same extent throughout the titration (Figs. 1 and 2). The linkers connecting the various metal-binding domains instead did not experience appreciable chemical shift changes calculated through the following equation (Fig. 3).
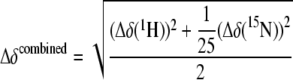 |
(Eq. 1) |
FIGURE 1.
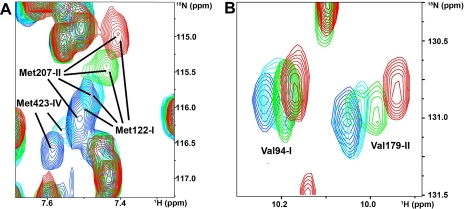
Formation of metal-mediated adducts between copper(I)-HAH1 and some MBDs of WLN1-6. In panel A and B selected peaks from the first, second, and fourth MBDs of WLN1-6 are shown as a function of the copper(I)-HAH1/WLN1-6 ratio. MBDs numbers are indicated in roman numerals. Blue, 0:1; cyan, 1.5:1; green, 3:1; red, 6:1 copper(I)-HAH1/WLN1-6 ratio.
FIGURE 2.
Metallation of the third, fifth, and sixth domains of WLN1-6 by copper(I)-HAH1. Panels A–D show an overlay of a selected region of the 1H-15N HSQC spectra of apo-WLN1-6 (blue) and of apo-WLN1-6 in the presence of 1.5 (cyan), 3.0 (green), and 6.0 (red) equivalents of unlabeled Cu(I)-HAH1. MBDs numbers are indicated in roman numerals.
FIGURE 3.
Interaction of WLN1-6 with copper(I)-HAH1 (viewed from the side of WLN1-6). Variations in the position of signals between WLN1-6 alone and in the presence of 3.0 eq (A) and 6.0 eq (B) of copper(I)-HAH1. C, variations in the position of signals between C1,2,4WLN1-6 mutant alone and in the presence of 6.0 eq of copper(I)-HAH1. These variations are quantitated through the combined chemical shift (52), which is calculated from the experimental 1H and 15N chemical shift changes (Δδ(1H) and Δδ(15N), respectively) measured between corresponding peaks in the two forms, through Equation 1. The red bars indicate the peaks that became broad beyond detection upon interaction. The boundaries of the six MBDs along the sequence are shown by blue boxes.
The signals of domains 1, 2, and 4 always appeared in a single form, and the corresponding peaks had a position in the spectrum that depended on the copper(I)-HAH1/WLN1-6 ratio (Figs. 1 and 2). The chemical shift changes observed for corresponding residues in these domains were similar throughout the titration (Fig. 3). The perturbed signals of domains 1, 2, and 4 belonged to both the metal-binding loop and the loop between the second α-helix and the fourth β-strand of each domain (loop 5). Loop 5 is located at the interface of the metal-bridged adduct formed by the homologous yeast proteins (35). Finally, some signals of the residues of the MBD of domain 4 experienced significant line broadening (e.g. Met-423 in Fig. 1), which led to them becoming undetectable. Eventually, signals from the copper(I) form could be detected.
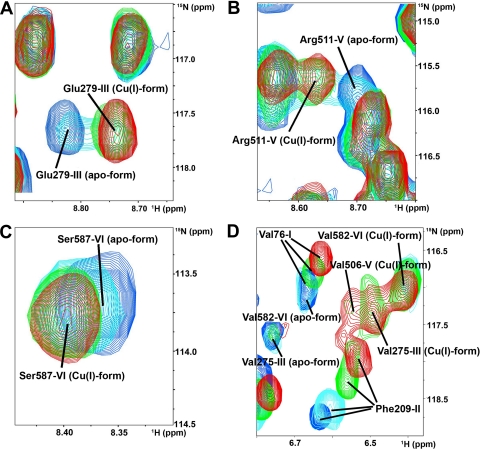
For domains 3, 5, and 6, the chemical shift changes were mainly limited to the metal-binding loop, and loop 5 was not affected by interaction with the partner (Fig. 3). The signals of the apo form disappeared with a similar dependence on the concentration of copper(I)-HAH1. However, not all of the corresponding signals of the copper(I) form reappeared at the same protein ratio (see the signals from Val-275, Val-506, and Val-582 in Fig. 2). Domains 3, 5, and 6 were essentially fully metallated at the end of the titration (6:1 copper(I)-HAH1/WLN1-6 ratio).
Looking at 15N-enriched copper(I)-HAH1 when interacting with apo-WLN1-6, it was observed that the interaction caused a change in the position of various peaks in the 1H-15N HSQC. In addition, essentially all of the signals of the residues around the metal-binding loop (Fig. 4) became broad beyond detection. In order to evaluate whether this was the result of the formation of an intermolecular adduct involving HAH1 and WLN1-6, we measured the correlation time (τR) for reorientation in solution of copper(I)-HAH1 alone and in the presence of equimolar WLN1-6. An increase of the τR of HAH1 in the latter system up to 7.4 ns from 4.9 ns of the isolated protein (38) was determined, which resulted from the generalized increase of 15N R2 relaxation rates and the concomitant decrease of 15N R1 relaxation rates. This increase in the τR of HAH1 indicates that the latter protein formed an intermolecular adduct with one or more of the domains of WLN1-6.
FIGURE 4.
Interaction of WLN1-6 with copper(I)-HAH1 (viewed from the side of HAH1). Variations in the position of signals between copper(I)-HAH1 alone and in the presence of 1 eq of WLN1-6 are quantitated through the combined chemical shift variation (see legend to Fig. 3). The white bars indicate the peaks that became broad beyond detection upon interaction.
The overall behavior indicates for WLN1-6 a clear cut differentiation between the signals from domains 1, 2, and 4 on the one hand and 3, 5, and 6 on the other. From all of these data and also on the basis of the similar behavior of other systems, such as human Menkes and yeast Ccc2, it can be concluded that (i) all of the six domains of WLN1-6 were metallated by copper(I)-HAH1; (ii) the latter protein formed metal-bridged adducts with domains 1, 2, and 4 at levels detectable by NMR spectroscopy, which were in fast (on the NMR time scale) exchange with the single species; and (iii) the copper(I) form of domains 3, 5, and 6 exchanged with the apo form in an intermediate to slow regime.
To discriminate whether domains 3, 5, and 6 were directly metallated by copper(I)-HAH1 or if an intramolecular copper transfer occurred, we studied the interaction of copper(I)-HAH1 with a mutant of WLN1-6, where the metal-binding cysteines of domains 1, 2, and 4 had been mutated to alanine (C1,2,4WLN1-6), thus making these domains unable to bind copper. The 1H-15N HSQC spectrum of C1,2,4WLN1-6 was essentially superimposable to that of wild type WLN1-6 recorded under the same experimental conditions, with the exception of the mutated amino acids and of their neighboring residues in sequence. Upon the addition of copper(I)-HAH1 to the mutant, we observed that domains 3, 5, and 6 acquired copper(I) from HAH1, since they experienced the same spectral changes that were observed in native WLN1-6 (Fig. 3). The new sets of signals of the metallated domains were in slow exchange with the apo form (Fig. 5). On the contrary, the signals of residues from domains 1, 2, and 4 did not experience significant chemical shift changes, indicating that no adducts were formed with the partner (Fig. 3).
FIGURE 5.
Copper(I) transfer from HAH1 to C1,2,4WLN1-6. Overlay of a selected region of the 1H-15N HSQC spectra of apo-C1,2,4WLN1-6 (black) and of apo-C1,2,4WLN1-6 in the presence of 3 eq of unlabeled Cu(I)-HAH1 (light gray).
DISCUSSION
When the WLN1-6 construct was presented with copper(I)-HAH1, the behavior of its six MBDs differentiated. In fact, the NMR signals of the backbone amide moieties from three domains (the first, second, and fourth) experienced a variation of their position that was dependent on the copper(I)-HAH1/WLN1-6 ratio (Fig. 1). Instead, for the other three domains (the third, fifth, and sixth), the addition of copper(I)-HAH1 gave rise to another set of signals in slow or intermediate exchange with the apo form (Fig. 2).
It has been extensively demonstrated in the available literature for related systems that the present behavior of the signals of the amino acids in domains 1, 2, and 4 results from the formation of an intermolecular, copper(I)-bridged adduct with HAH1 (14, 15, 28, 35, 43). In the present work, the formation of this adduct was experimentally shown by the increase of the τR of HAH1. Instead, the behavior shown by domains 3, 5, and 6 is attributable to the occurrence of metal transfer from the metallochaperone without accumulation of the protein-protein adduct. Indeed, in a closely related study of a two-domain construct of WLN that comprised only domains 3 and 4, it was demonstrated that the addition of copper(I)-HAH1 to apo-WLN3-4 induced a significant increase of the tumbling rate of domain 4, consistent with formation of an intermolecular copper(I)-bridged adduct, but did not affect the tumbling rate of domain 3 (15).
Within each group of domains (1, 2, and 4 on one hand, 3, 5, and 6 on the other), the signals from corresponding residues of the various domains did not all behave in the same way. Regarding domains 1, 2, and 4, the features of the signals of the fourth MBD suggested that the complex between this domain and copper(I)-HAH1 had a slower dissociation rate than in the case of the first and second MBDs. For domains 3, 5, and 6, the signals of the copper(I) form became detectable at different copper(I)-HAH1/WLN1-6 ratios, despite the similar affinity for the metal ion of the three domains (29). This is presumably due to the diverse dynamics of this loop in each MBD, related to conformational equilibria involving the apo and copper(I) forms, resulting in different broadening of the signals.
We then investigated whether the metallation of domains 3, 5, and 6 could be carried out by HAH1 alone or required the prior formation of an adduct involving one among domains 1, 2, or 4. As shown in Figs. 3 and 5, the C1,2,4WLN1-6 mutant could be metallated by copper(I)-HAH1. In addition, there was no significant perturbation of the backbone signals of domains 1, 2, and 4 (Fig. 3). The presence of the metal-binding cysteines is thus required for the formation of the intermolecular adduct. This confirms that the adduct is metal-mediated, analogously to the yeast Atx1/Ccc2a system (35). Furthermore, it appears that the metal transfer process from copper(I)-HAH1 to the third, fifth, or sixth MBDs can take place even without any prior intermolecular interaction with the other domains. In a previously reported study (14), a double domain construct comprising the fifth and sixth metal-binding domains of the WLN protein was unable to receive directly the metal from copper(I)-HAH1. This difference from our results is most likely linked to the different protein as well as to the different experimental conditions employed in the two studies and is not unprecedented. Indeed, for the third domain of the MNK protein, a difference in metal-binding capability has been reported when comparing the isolated domain to the six-domain construct (28, 44). In addition, the isolated second WLN MBD required somewhat higher concentrations of the partner metallochaperone than the fourth to afford comparable chemical shift variations (14), whereas in the present WLN1-6 construct, the first, second, and fourth MBDs experienced similar chemical shift variations along the titration. Considering all of the available experimental data on the wild type and mutant WLN and MNK systems (28), it appears that copper(I) can be transferred directly by the metallochaperone to any of the six MBDs. A mechanism in which the metal ion is first transferred to a domain and then relocates to another domain (to be ultimately pumped across the membrane) cannot be ruled out.
The different interaction of the MBDs of the MNK (28) and WLN proteins with copper(I)-HAH1 reflects different thermodynamic properties of the various species involved in the metal transfer pathway. In all known cases, including those of the yeast and bacterial homologues of the present proteins, it can be assumed that copper(I) transfer to a soluble MBD of the ATPase occurs according to the following scheme (35), where the metal ion bridges the proteins in the chaperone-Cu(I)-ATPase species, as it is coordinated by two cysteines of one protein and one cysteine of the other. For the yeast system, there are two forms of the chaperone-Cu(I)-ATPase species in equilibrium (35). The two forms differ in the identity of the protein providing the two cysteines, the form with the ATPase providing two cysteines being more stable (35). Scheme 1 is in agreement with a body of experimental evidence based on the combination of site-directed mutagenesis data and various spectroscopic techniques (32–35).
 |
Within the above general reaction scheme, the various systems experimentally investigated up to now feature a remarkable differentiation of the energetic and kinetic aspects of the process. In yeast, the affinity for copper(I) of the metallochaperone (Atx1) and of the first MBD of the Ccc2 ATPase is nearly the same (31). When 1 mm copper(I)-Atx1 is mixed with 1 mm Ccc2a, a copper(I)-bridged adduct forms, involving ∼70% of the protein molecules (45). The adduct is in fast exchange with the isolated proteins. The various MBDs of MNK (27, 46) and WLN (29) have a higher affinity for copper(I) than HAH1. In WLN, three MBDs form an adduct at detectable amounts with copper(I)-HAH1, whereas for the other three MBDs, the complex is not detectable, and only the final metallated form can be observed. In MNK, only two domains (the first and the fourth) form an adduct (28) at similar protein concentrations; the second MBD of MNK is metallated by copper(I)-HAH1 without the formation of detectable amounts of a protein-protein adduct. This behavior is observed both in the context of the entire tail (28) and for the isolated domain (46). The different behaviors described above correspond to different energy profiles for the copper transfer process, which are sketched in Fig. 6, for the reaction of Scheme 1. In Ccc2, the final state has a free energy only slightly lower than the initial state (Fig. 6A), whereas this difference is somewhat higher in MNK and WLN (Fig. 6B). Indeed, the HAH1-Cu(I)-WLN complex is more similar to the final product than the Atx1-Cu(I)-Ccc2 complex. In both cases, the occurrence of detectable amounts of the intermediate adduct is possible if its free energy is close to that of the final state of Scheme 1 but is independent of the relative affinity for copper(I) of the metallochaperone and the ATPase. When the adduct is formed, it is in fast equilibrium with the free proteins, implying low activation barriers. On the other hand, for those MBDs that do not form detectable amounts of adduct, the equilibrium between the apo and copper(I) forms is slow, regardless of the presence of HAH1. The latter, therefore, does not act as a catalyst in lowering the activation barrier between the two forms of the metal-binding domains (Fig. 6B).
FIGURE 6.
Energy profile along the copper(I) transfer reaction for different kinds of metallochaperone/ATPase pairs. A, the isolated proteins have very similar affinity for copper(I) and may (continuous line) or may not (dashed line) form a stable complex; B, the isolated MBD of the ATPase has higher affinity for copper(I) than the metallochaperone. The two proteins may (continuous line; e.g. MBD1, MBD2, and MBD4 of WLN1-6) or may not (dashed line; e.g. MBD3, MBD5, and MBD6 of WLN1-6) form a stable complex. In both cases, the complex and the isolated proteins may be in fast or slow (on the NMR time scale) exchange, depending on the height of the energetic barriers separating the various species. The two equilibria of Equation 1 do not need to be equally rapid; indeed, for the homologous system of Synechocystis PCC 6803, one of the two equilibria was fast, whereas the other was slow (53). G stands for Gibbs energy.
The N-terminal tail of P1B-type ATPases may have a regulatory role in the enzyme activity and/or trafficking (17, 20, 21). In particular, the tail could have an autoinhibitory role in the absence of copper(I) (18, 24). The structural model of CopA, a P1B-type ATPase from Archaeoglobus fulgidus, has suggested that in the absence of copper(I), the tail is in close contact with the other cytoplasmic domains (47). Both for WLN and MNK, the regulation of the ATPase activity and/or trafficking by the N-terminal tail could depend upon the interaction with copper(I)-HAH1, leading to a variation of the contacts of the tail with the other domains of the enzyme. Indeed, the present data, as well as those on MNK1-6 (28), suggested that the aforementioned rearrangements are linked to the formation, and presumably accumulation at high intracellular copper(I) concentrations, of adducts between the MBDs of the tail and copper(I)-HAH1. This view would also provide a rationale for the observed dependence of ATPase trafficking on the expression levels of HAH1 (48). Even when the equilibrium leading to the formation of the adduct with the metallochaperone or to the transfer of copper(I) is not fully shifted toward the products, every time one of these molecular events takes place, it can trigger the conformational rearrangements of the ATPase that ultimately lead to ATP hydrolysis. The latter reaction is effectively irreversible and thus pushes forward the catalytic cycle. The effective protein concentration in the cell is not known; the relatively high equilibrium constants for the protein-protein adducts observed here imply that the processes described above may occur also at micromolar concentrations.
The available literature points to the MBD or to the two MBDs closest to the transmembrane region of MNK and WLN as playing a role different than the most N-terminal four domains (49, 50). In this respect, it is noteworthy that for both human proteins, the fifth and sixth MBDs are metallated by the metallochaperone without forming detectable amounts of complex. The metallation of these two domains does not require the formation of copper(I)-mediated adducts with other metal-binding domains within the cytosolic tail, as shown by mutagenesis data for both MNK (28) and WLN (Fig. 3). This is at variance with what observed for yeast Ccc2. This ATPase has only two MBDs, of which at least one forms a complex with copper(I)-Atx1.
Of the first four MBDs, three domains in WLN and two in MNK can form detectable amounts of an adduct. The above differences are the consequence of subtle mutations of their surface properties, which define the energetic profile of the interaction, with copper(I)-HAH1 among those depicted in Fig. 6. This differentiation could contribute to tuning the regulation of the activity and/or trafficking of the ATPases. Specifically, for WLN and MNK, this could be a means to discriminate their properties in the tissues where they are coexpressed (51).
Supplementary Material
Acknowledgments
We thank David Huffman for many insightful discussions.
This work was supported by Ministero dell'Istruzione, dell'Università e della Ricerca (Fondo per gli Investimenti della Ricerca di Base project RBLA032ZM7), Ente Cassa di Risparmio di Firenze (Projects “Basi Molecolari di Patologie Umane Correlate a Disfunzioni della Catena Respiratoria” and “Relazione Varianti Proteiche Strutturali-malattie Genetiche”), and the European Commission (Project SPINE2-COMPLEXES-031220).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
Footnotes
The abbreviations used are: MBD, metal-binding domain; HSQC, heteronuclear single quantum coherence.
References
- 1.Petris, M. J., Mercer, J. F., Culvenor, J. G., Lockhart, P., and Camakaris, J. (1996) EMBO J. 15 6084-6095 [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer, M., Hopkins, R. G., Failla, M. L., and Gitlin, J. D. (1999) Am. J. Physiol. 276 G639-G646 [DOI] [PubMed] [Google Scholar]
- 3.Harrison, M. D., Jones, C. E., Solioz, M., and Dameron, C. T. (2000) Trends Biochem. Sci. 25 29-32 [DOI] [PubMed] [Google Scholar]
- 4.Puig, S., and Thiele, D. J. (2002) Curr. Opin. Chem. Biol. 6 171-180 [DOI] [PubMed] [Google Scholar]
- 5.Bull, P. C., Thomas, G. R., Rommens, J. M., Forbes, J. R., and Cox, D. W. (1993) Nat. Genet. 5 327-337 [DOI] [PubMed] [Google Scholar]
- 6.Petrukhin, K., Fischer, S. G., Pirastu, M., Tanzi, R. E., Chernov, I., Devoto, M., Brzustowicz, L. M., Cavanis, E., Vitale, E., and Russo, J. J. (1993) Nat. Genet. 5 338-343 [DOI] [PubMed] [Google Scholar]
- 7.Tanzi, R. E., Petrukhin, K., Chernov, I., Pellequer, J. L., Wasco W., Ross, B., Romano, D. M., Parano, E., Pavone, L., and Brzustowicz, L. M., Devoto, M., Peppercorn, J., Bush, A. I., Sternlieb, I., Pirastu, M., Gusella, J. F., Evgrafov, O., Penchaszadeh, G. K., Honig, B., Edelman, I. S., Soares, M. B., Scheinberg, I. H., and Gilliam, T. C. (1993) Nat. Genet. 5 344-350 [DOI] [PubMed] [Google Scholar]
- 8.Mercer, J. F., Livingston, J., Hall, B., Paynter, J. A., Begy, C., Chandrasekharappa, S., Lockhart, P., Grimes, A., Bhave, M., and Siemieniak, D. (1993) Nat. Genet. 3 20-25 [DOI] [PubMed] [Google Scholar]
- 9.Vulpe, C. D., Levinson, B., Whitney, S., Packman, S., and Gitschier, J. (1993) Nat. Genet. 3 7-13 [DOI] [PubMed] [Google Scholar]
- 10.Solioz, M., and Vulpe, C. (1996) Trends Biochem. Sci. 21 237-241 [PubMed] [Google Scholar]
- 11.Gitschier, J., Moffat, B., Reilly, D., Wood, W. I., and Fairbrother, W. J. (1998) Nat. Struct. Biol. 5 47-54 [DOI] [PubMed] [Google Scholar]
- 12.Jones, C. E., Daly, N. L., Cobine, P. A., Craik, D. J., and Dameron, C. T. (2003) J. Struct. Biol. 143 209-218 [DOI] [PubMed] [Google Scholar]
- 13.Banci, L., Bertini, I., Del Conte, R., D'Onofrio, M., and Rosato, A. (2004) Biochemistry 43 3396-3403 [DOI] [PubMed] [Google Scholar]
- 14.Achila, D., Banci, L., Bertini, I., Bunce, J., Ciofi-Baffoni, S., and Huffman, D. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5729-5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banci, L., Bertini, I., Cantini, F., Rosenzweig, A. C., and Yatsunyk, L. A. (2008) Biochemistry 47 7423-7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutsenko, S., Petrukhin, K., Cooper, M. J., Gilliam, C. T., and Kaplan, J. H. (1997) J. Biol. Chem. 272 18939-18944 [DOI] [PubMed] [Google Scholar]
- 17.Rice, W. J., Kovashilin, A., and Stokes, D. L. (2006) Biochem. Biophys. Res. Commun. 348 124-131 [DOI] [PubMed] [Google Scholar]
- 18.Hatori, Y., Hirata, A., Toyoshima, C., Lewis, D., Pilankatta, R., and Inesi, G. (2008) J. Biol. Chem. 283 22541-22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiDonato, M., Hsu, H. F., Narindrasorasak, S., Que, L. J., and Sarkar, B. (2000) Biochemistry 39 1890-1896 [DOI] [PubMed] [Google Scholar]
- 20.Cater, M. A., La Fontaine, S., and Mercer, J. F. (2007) Biochem. J. 401 143-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cater, M. A., Forbes, J. R., La Fontaine, S., Cox, D., and Mercer, J. F. (2004) Biochem. J. 380 805-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voskoboinik, I., Mar, J., Strausak, D., and Camakaris, J. (2001) J. Biol. Chem. 276 28620-28627 [DOI] [PubMed] [Google Scholar]
- 23.Payne, A. S., and Gitlin, J. D. (1998) J. Biol. Chem. 273 3765-3770 [DOI] [PubMed] [Google Scholar]
- 24.Huster, D., and Lutsenko, S. (2003) J. Biol. Chem. 278 32212-32218 [DOI] [PubMed] [Google Scholar]
- 25.Strausak, D., La Fontaine, S., Hill, J., Firth, S. D., Lockhart, P. J., and Mercer, J. F. (1999) J. Biol. Chem. 274 11170-11177 [DOI] [PubMed] [Google Scholar]
- 26.Voskoboinik, I., Strausak, D., Greenough, M., Brooks, H., Petris, M., Smith, S., Mercer, J. F., and Camakaris, J. (1999) J. Biol. Chem. 274 22008-22012 [DOI] [PubMed] [Google Scholar]
- 27.Banci, L., Bertini, I., Cantini, F., Migliardi, M., Rosato, A., and Wang, S. (2005) J. Mol. Biol. 352 409-417 [DOI] [PubMed] [Google Scholar]
- 28.Banci, L., Bertini, I., Cantini, F., Della Malva, N., Migliardi, M., and Rosato, A. (2007) J. Biol. Chem. 282 23140-23146 [DOI] [PubMed] [Google Scholar]
- 29.Yatsunyk, L. A., and Rosenzweig, A. C. (2007) J. Biol. Chem. 282 8622-8631 [DOI] [PubMed] [Google Scholar]
- 30.Bunce, J., Achila, D., Hetrick, E., Lesley, L., and Huffman, D. L. (2006) Biochim. Biophys. Acta 1760 907-912 [DOI] [PubMed] [Google Scholar]
- 31.Huffman, D. L., and O'Halloran, T. V. (2000) J. Biol. Chem. 275 18611-18614 [DOI] [PubMed] [Google Scholar]
- 32.Larin, D., Mekios, C., Das, K., Ross, B., Yang, A. S., and Gilliam, C. T. (1999) J. Biol. Chem. 274 28497-28504 [DOI] [PubMed] [Google Scholar]
- 33.Strausak, D., Howie, M. K., Firth, S. D., Schlicksupp, A., Pipkorn, R., Multhaup, G., and Mercer, J. F. (2003) J. Biol. Chem. 278 20821-20827 [DOI] [PubMed] [Google Scholar]
- 34.van Dongen, E. M., Klomp, L. W., and Merkx, M. (2004) Biochem. Biophys. Res. Commun. 323 789-795 [DOI] [PubMed] [Google Scholar]
- 35.Banci, L., Bertini, I., Cantini, F., Felli, I. C., Gonnelli, L., Hadjiliadis, N., Pierattelli, R., Rosato, A., and Voulgaris, P. (2006) Nat. Chem. Biol. 2 367-368 [DOI] [PubMed] [Google Scholar]
- 36.Alzari, P. M., Berglund, H., Berrow, N. S., Blagova, E., Busso, D., Cambillau, C., Campanacci, V., Christodoulou, E., Eiler, S., Fogg, M. J., Folkers, G., Geerlof, A., Hart, D., Haouz, A., Herman, M. D., Macieira, S., Nordlund, P., Perrakis, A., Quevillon-Cheruel, S., Tarandeau, F., Van Tilbeurgh, H., Unger, T., Luna-Vargas, M. P., Velarde, M., Willmanns, M., and Owens, R. J. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 1103-1113 [DOI] [PubMed] [Google Scholar]
- 37.Golovanov, A. P., Hautbergue, G. M., Wilson, S. A., and Lian, L. Y. (2004) J. Am. Chem. Soc. 126 8933-8939 [DOI] [PubMed] [Google Scholar]
- 38.Anastassopoulou, J., Banci, L., Bertini, I., Cantini, F., Katsari, E., and Rosato, A. (2004) Biochemistry 43 13046-13053 [DOI] [PubMed] [Google Scholar]
- 39.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., III, and Skelton, N. J. (1996) Protein NMR Spectroscopy: Principles and Practice, pp. 478-528, Academic Press, Inc., San Diego
- 40.Schanda, P., Kupce, E., and Brutscher, B. (2005) J. Am. Chem. Soc. 33 199-211 [DOI] [PubMed] [Google Scholar]
- 41.Ishima, R., and Torchia, D. A. (2000) Nat. Struct. Biol. 7 740-743 [DOI] [PubMed] [Google Scholar]
- 42.Keller, R. (2004) The Computer Aided Resonance Assignment Tutorial, CANTINA Verlag, Goldau, Switzerland
- 43.Banci, L., Bertini, I., Cantini, F., Chasapis, C., Hadjiliadis, N., and Rosato, A. (2005) J. Biol. Chem. 280 38259-38263 [DOI] [PubMed] [Google Scholar]
- 44.Banci, L., Bertini, I., Cantini, F., Della Malva, N., Rosato, A., Herrmann, T., and Wüthrich, K. (2006) J. Biol. Chem. 281 29141-29147 [DOI] [PubMed] [Google Scholar]
- 45.Arnesano, F., Banci, L., Bertini, I., Cantini, F., Ciofi-Baffoni, S., Huffman, D. L., and O'Halloran, T. V. (2001) J. Biol. Chem. 276 41365-41376 [DOI] [PubMed] [Google Scholar]
- 46.Banci, L., Bertini, I., Chasapis, C., Ciofi-Baffoni, S., Hadjiliadis, N., and Rosato, A. (2005) FEBS J. 272 865-871 [DOI] [PubMed] [Google Scholar]
- 47.Wu, C. C., Rice, W. J., and Stokes, D. L. (2008) Structure 16 976-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamza, I., Prohaska, J., and Gitlin, J. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1215-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsivkovskii, R., MacArthur, B. C., and Lutsenko, S. (2001) J. Biol. Chem. 276 2234-2242 [DOI] [PubMed] [Google Scholar]
- 50.Mercer, J. F., Barnes, N., Stevenson, J., Strausak, D., and Llanos, R. M. (2003) Biometals 16 175-184 [DOI] [PubMed] [Google Scholar]
- 51.Linz, R., and Lutsenko, S. (2007) J. Bioenerg. Biomembr. 39 403-407 [DOI] [PubMed] [Google Scholar]
- 52.Garrett, D. S., Seok, Y. J., Peterkofsky, A., Clore, G. M., and Gronenborn, A. M. (1997) Biochemistry 36 4393-4398 [DOI] [PubMed] [Google Scholar]
- 53.Banci, L., Bertini, I., Ciofi-Baffoni, S., Kandias, N. G., Spyroulias, G. A., Su, X. C., Robinson, N. J., and Vanarotti, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.