Abstract
Arylsulfatase A (ASA) catalyzes the intralysosomal desulfation of 3-O-sulfogalactosylceramide (sulfatide) to galactosylceramide. The reaction requires saposin B (Sap B), a non-enzymatic proteinaceous cofactor which presents sulfatide to the catalytic site of ASA. The lack of either ASA or Sap B results in a block of sulfatide degradation, progressive intralysosomal accumulation of sulfatide, and the fatal lysosomal storage disease metachromatic leukodystrophy. We studied the coupled Sap B-ASA reaction in vitro using detergent-free micellar and liposomal assay systems and in vivo using cell culture models of metachromatic leukodystrophy. Under in vitro conditions, the reaction had a narrow pH optimum around pH 4.3 and was inhibited by mono- and divalent cations, phosphate and sulfite. Bis(monoacylglycero) phosphate and phosphatidic acid were activators of the reaction, underscoring a significant role of acidic phosphoglycerolipids in sphingolipid degradation. Desulfation was negligible when Sap B was substituted by Sap A, C, or D. Up to a molar ratio between Sap B and sulfatide of 1:5, an elevation of Sap B concentrations caused a sharp increase of sulfatide hydrolysis, indicating the requirement of unexpected high Sap B levels for maximum turnover. Feeding of ASA-deficient, sulfatide-storing primary mouse kidney cells with ASA caused partial clearance of sulfatide. Co-feeding of Sap B or its precursor prosaposin resulted in the lysosomal uptake of the cofactor but did not promote ASA-catalyzed sulfatide hydrolysis. This suggests that Sap B is not a limiting factor of the coupled Sap B-ASA reaction in mouse kidney cells even if sulfatide has accumulated to unphysiologically high levels.
Glycosphingolipids (GSLs)2 are membrane components assembled from a hydrophobic ceramide moiety and a hydrophilic carbohydrate head group which contains one or several occasionally chemically modified monosaccharide units (1, 2). Degradation in the endosomal/lysosomal compartment is catalyzed by a series of acidic exohydrolases which remove modifying groups from the carbohydrate, stepwise release monosaccharide units from the non-reducing end of oligomeric head groups, eventually cleave off the basal monosaccharide from the ceramide moiety, and subsequently hydrolyze ceramide into sphingosine and fatty acid. The lysosomal degradation of GSLs with short hydrophilic head groups also depends on small, non-enzymatic polypeptides designated as sphingolipid activator proteins (1–4). This group of lysosomal cofactors comprises five polypeptides; (i) the four saposins (Saps) A, B, C, and D, which are generated by proteolytic cleavage of a common precursor, prosaposin, and (ii) the GM2 activator protein, which is encoded by another gene. Each sphingolipid activator protein binds and presents distinct target lipids to corresponding hydrolases which themselves are barely active on membrane-bound substrate molecules. The physiological significance of activator proteins is underscored by the occurrence of sphingolipid storage diseases due to mutations of individual activators and the lethal phenotype of complete prosaposin deficiency in humans and mice (3–5).
Arylsulfatase A (ASA) is one of nearly a dozen enzymes that are involved in the lysosomal degradation of GSLs (6). It catalyzes the first step in the catabolism of sulfatide, an anionic GSL with a sulfated galactosyl residue as head group, by removing the sulfate group. The desulfation also depends on Sap B, which binds membrane-bound sulfatide in a 1:1 complex and presents it to the substrate binding site of ASA (1–4). In the current view, presentation involves the shielding of the lipid tail by accommodating it in a hydrophobic cavity in the dimeric protein shell of Sap B and the full extraction of the lipid from the membrane leaflet possibly by a conformational change of the activator (7). Desulfation of sulfatide yields galactosylceramide, which is presented and hydrolyzed by the next saposin (Sap A) and lysosomal enzyme (galactocerebrosidase) of the catabolic cascade, respectively. The lack of ASA activity results in the glycosphingolipidosis metachromatic leukodystrophy (MLD), which is characterized by progressive accumulation of sulfatide in oligodendrocytes, Schwann cells, and other cell types, the loss of myelinating cells, progressive neurological symptoms, and premature death (6). Likewise, Sap B deficiency causes intralysosomal deposition of sulfatide. In this case additional GSLs, globotriaosylceramide and digalactosylceramide, accumulate that are not substrates of ASA and do not accumulate in classical MLD (3). It is likely that in normal cells these galactosylceramides are presented to the lysosomal α-galactosidase A by Sap B. Thus, differences in the substrate specificities of ASA and Sap B, which may also include different preferences for hydroxylated and non-hydroxylated sulfatide species (8), may explain why Sap B deficiency causes a disease in which the clinical picture is very similar but not identical to classical MLD.
To mimic the in vivo conditions of sphingolipid degradation in the endosomal/lysosomal compartment, micellar or liposomal in vitro systems have been established. They employ substrate-bearing micelles or liposomes exposed to the soluble enzymes, sphingolipid activator proteins, and possibly other factors (for review, see Ref. 1). Such in vitro systems expanded our knowledge about the lysosomal biogenesis and function substantially. One finding was that hydrolysis of many membrane-bound sphingolipids is stimulated by bis(monoacylglycero)phosphate (BMP; erroneously also called lysobisphosphatidic acid, LBPA), an acidic phosphoglycerolipid which accumulates in intraendosomal and intralysosomal membrane structures during the endocytic lipid sorting process. According to a recently proposed model these inner vesicular membrane structures descend from invaginations of the limiting lysosomal membrane and represent the main site of GSL breakdown (2).
Enzyme replacement therapy (ERT) using repeated intravenous injection of recombinant human ASA reduces sulfatide storage in the peripheral and central nervous system of ASA knock-out mice and, therefore, represents a new and promising treatment option for MLD (9). A better understanding of the intraendosomal/intralysosomal processes leading to substrate clearance might be critical for the optimization of this treatment regimen. For this purpose, we established detergent-free micellar and liposomal in vitro systems and studied the hydrolysis of radioactive and nonradioactive sulfatide in the Sap B-ASA concerted reaction. The role of Sap B in sulfatide hydrolysis was further investigated in cell culture models of MLD.
EXPERIMENTAL PROCEDURES
Materials—Unless otherwise indicated recombinant human ASA (rhASA) isolated from secretions of transfected baby hamster kidney cells was used (10). The baby hamster kidney rhASA had a specific activity of 31 units/mg. Five rhASA batches purified from the secretions of transfected Chinese hamster ovary cells were kindly provided by Zymenex A/S (Hillerød, Denmark) (9). Native Sap B was purified from pig kidney and human urine, respectively (11). Recombinant human Saps A, B, C, and D were produced using the Pichia pastoris expression system (12, 13). Prosaposin was purified as described (14). Rabbit antisera against human ASA, Sap B, and Sap D were as described (15, 16). BMP, dioleyl phosphatidylcholine, and dolichol were from Avanti Polar Lipids (Alabaster, AL). All other chemicals were obtained from Sigma-Aldrich, Merck, or Calbiochem-Novabiochem. Chemicals and solvents were of analytical grade.
Radiolabeling—Radiocarbon-labeled sulfatide was synthesized from lyso-sulfatide (17) and [1-14C]stearic acid (2.15 GBq/mmol obtained from GE Healthcare) following procedures as outlined (18). Native Sap B (11) was labeled with tritiated propionic acid N-succinimidyl ester essentially as described (19). Briefly, [2,3-3H]propionic acid N-succinimidyl ester (3.59 TBq/mmol obtained from GE Healthcare) was diluted with the unlabeled reagent to 18.5 GBq/mmol. The unlabeled ester was synthesized as described (20). The amount of the labeling reagent was chosen so as to achieve the conversion of one to two ε-amino groups of the peripheral lysine residues of the protein into propionamide residues. The degree of labeling was determined by measuring protein content and radioactivity. The conversion of nearly two lysine residues had no effect on the ability of Sap B to stimulate the degradation of radiolabeled sulfatide by ASA (see Fig. 5, A and B). Excess reagent was removed by exhaustive dialysis against phosphate buffer, pH 4.5, at 4 °C.
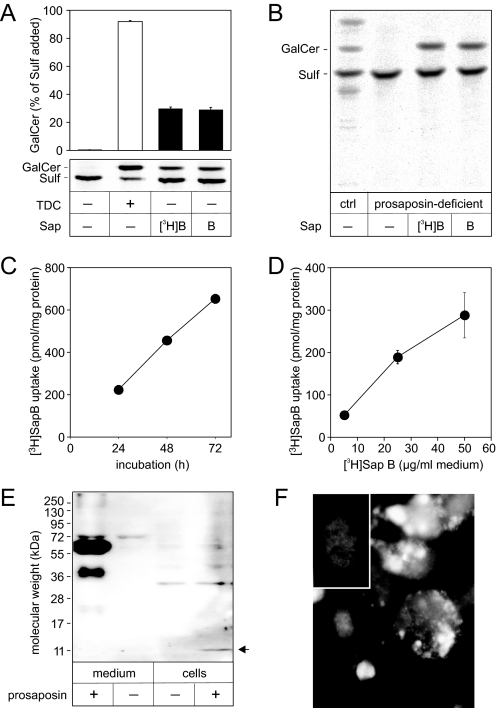
FIGURE 5.
Cellular uptake of [3H]Sap B and prosaposin. A, activity of [3H]Sap B tested by hydrolysis of [14C]sulfatide in a liposomal in vitro assay under standard conditions. Lipids were separated by TLC and quantified. Bars representing the means ± S.E. of n = 2 independent measurements are indicated. B, effect of [3H]Sap B and native Sap B on the degradation of [14C]sulfatide in prosaposin-deficient and healthy control (ctrl) fibroblasts. Cells were preincubated for 24 h with Sap B (25 μg/ml medium) and subsequently fed additionally with [14C]sulfatide (0.66 nmol/ml medium) for 48 h. Cellular lipids were separated by TLC and 14C-labeled compounds were visualized by autoradiography. C, uptake of [3H]Sap B by prosaposin-deficient human fibroblasts. Cells were incubated with [3H]Sap B (25 μg/ml) for 24, 48, and 72 h. The means ± S.E. of n = 2 independent experiments are indicated. D, uptake of [3H]Sap B by ASA-deficient murine kidney cells. Cells were incubated with different concentrations of [3H]Sap B for 24 h. The means ± S.E. of n = 3 independent experiments are indicated. E, prosaposin and Sap C in media and homogenates of ASA-deficient murine kidney cells fed with human prosaposin (2 μg/ml medium). An antiserum which specifically recognizes human prosaposin and human Sap C (arrow) was used for immunodetection. F, immunofluorescence staining of human Sap C in cells incubated with 2 μg/ml human prosaposin for 24 h. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole and appear diffuse gray in the black-and-white image. Bright signals for Sap C are mainly detectable in the perinuclear region. Signals are absent from cells which were cultured without human prosaposin (inset).
p-Nitrocatechol sulfate (pNCS) Assay—The specific activity of rhASA was measured with a cofactor-independent assay using the water-soluble sulfate ester (pNCS) as a substrate (21).
Sap B-dependent Sulfatide Hydrolysis—In initial experiments, 5 nmol of sulfatide was dried and suspended in 25 μl of reaction buffer (10 mm sodium acetate, 100 mm NaCl, pH 5.0) by ultrasonication. Then, 25 μl of reaction buffer containing 10 μg of bovine serum albumin, 40 milliunits rhASA, and 100 nmol taurodeoxycholate (Sigma-Aldrich) or up to 0.3 nmol Sap B was added. Under these conditions the sulfatide concentration is above the critical micellar concentration (22), and pure sulfatide micelles are formed. The reaction was stopped after 4 h at 37 °C. Reaction products were extracted according to Folch et al. (23) separated by high performance TLC using chloroform/methanol/water (70/30/4, v/v/v) as a solvent system and visualized by CuSO4 staining (24). Quantification was done using reference standards by densitometry scanning and the image analysis software AIDA (Raytest, Straubenhardt, Germany).
In later experiments liposomes containing 10 mol % sulfatide, 10 mol % cholesterol, and 80 mol % phosphatidylcholine were used as a substrate. For the preparation of liposomes, lipids dissolved in chloroform/methanol (1/1, v/v) were mixed, and the solvent was evaporated first under a stream of dry nitrogen and subsequently in a vacuum desiccator. After hydration of the dried lipids in 10 mm sodium acetate, pH 4.5, the mixture was sonicated in a Branson 1510 bath sonicator for 10 min (Branson Ultrasonics Corp., Danbury, CT). Under these conditions the lipids assemble into bilayered liposomes or liposome-like structures (25). The physical state of the structures was analyzed by electron transmission microscopy which revealed liposomes with a maximum diameter of ∼110 nm (not shown).
The highest rate of sulfatide hydrolysis was achieved with liposomes containing 5 nmol of sulfatide (10 mol %), 5 nmol of cholesterol (10 mol %), 15 nmol of BMP (30 mol %), and 25 nmol of phosphatidylcholine (50 mol %) suspended in 50 μl of 10 mm sodium acetate, pH 4.5, containing 10 μg bovine serum albumin, 3 nmol of SapB, and 30 milliunits of rhASA. Under standard conditions, however, only 0.3 nmol of Sap B was added, and no BMP was added.
For radioactive micellar assays, pure sulfatide micelles containing 4.5 nmol of sulfatide were augmented with 0.5 nmol of [14C]sulfatide (58.6 Ci/mol) and reacted under standard conditions. After the reaction, lipids were dried and resolved in 40 μl of chloroform/methanol (1/1, v/v), separated by TLC in chloroform, methanol, 0.22% CaCl2 (60/35/8, v/v/v), and visualized with a BioImaging Analyser1000 (Fuji Europe, Düsseldorf, Germany).
For radioactive liposomal assays, liposomes containing 0.5 nmol of [14C]sulfatide plus 4.5 nmol of sulfatide (10 mol %), cholesterol (10 mol %), BMP (0–30 mol %), and phosphatidylcholine (80–50 mol %) were prepared by established methods (12). After reacting under standard conditions, the assay was terminated by the addition of 0.5 ml of chloroform/methanol (1/1, v/v). The separation, visualization, and quantification of radiolabeled sulfatide and galactosylceramide was done as described above.
Cell Culture Experiments—Before feeding, primary kidney cells from ASA knock-out mice (26, 27) and prosaposin-deficient human fibroblasts (28) were grown for 24 h in Opti-MEM I and Dulbecco's modified Eagle's medium, respectively, containing 0.3% fetal bovine serum. For uptake studies [3H]Sap B was added subsequently. After incubation, fibroblasts were washed 3 times with PBS, detached with 0.25% trypsin, and collected by centrifugation. The cell pellets were washed 3 times with PBS and homogenized in water by sonication. Kidney cells were washed 3 times with PBS, once with 0.05% trypsin, 3 times with PBS, and then scraped off the plates with a rubber policeman. The radioactivity of the suspensions was determined in triplicate. Metabolic studies with prosaposin-deficient fibroblasts were performed as described (12). For metabolic studies with murine kidney cells, different combinations of rhASA, Sap B, and prosaposin were added. After feeding, cells were thoroughly washed with PBS, harvested by trypsinization, and suspended in PBS. Aliquots of the cell suspension were used for the determination of rhASA and lipid concentrations by enzyme-linked immunosorbent assay and TLC, respectively (9). Total protein was determined using the Dc assay from Bio-Rad. Loading volumes for TLC were normalized on protein concentrations, and relevant lipids were densitometrically quantified. Sulfatide levels were normalized on cholesterol levels by dividing the respective scan signals after background normalization. Immunofluorescence and immunoblots with antibodies for ASA, Sap B, or Sap C was performed according to established techniques (27).
RESULTS
Standard Assay for the Measurement of Sap B-activated Sulfatide Desulfation—Desulfation of sulfatide to galactosylceramide was successfully reconstituted by incubating pure sulfatide micelles with rhASA and either detergent (sodium taurodeoxycholate) or Sap B under pH and salt conditions described for the hydrolysis of the water-soluble sulfate ester p-nitrocatechol sulfate (22, 29) (Fig. 1). Lysosomal membranes, representing the natural target for Sap B, are rich in phosphoglycerolipids and contain comparably low levels of cholesterol. To mimic the composition of lysosomal membranes, liposomes composed of 10 mol % sulfatide, 10 mol % cholesterol, and 80 mol % phosphatidylcholine were used as a substrate in subsequent experiments. Up to a relative concentration of 30 mol % (cholesterol) or 90 mol % (phosphatidylcholine) the two lipids did not significantly affect sulfatide hydrolysis (not shown).
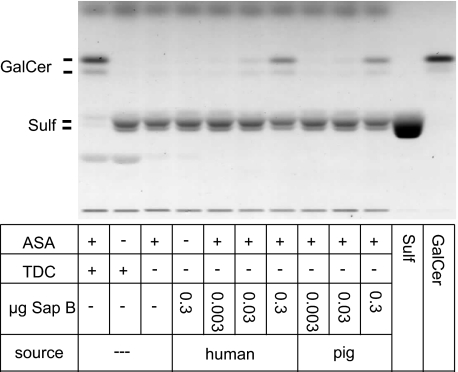
FIGURE 1.
Sap B-dependent hydrolysis of sulfatide in a non-radioactive assay using pure sulfatide micelles as a substrate. Micelles were incubated with 40 milliunits of rhASA and increasing concentrations of human or porcine Sap B for 4 h. Lipids were separated by high performance TLC. For a positive control, Sap B was substituted by taurodeoxycholate (TDC). For a negative control, reactions were conducted without ASA or without detergent and Sap B.
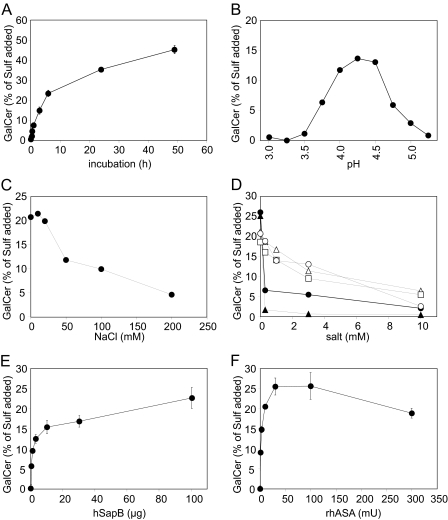
Investigation of the Sap B-dependent Sulfatide Hydrolysis at Liposomal Surfaces—Liposomes were used to evaluate the influence of various parameters on the Sap B-dependent hydrolysis of sulfatide (Fig. 2). In a first series of experiments the (i) incubation time, (ii) pH value, (iii) sodium chloride concentration, or (iv) concentration of different divalent cations and anions was varied. The reaction increased steadily up to a sulfatide turnover of ∼25%, which was reached after 4–6 h under standard conditions (Fig. 2A). Because of the slow decline of the reaction velocity within this time interval, the galactosylceramide concentration determined after 4 h was taken as an estimate of the reaction rate in subsequent experiments. In experiments in which the reaction rate was increased, for example by the addition of high Sap B concentrations (see below), the incubation time was reduced to keep the sulfatide consumption below 25% and to avoid substrate shortage. When tested at different pH values, the desulfation was highest in a narrow pH interval between pH 4.0 and 4.5. It was less than half-maximum at pH values below pH 3.75 and above pH 4.75 (Fig. 2B). Thus, the pH optimum of the Sap B-dependent sulfatide hydrolysis closely resembled the pH of the lysosomal compartment, which ranges between 4.3 and 4.5 in human fibroblasts (30). The hydrolysis was inhibited at NaCl concentrations exceeding 20 mm (Fig. 2C). At 50 and 200 mm the rate of sulfatide hydrolysis was diminished by ∼50 and 75%, respectively. In contrast to sodium chloride, MgCl2, CaCl2, and MnCl2 reduced the reaction rate in the low millimolar range, suggesting a profound inhibitory effect of divalent cations (Fig. 2D). At a concentration of 3 mm, these salts inhibited the hydrolysis by ∼30–50%. Phosphate and sulfite, two known competitive inhibitors of ASA (31), were even more effective in inhibiting the hydrolysis of sulfatide (Fig. 2D).
FIGURE 2.
Sulfatide hydrolysis under different experimental conditions using liposomes containing 10 mol % sulfatide, 10 mol % cholesterol, and 80 mol % phosphatidylcholine as a substrate. Unless otherwise indicated, the reaction conditions were 10 mm NaAc, pH 4.5, 30 milliunits of rhASA, 5 nmol of sulfatide, and 0.3 nmol of Sap B. Either 20 mm NaCl (B) or no NaCl (A, D–F) was added. Reaction times were 4 h (B–D, F) or 2 h (E). Experiments shown in B–D were done in single assays. Data points and bars shown in A, E, and F represent the means ± S.D. of n = 3 independent experiments per condition. Please note that in some cases error bars are hidden by the symbols. Shown are variation of incubation time (A), pH value (B), NaCl concentration (C), concentration of MgCl2 (open triangles), CaCl2 (open squares), MnCl2 (open circles), NaH2PO4 (closed circles), and Na2SO3 (closed triangles) (E), concentration of native human Sap B (D), and activity of rhASA (F).
In a second set of experiments, the Sap B or rhASA concentration was varied. To evaluate the dose-dependent effects of Sap B, the rhASA concentration was kept constant, and 0–100 μg of native human Sap B (0–200 μm) was added. At the highest concentration, Sap B was present in a 2-fold molar excess upon sulfatide. The rate of sulfatide hydrolysis increased continuously with increasing Sap B concentrations (Fig. 2E). The most prominent increase was visible between 0 and 10 μg of Sap B, i.e. up to a molar ratio between Sap B and sulfatide of 1:5. A further elevation of the Sap B concentration to a molar ratio around 2:1 (100 μg Sap B) increased the mean sulfatide hydrolysis only by another ∼50%. Effects of increasing rhASA concentrations were measured at a constant Sap B concentration of 3 μg per 50 μl (6 μm). Up to a total activity of 30 milliunits (0.3 μm) the reaction rate increased almost linearly. It leveled off at 100 milliunits and decreased by ∼25% at 300 milliunits (Fig. 2F).
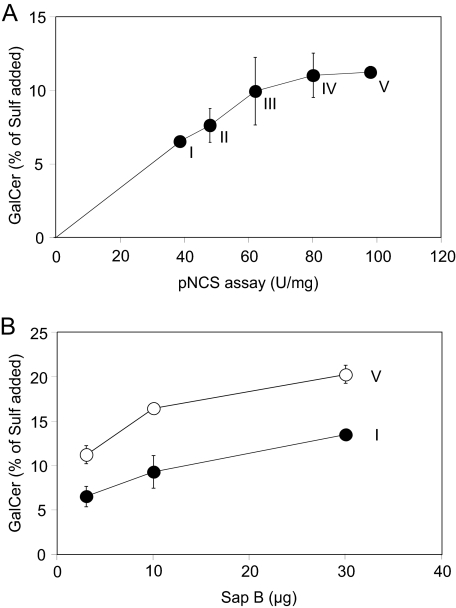
Comparison of pNCS and Sulfatide Hydrolyzing Activities of Different ASA Batches—The conventional assay for the determination of the ASA activity utilizes the artificial sulfate ester pNCS as a substrate. Because pNCS is water-soluble, the assay is cofactor-independent and does not require Sap B or sodium taurodeoxycholate. To compare the hydrolysis of pNCS and sulfatide, we used five rhASA preparations with widely differing specific activities. The rhASA batches were prepared from the secretions of a stably transfected Chinese hamster ovary cell line, which had been cultured under different conditions. This led to a more or less efficient modification of the active site cysteine to formylglycine, which is essential for catalytic activity of ASA (32). In the pNCS assay, the five Chinese hamster ovary-rhASA preparations displayed 39–98 units/mg, i.e. 1 mg of rhASA hydrolyzed 39–98 μmol of pNCS per hour (Fig. 3A). In the Sap B-dependent assay, 2 μg of the 5 rhASA batches cleaved between 6.5 and 11.2% of the input sulfatide within 20 min. This is equivalent to a specific activity of 0.49–0.84 μmol of sulfatide/h/mg. Thus, the rate of sulfatide cleavage was 80–120-fold lower than the rate of pNCS cleavage. Notably, the rate of sulfatide and pNCS hydrolysis failed to correlate for preparations with high specific activities. Thus, between 80 and 100 units/mg virtually no difference in the rate of sulfatide turnover was discernible.
FIGURE 3.
Activities of five different rhASA preparations (I–V) purified from secretions of Chinese hamster ovary cells. The activities were measured in a cofactor-independent assay using the water-soluble substrate pNCS (21) and in a Sap B-dependent assay using pure sulfatide micelles as a substrate. For the sulfatide hydrolyzing activity, the means ± S.D. of n = 3 independent measurements are indicated. A, correlation between pNCS hydrolytic activity (abscissa) and sulfatide hydrolytic activity (ordinate). B, effect of increasing concentrations of native human Sap B on sulfatide hydrolyzing activity of preparation I and V.
To investigate in how far Sap B limits sulfatide hydrolysis, the activity of the two rhASA preparations with the highest and the lowest pNCS hydrolyzing activity was retested in the presence of increasing Sap B concentrations. A 10-fold increase of the Sap B concentration stimulated the sulfatide turnover of each of the two batches around 2-fold (Fig. 3B). Notably, in the presence of 30 μg of Sap B, the rhASA preparation with the low specific activity hydrolyzed more sulfatide than the preparation with the high activity at 3 μg of Sap B. Thus, a 2.5-fold lower enzymatic activity could be overcompensated by a 10-fold increase of Sap B.
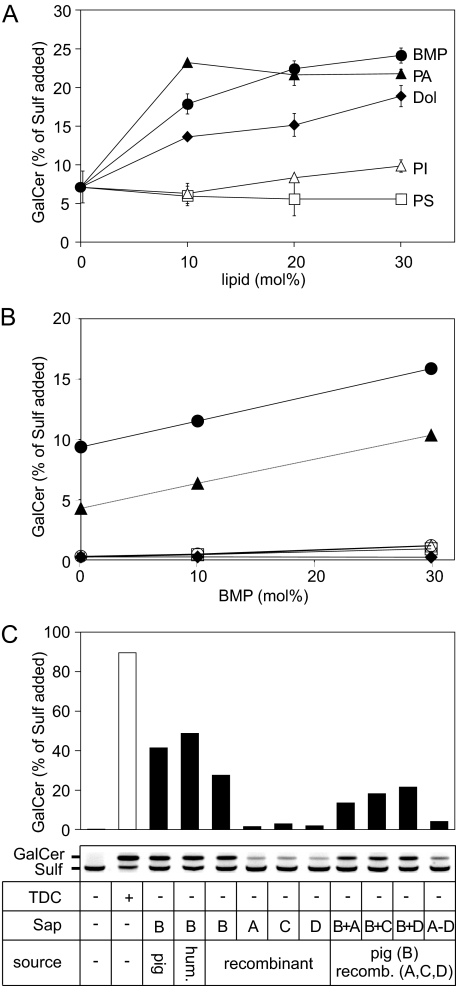
Effect of Different Lipid Additives on Sap B-dependent Sulfatide Hydrolysis—The hydrolysis of various sphingolipids in micellar and liposomal in vitro systems is influenced by the lipid composition of the latter (2). To detect the possible effects of acidic phosphoglycerolipids or the isoprenoid dolichol on sulfatide hydrolysis, sulfatide-containing liposomes were augmented with respective lipids and reacted under standard conditions. In detail, 10 mol % sulfatide was mixed with 10 mol % cholesterol, 0–30 mol % of either BMP, phosphatidylinositol, phosphatidylserine, phosphatidic acid, or dolichol, and the lipid mixtures were complemented with 80–50 mol % phosphatidylcholine. High performance TLC revealed that the individual lipid additives affected sulfatide hydrolysis to different extents. BMP, phosphatidic acid, and dolichol were moderate activators of sulfatide cleavage. At a relative concentration of 30 mol %, these lipids stimulated the reaction rate by a factor of 3.4 (BMP), 3.1 (phosphatidic acid), and 2.7 (dolichol), respectively (Fig. 4A). Phosphatidylinositol and phosphatidylserine, on the contrary, had virtually no effect on the rate of sulfatide hydrolysis.
FIGURE 4.
Effect of acidic lipids and saposin A, C, and D on sulfatide hydrolysis. A, stimulatory effects of different lipid additives. Liposomes composed of 10 mol % of sulfatide, 10 mol % of cholesterol, and 80–50 mol % of phosphatidylcholine were mixed with 0–30 mol % of one of the following lipids: BMP (closed circles;), phosphatidic acid (closed triangles, PA), dolichol (closed diamonds, dol), phosphatidylinositol (open triangles, PI), phosphatidylserine (open squares, PS). Reaction conditions are as outlined in the legend of Fig. 2. For each condition the mean ± S.D. of n = 3 independent measurements are indicated. B, saposin- and BMP-dependent sulfatide hydrolysis. Liposomes consisting of 10 mol % [14C]sulfatide, 10 mol % cholesterol, 0–30 mol % BMP, and 80–50 mol % phosphatidylcholine were reacted in the presence of one of the following Saps (0.3 nmol): native human Sap B (closed circles), native porcine Sap B (closed triangles), recombinant human Sap A (open circles), recombinant human Sap C (open triangles), recombinant human Sap D (open squares). As a negative control, no Sap was added (closed diamonds). The graphs for the negative control, Sap A, C, and D are hardly discernible due to substantial overlap close to the base line. Data are from single assays. C, saposin-dependent sulfatide hydrolysis in pure sulfatide micelles containing [14C]sulfatide. Different Saps were added alone or in combination as indicated. After TLC, radioactive Sulf and GalCer was visualized (inset) and densitometrically quantified (histogram). The histogram bar upper limits represent the means of two independent experiments. TDC, taurodeoxycholate.
For a more precise evaluation of the stimulatory effect of BMP, liposomes containing [14C]sulfatide were used. After carrying out the reaction under standard conditions, the densitometric quantification of radioactive galactosylceramide revealed a 1.7- and 2.4-fold increase of the sulfatide turnover in the presence of 30 mol % BMP for human and porcine Sap B, respectively (Fig. 4B).
Substitution of Sap B by Other Saposins—Liposomes containing radiolabeled sulfatide and different concentrations of BMP were also used to determine whether and to which extent Sap A, C, and D can substitute for Sap B in presenting sulfatide to ASA. In this experiment human and porcine Sap B afforded hydrolysis of up to ∼10 and ∼16% of sulfatide, whereas less than 1.2% of sulfatide was hydrolyzed when recombinant human Sap A, C, or D was added instead of Sap B (Fig. 4B).
The inability of Sap A, C, and D to efficiently present sulfatide to ASA was confirmed by radioactive assays using pure sulfatide micelles as a substrate. Also under these conditions Sap A, C, and D did not allow a significant turnover of sulfatide, and only 1.5, 2.9, and 1.9% of [14C]sulfatide was hydrolyzed, respectively (Fig. 4C). On the contrary, three different Sap B preparations, which were used as positive controls, hydrolyzed 28–49% of the sulfatide added. It has to be mentioned that in this experiment the reaction conditions were chosen to allow detection of also minor stimulatory effects of Sap A, C, and D. The rates of the Sap B-promoted reactions may have been underestimated due to a high sulfatide consumption and resulting substrate shortage.
To evaluate possibly synergistic effects of Sap A, C, or D on the Sap B-promoted sulfatide hydrolysis, combinations of Saps were also tested (0.3 nmol each). Compared with Sap B alone, combinations with any of the three other Saps led to a substantial decrease of the sulfatide turnover (Fig. 4C). The inhibitory effect of unspecific Saps on sulfatide hydrolysis was confirmed by a combination of all four cofactors. Under this condition only 4.1% of the sulfatide was hydrolyzed.
Co-feeding of rhASA and Activator in a Cell Culture Model of MLD—The in vitro data indicated that a low ASA activity can be overcompensated for by an increase in the concentration of Sap B (Fig. 3B) and that unexpected high levels of Sap B are required for maximum sulfatide turnover (Fig. 2E). We, therefore, hypothesized that Sap B expression might be a limiting factor of sulfatide hydrolysis also in vivo. To test this notion we used an ERT cell culture model consisting of primary murine ASA-deficient kidney cells. Because of the lack of endogenous ASA, these cells are unable to hydrolyze sulfatide and, as a consequence, develop intralysosomal sulfatide deposits (27). As shown previously, feeding of the cells with ASA leads to a dose-dependent reduction of sulfatide storage (33). To allow for the detection of possible synergistic effects of co-feeding with the activator in the present experiments, rhASA was used in a low concentration (2.5–6 μg/ml), leading only to a partial decline of storage. The cells were either incubated with rhASA alone or in combination with Sap B. Alternatively to Sap B, the Sap B precursor prosaposin was added.
First, the cellular uptake of rhASA, Sap B, and prosaposin was analyzed. Endocytosis and lysosomal targeting of rhASA is clearly demonstrated by the capacity of externally added rhASA to compensate the catabolic defect of ASA-deficient kidney cells (see below). Uptake and lysosomal delivery was confirmed by immunofluorescence staining using antibodies to human ASA and the lysosomal marker lamp-2 (not shown; for previous experiments, see Ref. 33). Quantification of the internalized rhASA by enzyme-linked immunosorbent assay revealed a concentration of 0.39 ± 0.09 ng (mean ± S.D., n = 3) or ∼7 fmol of rhASA/mg of cell protein 24 h after feeding of 2.5 μg/ml rhASA (not shown).
Uptake studies for Sap B were complicated by the lack of antibodies which recognize Sap B with sufficient specificity and sensitivity. To circumvent this limitation, Sap B was radioactively labeled using [2,3-3H]propionic acid N-succinimidyl ester as a radiolabeling reagent. Possible adverse effects of the modification on the sulfatide-presenting activity of the activator were tested in a liposomal in vitro assay and cell culture experiments. In the in vitro assay, unmodified Sap B and [3H]Sap B promoted the hydrolysis of [14C]sulfatide to the same extent (Fig. 5A). In the cell culture assay the ability of externally added Sap B (25 μg/ml) to compensate the defective sulfatide catabolism of prosaposin-deficient human fibroblasts was exploited (12). Feeding of prosaposin-deficient cells revealed no difference in the corrective capacity of unmodified Sap B and [3H]Sap B as shown by the complete restoration of the sulfatide catabolism under both conditions (Fig. 5B). Thus, Sap B is not inactivated by the labeling reaction and is taken up in functionally relevant amounts. Feeding of 25 μg/ml [3H]Sap B to prosaposin-deficient human fibroblasts and primary murine kidney cells revealed similar uptake rates of 220- and 190-pmol cofactor/mg of cellular protein and 24 h (Fig. 5, C and D). As tested in murine kidney cells, uptake of Sap B was not saturable up to a concentration of 50 μg/ml (5 μm) (Fig. 5D).
Uptake of prosaposin was investigated with an antiserum to human Sap C, which detects human Sap C and human prosaposin but not their murine homologues. Therefore, uptake of human prosaposin by primary murine kidney cells could be specifically investigated by immunoblotting. Upon feeding of ASA-deficient murine kidney cells with 2 μg/ml prosaposin for 24 h, prosaposin (62 kDa) is detectable in the medium but not in the cell homogenate. However, a 10-kDa intracellular Sap C signal is discernible in fed cells, demonstrating lysosomal delivery and intralysosomal proteolysis of the precursor to the mature Saps (Fig. 5E). Endocytosis and lysosomal targeting of prosaposin was confirmed by immunofluorescence staining with α-Sap C antiserum (Fig. 5F).
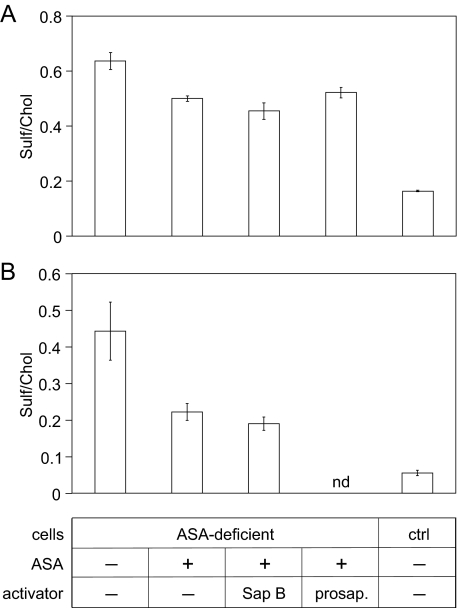
After demonstration of uptake of rhASA, Sap B and prosaposin, possible effects of activator co-feeding on the catabolism of ASA-deficient kidney cells, were analyzed. In an initial experiment, 2.5 μg/ml rhASA was added in combination with either 25 μg/ml Sap B or 2 μg/ml prosaposin (Fig. 6A). Feeding with rhASA alone reduced the mean sulfatide levels by ∼20%. However, neither Sap B nor prosaposin significantly increased the sulfatide turnover. In other co-feeding experiments the incubation time and/or the molar ratio between ASA and activator was varied. Cells were fed, for example, for 48 h with 6 μg/ml rhASA alone or in combination with 4.5 μg/ml Sap B (Fig. 6B). Around 50% (rhASA-feeding) and 58% of sulfatide (rhASA-Sap B co-feeding) were lost from the cells, confirming that activator co-feeding has no significant stimulatory effect on sulfatide hydrolysis (Fig. 6B). Also, preincubation of cells with Sap B or prosaposin alone for 24 h and subsequent co-feeding of cofactor and rhASA for another 24 h failed to increase the rate of sulfatide hydrolysis (not shown).
FIGURE 6.
Effect of Sap B and prosaposin co-feeding on sulfatide storage reduction in primary murine ASA-deficient kidney cells incubated with rhASA. Wild type primary kidney cells were used as a control (ctrl). Sulfatide levels were normalized on cholesterol (chol) levels. Bars indicate the means ± S.D. of n = 3 experiments per condition. nd, not determined. A, cells were treated with 2.5 μg/ml rhASA and 25 μg/ml native human Sap B or 2 μg/ml prosaposin as indicated in the table below (B). After 24 h cells were harvested for lipid analysis. B, sulfatide levels of cells after treatment with 6 μg/ml rhASA and 4.5 μg/ml native human Sap B for 48 h.
DISCUSSION
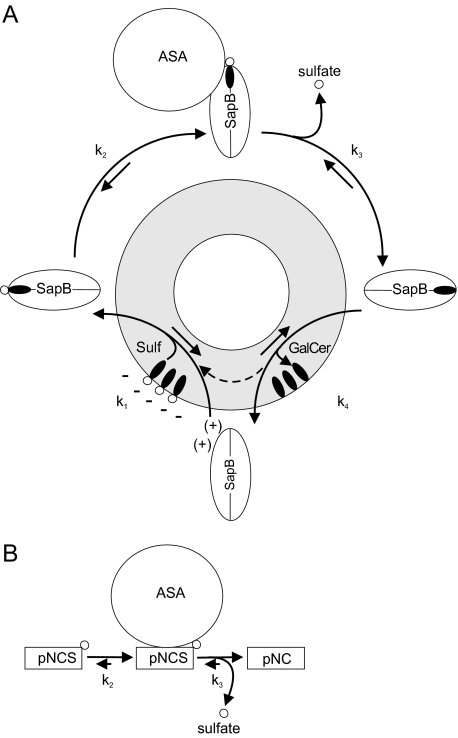
We have established micellar and liposomal in vitro assays to examine the ASA-catalyzed desulfation of sulfatide in a Sap B-dependent manner. These assays revealed that none of the Saps A, C, or D can substitute for Sap B in presenting sulfatide to ASA (Fig. 4, B and C). Conventional ASA activity assays circumvent the need for Sap B by using either a detergent which mimics the mobilization function of Sap B (9, 34) or by using water-soluble substrates like the sulfate ester pNCS (21, 30). In both experimental set-ups the reaction rate is determined by the catalytic rate of ASA (Fig. 7B). Sulfatide hydrolysis in the lysosomal compartment, however, also depends on the cofactor Sap B, which binds and solubilizes membrane-bound sulfatide in a 1:1 complex and presents it to the substrate binding site of ASA (1–4). The availability and activity of the cofactor rather than the catalytic rate of ASA might, therefore, be rate-limiting under in vivo conditions. This notion is supported by our in vitro data, which demonstrate that unexpected high concentrations of Sap B are required for a maximum turnover of sulfatide. Thus, the rate of sulfatide hydrolysis increases substantially up to a molar ratio between Sap B and sulfatide of 1:5 (Figs. 2E and 3B). Furthermore, at a constant Sap B concentration, a saturation of the sulfatide hydrolysis is rapidly obtained when the specific ASA activity is increased (Fig. 3A).
FIGURE 7.
Kinetic models of ASA-catalyzed reactions. A, kinetic model of the coupled Sap B-ASA reaction. The reaction involves binding of Sap B to the lysosomal target membrane and extraction of membrane-bound sulfatide (k1). The sensitivity of the mobilization process to anionic lipids, pH value, and ionic strength (12) suggests that the Sap B-membrane interaction is based on weak ionic attraction of the negatively charged membrane surface and Sap B, which, due to its isoelectric point close to the lysosomal pH, has only a small positive net charge. Anionic lipids, particularly BMP, might facilitate sulfatide extraction by providing negative surface charges and perturbing the membrane structure. The mobilized sulfatide is recognized by ASA (k2) and hydrolyzed to galactosylceramide and sulfate (k3). Galactosylceramide is then released from the lipid binding site of Sap B (k4), allowing another round of the reaction cycle. The requirement of high concentrations of Sap B for maximum hydrolysis suggests that the product-substrate exchange (determined by k4 and k1) is the rate-limiting step of the coupled Sap B-ASA reaction. B, kinetic model of ASA-catalyzed hydrolysis of the sulfate ester pNCS. This artificial substrate is conventionally used to measure ASA activity. Under standard conditions ASA molecules are saturated by an excess of pNCS, and the catalytic rate of ASA (k3) is rate-limiting.
The function of Sap B includes binding of sulfatide, its presentation to ASA, and the release of the reaction product galactosylceramide (Fig. 7A). As proposed for lipid binding enzymes, sulfatide presentation might be described by a two-step model following a surface dilution kinetics (35). In a first step, Sap B associates with membranes, micelles, or liposomes. This reaction is of second order and will be greatly accelerated by high activator concentrations. Sequestration of Sap B at the two-dimensional lipid interface may then facilitate detection and binding of a sulfatide molecule by a surface scanning mechanism. In the in vitro assay, Sap B and micelles or liposomes are freshly mixed, and surface association might, therefore, determine the rate of the two-step reaction. In lysosomes, however, the bulk of Sap B may already be attached to target membranes. The in vitro assay may, therefore, overestimate the requirement of Sap B. Several observations argue against this view. Surface plasmon resonance spectroscopy indicated that Sap B binds fast but not stably to immobilized liposomes, resulting in a rapid equilibrium between surface-bound and -unbound Sap B (12). Fast association/dissociation rates explain why Sap B is able to efficiently transport lipids from donor to acceptor liposomes (36) and seem to be mandatory for sulfatide presentation in the lysosomal microenvironment. Thus, a Sap B mutant which binds more stably to membranes due to the lack of N-glycosylation is unable to maintain normal sulfatide turnover and causes MLD (12). As supported by structural considerations (7), it is, therefore, very likely that the function of Sap B involves full extraction and presentation of sulfatide in a soluble Sap B-lipid complex. Association reactions as required for in vitro activity will, therefore, be an inherent part of the Sap B function also in vivo.
Kinetic data on the release of the reaction product from the lipid binding site and the reentry of a new substrate molecule are scarce. A comparative binding study in which lipids competed for binding to Sap B revealed that although no simple “binding rule” can be established, ligands with longer and/or more complex lipoidal and polar adducts appear to be favored (29, 37). The displacement of galactosylceramide by sulfatide might, therefore, be driven by only minor differences in the affinity of Sap B to sulfatide and galactosylceramide. Hence, a slow product-substrate exchange may be the rate-limiting step of the coupled Sap B-ASA reaction. Although saposins other than Sap B do not present sulfatide to ASA (Fig. 4, B and C), they might have an indirect stimulatory effect on the hydrolysis of sulfatide. Thus, Sap A, which has to present galactosylceramide to the next hydrolytic enzyme in the catabolic cascade, galactocerebrosidase, might extract the desulfation product galactosylceramide from Sap B, thereby enabling the reentry of a new sulfatide molecule. To test this notion, we combined Sap B with Sap A or other saposins. However, compared with Sap B alone, none of the combinations increased the rate of sulfatide desulfation (Fig. 4C). On the contrary, all combinations reduced the hydrolytic rate, suggesting that competition for galactosylceramide is not a driving force for the degradation of sulfatide but, rather, that competition for sulfatide leads to the sequestration of sulfatide molecules.
It has been shown previously that acidic phosphoglycerolipids, some of which accumulate in intraendosomal/intralysosomal vesicles during the endosomal lipid sorting process, enhance the hydrolysis of various sphingolipids in micellar and liposomal in vitro systems (for review, see Ref. 2). Acidic phosphoglycerolipids might, therefore, act as physiological promoters of sphingolipid presentation. The most potent promoter, the acidic phosphoglycerolipid BMP, accelerates in vitro hydrolysis of sphingomyelin, ceramide, and gangliosides by the respective lysosomal hydrolases up to 100-fold (2). Also, phosphatidylinositol and phosphatidylserine enhance hydrolysis in some in vitro systems. When tested on the SapB-dependent hydrolysis of sulfatide, phosphatidylinositol and phosphatidylserine had virtually no effect (Fig. 4A). BMP, phosphatidic acid, and the isoprenoid dolichol, on the contrary, activated the reaction to some extent (Fig. 4, A and B). However, even for BMP the stimulation was comparably low, and only around 2–3-fold more sulfatide was hydrolyzed when the BMP concentration was increased to 30 mol % (Fig. 4, A and B). The differential effect of anionic phosphoglycerolipids on the hydrolysis of sulfatide and other sphingolipids might be most easily explained by the fact that sulfatide itself is an acidic molecule whose negative charge makes any accessory function of other negatively charged membrane components dispensable. Negative surface charges might be particularly important to favor the contact of Saps to target membranes because Saps have an isoelectric point close to the lysosomal pH and, therefore, carry only weak net positive charges. A weak affinity of Sap B to membranes may also explain why the coupled Sap B-ASA reaction is much more sensitive to changes in the pH and ionic strength (Fig. 2, B and C) than Sap B-independent reactions accomplished by ASA alone (38). The notion that pH value, salt concentration, and acidic lipids are critical for the function of Sap B rather than ASA is supported by the impact of these factors on the Sap B-mediated extraction of sulfatide from immobilized liposomes (12).
The requirement of unexpectedly high Sap B concentrations for the maximum rate of sulfatide hydrolysis also raised the question of whether Sap B may be a limiting factor of sulfatide desulfation in vivo. The phenotype of mice expressing reduced levels of prosaposin lends support to this notion. Thus, a ∼60% reduced prosaposin mRNA level causes accumulation and intralysosomal deposition of sulfatide in the brain stem and kidney of mice (39). Moreover, changes of the lipid pattern are detectable in mice which are heterozygous for a disrupted prosaposin allele (40). In contrast to lysosomal enzymes, which are able to maintain lysosomal homeostasis at residual levels of only 5% of normal (6), the Sap B expression might, therefore, be regulated to a level much closer to the threshold required for the normal lysosomal function. The question of whether Sap B can delimit sulfatide hydrolysis in vivo is particularly important in view of ongoing ERT studies which investigate the therapeutic efficacy of ERT in children with inborn ASA-deficiency (see clinicaltrials.gov/ct2/show/NCT00418561). ERT is based on the ability of patient cells to endocytose systemically administered rhASA via mannose 6-phosphate receptors. Although these receptors are believed to be ubiquitously expressed, the rate of endocytosis seems to be cell type-specific, and not all cells profit equally from treatment. Our observation that an increase of the Sap B concentration can overcompensate low ASA activities (Fig. 3B) might, therefore, be exploited to subsidize cell types with limited access to ASA by providing Sap B in addition. Primary ASA-deficient murine kidney cells were used to analyze the effects of Sap B uptake on the sulfatide hydrolysis in an ERT model of classical MLD. ASA was fed in a concentration which was considerably below the concentration required for the saturation of mannose 6-phosphate receptors (41). Therefore, only partial clearance of sulfatide storage was achieved (Fig. 6A), and a possible synergistic effect of Sap B co-feeding on sulfatide clearance could be analyzed. However, no significant stimulatory effect of activator co-feeding was detectable under different conditions (Fig. 6). Also prosaposin, which is efficiently endocytosed and releases the mature Saps in the lysosome (Fig. 5, E and F), did not have the potential to promote sulfatide hydrolysis in this system (Fig. 6A). The reason for the failure of activator cosubstitution to stimulate sulfatide hydrolysis is not clear. Because it is unknown how much Sap B is expressed by murine kidney cells, the contribution of internalized Sap B to the endogenous pool can only be roughly estimated. Fluharty et al. (11) isolated around 100 mg of Sap B from 1 kg of pig kidney representing 2 nmol of Sap B/mg of extractable protein. In the present study mouse kidney cells internalized around 0.2 nmol of Sap B/mg of extractable protein (Fig. 5D), i.e. 10% of the amount isolated from pig kidney. Although mouse and pig kidney cannot be directly compared, the data suggest that kidney cells express very high levels of Sap B so that uptake of externally substituted Sap B does not substantially increase the intralysosomal concentration of the cofactor. In this case, an MLD cell culture model with low Sap B expression would be more appropriate to detect possible synergistic effects.
The efficient uptake of Sap B by cultured cells (Fig. 5, C and D) was unexpected, as the activator is believed to possess no specific endocytosis signal. The mechanism which drives this rapid uptake is, therefore, unknown. In co-feeding experiments, in which Sap B was present in a high molar excess upon rhASA, ASA uptake was not significantly reduced (not shown). This suggests that Sap B does not compete for components of the mannose 6-phosphate receptor pathway. Furthermore, internalization of Sap B was not saturated up to concentrations of 5 μm (Fig. 5E). This may indicate that Sap B is internalized via a receptor-independent pathway such as fluid or adsorptive endocytosis (pinocytosis). How much Sap B actually reaches the lysosomal compartment cannot be decided from the present experiments. The rate of lysosomal delivery is, however, sufficient to completely correct the defective sulfatide catabolism of prosaposin-deficient cells (Fig. 5B). Analogous to ERT of classical MLD (9), replacement therapy with Sap B might, therefore, represent a viable option for the treatment of patients suffering from an inherited Sap B deficiency.
Acknowledgments
We thank Dieter Hartmann (Anatomical Institute, Bonn, Germany), Jens Fogh (Zymenex A/S, Hillerød, Denmark), and Nicole Brunett (Kekulé-Institute, Bonn, Germany) for microscopic analysis of liposomes and providing rhASA and recombinant Saps, respectively.
This work was supported by research grants from the European Leukodystrophy Association, the Bundesministerium für Bildung und Forschung (Leukonet), Deutsche Forschungsgemeinschaft Grants Sa 257/21 and SFB 645, and the seventh framework program of the EU-funded “Lipidomic-Net” (proposal 202272).
Footnotes
The abbreviations used are: GSL, glycosphingolipid; ASA, arylsulfatase A; rhASA, recombinant human ASA; BMP, bis(monoacylglycero)phosphate; ERT, enzyme replacement therapy; MLD, metachromatic leukodystrophy; pNCS, p-nitrocatechol sulfate; Sap, saposin; GalCer, galactosylceramide; Sulf, sulfatide; PBS, phosphate-buffered saline.
References
- 1.Sandhoff, K., and Kolter, T. (2003) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358 847-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolter, T., and Sandhoff, K. (2005) Annu. Rev. Cell Dev. Biol. 21 81-103 [DOI] [PubMed] [Google Scholar]
- 3.Sandhoff, K., Kolter, T., and Harzer, K. (2001) in The Metabolic and Molecular Basis of Inherited Disease (Scriver, C. R., Beaudet, A. L., Sly, W. S., and Valle, D., eds) pp. 3371-3388, McGraw-Hill, New York
- 4.Kishimoto, Y., Hiraiwa, M., and O'Brien, J. S. (1992) J. Lipid Res. 33 1255-1267 [PubMed] [Google Scholar]
- 5.Fujita, N., Suzuki, K., Vanier, M. T., Popko, B., Maeda, N., Klein, A., Henseler, M., Sandhoff, K., Nakayasu, H., and Suzuki, K. (1996) Hum. Mol. Genet. 5 711-725 [DOI] [PubMed] [Google Scholar]
- 6.von Figura, K., Gieselmann, V., and Jaeken, J. (2001) in The Metabolic and Molecular Basis of Inherited Disease (Scriver, C. R., Beaudet, A. L., Sly, W. S., and Valle, D., eds) pp. 3695-3724, McGraw-Hill, New York
- 7.Ahn, V. E., Faull, K. F., Whitelegge, J. P., Fluharty, A. L., and Privé, G. G. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 38-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norris, A. J., Whitelegge, J. P., Yaghoubian, A., Alattia, J. R., Privé, G. G., Toyokuni, T., Sun, H., Brooks, M. N., Panza, L., Matto, P., Compostella, F., Remmel, N., Klingenstein, R., Sandhoff, K., Fluharty, C., Fluharty, A., and Faull, K. F. (2005) J. Lipid Res. 46 2254-2264 [DOI] [PubMed] [Google Scholar]
- 9.Matzner, U., Herbst, E., Hedayati, K. K., Lüllmann-Rauch, R., Wessig, C., Schröder, S., Eistrup, C., Möller, C., Fogh, J., and Gieselmann, V. (2005) Hum. Mol. Genet. 14 1139-1152 [DOI] [PubMed] [Google Scholar]
- 10.Sommerlade, H. J., Hille-Rehfeld, A., von Figura, K., and Gieselmann, V. (1994) Biochem. J. 297 123-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluharty, A. L., Katona, Z., Meek, W. E., Frei, K., and Fowler, A. V. (1992) Biochem. Med. Metab. Biol. 47 66-85 [DOI] [PubMed] [Google Scholar]
- 12.Remmel, N., Locatelli-Hoops, S., Breiden, B., Schwarzmann, G., and Sandhoff, K. (2007) FEBS J. 274 3405-3420 [DOI] [PubMed] [Google Scholar]
- 13.Locatelli-Hoops, S., Remmel, N., Klingenstein, R., Breiden, B., Rossocha, M., Schoeniger, M., Koenigs, C., Saenger, W., and Sandhoff, K. (2006) J. Biol. Chem. 281 32451-32460 [DOI] [PubMed] [Google Scholar]
- 14.Gopalakrishnan, M. M., Grosch, H. W., Locatelli-Hoops, S., Werth, N., Smolenová, E., Nettersheim, M., Sandhoff, K., and Hasilik, A. (2004) Biochem. J. 383 507-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schierau, A., Dietz, F., Lange, H., Schestag, F., Parastar, A., and Gieselmann, V. (1999) J. Biol. Chem. 274 3651-3658 [DOI] [PubMed] [Google Scholar]
- 16.Henseler, M., Klein, A., Glombitza, G. J., Suziki, K., and Sandhoff, K. (1996) J. Biol. Chem. 271 8416-8423 [DOI] [PubMed] [Google Scholar]
- 17.Koshy, K. M., and Boggs, J. M. (1982) Lipids 17 998-1000 [DOI] [PubMed] [Google Scholar]
- 18.Schwarzmann, G., and Sandhoff, K. (1987) Methods Enzymol. 138 319-341 [DOI] [PubMed] [Google Scholar]
- 19.Dolly, J. A., Nockles, E. A. V., Lo, M. M. S., and Barnard, E. A. (1981) Biochem. J. 193 919-923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapidot, Y., Rappoport, S., and Wolman, Y. (1967) J. Lipid Res. 8 142-145 [PubMed] [Google Scholar]
- 21.Baum, H., Dodgson, K. S., and Spencer, B. (1959) Clin. Chim. Acta 4 453-455 [DOI] [PubMed] [Google Scholar]
- 22.Jeffrey, H. J., and Roy, A. B. (1977) Aust. J. Exp. Biol. Med. Sci. 55 339-346 [DOI] [PubMed] [Google Scholar]
- 23.Folch, J., Lees, M., and Sloane Stanley, G. H. (1957) J. Biol. Chem. 226 497-509 [PubMed] [Google Scholar]
- 24.Yao, J. K., and Rastetter, G. M. (1985) Anal. Biochem. 150 111-116 [DOI] [PubMed] [Google Scholar]
- 25.Cestaro, B., Marchesini, S., Cervato, G., Viani, P., and Vesely, S. (1984) Ital. J. Biochem. 33 381-391 [PubMed] [Google Scholar]
- 26.Hess, B., Saftig, P., Hartmann, D., Coenen, R., Lüllmann-Rauch, R., Goebel, H. H., Evers, M., von Figura, K., D'Hooge, R., Nagels, G., De Deyn, P., Peters, C., and Gieselmann, V. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14821-14826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, D., Büssow, H., Fewou, S. N., and Gieselmann V. (2005) Biochem. Biophys. Res. Commun. 327 663-667 [DOI] [PubMed] [Google Scholar]
- 28.Harzer, K., Paton, B. C., Poulos, A., Kustermann-Kuhn, B., Roggendorf, W., Grisar, T., and Popp, M. (1989) Eur. J. Pediatr. 149 31-39 [DOI] [PubMed] [Google Scholar]
- 29.Vogel, A., Schwarzmann, G., and Sandhoff, K. (1991) Eur. J. Biochem. 200 591-597 [DOI] [PubMed] [Google Scholar]
- 30.Bach, G., Chen, C. S., and Pagano, R. E. (1999) Clin. Chim. Acta 280 173-179 [DOI] [PubMed] [Google Scholar]
- 31.Kolodny, E. H., and Fluharty, A. L. (1995) in The Metabolic and Molecular Basis of Inherited Disease (Scriver, C. R., Beaudet, A. L., Sly, W. S., and Valle, D., eds) pp. 2693-2740, McGraw-Hill, New York
- 32.Dierks, T., Dickmanns, A., Preusser-Kunze, A., Schmidt, B., Mariappan, M., von Figura, K., Ficner, R., and Rudolph, M. G. (2005) Cell 121 541-552 [DOI] [PubMed] [Google Scholar]
- 33.Matzner, U., Matthes, F., Weigelt, C., Andersson, C., Eistrup, C., Fogh, J., and Gieselmann V. (2008) J. Mol. Med. 86 433-442 [DOI] [PubMed] [Google Scholar]
- 34.Porter, M. T., Fluharty, A. L., De la Flor, S. D., and Kihara, H. (1972) Biochim. Biophys. Acta 258 769-778 [DOI] [PubMed] [Google Scholar]
- 35.Carman, G. M., Deems, R. A., and Dennis E. A. (1995) J. Biol. Chem. 270 18711-18714 [DOI] [PubMed] [Google Scholar]
- 36.Ciaffoni, F., Tatti, M., Boe, A., Salvioli, R., Fluharty, A., Sonnino, S., and Vaccaro A. M. (2006) J. Lipid Res. 47 1045-1053 [DOI] [PubMed] [Google Scholar]
- 37.Fluharty, C. B., Johnson, J., Whitelegge, J., Faull, K. F., and Fluharty A. L. (2001) J. Neurosci. Res. 63 82-89 [DOI] [PubMed] [Google Scholar]
- 38.Stinshoff, K., and Jatzkewitz, H. (1975) Biochim. Biophys. Acta 377 126-138 [DOI] [PubMed] [Google Scholar]
- 39.Sun, Y., Qi, X., Witte, D. P., Ponce, E., Kondoh, K., Quinn, B., and Grabowski, G. A. (2002) Mol. Genet. Metab. 76 271-286 [DOI] [PubMed] [Google Scholar]
- 40.Doering, T., Holleran, W. M., Potratz, A., Vielhaber, G., Elias, P. M., Suzuki, K., and Sandhoff, K. (1999) J. Biol. Chem. 274 11038-11045 [DOI] [PubMed] [Google Scholar]
- 41.Klein, D., Yaghootfam, A., Matzner, U., Koch, B., Braulke, T., and Gieselmann V. (2009) Biol. Chem. 390 41-48 [DOI] [PubMed] [Google Scholar]