Abstract
Calcineurin phosphatase plays a crucial role in T cell activation. Dephosphorylation of the nuclear factors of activated T cells (NFATs) by calcineurin is essential for activating cytokine gene expression and, consequently, the immune response. Current immunosuppressive protocols are based mainly on calcineurin inhibitors, cyclosporine A and FK506. Unfortunately, these drugs are associated with severe side effects. Therefore, immunosuppressive agents with higher selectivity and lower toxicity must be identified. The immunosuppressive role of the family of proteins regulators of calcineurin (RCAN, formerly known as DSCR1) which regulate the calcineurin-NFAT signaling pathway, has been described recently. Here, we identify and characterize the minimal RCAN sequence responsible for the inhibition of calcineurin-NFAT signaling in vivo. The RCAN-derived peptide spanning this sequence binds to calcineurin with high affinity. This interaction is competed by a peptide spanning the NFAT PXIXIT sequence, which binds to calcineurin and facilitates NFAT dephosphorylation and activation. Interestingly, the RCAN-derived peptide does not inhibit general calcineurin phosphatase activity, which suggests that it may have a specific immunosuppressive effect on the calcineurin-NFAT signaling pathway. As such, the RCAN-derived peptide could either be considered a highly selective immunosuppressive compound by itself or be used as a new tool for identifying innovative immunosuppressive agents. We developed a low throughput assay, based on the RCAN1-calcineurin interaction, which identifies dipyridamole as an efficient in vivo inhibitor of the calcineurin-NFAT pathway that does not affect calcineurin phosphatase activity.
Calcineurin (Cn)3 is a calcium/calmodulin-dependent serine/threonine protein phosphatase and a key enzyme in many cellular processes, such as T cell activation (1). Activated Cn dephosphorylates many substrates, including the nuclear factor of activated T cells (NFAT) 1–4 transcription factors, thereby inducing their translocation to the cell nucleus. Nuclear NFAT is a key component of the cytokine gene expression stimulation that triggers T cell activation (2). Cn-induced NFAT dephosphorylation is regulated by a Cn-NFAT protein-protein interaction that involves the NFAT PXIXIT motif, which is crucial for NFAT activation and signaling (3).
Current immunosuppressive protocols in transplantation therapies and in the treatment of autoimmune diseases include the administration of cyclosporine A (CsA) or FK506. These drugs are potent inhibitors of Cn phosphatase activity (4), and their continued administration has been associated with severe side effects. The binding of these drugs to intracellular immunophilin receptors, before the interaction with calcineurin, induces complete suppression of Cn phosphatase activity (5). Therefore, a deeper characterization of the molecular mechanisms that govern the interaction between endogenous Cn regulators and Cn in T cell activation would provide useful information for the development of novel immunosuppressive drugs.
Several endogenous Cn inhibitors have been identified in the literature (6), including the protein family of regulators of Cn (RCAN, previously known as calcipressins or DSCR1) (7), which interact physically with Cn and modulate its phosphatase activity (8–11). In humans, this family has three members: RCAN1, RCAN2, and RCAN3. RCAN1 has two different and independent Cn-interacting motifs: CIC (Cn inhibitor of Calcipressin) and PXIXXT. In contrast, the interaction between RCAN3 and Cn appears to be driven solely by the CIC motif. Although RCAN proteins can contain both of these Cn-binding sites, only the RCAN CIC motif (consensus sequence in mammals is LGPGEKYELHA(G/A)T(D/E)(S/T)TPSVVV HVC(E/D)S) is responsible for inhibiting Cn-NFAT-dependent signaling in human T cells and, as a result, T cell activation (12, 13).
In the present study, we characterized a minimal RCAN-derived sequence (part of the RCAN CIC motif) that retains the ability to inhibit Cn-NFAT signaling in vivo. The peptides spanning this sequence, RCAN1198–218 from RCAN1 and RCAN3183–203 from RCAN3, were used to conduct a detailed analysis of the RCAN-Cn-binding mechanism. In vitro, these RCAN-derived peptides bind to Cn with high affinity and selectively inhibit the interaction of Cn with NFAT. Our results also suggest that the RCAN-derived peptides impede Cn interaction with the NFAT PXIXIT motif but not with the NFAT LXVP motif. Interestingly, the RCAN-derived peptides characterized here are specific inhibitors of the Cn-NFAT signaling pathway but do not inhibit general Cn phosphatase activity in vitro. Consequently, these peptides could have a favorable profile for immunosuppressive therapy. We hypothesized that these peptides could serve as valuable pharmacological tools for identifying new immunosuppressant compounds. By screening chemical and peptide libraries, we identified dipyridamole to be a regulator of the RCAN-Cn interaction that inhibits the Cn-NFAT signaling pathway in vivo.
EXPERIMENTAL PROCEDURES
Plasmid Construct—Overlapping EGFP·RCAN fusion proteins for RCAN1-1 (NCBI accession number AAP96743) and RCAN3-2 (NCBI accession number AF176116) were designed and expressed using standard techniques (12). All DNA sequences were confirmed by DNA sequencing. The pNFAT1-EYFP plasmid construct was described previously (12). The pFLAG-NFAT1 (14), human pGEX-6P-1-CnAα (amino acids 2–347) (15) and 3XNFAT-luc (16) plasmid constructs were kindly provided by Dr. Gerald Crabtree, Dr. Patrick Hogan, and Dr. José Aramburu, respectively.
Peptides and Reagents—The peptides RCAN1198–218, KYELHAATDTTPSVVVHVCES; RCAN3183–203, KYELHAGTESTPSVVVHVCES; scrambled RCAN1, SAVTHKLESVDPATVYCETHV; unrelated peptide, RGKWTYNGYTIEGR; PXIXIT (motif sequence from NFAT1), ASGLSPRIEITPSHEL; LXVP (motif sequence from NFAT2), DQYLAVPQHPYQWAK; mutLXVP, DQYAAAAQHPYQWAK; and phosphorylated RII peptide, DLDVPIPGRFDRRV-phosphorylatedS-VAAE were synthesized as acetylated N-terminal and C-terminal amides using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry on a 433A peptide synthesizer (Applied Biosystems). Similar procedures were used to synthesize the N-terminal carboxyfluorescein derivatives. Fully deprotected cleaved peptides were precipitated in cold t-butyl methyl ether, centrifuged, dissolved in acetic acid, lyophilized, and purified by reversed phase-high pressure liquid chromatography (17). MALDI-TOF mass spectrometry was used to confirm peptide identity. FK506 was kindly provided by Fujisawa. Dipyridamole and FKBP12 were purchased from Sigma-Aldrich. Pyrimidopyrimidine was synthesized at IIQAB (CSIC, Barcelona, Spain).
In Vivo Cellular Assays—COS-7 and U-2 OS cells were transiently transfected with pFLAG-NFAT1 or pNFAT1-EYFP and different EGFP·RCAN plasmids to analyze the inhibition of NFAT nuclear import. HEK 293T cells were transiently transfected with EGFP·RCAN1 fusion proteins to analyze their interaction with endogenous Cn in co-immunoprecipitation assays. Jurkat cells were electroporated with different EGFP· RCAN plasmids and the 3xNFAT-luciferase reporter to analyze NFAT activity. Endogenous cytokine NFAT-dependent gene expression of Jurkat cells, either untreated or treated with dipyridamole, was analyzed by reverse transcription-PCR. All protocols were performed as described previously (12, 13) with some modifications, as indicated in the supplemental data.
Fluorescence Polarization Measurement and Library Screening—Binding and competition assays were performed on a Wallac VICTOR2 1420 multilabel high throughput screening counter (PerkinElmer Life Sciences). For a detailed description of the assay, see the supplemental data. The truncated human CnAα-(2–347) protein (hereafter referred to as CnA), which contains the catalytic domain and the linker region (see supplemental Fig. 2A for a schematic representation), was used to characterize protein-peptide interactions due to its proven ability to bind NFAT (18, 19). The Prestwick Chemical Library® was initially screened at 100 μm, and compounds promoting a >45% displacement of the RCAN1198–218-CnA interaction were evaluated in dose-dependent assays.
Calcineurin Phosphatase Activity—Cn phosphatase activity was analyzed using the calcineurin cellular assay kit PLUS (BIOMOL International LP) according to the manufacturer's instructions, with the modifications noted in the supplemental data.
RESULTS
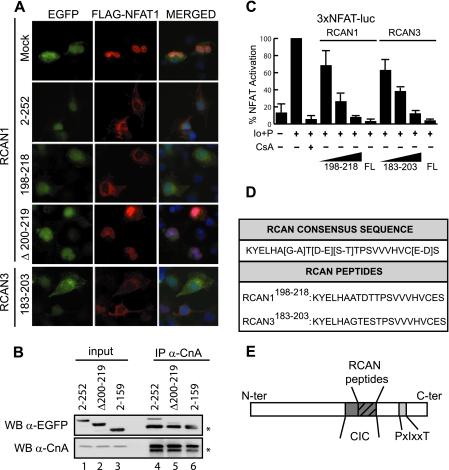
Identification of the Minimal RCAN Sequence Responsible for Inhibiting the Cn-NFAT Signaling Pathway in Vivo—Immunosupression of the Cn-NFAT signaling pathway in vivo by RCAN proteins is caused by the RCAN CIC motif, which spans amino acids 191–218 for RCAN1 and 178–203 for RCAN3 (12, 13). In this report, we present the minimal RCAN CIC inhibitor consensus sequence, spanning amino acids 198–218 for RCAN1 and 183–203 for RCAN3, that retains this immunosuppressive role (Fig. 1 and supplemental Fig. 1). These RCAN sequences are directly responsible for inhibiting NFAT nuclear translocation in calcium-stimulated cells (Fig. 1A, compare EGFP·RCAN1-(2–252) full-length protein and EGFP· RCAN1-(198–218) and EGFP·RCAN3-(183–203) proteins with mock vector, FLAG-NFAT1, red panels). In contrast, a full-length RCAN1 mutant lacking this specific sequence fails to inhibit Cn-regulated NFAT nuclear translocation (Fig. 1A, EGFP·RCAN1 (Δ200–219); FLAG-NFAT1, red panel). However, this RCAN1 mutant retains the ability to bind in vivo to endogenous Cn (Fig. 1B, IP α-CnA, lane 5) due to the presence of the PXIXXT motif, which is an additional Cn-interacting motif in RCAN1 (12). Furthermore, both EGFP·RCAN1-(198–218) and EGFP·RCAN3-(188–203) fusion proteins inhibit NFAT-driven promoters in a dose-dependent manner, with activity comparable with that of the full-length proteins (Fig. 1C). These results suggest that the inhibitory activity of RCAN proteins on the Cn-NFAT signaling pathway is specifically dependent on the minimal RCAN inhibitory consensus sequence identified here. On the strength of these data, we synthesized a series of RCAN-derived peptides, RCAN1198–218 and RCAN3183–203 (Fig. 1, D and E), which do not contain the RCAN PXIXXT motif, and analyzed in vitro the molecular basis of their binding to Cn.
FIGURE 1.
The RCAN1-(198–218) and RCAN3-(183–203) sequences inhibit Cn-dependent NFAT signaling. A, immunofluorescence microscopy of COS-7 cells transiently co-transfected with pFLAG-NFAT1 and different pEGFP·RCAN1 and pEGFP·RCAN3 plasmid constructs. Cells were stimulated with 1 μm ionomycin and 10 mm CaCl2. FLAG-NFAT1 (in red) was immunodetected using a secondary antibody conjugated to the Alexa Fluor® 568 fluorochrome, whereas RCAN proteins (in green) were visualized by the presence of EGFP in the fusion protein. 4′,6-Diamidino-2-phenylindole staining (in blue) confirmed cell integrity. B, co-immunoprecipitation (IP) of CnA and RCAN1 proteins in HEK 293T cellular extracts transiently transfected with different EGFP·RCAN1 fusion proteins. Proteins were co-immunoprecipitated using an anti-CnA antibody. Asterisks correspond to the immunoglobulin heavy chain. WB, Western blotting. C, RCAN proteins inhibit NFAT-driven gene expression. Jurkat cells (20 × 106) were transiently co-transfected with 10 μg of pEGFP·RCAN1, pEGFP·RCAN3 full-length (FL) plasmids, or increasing concentrations (100 ng, 1 μg, and 10 μg) of pEGFP·RCAN1-(198–218) or pEGFP·RCAN3-(183–203), together with 3xNFAT-luc (20 μg). Renilla-luc (pGL4.75(hRluc/CMV), 0.2 μg) was used as an internal transfection control. Cells were stimulated with 1 μm ionomycin (Io) and 20 nm phorbol 12-myristate 13-acetate (indicated by P in the figure) for 4 h. Results from promoter fold activation of the luciferase reporter plasmid (luciferase/Renilla units) are given as percentages of NFAT activation. Values from Io + phorbol 12-myristate 13-acetate (Io+P)-stimulated cells transfected with EGFP, 3xNFAT-luc, and Renilla-luc were considered to be 100% NFAT activation. The graph shows the mean ± S.D. of three independent experiments performed in triplicate. D, RCAN-derived peptide consensus sequence in mammalian and human RCAN1198–218 and RCAN3183–203 peptide sequences. E, schematic diagram showing the Cn-binding RCAN1 motifs characterized to date. The light gray box corresponds to the RCAN PXIXXT motif, the dark gray box corresponds to the RCAN CIC motif, and the striped box indicates the localization of the RCAN-derived peptides. N-ter, N-terminal; C-ter, C-terminal.
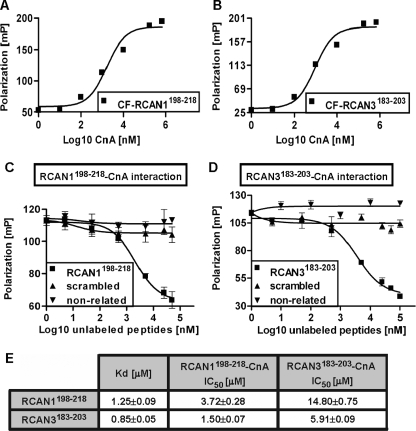
RCAN1198–218 and RCAN3183–203 Peptides Bind to CnA with High Affinity—The protein-protein interaction between RCAN1 and Cn is well established in the literature (8–11). However, the only biochemical characterization reported for this interaction involves Cn and a long polypeptide spanning the RCAN1-(196–252) C-terminal amino acids. This RCAN1 region contains both the CIC and the PXIXXT Cn-binding motifs (20). However, our in vivo results show that the RCAN PXIXXT motif is not essential for exerting a functional inhibitory effect on the Cn-NFAT signaling pathway (Fig. 1 and supplemental Fig. 1). The RCAN1198–218 and RCAN3183–203 peptides, whose sequences include part of the RCAN CIC but not the RCAN PXIXXT motif (Fig. 1E), were used to obtain a more detailed characterization of the Cn-RCAN interaction. Importantly, this study is the first biochemical analysis of the recently identified Cn-RCAN3 interaction (13). We used human CnA and the carboxyfluorescein (CF)-labeled RCAN1198–218 and RCAN3183–203 peptides (CF-RCAN1198–218 and CF-RCAN3183–203, respectively) to explore the protein-peptide interactions by fluorescence anisotropy spectroscopy (21). Both the CF-RCAN1198–218 and the CF-RCAN3183–203 peptide bind to CnA (Fig. 2, A and B, respectively) with apparent dissociation constants (Kd) in the low physiological micromolar range (Fig. 2E). The interaction between CnA and the RCAN-derived peptides was also confirmed qualitatively by peptide-protein cross-linking followed by mass spectrometry (MALDI-TOF) analysis (data not shown). The specificity of the CF-RCAN1198–218-CnA interaction was assessed by conducting competition experiments with the unlabeled peptide RCAN1198–218, which competed effectively with the interaction, and with a non-related and a scrambled peptide (see “Experimental Procedures”), both of which were unable to compete with the interaction (Fig. 2C). Similar experiments demonstrated the specificity for the binding of CF-RCAN3183–203 to CnA (Fig. 2D). As expected, due to the high amino acid conservation in both RCAN-derived peptides, cross-competition experiments showed cross-affinity for the CnA-binding site (Fig. 2E, see corresponding apparent IC50 values), which suggests that both peptides could interact with the same CnA region. In conclusion, our results show that both RCAN-derived peptides interact specifically and with high affinity with CnA.
FIGURE 2.
RCAN1198–218 and RCAN3183–203 peptides bind with high affinity to human CnA. A and B, binding of 30 nm CF-labeled RCAN peptide CF-RCAN1198–208 and CF-RCAN3183–203, respectively, to increasing amounts of CnA. Data, given as millipolarization (mP) units, were analyzed with the GraphPad PRISM® 4.0 program and fitted using a non-linear regression model to a sigmoidal dose-response curve. C and D, RCAN1198–218-CnA and RCAN3183–203-CnA interactions were established using 30 nm CF-RCAN peptides and 1.5 μm CnA and competed with increasing amounts of unlabeled peptides. A RCAN1198–218 scrambled peptide and a non-related peptide were included as controls. Data were fitted to a one-site competition model. E, summary of apparent Kd and IC50 values obtained for the RCAN1198–218 and RCAN3183–203 peptides in the interaction of CnA.
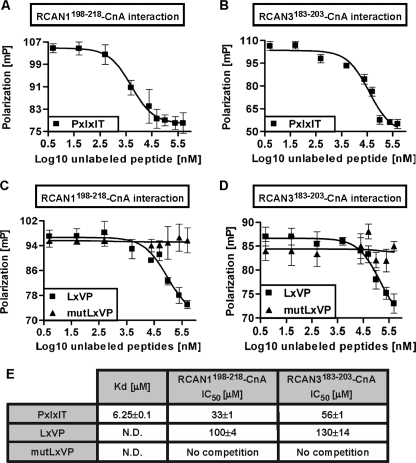
The CnA-binding PXIXIT Sequence in NFAT Interferes with RCAN-CnA Interaction—Previous reports have demonstrated that two independent NFAT motifs participate in the NFAT-CnA interaction: the PXIXIT motif, which binds to Cn and facilitates NFAT dephosphorylation and activation (3), and the LXVP motif (22) (see supplemental Fig. 2B for a schematic representation). We wanted to determine whether the RCAN-derived peptides could inhibit NFAT dephosphorylation by interfering with the Cn-binding sequence motifs in NFAT (Fig. 3). In fact, a peptide derived from the human NFAT1 PXIXIT motif (called PXIXIT) competed strongly with the interactions between CnA and the RCAN1198–218 and RCAN3183–203 peptides (Fig. 3, A and B; apparent IC50 values are shown in Fig. 3E). Similar results (data not shown) were observed when competition experiments were performed with the previously reported VIVIT peptide (16), which is a high affinity Cn-binding peptide optimized from the NFAT PXIXIT natural sequence. In contrast, a peptide derived from the NFAT2 LXVP motif (called LXVP) only showed competition at high peptide concentrations (Fig. 3, C and D). An analog peptide in which alanine residues replace the key amino acids of the LXVP peptide (13) (called mutLXVP, see “Experimental Procedures”) showed no competition (Fig. 3, C and D; apparent IC50 values are summarized in Fig. 3E). As a control, we confirmed that the NFAT peptides PXIXIT and LXVP, but not mutLXVP, were able to compete with the NFAT PXIXIT-CnA interaction in vitro, which suggests that these peptides bind effectively to CnA (data not shown). Overall, these results suggest that RCAN proteins disrupt NFAT binding to CnA. This is caused by the interference between either RCAN1198–218 or RCAN3183–203 and the NFAT PXIXIT peptide, which is crucial in the recognition and dephosphorylation of NFAT by Cn and, by extension, in NFAT activation (3). Interestingly, it has recently been reported that the NFAT PXIXIT sequence motif interacts with CnA through the CnA catalytic domain (amino acids 70–333) (19, 23, 24). Our competition experiments strongly support the hypothesis that the RCAN-derived peptides interact with CnA through the Cn catalytic domain, as described for the NFAT PXIXIT sequence motif (19, 23).
FIGURE 3.
The NFAT PXIXIT motif interferes with the binding of the RCAN-derived peptides to CnA. A and B, competition of the RCAN1198–218-CnA and RCAN3183–203-CnA interactions, respectively, with increasing concentrations of unlabeled NFAT PXIXIT peptide. mP, millipolarization units. C and D, competition experiments of the RCAN1198–218-CnA and RCAN3183–203-CnA interactions, respectively, in the presence of the unlabeled LXVP or mutLXVP peptides. E, summary of apparent Kd and IC50 values for NFAT PXIXIT, LXVP, and mutLXVP peptides. N.D. = not determined.
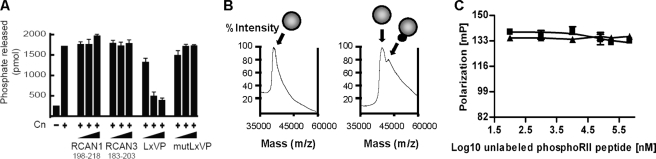
The RCAN-derived Peptides Do Not Inhibit General Cn Phosphatase Activity—The RCAN1 C-terminal region (which includes the CIC and the PXIXXT sequences that bind Cn) has been found to inhibit Cn phosphatase activity on the synthetic substrate p-nitrophenyl phosphate (20, 25). An in vitro enzyme assay was performed to evaluate the effect of the RCAN-derived peptides on Cn phosphatase activity (Fig. 4A). The RCAN1198–218 and RCAN3183–203 peptides do not inhibit the enzymatic activity of Cn toward the phosphorylated RII peptide substrate (Fig. 4A). As a control, the NFAT LXVP peptide (but not the mut-LXVP) (24) inhibited Cn phosphatase activity in a dose-dependent manner (Fig. 4A). Furthermore, protein-peptide cross-linking and mass spectrometry analyses showed that the phosphorylated RII peptide is able to bind CnA under our experimental conditions (Fig. 4B). Finally, in agreement with the results shown in Fig. 4A, the phosphorylated RII peptide does not compete with the RCAN1198–218-CnA or RCAN3183–203-CnA interactions (Fig. 4C). Taken together, these results suggest that both RCAN-derived peptides bind to the catalytic domain of CnA without directly affecting the catalytic activity of Cn. This contrasts with CsA and FK506 immunosuppressant drugs, which totally abrogate general Cn phosphatase activity. Therefore, from a therapeutic perspective, the RCAN1198–218 and RCAN3183–203 peptides could be considered novel, highly specific compounds with which to define new specific immunosuppressive drugs.
FIGURE 4.
The RCAN-derived peptides do not inhibit Cn phosphatase activity. A, the in vitro phosphatase activity of Cn on the phosphorylated RII substrate was measured in the absence or presence of different peptides (RCAN1198–218, RCAN3183–203, LXVP, and mutLXVP) at concentrations of 10, 100, and 500 μm. Each reaction was performed in the presence of 40 units of human recombinant Cn (Calbiochem) and at the indicated concentrations of the three peptides for 1 h at 30 °C. The specific activity of Cn was 2.8 × 10 units/mg of protein. The picomoles of free phosphate released during the reaction time were measured. The data shown represent the mean ± S.D. of three independent experiments. B, MALDI-TOF mass spectrometric analysis of CnA (41 kDa) cross-linked with the phosphorylated RII peptide (2239 Da) indicated by the gray and black spheres, respectively. C, competition of the RCAN1198–218-CnA (▪) and the RCAN3183–203-CnA (▿) interactions with increasing amounts of unlabeled phosphorylated RII (phosphoRII) peptide.
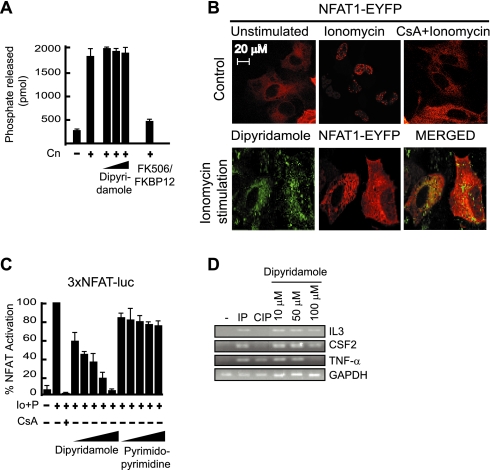
Dipyridamole Interferes with in Vivo Cn-NFAT Signaling—Based on the specific immunosuppressive potential of the RCAN-derived peptides toward the Cn-NFAT signaling pathway, we screened several chemical libraries to identify molecules capable of disrupting the interaction between CnA and the RCAN1198–218 peptide. We first screened a hexapeptide-based library (26, 27) and a diversity-oriented positional scanning library of N-alkylglycine trimers, both of which had previously provided lead compounds for different biological assays (28–31); however, no suitable hit was found. We then screened the Prestwick Chemical Library®, which comprises 880 biologically active molecules with high chemical and pharmacological diversity; dipyridamole was identified as a compound that competes with CnA for binding to fluorescently labeled RCAN1198–218 peptide.
Dipyridamole has been found to inhibit tumor necrosis factor-α production in activated human peripheral blood leukocytes (32), but the mechanism responsible for this activity has not yet been described. Moreover, it is difficult to establish a molecular characterization of the mechanism by which dipyridamole binds to CnA due to the intrinsic fluorescence of the molecule. To address this problem, we performed a functional evaluation of the inhibitory effect of dipyridamole on Cn-NFAT signaling (Fig. 5). First, we analyzed the in vitro effect of dipyridamole on Cn phosphatase activity. Our results show that dipyridamole does not inhibit general Cn phosphatase activity (Fig. 5A), which is also the case of RCAN-derived peptides (Fig. 4A). Next, we performed several experiments to determine the in vivo effect of dipyridamole on Cn-NFAT signaling. Dipyridamole inhibits NFAT nuclear translocation in vivo at 50 μm (Fig. 5B). In addition, dipyridamole, but not its synthetic analog pyrimidopyrimidine, specifically inhibits an NFAT-driven promoter in a dose-dependent manner (Fig. 5C). Consequently, cytokine NFAT-dependent transcription is affected in a dose-dependent manner in human T cells (Fig. 5D). Our in vitro and in vivo experiments demonstrate the dipyridamole-dependent inhibition of the Cn-NFAT signaling pathway. This inhibitory effect on Cn seems to be specific to the NFAT substrates because dipyridamole does not affect general Cn phosphatase activity. These data constitute the first experimental identification of a disrupter of the interaction between Cn and RCAN in vitro that affects Cn-NFAT dependent signaling in vivo.
FIGURE 5.
Dipyridamole interferes with Cn-NFAT cellular signaling in vivo. A, the in vitro phosphatase activity of Cn on the phosphorylated RII substrate was measured in the absence or presence of dipyridamole (10, 50, and 500 μm). As an inhibitory control, calcineurin activity was measured in the presence of 10 μm FK506 and 10 μm FKBP12. The picomoles of free phosphate released during the reaction were measured, and the data obtained from three independent experiments was expressed as the mean ± S.D. B, U-2 OS cells were transfected with NFAT1·EYFP (red) and unstimulated, stimulated with 2 μm Io and 10 mm CaCl2 for 25 min in the presence of vehicle or dipyridamole (green; added previously at 50 μm for 16 h), or inhibited with 1 μm CsA for 30 min before administration of the stimuli. The images represent three independent experiments. C, Jurkat cells were transfected with 3xNFAT-luc and Renilla-luc (pGL4.75(hRluc/CMV)), as an internal transfection control, and treated 12 h after transfection with either 1 μm CsA, as a control, or increasing concentrations of either dipyridamole or pyrimidopyrimidine (1, 10, 20, 50, and 100 μm) for 3 h at 37 °C and 5% CO2. Cells were then either not treated or treated with 1 μm Io and 20 nm phorbol 12-myristate 13-acetate (P) for 4 h. Luciferase/Renilla unit results are shown as in Fig. 1C. The values represent the average of three independent experiments. D, Jurkat cells were treated with vehicle or dipyridamole at different concentrations, as shown in panel A. The cells were then left unstimulated (-), stimulated with 1 μm Io and 20 nm phorbol 12-myristate 13-acetate for 4 h (IP), or inhibited with 1 μm CsA for 30 min before administration of the stimuli (CIP). NFAT-dependent gene expression was analyzed by semiquantitative reverse transcription-PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcription level was used as a control. IL3, interleukin-3; CSF2, colony stimulating factor 2; TNF-α, tumor necrosis factor-α.
DISCUSSION
RCAN1198–218 and RCAN3183–203 Are High Affinity CnA-binding Peptides—We have identified the minimal RCAN region, RCAN1-(198–218) and RCAN3-(183–203), responsible for in vivo inhibition of the Cn-NFAT signaling pathway (Fig. 1 and supplemental Fig. 1). More importantly, biochemical characterization of the interaction between two RCAN-derived peptides spanning these sequences (RCAN1198–218 and RCAN3183–203) and CnA has revealed that both RCAN-derived peptides interact with high affinity with CnA through a region located between amino acids 2 and 347. This region includes the CnA catalytic domain (residues 70–333) and the linker region (residues 335–347) (Fig. 2, A and B, and supplemental Fig. 2A). These results reinforce previous evidence indicating a putative role of the CnA catalytic domain and linker region in binding to RCAN1 (10, 11). These regions have also been implicated in CnA binding to NFAT and other Cn inhibitor proteins such as Cabin1 and AKAP79 (5, 33). Taken together, these data suggest that different Cn endogenous inhibitors may interact with Cn through closely related regions.
The NFAT PXIXIT Sequence Interferes with the Interaction between RCAN-derived Peptides and CnA—The regulatory mechanism by which Cn binds to NFAT or RCAN proteins is a matter of speculation due to the presence of different Cn-binding sites in NFAT (the PXIXIT and LXVP motif sequences) and in RCAN (the CIC and PXIXXT motif sequences). RCAN proteins have been found in different cell systems to interfere in vivo with Cn-NFAT signaling (10, 20, 34). In addition, the RCAN1-4 protein isoform has been found to compete for Cn binding with the VIVIT peptide, which is a peptide derivative of the NFAT PXIXIT sequence motif (35). However, a detailed identification of the RCAN sequence responsible for these molecular events was not carried out. The competition experiments presented here describe for the first time the existence of direct competition between RCAN-derived peptides and the NFAT1 PXIXIT peptide for binding to CnA. Therefore, these results could potentially be extrapolated to full-length proteins. The functional consequence of this competition is to prevent NFAT activation and, subsequently, T cell activation. Our results also suggest that the RCAN-derived peptides could bind to CnA through a region that either partially overlaps or is proximal to where the NFAT PXIXIT motif binds. However, we cannot rule out that binding of one peptide (the RCAN-derived peptides) to any other calcineurin domain could affect the enzyme conformation and binding to the other peptide (NFAT PXIXIT peptide). We hypothesized that both RCAN peptides would interact within the catalytic domain of CnA through a region located outside the CnA active site and probably without direct participation of the linker region. This hypothesis is further supported by the recent structural characterization of the protein-protein surface interaction, defined by binding of the VIVIT peptide (16) to CnA (19, 23). The surface is defined by direct interaction of an exposed short β-strand of CnA and by the β-sheet conformation adopted by the VIVIT peptide. The structure of the complex did not reveal any interactions between the CnA linker region and the peptide. Finally, taking into consideration our data here presented (Fig. 3E), we cannot discard that the NFAT LXVP peptide could also play some role in the competition of the RCAN-derived peptides interaction with CnA. Nevertheless, this competitive effect would be expected to be in a lesser extent than the one mediated by the NFAT PXIXIT peptide. Further experiments would need to be addressed to clarify this point.
The RCAN-derived Peptides Do Not Inhibit General Cn Phosphatase Activity—One of the main findings of the present study is that the RCAN-derived peptides do not inhibit Cn phosphatase activity toward phosphorylated RII peptide substrate (Fig. 4A). In addition, the LXVP peptide, which significantly blocks RII dephosphorylation by Cn (24), only competed with the RCAN-derived peptides for binding to CnA at high concentrations (Fig. 3, C and D). Neither CsA-cyclophilin A nor FK506-FKBP12 complexes competed with the CnA-RCAN-derived peptide interactions, although both interact with CnA under our experimental conditions (data not shown). Overall, these results suggest that the interaction between RCAN-derived peptides and CnA does not involve the direct participation of the CnA active site. These data differ from those of previous reports, in which the complete C-terminal region of RCAN1 was shown to be a competitive inhibitor of Cn in a phosphatase activity assay using the p-nitrophenyl phosphate or synthetic RII peptide as substrates (20, 25). Here, we used well defined C-terminal RCAN-derived peptides, which do not include the RCAN PXIXXT motif and enabled fine characterization of the RCAN motif that inhibits Cn-NFAT signaling in vivo. Interestingly, the RCAN-derived peptides do not abolish Cn phosphatase activity in vitro, unlike CsA or FK506. This suggests that the Cn inhibitory effect of these peptides might be specific to NFAT dephosphorylation.
RCAN1198–218 and RCAN3183–203 Are Potential Immunosuppressive Peptides That Could Be Used as Screening Tools—Our data suggest a potential model for the mechanism by which the RCAN-derived peptides bind to Cn via its catalytic domain, through a region located outside the active site. In this position, the RCAN-derived peptides would not inhibit Cn phosphatase activity but would selectively interfere with the Cn-binding PXIXIT motif of NFAT. This effect would, in turn, prevent NFAT dephosphorylation and, as a result, T cell activation. RCAN-derived peptides might also interfere with CnA-binding sites for other proteins, which could be located close to the RCAN-binding site, although this hypothesis demands further examination. The RCAN-derived peptides identified here could constitute novel, selective immunosuppressive lead candidates and potential therapeutic alternatives to CsA or FK506, presumably with fewer side effects (16, 36). However, given the multiple effects of Cn-dependent NFAT activation on cell regulation, we cannot rule out the possibility that NFAT inhibitors that do not affect the Cn active site could also have some toxic side effects. Specific experiments must be performed to address this question. Nevertheless, these RCAN-derived peptides could be used to identify new, more specific immunosuppressant drugs through the exploration of alternative Cn-binding sites that may inhibit the Ca2+-Cn-NFAT signaling pathway (15, 37, 38). Therefore, the binding site in CnA for the RCAN-derived peptides may be a new target site for the development of such drugs. Based on this hypothesis, we screened chemical libraries under restrictive assay conditions. By imposing screening constraints, we limited the putative hits to molecules that could compete with CF-RCAN1198–218, bind to its binding site in CnA, and inhibit the protein-protein interaction that produces the active CnA-NFAT complex. The screening identified dipyridamole to be a hit compound for the development of this new class of inhibitors. It competed efficiently with CF-RCAN1198–218 in in vitro CnA binding assays. Dipyridamole also inhibited Cn-dependent NFAT nuclear translocation (Fig. 5B), NFAT activity (Fig. 5C), and NFAT-dependent cytokine gene expression in human T cells (Fig. 5D), without affecting general phosphatase activity (Fig. 5A). Although dipyridamole is widely used in clinical practice as an antiplatelet agent to prevent recurrence of ischemic stroke (39), in recent years, it has also been suggested that this drug could have an immunomodulatory effect in vivo (32, 40). However, the molecular mechanism responsible for this novel role remains unknown. Interestingly, our results suggest that dipyridamole may play a role in immunosuppression and that this role could be regulated through the inhibition of the Cn-NFAT signaling pathway.
In summary, we have gained crucial insights into the underlying mechanism of Cn binding to RCAN1 and to RCAN3. When overexpressed in cells, the amino acid sequences of RCAN1-(198–218) and RCAN3-(183–203) inhibit Cn-NFAT signaling, thereby preventing activation of NFAT without affecting Cn phosphatase activity. We have also defined two novel sequences, the RCAN1198–218 and RCAN3183–203 peptides, that could be valuable pharmacological tools. Once the current difficulties of intracellular peptide delivery have been overcome, these synthetic peptides could also prove beneficial in immunosuppressive therapy. Using these peptides as screening tools, we identified dipyridamole to be a hit compound for the development of new immunosuppressant molecules, providing advantages over the currently used drugs CsA and FK506.
Supplementary Material
Acknowledgments
We thank Jordi Bujons, Miguel Hueso, Lluis Espinosa, and Anna Bigas for helpful discussions, Carina Delpiccolo for pyrimidopyrimidine synthesis, and Ana Giménez, Laura Cascales, the Proteomic Unit of the Centro de Investigación Príncipe Felipe, and Raquel Garcia from the Barcelona Science Park for their skillful technical assistance.
This work was supported by grants from the Fundació La Marató de TV3 (Reference 030830), the Generalitat de Catalunya (Reference 2006 BE 00051), the Spanish Ministry of Education and Science (Grants SAF2006-04815, BIO2004-00998, BIO2007-60066, CTQ2005-00995/BQU), and the Fundación Mutua Madrileña 2007. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and two supplemental figures.
Footnotes
The abbreviations used are: Cn, calcineurin; CnA, catalytic subunit of Cn; RCAN, regulator of Cn; CIC, Cn inhibitor of calcipressin; CF, carboxyfluorescein; NFAT, cytosolic nuclear factor of activated T cells 1–4; CsA, cyclosporine A; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein; Io, ionomycin; luc, luciferase.
References
- 1.Winslow, M. M., Neilson, J. R., and Crabtree, G. R. (2003) Curr. Opin. Immunol. 15 299-307 [DOI] [PubMed] [Google Scholar]
- 2.Macian, F. (2005) Nat. Rev. Immunol. 5 472-484 [DOI] [PubMed] [Google Scholar]
- 3.Aramburu, J., Garcia-Cozar, F., Raghavan, A., Okamura, H., Rao, A., and Hogan, P. G. (1998) Mol. Cell 1 627-637 [DOI] [PubMed] [Google Scholar]
- 4.Fruman, D. A., Klee, C. B., Bierer, B. E., and Burakoff, S. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 3686-3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke, H., and Huai, Q. (2003) Biochem. Biophys. Res. Commun. 311 1095-1102 [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Martinez, S., and Redondo, J. M. (2004) Curr. Med. Chem. 11 997-1007 [DOI] [PubMed] [Google Scholar]
- 7.Davies, K. J., Ermak, G., Rothermel, B. A., Pritchard, M., Heitman, J., Ahnn, J., Henrique-Silva, F., Crawford, D., Canaider, S., Strippoli, P., Carinci, P., Min, K. T., Fox, D. S., Cunningham, K. W., Bassel-Duby, R., Olson, E. N., Zhang, Z., Williams, R. S., Gerber, H. P., Perez-Riba, M., Seo, H., Cao, X., Klee, C. B., Redondo, J. M., Maltais, L. J., Bruford, E. A., Povey, S., Molkentin, J. D., McKeon, F. D., Duh, E. J., Crabtree, G. R., Cyert, M. S., de la Luna, S., and Estivill, X. (2007) FASEB J. 21 3023-3028 [DOI] [PubMed] [Google Scholar]
- 8.Gorlach, J., Fox, D. S., Cutler, N. S., Cox, G. M., Perfect, J. R., and Heitman, J. (2000) EMBO J. 19 3618-3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kingsbury, T. J., and Cunningham, K. W. (2000) Genes Dev. 14 1595-1604 [PMC free article] [PubMed] [Google Scholar]
- 10.Fuentes, J. J., Genesca, L., Kingsbury, T. J., Cunningham, K. W., Perez-Riba, M., Estivill, X., and de la Luna, S. (2000) Hum. Mol. Genet 9 1681-1690 [DOI] [PubMed] [Google Scholar]
- 11.Rothermel, B., Vega, R. B., Yang, J., Wu, H., Bassel-Duby, R., and Williams, R. S. (2000) J. Biol. Chem. 275 8719-8725 [DOI] [PubMed] [Google Scholar]
- 12.Aubareda, A., Mulero, M. C., and Perez-Riba, M. (2006) Cell. Signal. 18 1430-1438 [DOI] [PubMed] [Google Scholar]
- 13.Mulero, M. C., Aubareda, A., Schluter, A., and Perez-Riba, M. (2007) Biochim. Biophys. Acta 1773 330-341 [DOI] [PubMed] [Google Scholar]
- 14.Northrop, J. P., Ho, S. N., Chen, L., Thomas, D. J., Timmerman, L. A., Nolan, G. P., Admon, A., and Crabtree, G. R. (1994) Nature 369 497-502 [DOI] [PubMed] [Google Scholar]
- 15.Roehrl, M. H., Kang, S., Aramburu, J., Wagner, G., Rao, A., and Hogan, P. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7554-7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aramburu, J., Yaffe, M. B., Lopez-Rodriguez, C., Cantley, L. C., Hogan, P. G., and Rao, A. (1999) Science 285 2129-2133 [DOI] [PubMed] [Google Scholar]
- 17.Vilar, M., Esteve, V., Pallas, V., Marcos, J. F., and Perez-Paya, E. (2001) J. Biol. Chem. 276 18122-18129 [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Cozar, F. J., Okamura, H., Aramburu, J. F., Shaw, K. T., Pelletier, L., Showalter, R., Villafranca, E., and Rao, A. (1998) J. Biol. Chem. 273 23877-23883 [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi, K., Roehrl, M. H., Sun, Z. Y., and Wagner, G. (2007) Structure (Camb.) 15 587-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan, B., Greenan, G., McKeon, F., and Ellenberger, T. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13075-13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Paya, E., Dufourcq, J., Braco, L., and Abad, C. (1997) Biochim. Biophys. Acta 1329 223-236 [DOI] [PubMed] [Google Scholar]
- 22.Park, S., Uesugi, M., and Verdine, G. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7130-7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H., Zhang, L., Rao, A., Harrison, S. C., and Hogan, P. G. (2007) J. Mol. Biol. 369 1296-1306 [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Martinez, S., Rodriguez, A., Lopez-Maderuelo, M. D., Ortega-Perez, I., Vazquez, J., and Redondo, J. M. (2006) J. Biol. Chem. 281 6227-6235 [DOI] [PubMed] [Google Scholar]
- 25.Ryeom, S., Greenwald, R. J., Sharpe, A. H., and McKeon, F. (2003) Nat. Immunol. 4 874-881 [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Garcia, B., Perez-Paya, E., and Marcos, J. F. (2002) Appl. Environ. Microbiol. 68 2453-2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canela, N., Orzaez, M., Fucho, R., Mateo, F., Gutierrez, R., Pineda-Lucena, A., Bachs, O., and Perez-Paya, E. (2006) J. Biol. Chem. 281 35942-35953 [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Martinez, C., Humet, M., Planells-Cases, R., Gomis, A., Caprini, M., Viana, F., De La Pena, E., Sanchez-Baeza, F., Carbonell, T., De Felipe, C., Perez-Paya, E., Belmonte, C., Messeguer, A., and Ferrer-Montiel, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2374-2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humet, M., Carbonell, T., Masip, I., Sanchez-Baeza, F., Mora, P., Canton, E., Gobernado, M., Abad, C., Perez-Paya, E., and Messeguer, A. (2003) J. Com. Chem. 5 597-605 [DOI] [PubMed] [Google Scholar]
- 30.Mora, P., Masip, I., Cortes, N., Marquina, R., Merino, R., Merino, J., Carbonell, T., Mingarro, I., Messeguer, A., and Perez-Paya, E. (2005) J. Med. Chem. 48 1265-1268 [DOI] [PubMed] [Google Scholar]
- 31.Malet, G., Martin, A. G., Orzaez, M., Vicent, M. J., Masip, I., Sanclimens, G., Ferrer-Montiel, A., Mingarro, I., Messeguer, A., Fearnhead, H. O., and Perez-Paya, E. (2006) Cell Death Differ. 13 1523-1532 [DOI] [PubMed] [Google Scholar]
- 32.Borisy, A. A., Elliott, P. J., Hurst, N. W., Lee, M. S., Lehar, J., Price, E. R., Serbedzija, G., Zimmermann, G. R., Foley, M. A., Stockwell, B. R., and Keith, C. T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7977-7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez, A., Martinez-Martinez, S., Lopez-Maderuelo, M. D., Ortega-Perez, I., and Redondo, J. M. (2005) J. Biol. Chem. 280 9980-9984 [DOI] [PubMed] [Google Scholar]
- 34.Hesser, B. A., Liang, X. H., Camenisch, G., Yang, S., Lewin, D. A., Scheller, R., Ferrara, N., and Gerber, H. P. (2004) Blood 104 149-158 [DOI] [PubMed] [Google Scholar]
- 35.Vega, R. B., Yang, J., Rothermel, B. A., Bassel-Duby, R., and Williams, R. S. (2002) J. Biol. Chem. 277 30401-30407 [DOI] [PubMed] [Google Scholar]
- 36.Noguchi, H., Matsushita, M., Okitsu, T., Moriwaki, A., Tomizawa, K., Kang, S., Li, S. T., Kobayashi, N., Matsumoto, S., Tanaka, K., Tanaka, N., and Matsui, H. (2004) Nat. Med. 10 305-309 [DOI] [PubMed] [Google Scholar]
- 37.Venkatesh, N., Feng, Y., DeDecker, B., Yacono, P., Golan, D., Mitchison, T., and McKeon, F. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8969-8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang, S., Li, H., Rao, A., and Hogan, P. G. (2005) J. Biol. Chem. 280 37698-37706 [DOI] [PubMed] [Google Scholar]
- 39.De Schryver, E. L., Algra, A., and van Gijn, J. (2003) Cochrane Database Syst. Rev. 1 CD001820. [DOI] [PubMed] [Google Scholar]
- 40.Weyrich, A. S., Denis, M. M., Kuhlmann-Eyre, J. R., Spencer, E. D., Dixon, D. A., Marathe, G. K., McIntyre, T. M., Zimmerman, G. A., and Prescott, S. M. (2005) Circulation 111 633-642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.