Abstract
Despite central roles of egg coat proteins in gamete recognition, their functions and composition are poorly understood. Here, we report that the proteome of the egg coat in the solitary ascidian Ciona intestinalis, called vitelline coat (VC) fraction, contains more than 800 proteins identified by mass spectrometry-based analyses. Over 100 proteins were enriched in the VC fraction compared with the VC-free egg proteome. The most abundant component in the VC was an apolipoprotein-like protein. The VC contained multiple homologs of mammalian zona pellucida (ZP) proteins, the number of which was unexpectedly large and most of which possessed epidermal growth factor-like repeats. Furthermore, the present study revealed that two fibrinogen-like proteins, v-Themis-A and -B, both of which are expressed in the VC, are the molecules responsible for the two self-sterility loci that were identified by our previous genetic study in this species.
Successful fertilization is a crucial process for sexual reproduction in living organisms. One of the key events in fertilization is a sperm-egg recognition step that takes place on the egg coat, an extracellular matrix of the egg. The egg coat provides both an adhesive scaffold and a formidable barrier to sperm so that only a single, congeneric and compatible sperm can penetrate through it. This recognition process mostly depends on a set of finely prearranged molecules that reside on the egg coat and sperm plasma membrane. Therefore comprehensive identification and characterization of such biomolecules are very important to understand the molecular mechanism of gamete recognition. Although several proteomic analyses have been performed on the sperm proteins in various organisms (1-3), proteomic studies on the egg coat proteins have been scarcely conducted because of technical difficulties in the sample preparation.
A cosmopolitan ascidian, Ciona intestinalis, provides an ideal animal system for fertilization studies. The ascidian is a closest relative of vertebrates. Its egg coat is called vitelline coat (VC).4 C. intestinalis undergoes external fertilization. Compared with mammalian eggs, a large quantity of readily fertilizable eggs can be easily obtained. Moreover, draft genome sequences and a large set of transcriptomic data are available (4, 5).
Another interesting feature of the C. intestinalis VC is its allorecognizable ability. Nearly all ascdians are hermaphroditic, releasing both sperm and eggs nearly simultaneously, but many species, including C. intestinalis, exhibit self-sterility. It is shown that sperm fertility is closely related to its capability to bind to the VC: self-sperm do not bind as firmly to the VC as nonself-sperm (6). This self/nonself-discriminating activity is obvious even between isolated (dead) VC and intact sperm, indicating that the VC contains female-side self-sterility factors.
In this report, we identified more than 800 proteins in the VC fraction of C. intestinalis by mass spectrometry (MS)-based proteomic analyses. Over 100 proteins were enriched in the VC fraction compared with the VC-free egg proteome. To the best of our knowledge, this is the most comprehensive proteome of the egg coat in animals.
We previously performed a positional cloning of two loci responsible for the self-sterility system of this species (7). Combination of the genetic studies and the present proteomic analyses enabled us to present robust evidence that two fibrinogen-like proteins (v-Themis-A and -B) are responsible for the female-side self-recognition molecules.
EXPERIMENTAL PROCEDURES
Biological Materials—Adult C. intestinalis animals were collected in Mikawa Bay, Japan. Eggs were surgically obtained from the gonoduct. Follicle cells surrounding the egg were removed by shaking in Ca2+/Mg2+-free seawater. Eggs were gently homogenized in 0.2 × Ca2+/Mg2+-free artificial seawater (4 mm EPPS (pH 8.0), 92 mm NaCl, and 2 mm KCl) containing a protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and Complete Mini (Roche)) with a Teflon homogenizer. The homogenate was filtered through a nylon mesh (40 μm). The flow-through (FT) was saved for use as the VC-free egg extract fraction. The vitelline coat (VC) that remained on the mesh was washed by pipetting, using 0.2 × Ca2+/Mg2+-free artificial seawater containing 0.005% Triton X-100 and further purified manually under a binocular microscope. The acid extract (AE) sample was prepared by immersing the defolliculated eggs in 10 mm HCl-containing artificial seawater for 15 min, and the supernatant fraction was used. All samples were extracted with Laemmli SDS-PAGE sample buffer containing 5% 2-mercaptoethanol and were denatured by boiling for 5 min prior to SDS-PAGE.
Gel Electrophoresis and In Gel Digestion—Samples of VC (100 μg), FT (40 μg), and AE (40 μg) were used for SDS-PAGE (10% gel). After electrophoresis, the gel was stained with Coo-massie Brilliant Blue and then cut into 48 (VC), 29 (FT), and 29 (AE) slices. Each gel piece was subjected to in-gel digestion with trypsin as described previously (8). Briefly, each gel piece was destained with 25 mm ammonium bicarbonate (AB) in 50% acetonitrile (MeCN). After dehydration with MeCN and drying in a vacuum concentrator, the gel piece was re-hydrated with 10 mm dithiothreitol in 25 mm AB and incubated for 1 h at 56 °C for reduction of disulfide bonds. The resulting free thiol groups of proteins were subsequently alkylated by incubating with 55 mm iodoacetamide in 50 mm AB for 45 min at 25 °C in the dark. The gel piece was washed twice with 25 mm AB in 50% MeCN, dehydrated with MeCN, and dried up. Then each gel piece was re-hydrated in 50 μl of 50 mm AB containing 10 μg/ml trypsin (sequence-grade modified trypsin, Promega) for 30 min on ice. After removal of the excess trypsin solution, each gel piece was incubated at 37 °C overnight. The trypsin-digested peptides were extracted from the gel by incubating with 70 μl of 50% MeCN containing 3% formic acid (FA) three times. The extracts were combined and evaporated. The digested peptide mixture from each gel piece was dissolved in 6 μl of MeCN containing 0.1% FA.
LC/MS/MS Analysis—The digested peptides were analyzed using a capillary liquid chromatography system (CapLC; Waters/Micromass, MA) connected online to a tandem mass spectrometer (QToF-2, Waters/Micromass) equipped with a nanoelectrospray source (Z spray) operated in the positive ion mode. Extracted peptides were injected onto a trapping column (μ-Precolumn cartridge filled with PepMap C18, 5 μm, 100 Å, 300 μm in inner diameter, 5 mm in length (LC Packings, Sunny-vale, CA)) and washed with aqueous 0.1% FA at a flow rate of 25 μl/min for 3 min. The trap column was then connected to a C18 reversed-phase analytical column (filled with PepMap C18, 5 μm, 300 Å, 75 μm in inner diameter, 150 mm in length (LC Packings)) by valve switching. The analytical column was pre-equilibrated with 5% solvent B (see below) and eluted at a flow rate of 200 nl/min with a 100-min MeCN gradient (see below). For gradient elution, the 2 μl/min combined flow from pumps A (solvent A: 5% MeCN, 0.1% FA) and B (solvent B: 95% MeCN, 0.1% FA) was reduced to ∼200 nl/min by splitting. Gradients (starting with sample injection) were: 5% B from 0 to 30 min, linear increase to 22% B over the next 55 min, increase to 50% B over the next 31 min, increase to 95% B over the next 5 min, maintenance at 95% B for the next 2 min, and decrease to 5% B over the next 2 min.
Acquisition of MS/MS spectra was performed with the following parameters. Electrospray ionization was performed with the capillary voltage set at 2,200 V, the cone set at 40 V, and the extractor set at 0 V. The collision cell was set at 10 V as a default setting, the source block temperature was 80 °C, and the multichannel plate detector was set at 2,000-2,200 V. The mass spectrometer was operated in the survey scan mode, with up to two of the most intense precursor ions automatically selected. Singly, doubly, or triply charged ions were targeted for collision-induced dissociation fragmentation. Survey scans were recorded from 400 to 1,700 m/z for 0.95 s with an interscan time of 0.05 s, whereas MS/MS scans, performed sequentially for the selected precursor ions, were recorded for 0.95 s from 50 to 1,600 m/z. After total ion counts of MS/MS spectra had risen above 1,200 or MS/MS spectra had been collected for 6 s, the MS/MS scan was switched back to the survey scan. Target ions already selected for MS/MS were dynamically excluded during the subsequent period of 200 s.
After acquisition of the LC/MS/MS data, the raw data were automatically processed using MassLynx version 4.0 (Waters) software to extract peak lists, using the following parameters: for background subtraction, polynomial order 1 and below curve, 40%; for smoothing, smooth window, 3.00, number of smoothings, 2 with smoothing mode Savis Golay; for centroid, minimum peak width at half-height, 4, and centroid mode, centroid top 80%.
Data Base Search and Data Analysis—The obtained peak lists were subjected to a protein data base search using MASCOT version 2.1 (Matrix Science, London, UK) (9). The protein data base used for the present analyses, named “Ciona+NCBInr” (4,225,027 sequences, 1,466,783,644 residues), included Ciona-derived sequences, “Ciona” (125,201 sequences, 52,793,543 residues), and excess decoy sequences (NCBI non-redundant sequences; 4,099,826 sequences, 1,413,990,101 residues; release 156.0, October 15, 2006; ncbi.nih.gov/blast/db/FASTA/nr.gz). The following sequences of predicted proteins were combined for Ciona: KYOTOGRAIL2005 gene models and full-length cDNA sequences (ghost.zool.kyoto-u.ac.jp/indexr1.html) (5), JGI versions 1.0 and 2.0 filtered gene models (genome.jgi-psf.org/Cioin2/Cioin2.home.html), Ensemble ab-initio peptide version 2.0 (ensembl.org/Ciona_intestinalis/index.html), and all Themis protein sequences (7). The following criteria were used to obtain confident results: 1) the hits of peptide derived from Ciona sequences were selected; 2) each ion score should be more than 20; and 3) the protein score, which is the sum of peptide scores, must be greater than 40. Ratio of false identifications was estimated to be about 1.0% by a data base search with the same query peak list and the same criteria against a decoy data base containing reversed Ciona sequences (Fig. 1D). In the present analyses, proteins that had more than three identified peptides were selected, and their protein scores were employed for approximate quantification of abundance of each protein and comparison of relative abundances between different samples (10-12). Protein score in each sample was normalized on the basis of protein score with linear normalization. A linear normalization factor was computed per sample by setting the geometric mean of protein score to 300. The normalization values were 1.07, 1.51, and 1.23 for VC, FT, and AE samples, respectively. VC-enriched proteins were selected by the following criteria: 1) the protein should have more than three peptides identified in the VC sample and 2) relative values (“ratio” in supplemental Table S3), which were calculated by dividing the normalized protein score in the VC by that in FT, were more than 10, or the protein was not detected in FT. All proteins identified in the VC were categorized into several groups according to their functions that were predicted from BLASTP results and/or domain architectures determined by SMART (smart.embl-heidelberg.de/) program (supplemental Table S2). The functional classification was performed as previously described (13).
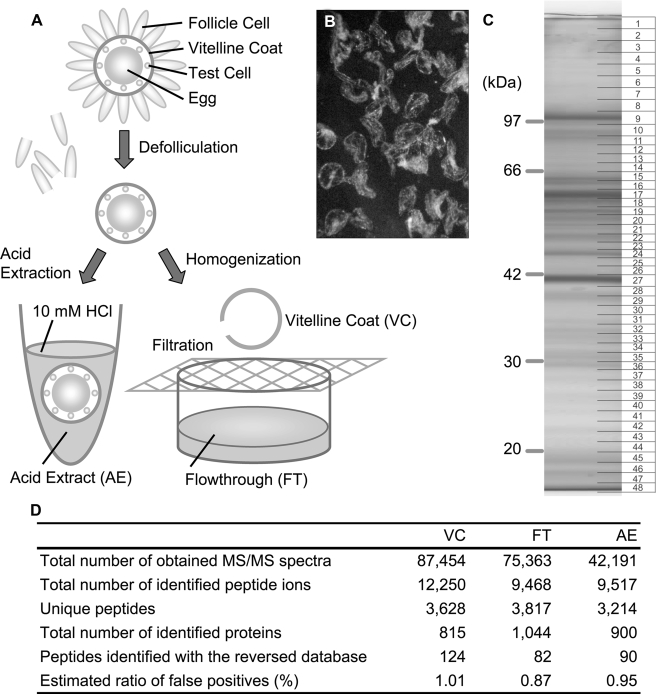
FIGURE 1.
Proteomic analysis of the VC proteins. A, procedure for isolation of VC, FT, and AE samples. B, manually purified VC. C, one-dimensional PAGE of the VC fraction. Numbers at right indicate the slices used for in-gel digestion. D, summary of the proteomic analyses. All MS/MS spectra that hit the forward data base were subjected to a search against the reverse data base and used for the estimation of false-positive rate. Formula for the false-positive rate (%): 100 × (number of reverse identifications) per (number of forward identifications).
Immunological Procedures—A mouse antiserum was raised against a glutathione S-transferase fusion protein of a Ci-ApoBL fragment containing the region corresponding from KLYREDWL to AGRVNLKF.
To examine the spatial distribution of Ci-ApoBL, defolliculated eggs were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) on ice for 1 h and then washed with PBS. Saturation of nonspecific binding sites was achieved by incubation with 3% bovine serum albumin in PBS for 1 h. After incubation with the anti-Ci-ApoBL polyclonal antibody in PBS for 3 h, the specimen was washed with PBS. Signals were detected with a rhodamine-conjugated anti-mouse IgG.
Far Western analysis was performed as previously reported (14). Sperm homogenate was subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. After a 1-h incubation in a blocking buffer (1% skim milk in 1 × PBS), Ci-ApoBL-containing egg homogenate in the blocking buffer was overlaid on the membrane, allowing Ci-ApoBL to bind to its potential binding partners on the membrane overnight at room temperature. The Ci-ApoBL-interacting proteins were detected by the anti-Ci-ApoBL antiserum.
Single Nucleotide Polymorphism Analysis—Gonad RNA and genomic DNA samples were individually prepared from wild animals. cDNAs were synthesized using a SuperScript III first-strand synthesis system (Invitrogen) primed with oligo(dT). cDNA and genomic fragments were amplified with gene-specific primers (v-Themis-like1-fwd, 5′-GAGCGGAATGTACAAAATCTGGTTGG-3′; v-Themis-like1-rev, 5′-TACAAATCGCAGGTCTGGGTTAGCTG-3′; kyotograil2005.78.4.1-fwd, 5′-AAAGTTTGGCAGCGTGTACCTTGCT-3′; kyotograil2005.78.4.1-rev, 5′-AAGTCGGTGTCGTGGTTCCAACTTT-3′; kyotograil2005.726.5.1-fwd1, 5′-GCTCCCTTCATTGCAAAACGAAGAA-3′; kyotograil2005.726.5.1-rev1, 5′-CACTCGTAAAAGCAATTGGCACGAG-3′; kyotograil2005.726.5.1-fwd2, 5′-TGGAAAGCCAAGAATTGTCAACCAA-3′; kyotograil2005.726.5.1-rev2, 5′-TGAAATGATGAGAATCCTGTGCAAGC-3′; kyotograil2005.739.4.1-fwd1, 5′-AAGATGAAGACAACGACGATGCAATA-3′; kyotograil2005.739.4.1-fwd2, 5′-TATTGGTGACATAGAAGCGAGGTTGA-3′; kyotograil2005.739.4.1-rev1, 5′-AACATGATGGTTCGTATTCACTGACG-3′; kyotograil2005.739.4.1-rev2, 5′-CATGTACACGACCAAATTCCACGATA-3′; kyotograil2005.112.19.1-fwd, 5′-ACACAACTGTATCAGGGGTGGCTCA-3′; kyotograil2005.112.19.1-rev, 5′-GCCTCTTGTTCTGCCTGGTTTGTTT-3′). The amplified fragments were sequenced directly using the same primers as those used for amplification.
RESULTS AND DISCUSSION
Proteomic Analysis of the VC in C. intestinalis—The mature egg of C. intestinalis is surrounded by test cells, the VC, and a layer of follicle cells covering the outer surface of the VC (Fig. 1A). Defolliculated eggs were homogenized and filtered through a nylon mesh. Homogenates of the egg and test cell passed through the mesh, whereas the VC was retained. To reduce the contaminating egg fragments or test cell-derived materials, the VC was further purified manually under a binocular microscope (Fig. 1B). The flow-through specimen (FT) was also saved for later use as the VC-free egg extract.
The VC fraction was subjected to SDS-PAGE and the gel was cut into small pieces (Fig. 1C). Each gel slice was treated with trypsin, followed by reverse-phased high performance liquid chromatography/electrospray ionization tandem mass spectrometry (LC/MS/MS). Each sample was measured twice. Fig. 1D (left column) shows a summary of proteomic analyses of the VC fraction. More than 80,000 spectra, each of which was derived from a trypsin-digested peptide ion, were detected in LC/MS/MS analysis. A data base search against a combined data base of the three predicted gene models in this species (e.g. Refs. 4 and 5) (ghost.zool.kyoto-u.ac.jp/indexr1.html, genome.jgi-psf.org/Cioin2/Cioin2.home.html, and www.ensembl.org/Ciona_intestinalis/index.html) using the MASCOT search engine (9) yielded 815 proteins. A comprehensive list of the proteins is available in supplemental Table S1. The rate of false identifications was estimated to be around 1.0% by searching the same data set against a decoy data base consisting of reversed protein sequences of the same gene model set (15). Abundance of each protein can be roughly estimated with the protein score presented by the MASCOT search engine, which is the sum of peptide scores, and spectral counting (supplemental Table S1 and Refs. 10-12).
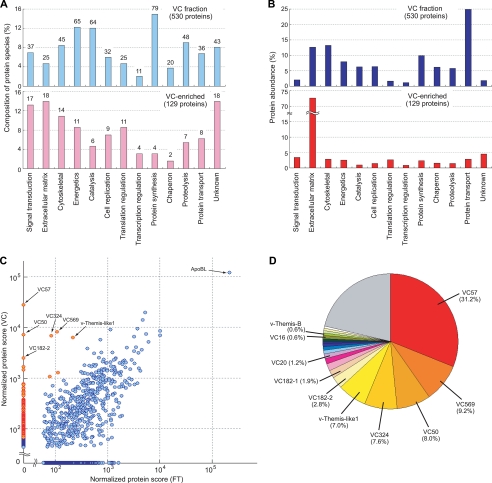
Among the 815 proteins, 530 identified with more than three peptides were chosen for the following functional classification. Thirteen categories used for this analysis are shown in supplemental Table S2. Based on the number of protein species, the VC fractions, thus prepared, contained many proteins involved in protein synthesis (79 proteins, 14.9%) as well as many proteins for energy reproduction (65, 12.3%), catalysis (64, 12.1%), and proteolysis (48, 9.1%) (Fig. 2A, upper panel). On the other hand, based on the protein amounts, proteins involved in protein transport were the most abundant (24.9%) in the VC, followed by extracellular matrix proteins (14.7%) and cytoskeletal proteins (13.3%) (Fig. 2B, upper panel). Several proteins that were predicted to be localized intracellularly, such as metabolic enzymes and mitochondrial proteins, were also detected. These results suggest that our VC fraction is still contaminated with the egg- or test cell-derived proteins.
FIGURE 2.
Protein composition of the VC fraction. A and B, functional classification of the total VC fraction (upper panel) and the VC-enriched proteins (lower panel). The bar height is proportional to the number of protein species (A) or the protein abundances (B). The number of protein species classified in each category is shown above each bar in A. C, scatter plot showing a comparison of the relative abundance of each protein identified in the VC and FT fractions. Red spots represent proteins enriched in the VC fraction. The protein scores are plotted in logarithmic scale. D, relative abundances of the 20 most abundant proteins enriched in the VC fraction. The top 20 abundant proteins are shown separately and the remaining proteins are combined in a single gray area.
To distinguish “true” VC-specific components from such putative contaminants, we also analyzed the protein composition of the mesh FT fraction, which contains egg cell/test cell homogenates and fluid in the perivitelline space (Fig. 1A). By using the same strategy and criteria as those used for the VC fraction, 1,044 proteins were identified in the FT fraction (Fig. 1D, middle, and supplemental Table S3). Then we compared the protein abundance of the VC fraction and those of the FT fraction (Fig. 2C). We identified 129 proteins that are abundant in the VC fraction but almost absent in the FT fraction, which are defined as “VC-enriched” components. Table 1 and supplemental Table S4 show lists of the VC-enriched components, of which relative abundances in the VC are more than 10 times higher than those in the FT. Functional classification of the VC-enriched proteins revealed that the percentages of protein species involved in catalysis (6 proteins, 4.7%) and protein synthesis (4, 3.1%) were clearly decreased, whereas the percentages of protein species involved in extracellular matrix formation (18, 14.0%) and signal transduction (17, 13.2%) were increased (Fig. 2A, lower panel). This tendency became more apparent when protein abundance was considered: 72.7% of the VC-enriched proteins were classified as extracellular matrix proteins (Fig. 2B, lower panel). We concluded that the comparative analysis could reveal the true VC-specific proteins.
TABLE 1.
VC-enriched proteins
|
Vitelline coat
|
Flow-through
|
Acid extract
|
|||||
|---|---|---|---|---|---|---|---|
| Representative gene model IDa | Product | Normalized protein score | Count of spectra | Normalized protein score | Count of spectra | Normalized protein score | Count of spectra |
| kyotograil2005.57.4.1 | CiVC57 | 27641.73 | 570 | 0 | 0 | 0 | 0 |
| kyotograil2005.569.7.1 | CiVC569 | 8135.03 | 158 | 110.64 | 1 | 0 | 0 |
| kyotograil2005.50.29.1 | CiVC50 | 7102.68 | 158 | 0 | 0 | 547.17 | 9 |
| kyotograil2005.324.11.1 | CiVC324 | 6773.99 | 146 | 85.44 | 2 | 79.89 | 2 |
| kyotograil2005.78.35.1 | v-Themis-like 1 | 6234.18 | 129 | 221.93 | 4 | 1224.99 | 21 |
| Cioin2|estExt_fgenesh3_pg.C_chr_04q0435 | CiVC182-2 | 2490.85 | 55 | 0 | 0 | 0 | 0 |
| kyotograil2005.182.31.1 | CiVC182-1 | 1650.86 | 34 | 0 | 0 | 0 | 0 |
| kyotograil2005.131.13.1 | Isoleucyl-tRNA synthetase | 1260.94 | 33 | 114.44 | 2 | 0 | 0 |
| Cioin2|estExt_fgenesh3_kg.C_1060014 | Vesicle docking protein | 1447.09 | 28 | 0 | 0 | 0 | 0 |
| kyotograil2005.20.78.1 | CiVC20 | 1047.20 | 26 | 79.55 | 2 | 0 | 0 |
| kyotograil2005.739.4.1 | Fibronectin type1 domain-containing protein | 638.59 | 17 | 0 | 0 | 77.33 | 1 |
| kyotograil2005.60.2.1 | EGF-like and transforming growth factor-β-like domain-containing protein | 734.51 | 16 | 0 | 0 | 0 | 0 |
| kyotograil2005.250.16.1 | Complement component C3 | 767.56 | 14 | 0 | 0 | 0 | 0 |
| kyotograil2005.2.149.1 | Ci-DNAJA1/2/4 | 625.56 | 12 | 0 | 0 | 0 | 0 |
| kyotograil2005.16.45.1 | CiVC16 | 550.05 | 12 | 0 | 0 | 0 | 0 |
| SNAP_CIONA00000044790 | vWA domain-containing protein | 531.95 | 12 | 0 | 0 | 0 | 0 |
| Cioin2|e_gw1.8117.2.1 | v-Themis-B (8117) | 537.06 | 11 | 0 | 0 | 0 | 0 |
| kyotograil2005.12.5.1 | Piwi-like 1 | 534.24 | 11 | 0 | 0 | 0 | 0 |
| kyotograil2005.247.2.1 | Similar to sea urchin XP_785293 (no putative domain) | 448.72 | 11 | 0 | 0 | 50.07 | 1 |
| kyotograil2005.181.21.1 | α3 Collagen | 348.31 | 10 | 0 | 0 | 0 | 0 |
| ciad011b02 | Ribosomalprotein L30 | 498.58 | 9 | 0 | 0 | 0 | 0 |
| Cioin2|e_gw1.01p.326.1 | NADH:ubiquinone oxidoreductase | 354.44 | 9 | 0 | 0 | 0 | 0 |
| ci0100149229 | Thrombospondin type1 repeat-containing protein | 468.78 | 8 | 0 | 0 | 0 | 0 |
| kyotograil2005.26.14.1 | NAD-dependent alcohol dehydrogenase | 375.66 | 8 | 0 | 0 | 0 | 0 |
| kyotograil2005.459.8.1 | Heterogeneous nuclear ribonucleoprotein | 342.78 | 8 | 0 | 0 | 0 | 0 |
| kyotograil2005.117.14.1 | NADPH-dependent retinol dehydrogenase | 336.22 | 8 | 0 | 0 | 0 | 0 |
| Cioin2|e_gw1.02q.956.1 | Small inducible cytokine subfamily E | 313.85 | 8 | 0 | 0 | 0 | 0 |
| Cioin2|e_gw1.07q.1362.1 | Small nuclear ribonucleoprotein | 296.25 | 8 | 0 | 0 | 0 | 0 |
| kyotograil2005.400.6.1 | CUB and Sushi multiple domains protein | 269.75 | 8 | 0 | 0 | 0 | 0 |
| kyotograil2005.404.4.1 | (No putative domain and homology) | 362.04 | 7 | 0 | 0 | 0 | 0 |
| kyotograil2005.247.1.1 | sImilar to sea urchin XP_801877 (no putative domain) | 321.83 | 7 | 0 | 0 | 482.87 | 9 |
| kyotograil2005.124.19.1 | Endothelin converting enzyme | 319.13 | 7 | 0 | 0 | 0 | 0 |
| Cioin2|fgenesh3_pg.C_chr_02q000529 | v-Themis-A (JGI) | 305.80 | 7 | 0 | 0 | 0 | 0 |
| kyotograil2005.40.5.1 | Transglutaminase | 303.62 | 7 | 0 | 0 | 0 | 0 |
| kyotograil2005.607.1.1 | Lectin-associated matrix protein | 271.85 | 7 | 0 | 0 | 0 | 0 |
| Cioin2|gw1.902.1.1 | v-Themis-B (902) | 252.19 | 7 | 0 | 0 | 0 | 0 |
| kyotograil2005.145.31.1 | Tyrosyl-tRNA synthetase | 248.96 | 7 | 0 | 0 | 0 | 0 |
| GENEFINDER00000092366 | (No putative domain and homology) | 200.81 | 7 | 0 | 0 | 0 | 0 |
| kyotograil2005.75.28.1 | Rhamnose binding lectin | 337.69 | 6 | 0 | 0 | 0 | 0 |
| kyotograil2005.51.66.1 | Similar to mannose receptor | 295.14 | 6 | 0 | 0 | 0 | 0 |
| kyotograil2005.20.56.1 | Ci-Vanabin1 | 284.65 | 6 | 0 | 0 | 0 | 0 |
| kyotograil2005.19.63.1 | Mini chromosome maintenance complex component 2 | 233.94 | 6 | 0 | 0 | 0 | 0 |
| kyotograil2005.147.19.1 | Laminin, gamma1 | 175.27 | 6 | 0 | 0 | 0 | 0 |
| kyotograil2005.25.22.1 | (No putative domain and homology) | 282.47 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.18.43.1 | Ras suppressor protein 1 | 249.33 | 5 | 0 | 0 | 0 | 0 |
| ci0100131162 | Cleavage and polyadenylation factorI | 239.92 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.670.5.1 | Mannose 6-phosphate protein | 232.88 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.1612.2.1 | β-Catenin | 221.71 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.35.23.1 | Nuclear matrix protein SNEV | 218.50 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.90.22.1 | Replication factor Cp37 subunit | 203.87 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.73.19.1 | Ci-HMG1/2 | 197.72 | 5 | 0 | 0 | 0 | 0 |
| SNAP_CIONA00000035881 | cbb3-type cytochrome c oxidase subunit | 181.76 | 5 | 0 | 0 | 0 | 0 |
| kyotograil2005.899.7.1 | Ci-BTF3 | 181.32 | 5 | 0 | 0 | 0 | 0 |
| GENEFINDER00000088127 | Similar to acetolactate synthase | 153.50 | 5 | 0 | 0 | 0 | 0 |
kyotograil2005x, from Kyotograil 2005 gene models (ghost.zool.kyoto-u.ac.jp/indexr1.html); e.g. ciad011b02, from full-length cDNA sequences (ghost.zool.kyoto-u.ac.jp/indexr1.html); ci0100x and Cioin2|x, from JGI version 1.0 and 2.0 filtered gene models (genome.jgi-psf.org/Cioin2/Cioin2.home.html), respectively; Remaining gene models correspond to Ensemble ab-initio peptide version 2.0 (www.ensembl.org/Ciona_intestinalis/index.html).
A Putative Egg Yolk Protein Abundantly Resides on the VC—The most abundant component found in the VC was an apolipoprotein-B-like protein (Ci-ApoBL). Because of its abundance in the FT fraction, it was not classified as a VC-enriched protein. This protein probably represents the major yolk protein in the egg. To determine whether Ci-ApoBL is a true VC component, we raised a mouse polyclonal antibody against a glutathione S-transferase fusion protein of Ci-ApoBL. The antiserum detected a distinct 100-kDa band in both of the VC and the VC-free FT lanes (supplemental Fig. S1A). Immunocytochemistry showed that Ci-ApoBL resides on the VC as well as in the egg cytoplasm (supplemental Fig. S1B).
To investigate a possible role of the VC-resident Ci-ApoBL protein in fertilization, we explored sperm-borne binding partners for Ci-ApoBL by Far Western analysis. The results showed that Ci-ApoBL specifically recognizes a 70-kDa sperm protein (supplemental Fig. S1F). This suggests that Ci-ApoBL on the VC may be involved in the interaction between sperm and the VC of the egg.
Abundance of ZP Domain-containing Proteins in the VC—The egg coat of mammals, called the zona pellucida (ZP), has a relatively simple molecular composition (16). For example, in mouse, the ZP is composed of three major glycoproteins, designated ZP1, ZP2, and ZP3, all of which contain a ZP domain at the C terminus and are highly glycosylated with N- and O-linked carbohydrate chains. It is thought that ZP2 and ZP3 dimerize to form long filaments, which are cross-linked by ZP1. The ZP domain has been shown to be responsible for polymerization of ZP-domain-containing proteins (17). It has been suggested that ZP3 is a primary species-selective adhesion molecule for sperm on the ZP (16). ZP2 is thought to play a role in the secondary binding of sperm to the ZP after acrosome reaction and during ZP penetration (18). In addition to mammals, ZP-domain-containing proteins are found in the egg coat of nearly all vertebrates (19) and some invertebrates (20), suggesting that the molecular composition of the egg coat is evolutionally conserved.
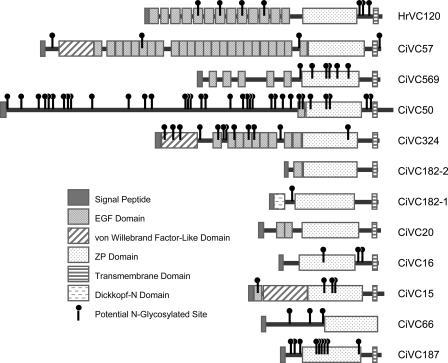
In C. intestinalis, the identified VC components included 11 proteins that harbor a ZP domain, four of which have already been reported (CiVC16, CiVC20, CiVC182-1, and CiVC569) (21, 22) but the others being novel (supplemental Table S1). Following the nomenclature proposed in Ref. 22, we designated the seven novel proteins as CiVC15 (encoded in scaffold 15 of the genome assemble in JGI version 1 (Ref. 4)), CiVC50, CiVC57, CiVC66, CiVC182-2, CiVC187, and CiVC324, respectively. CiVC182 originally reported in Ref. 21 was renamed CiVC182-1 because scaffold 182 encodes two ZP-domain-containing VC proteins. All of the proteins except for CiVC66 and CiVC187 were highly enriched in the VC (Table 1 and Fig. 2D). Moreover, the seven CiVC proteins each contain a single or multiple epidermal growth factor (EGF)-like domains (Fig. 3). This architectural feature seems to be characteristic of ascidian VC proteins. In another ascidian, Halocynthia roretzi, it has been shown that a 70-kDa glycoprotein, HrVC70, is the most major component of the VC and that it is processed from its precursor HrVC120, containing 13 EGF-like repeats and a ZP domain (23). In Halocynthia aurantium, HaVC130, the ortholog of HrVC120, was isolated (24). In H. roretzi, two more proteins that contain both EGF and ZP domains, designated as HrVLP-1 and -2, were identified as HrVC70-interacting proteins by yeast two-hybrid screening baited with HrVC70 and shown to be highly expressed in the ovary (25). In addition to EGF-like domains, several CiVC proteins contain other functional domains such as a von Willebrand factor domain or a Dick-kopf-N domain in the N-terminal side of the ZP domain.
FIGURE 3.
CiVC proteins. Domain architectures of CiVC proteins.
Close inspection of the proteomic data provided some insights into putative post-translational modifications on CiVC proteins. For example, the observed high molecular masses of CiVC15 and CiVC66 suggest that they are highly N-glycosylated, as they have a number of predicted N-glycosylation sites (supplemental Fig. S2A). CiVC57 was detected in almost all of the gel slices with a wide range of molecular masses. The profile of CiVC57-derived peptide ions detected in each gel slice predicts its possible processing pattern on the VC (supplemental Fig. S2B). Among the characterized CiVC proteins, CiVC57 is a main component and seems to be an ortholog of HrVC120 and HaVC130 (Fig. 3).
Functions of ZP proteins have been most extensively studied in mouse. Although ZP3 has been shown to play an important role in initial gamete adhesion (16), an alternative binding mechanism has been proposed (26). In mouse, ovulated ZP but not ovarian ZP shows a ZP3-independent sperm binding activity. Although the molecular natures of ZP-borne and sperm surface-borne binding molecules involved in this alternative binding are not known, it has been proposed that interaction between these hypothetical molecules is mediated by a bridging protein named SED1, which contains EGF-like repeats and discoidin/F5/8 C domains (26). In fact, sperm from SED1-null males reduces the binding ability to the ZP, although the animal retains partial fertility.
The architecture of the CiVC proteins harboring both EGF-like repeats and a ZP domain raises a possibility that they have dual roles corresponding to ZP3-dependent and -independent binding molecules in mammals. In H. roretzi, the physiological role of EGF-like domains of HrVC120 (HrVC70) has already been demonstrated by isolation of its sperm-borne binding partner, which was designated as HrUrabin (14).
Identification of VC-resident Self-sterility Molecules—C. intestinalis is a hermaphroditic animal showing a highly evolved self-sterility system. We previously characterized the genetic scheme of this system, governed by two unlinked loci, designated as loci A and B (7). The candidate region for locus A in chromosome 2q was restricted to a region as narrow as 170-kbp in length, whereas that for locus B was mapped within a 1-Mbp region in chromosome 7q. Each self-sterility locus encodes a linked set of genes of male-side and female-side recognition molecules, which make up a single haplotype. Self/nonself discrimination takes place on the basis of the self-recognition between molecules from the same haplotype. The genetic proximity of male-side and female-side molecules ensures successful inheritance of the allo-specific interaction between them. Also, previous biochemical studies have shown that the female-side recognition molecules reside on the VC (6).
In the present study, we detected more than 800 proteins in the VC fraction (supplemental Table S1). This list is very comprehensive and should include almost all VC components that are soluble in the SDS sampling buffer. Within the candidate region for locus A, only one protein appears in the list (Table 2). This protein, which has already been designated as v-Themis-A, consisted of a fibrinogen-C-terminal domain. Similarly, locus B encoded five VC proteins (Table 2), one of which turned out to be a v-Themis-B, showing significant homology to v-Themis-A. Both v-Themis-A and -B are highly polymorphic proteins (7). We revealed that four VC-resident molecules other than v-Themis-B encoded in locus B have little or no polymorphisms among individuals (data not shown). Therefore we concluded that v-Themis-A and -B are the female-side responsible genes for the self-recognition of loci A and B, respectively. The results that both v-Themis proteins were enriched in the VC fraction (Table 1) also coincided with our above mentioned conclusion.
TABLE 2.
Identified VC proteins encoded within the candidate reagions of self-sterility loci
|
Vitelline coat
|
Flow-through
|
||||||
|---|---|---|---|---|---|---|---|
| Representative gene model IDa | Product | Normalized protein score | Count of spectra | Normalized protein score | Count of spectra | Chromosome | Location |
| Locus A (chr_02q:1945933-2117245) | |||||||
| Cioin2|fgenesh3_pg.C_chr_02q000529 | v-Themis-A (JGI) | 305.80 | 7 | 0 | 0 | chr_02q | 2096379-2095300(−) |
| Locus B (chr_07q:2455323-3487288) | |||||||
| kyotograil2005.78.4.1 | Aurora/lpl1p-related kinase | 76.91 | 2 | 0 | 0 | chr_07q | 2604185-2602744(−) |
| kyotograil2005.726.5.1 | Ferrochelatase | 250.79 | 6 | 348.08 | 6 | chr_07q | 2455320-2459929(+) |
| Cioin2|e_gw1.8117.2.1 | v-Themis-B (8117) | 537.06 | 11 | 0 | 0 | chr_07q | Undetermined |
| Cioin2|gw1.902.1.1 | v-Themis-B (902) | 252.19 | 7 | 0 | 0 | chr_07q | Undetermined |
| kyotograil2005.360.12.1 | v-Themis-B (JGI) | 169.11 | 3 | 0 | 0 | chr_07q | 2613145-2613657(−) |
| kyotograil2005.739.4.1 | Fibronectin type1 domain-containing protein | 638.59 | 17 | 0 | 0 | chr_07q | 2897429-2900664(+) |
| kyotograil2005.112.19.1 | Propionyl-CoA carboxylase | 112.84 | 2 | 0 | 0 | chr_07q | 3419144-3414011(−) |
See the caption of Table 1.
Our MS/MS analysis revealed several types of alleles for both v-Themis proteins in the VC fraction, which was derived from limited numbers of wild animals. This implies that sequence variations of v-Themis may not be random and that the numbers of their alleles are finite; otherwise we would have failed to identify the v-Themis-derived peptides in the protein data base because the degree of their sequence variations is extremely high. It is notable that observed molecular masses were always higher than the predicted one (∼37 kDa) and showed considerable variations among alleles (supplemental Fig. S3).
Other VC-enriched Proteins—Besides homologs of vertebrate ZP proteins and v-Themis proteins, a number of proteins were significantly enriched in the VC fraction. As shown in Table 1, a protein showing striking homology to v-Themis-A/B (kyotograil2005.78.35.1) was found in the VC-enriched protein list. We designated this protein v-Themis-like1. v-Themis-like1 is composed of a single fibrinogen C-terminal domain like v-Themis, and it shows not only structural similarity but also chromosomal proximity to v-Themis-B, although v-Themis-like1 itself is located just outside the candidate region for locus B. Its abundance in the VC was much higher than that of v-Themis-B. We performed reverse transcriptase-PCR experiments on the v-Themis-like1 gene for several individuals and found that it was expressed in all individuals and showed little sequence variations (data not shown). This indicates that v-Themis-like1 is not an allele of the v-Themis-B gene but an independent gene.
The VC-enriched proteins also include a protein containing numerous EGF-like and transforming growth factor-β-like domains (kyotograil2005.60.2.1), a von Willebrand factor-like domain-containing protein (SNAP_CIONA00000044790), a CUB and Sushi multiple domain-containing protein (kyotograil2005.400.6.1), and a thrombospondin type 1 repeat-containing protein (ci0100149229) (Table 1 and supplemental Table S4). Although functions of these proteins for gamete recognition are not known yet, their domain architectures seem to imply that they have functions in molecular interactions.
v-Themis Proteins Are Not Extracted from the VC with Acid Treatment—In ascidians, the VC barrier against self-fertilization is known to be sensitive to acid or protease treatment, and this was previously thought to be due to the loss of self/nonself-recognizable barrier molecules from the VC (27-29). We therefore performed proteomic analysis of the supernatant fraction extracted from non-homogenized and defolliculated eggs with 10 mm HCl-containing artificial seawater (AE, Fig. 1A) and identified 900 proteins (Fig. 1D, right, supplemental Table S5). However, neither v-Themis-A nor -B proteins were detected in the acid supernatant (Table 1). Moreover, CiVC57 and the other CiVC proteins were scarcely extracted from the VC by acid treatment under these conditions (Table 1).
In contrast to the classical views (27-29), allorecognition molecules (v-Themis proteins) were hardly extracted by weak acid. Although v-Themis itself is resistant to acid, certain auxiliary molecules that play essential roles for the function of v-Themis may be sensitive to weak acid or may be lost from the VC by the acid treatment. As shown in Table 1, the relatively small number of VC-enriched proteins was extracted with weak acid. Among the VC-enriched proteins, v-Themis-like1 was most abundant in the AE fraction. It would be an attractive hypothesis that v-Themis-like1, which also contains a fibrinogen C-terminal domain like v-Themis, interact and cooperate with v-Themis proteins on the VC, and its loss by acid treatment leads a breakage of the self-sterility barrier.
Conclusions—Fertilization is a choreographed biological reaction performed by prearranged molecules rather than by newly synthesized ones. This implies that the proteome and interactome involved in fertilization are relatively “static” compared with those in other biological reactions. Therefore, identifications of each molecule and molecular interaction are quite important for understanding the total mechanism of fertilization.
Comprehensiveness of our present “OMICS” analysis provides a firm foundation for studies on the VC function. For example, one of the prominent functions of the VC in this species is a barrier, acting specifically against homogenous sperm. Identification of v-Themis proteins as the VC-resident female-side self-sterility molecules, which had long been searched for since the discovery of this phenomenon by T. H. Morgan (30), was achieved by a combination of our previous genetic and present proteomic analyses. Particularly, in locus A, these lines of evidence clearly showed that v-Themis-A is a sole candidate for the self-sterility gene.
Each v-Themis gene is located within the first intron of the respective s-Themis gene, which encodes a polycystin 1-like cation channel, in the chromosome. s-Themis gene is highly expressed in the testis, and extracellular domains of s-Themis proteins show extreme polymorphisms (7). The inheritance mode of the self-sterility phenotype suggests that v-Themis ligand protein on the VC is specifically recognized by “self-” (encoded by the same haplotype) s-Themis receptor protein. The molecular mechanism of this v/s-Themis-mediated self-incompatibility system was exhaustively discussed elsewhere (7, 31). Our next goal is to determine the male-side self-sterility molecule by a similar proteomic analysis on the sperm surface proteins.
Supplementary Material
Acknowledgments
We thank Kazuko Hirayama and Maizuru Fisheries Research Station of Kyoto University for help in culturing of C. intestinalis, which was supported by the National Bioresource Project (NBRP), MEXT, Japan.
This work was supported by a research fellowship of the Japan Society for the Promotion of Science (JSPS) for young scientists (to L. Y.), the 21st COE program “Disease Proteomics” and Grants-in-aid for Scientific Research 19370041 and 19659080 by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (to H. T.), Grants-in-aid for Exploratory Research 16659021 and 19659018 from MEXT (to H. S.), and Grant-in-aid 18770198 for Young Scientists (B) from MEXT (to Y. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3 and Tables S1-S5.
Footnotes
The abbreviations used are: VC, vitelline coat; AE, acid extract; EGF, epidermal growth factor; EPPS, N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid; FT, flow-through; LC, liquid chromatography; MS, mass spectrometry; ZP, zona pellucida; AB, ammonium bicarbonate; FA, formic acid; PBS, phosphate-buffered saline.
References
- 1.Dorus, S., Busby, S. A., Gerike, U., Shabanowitz, J., Hunt, D. F., and Karr, T. L. (2006) Nat. Genet. 38 1440-1445 [DOI] [PubMed] [Google Scholar]
- 2.Stein, K. K., Go, J. C., Lane, W. S., Primakoff, P., and Myles, D. G. (2006) Proteomics 6 3533-3543 [DOI] [PubMed] [Google Scholar]
- 3.Baker, M. A., Hetherington, L., Reeves, G., Muller, J., and Aitken, R. J. (2008) Proteomics 8 2312-2321 [DOI] [PubMed] [Google Scholar]
- 4.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., Harafuji, N., Hastings, K. E., Ho, I., Hotta, K., Huang, W., Kawashima, T., Lemaire, P., Martinez, D., Meinertzhagen, I. A., Necula, S., Nonaka, M., Putnam, N., Rash, S., Saiga, H., Satake, M., Terry, A., Yamada, L., Wang, H. G., Awazu, S., Azumi, K., Boore, J., Branno, M., Chin-Bow, S., DeSantis, R., Doyle, S., Francino, P., Keys, D. N., Haga, S., Hayashi, H., Hino, K., Imai, K. S., Inaba, K., Kano, S., Kobayashi, K., Kobayashi, M., Lee, B. I., Makabe, K. W., Manohar, C., Matassi, G., Medina, M., Mochizuki, Y., Mount, S., Morishita, T., Miura, S., Nakayama, A., Nishizaka, S., Nomoto, H., Ohta, F., Oishi, K., Rigoutsos, I., Sano, M., Sasaki, A., Sasakura, Y., Shoguchi, E., Shin-i, T., Spagnuolo, A., Stainier, D., Suzuki, M. M., Tassy, O., Takatori, N., Tokuoka, M., Yagi, K., Yoshizaki, F., Wada, S., Zhang, C., Hyatt, P. D., Larimer, F., Detter, C., Doggett, N., Glavina, T., Hawkins, T., Richardson, P., Lucas, S., Kohara, Y., Levine, M., Satoh, N., and Rokhsar, D. S. (2002) Science 298 2157-2167 [DOI] [PubMed] [Google Scholar]
- 5.Satou, Y., Kawashima, T., Shoguchi, E., Nakayama, A., and Satoh, N. (2005) Zool. Sci. (Tokyo) 22 837-843 [DOI] [PubMed] [Google Scholar]
- 6.Rosati, F., and de Santis, R. (1978) Exp. Cell Res. 112 111-119 [DOI] [PubMed] [Google Scholar]
- 7.Harada, Y., Takagaki, Y., Sunagawa, M., Saito, T., Yamada, L., Taniguchi, H., Shoguchi, E., and Sawada, H. (2008) Science 320 548-550 [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi, M., Hatano, N., Yokota, S., Shimozawa, N., Imanaka, T., and Taniguchi, H. (2004) J. Biol. Chem. 279 421-428 [DOI] [PubMed] [Google Scholar]
- 9.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551-3567 [DOI] [PubMed] [Google Scholar]
- 10.Allet, N., Barrillat, N., Baussant, T., Boiteau, C., Botti, P., Bougueleret, L., Budin, N., Canet, D., Carraud, S., Chiappe, D., Christmann, N., Colinge, J., Cusin, I., Dafflon, N., Depresle, B., Fasso, I., Frauchiger, P., Gaertner, H., Gleizes, A., Gonzalez-Couto, E., Jeandenans, C., Karmime, A., Kowall, T., Lagache, S., Mahe, E., Masselot, A., Mattou, H., Moniatte, M., Niknejad, A., Paolini, M., Perret, F., Pinaud, N., Ranno, F., Raimondi, S., Reffas, S., Regamey, P. O., Rey, P. A., Rodriguez-Tome, P., Rose, K., Rossellat, G., Saudrais, C., Schmidt, C., Villain, M., and Zwahlen, C. (2004) Proteomics 4 2333-2351 [DOI] [PubMed] [Google Scholar]
- 11.Colinge, J., Chiappe, D., Lagache, S., Moniatte, M., and Bougueleret, L. (2005) Anal. Chem. 77 596-606 [DOI] [PubMed] [Google Scholar]
- 12.Heller, M., Schlappritzi, E., Stalder, D., Nuoffer, J. M., and Haeberli, A. (2007) Mol. Cell Proteomics 6 1059-1072 [DOI] [PubMed] [Google Scholar]
- 13.Lee, Y. H., Huang, G. M., Cameron, R. A., Graham, G., Davidson, E. H., Hood, L., and Britten, R. J. (1999) Development 126 3857-3867 [DOI] [PubMed] [Google Scholar]
- 14.Urayama, S., Harada, Y., Nakagawa, Y., Ban, S., Akasaka, M., Kawasaki, N., and Sawada, H. (2008) J. Biol. Chem. 283 21725-21733 [DOI] [PubMed] [Google Scholar]
- 15.Elias, J. E., Haas, W., Faherty, B. K., and Gygi, S. P. (2005) Nat. Methods 2 667-675 [DOI] [PubMed] [Google Scholar]
- 16.Wassarman, P. M., Jovine, L., and Litscher, E. S. (2004) Cytogenet. Genome Res. 105 228-234 [DOI] [PubMed] [Google Scholar]
- 17.Jovine, L., Qi, H., Williams, Z., Litscher, E., and Wassarman, P. M. (2002) Nat. Cell Biol. 4 457-461 [DOI] [PubMed] [Google Scholar]
- 18.Howes, E., Pascall, J. C., Engel, W., and Jones, R. (2001) J. Cell Sci. 114 4127-4136 [DOI] [PubMed] [Google Scholar]
- 19.Goudet, G., Mugnier, S., Callebaut, I., and Monget, P. (2008) Biol. Reprod. 78 796-806 [DOI] [PubMed] [Google Scholar]
- 20.Aagaard, J. E., Yi, X., MacCoss, M. J., and Swanson, W. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17302-17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurn, U., Sommer, F., Hemmrich, G., Bosch, T. C., and Khalturin, K. (2007) Dev. Comp. Immunol. 31 360-371 [DOI] [PubMed] [Google Scholar]
- 22.Kurn, U., Sommer, F., Bosch, T. C., and Khalturin, K. (2007) Dev. Comp. Immunol. 31 1242-1254 [DOI] [PubMed] [Google Scholar]
- 23.Sawada, H., Sakai, N., Abe, Y., Tanaka, E., Takahashi, Y., Fujino, J., Kodama, E., Takizawa, S., and Yokosawa, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1223-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ban, S., Harada, Y., Yokosawa, H., and Sawada, H. (2005) Dev. Biol. 286 440-451 [DOI] [PubMed] [Google Scholar]
- 25.Harada, Y., and Sawada, H. (2007) Mol. Reprod. Dev. 74 1178-1187 [DOI] [PubMed] [Google Scholar]
- 26.Ensslin, M. A., and Shur, B. D. (2003) Cell 114 405-417 [DOI] [PubMed] [Google Scholar]
- 27.Kawamura, K., Nomura, M., Kameda, T., Shimamoto, H., and Nakauchi, M. (1991) Dev. Growth Differ. 33 139-148 [DOI] [PubMed] [Google Scholar]
- 28.Marino, R., Pinto, M. R., Cotelli, F., Lamia, C. L., and De Santis, R. (1998) Development 125 899-907 [DOI] [PubMed] [Google Scholar]
- 29.Sawada, H., Tanaka, E., Ban, S., Yamasaki, C., Fujino, J., Ooura, K., Abe, Y., Matsumoto, K., and Yokosawa, H. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15615-15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan, T. H. (1910) Roux Arch. Entwicklungsmech 30 206-235 [Google Scholar]
- 31.Harada, Y., and Sawada, H. (2008) Int. J. Dev. Biol. 52 637-645 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.