Abstract
Endosialidases (endoNs), as identified so far, are tailspike proteins of bacteriophages that specifically bind and degrade the α2,8-linked polysialic acid (polySia) capsules of their hosts. The crystal structure solved for the catalytic domain of endoN from coliphage K1F (endoNF) revealed a functional trimer. Folding of the catalytic trimer is mediated by an intramolecular C-terminal chaperone domain. Release of the chaperone from the folded protein confers kinetic stability to endoNF. In mutant c(S), the replacement of serine 911 by alanine prevents proteolysis and generates an enzyme that varies in activity from wild type. Using soluble polySia as substrate a 3-times higher activity was detected while evaluation with immobilized polySia revealed a 190-fold reduced activity. Importantly, activity of c(S) did not differ from wild type with tetrameric sialic acid, the minimal endoNF substrate. Furthermore, we show that the presence of the chaperone domain in c(S) destabilizes binding to polySia in a similar way as did selective disruption of a polySia binding site in the stalk domain. The improved catalytic efficiency toward soluble polySia observed in these mutants can be explained by higher dissociation and association probabilities, whereas inversely, an impaired processivity was found. The fact that endoNF is a processive enzyme introduces a new molecular basis to explain capsule degradation by bacteriophages, which until now has been regarded as a result of cooperative interaction of tailspike proteins. Moreover, knowing that release of the chaperone domain confers kinetic stability and processivity, conservation of the proteolytic process can be explained by its importance in phage evolution.
All endosialidases (endo-N-acylneuraminidases (endoN))3 known so far are specialized tailspike proteins of bacteriophages infecting encapsulated Escherichia coli strains. Phages infecting the neuroinvasive bacterium E. coli K1, a major cause of meningitis and sepsis in neonates, have been intensively studied. The dense capsule of these bacteria consists of α2,8-linked polysialic acid (polySia) (1, 2) and provides an efficient infection barrier for coliphages lacking polySia-degrading tailspikes (3).
PolySia is widely expressed also in the vertebrate system, where it forms a post-translational modification on some cell surface proteins. The most prominent polySia carrier is the neural cell adhesion molecule (for review see Refs. 4 and 5). In vertebrates polySia is essential for regular ontogenesis and for maintenance of neuronal plasticity into adulthood (for review see Ref. 6). Because phage-borne endosialidases are the only known enzymes that specifically degrade polySia, they are widely applied in polySia research and particularly in neuroscience (for review see Ref. 4).
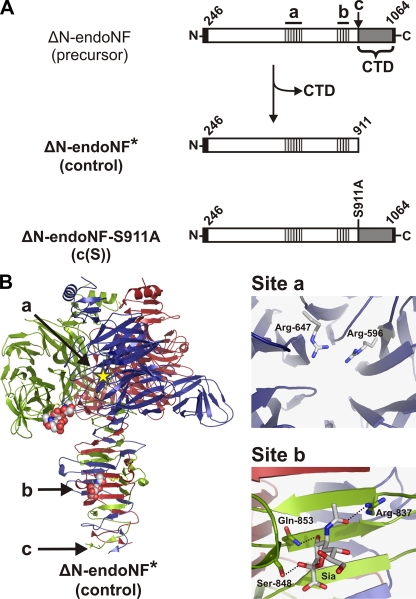
In contrast to exosialidases, which cleave terminal α-ketosidically linked sialic acid residues, endosialidases require α2,8-ketosidically linked sialic acid oligo- or polymers. Five endosialidase genes have been cloned from different E. coli K1 bacteriophages (7-13). The proteins encoded by these genes share a common architecture with three linearly organized domains as follows. (i) An N-terminal capsid binding domain is required to anchor the endosialidase tailspike to the phage particle. This domain varies in length in different phages and is dispensable for enzymatic activity (11, 12). (ii) A highly conserved central catalytic domain that comprises the polySia binding and cleavage activities (14). (iii) A short C-terminal domain (CTD) that functions as an intramolecular chaperone and is released from the matured enzyme (11, 15) (cf. Fig. 1A).
FIGURE 1.
Schematic representation of endoNF maturation and definition of polySia interaction sites in the ΔN-endoNF* structure (adapted from Ref. 14). A, schematic representation of endoNF variants lacking the N-terminal capsid binding domain (ΔN): the endosialidase F precursor (ΔN-endoNF), the processed catalytic part ΔN-endoNF* (Control), and the non-cleavable mutant ΔN-endoNF-S911A (c(S)). The catalytic part is shown as an open bar, the C-terminal chaperone domain (CTD), which is released from the mature protein, in gray. The N-terminal Strep-tag II and C-terminal His6 tag are given in black. The first and the last amino acid number of ΔN-endoNF are depicted diagonally. The active site, a, and the sialic acid binding site, b, are indicated as gray-black hatched boxes. The proteolytic cleavage site Ser-911 is indicated as c. The amino acid substitution S911A is written vertically. B, ribbon diagram of trimeric endoNF, with the monomers colored in red, green, and blue. The active site (a) of the blue subunit is schematically depicted with a yellow star. Spheres represent di-sialic acid and sialic acid bound in the β-barrel domain of the green subunit and the β-prism domain of the red subunit (site b), respectively. The proteolytic cleavage site (Ser-911) is indicated as c. Catalytic amino acids (site a, the view is along the pseudo 6-fold axis of the β-propeller) and residues involved in Sia binding in the stalk domain (Site b) are highlighted.
We have recently solved the crystal structure of the catalytic domain of endosialidase F isolated from the phage K1F (endoNF) (14). The functional enzyme forms a homotrimer with a mushroom-like outline (see also Fig. 1B). The “mushroom cap” is formed by three six-bladed β-propellers each harboring an active site “a.” Additionally, a lectin-like β-barrel domain extends from each β-propeller and contains a di-sialic acid binding site. The “mushroom stalk” is built by the intertwining C-terminal portions and is composed of a triple β-helix that is interrupted by a short triple β-prism domain. Both folds are found in other phage tailspike proteins and have been suggested to mediate the unusual stability of these protein complexes (16). Interestingly, a sialic acid binding site “b” was identified in the stalk domain, indicating that this domain functions not only in stabilizing the endoNF trimer but directly participates in substrate binding (14).
In contrast to the catalytic domain, no structural information for the CTD is available. This intramolecular chaperone is known to be essential for folding of the functional catalytic trimer (11). Interestingly, the CTD is conserved in a number of otherwise unrelated tailspike and fiber proteins (15). Furthermore, the CTD is proteolytically released in all cases at a highly conserved serine residue post-folding (11, 15) (“c” in Fig. 1). We have recently demonstrated that proteolytic release of the CTD confers kinetic stability to the trimeric enzyme (15).
In this study, we demonstrate that release of the CTD also fundamentally influences the mode of endosialidase activity. We show that the wild-type endoNF (control, Fig. 1A) is processive, meaning that the enzyme remains attached to the polymeric substrate for several rounds of cleavage after an initial association event. Introduction of mutations that either prevent the release of the CTD or destroy polySia-binding site b drastically interfere with processivity. Concordant with other processively active enzymes (17-19) the mutant forms of endoNF gained catalytic efficiency toward the soluble polySia substrate. With these data we provide new insight into the molecular basis underlying processivity of endoNF. Moreover, we indorse the notion that the proteolytic release of the chaperone CTD not only confers kinetic stability to the endoNF tailspike, but is also essential for an optimal balance of catalytic and binding functions of this receptor-destroying enzyme.
EXPERIMENTAL PROCEDURES
Materials—pBlueScript SK- was purchased from Stratagene. pET expression vectors were obtained from Novagen. Polysialic acid (polySia) for ELISA-based assays was purified from E. coli K1 membrane fractions according to Decher and coworkers (20). PolySia isolated according to this protocol has been shown to contain a lipid anchor at the reducing end (20), which allows efficient immobilization of polySia on microtiter plates (14). A recently developed protocol (21) was used to isolate lipid-free polySia (termed “soluble polySia” in this study) from the supernatants of E. coli K1 cultures. PolySia chains of this preparation contain >130 residues (21). Sialic acid and oligomeric α2,8-linked sialic acid (oligoSia) with a degree of polymerization of 2 to 4 were obtained from Nacalai Tesque (Kyoto, Japan).
Bacteria—The wild-type strain E. coli B2032/82 serotype K1 is an original clinical isolate obtained from the Dept. of Medical Microbiology of the Medizinische Hochschule Hannover (Hannover, Germany) (22). E. coli BL21-Gold(DE3) were purchased from Stratagene.
Site-directed Mutagenesis—Mutagenesis was performed by PCR using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's guidelines with the following primer pairs (mutated nucleotides are shown in uppercase): KS084 5′-c ccg gca ggg cag GCA atc ata ttt tgc ggg gg-3′ and KS085 5′-cc ccc gca aaa tat gat TGC ctg ccc tgc cgg g-3′ for endoNF-R837A; KS080 5′-t gaa ggc acc agt Gca acg act ggc gc-3′ and KS081 5′-gc gcc agt cgt tgC act ggt gcc ttc a-3′ for endoNF-S848A; KS082 5′-tca acg act ggc gca GCg att acg cta tat ggt gc-3′ and KS083 5′-gc acc ata tag cgt aat cGC tgc gcc agt cgt tga-3′ for endoNF-Q853A. Plasmids containing the corresponding sequence encoding for endoNF protein lacking the N-terminal capsid binding domain (ΔN-endoNF, cf. Ref. 15) were used as a template. The sequence identity of all PCR products was confirmed by sequencing. PCR products were ligated into BamHI/XhoI sites of the expression vector pET22b-Strep, a modified pET22b vector containing the sequence encoding an N-terminal Strep-tag II followed by a thrombin cleavage site (WSHPQFEKGALVPR'GS) and a C-terminal His6 tag. Enzymatically inactive endoNF variants were generated by exchanging the 1009-bp endoNF NdeI/XhoI fragment in pET22b-Strep containing the gene encoding for ΔN-endoNF-R596A/R647A (14) with the corresponding fragment from the respective endoNF binding site mutant.
Protein Expression and Purification—Proteins were expressed in E. coli BL21-Gold(DE3) in the presence of 100 μg/ml Carbenicillin. Bacteria were cultivated in PowerBroth (Athena Enzyme Systems) at 30 °C. Expression of endoNF was induced by adding 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at an optical density (A600) of 1.5, and bacteria were harvested 6-7 h after induction. For the analysis of soluble and insoluble proteins, bacteria were lysed by sonication. Soluble and insoluble fractions were obtained after centrifugation (22,000 × g, 20 min, 4 °C). Protein purification was performed as described previously (15).
Size-exclusion Chromatography—Size-exclusion chromatography was carried out on a Superdex 200 HR 10/30 column (Amersham Biosciences) equilibrated with 10 mm sodium phosphate buffer, pH 7.4. The column was calibrated using the Gel Filtration Molecular Weight Markers (MW-GF-200) from Sigma.
SDS-PAGE—SDS-PAGE was performed at 85 V and 20 °C. Proteins were incubated in the presence of 1% SDS for 5 min at 95 °C prior to loading to the gel. For the detection of SDS-resistant complexes, the incubation step was omitted. Proteins were stained using RotiBlue (Carl Roth GmbH) according to the manufacturer's guidelines. Scanning was performed with a LI-COR Odyssey Infrared imaging system.
NMR—All NMR experiments were performed on a Bruker Avance 600-MHz spectrometer, equipped with a 5-mm TXI probe with triple axis gradients. Deuterated phosphate buffer (10 mm sodium phosphate, pH 7.4, 150 mm NaCl) was used for all NMR experiments. The measurements were performed at 298 K without sample spinning. 1H NMR spectra were acquired with 32 scans and a 2-s relaxation delay over a spectral width of 6000 Hz. Solvent suppression of the residual semiheavy water (HDO) peak was achieved by low power presaturation during the relaxation delay. Data acquisition and processing were performed using XWINNMR software. Chemical shift assignment of DP4 oligoSia was achieved by COSY, TOCSY, NOESY, and HSQC NMR experiments. Digestion of 1 mg of DP4 oligoSia was monitored by acquiring 1H NMR spectra in 10-min time intervals over 16-h postincubation with 55 pmol of endoNF.
Analysis of OligoSia and PolySia Cleavage Products by Anion-exchange Chromatography—To monitor cleavage of DP3 or DP4 oligoSia 400 μg of the oligomer were incubated with 2 pmol of endoNF (control or mutants) in 100 mm sodium phosphate buffer, pH 5.1, for 16 h at 37 °C. To stop the reaction 1 volume of cold (-80 °C) absolute ethanol was added, and samples were frozen. Protein precipitates were removed by centrifugation (3 min at 19,000 × g), and supernatants were filtered (0.22 μm) before chromatographic separation. One-half volume of the supernatant was loaded onto anÄKTA design system equipped with a Mono Q HR 5/50 column (Amersham Biosciences) using 10 mm Tris-HCl, pH 8.0, as running buffer. Elution was performed using a segmented linear gradient: 2 ml at 0%, 2 ml at 0-8%, 7 ml at 8-20%, 8 ml at 20-28%, 1 ml at 28-100%, and 5 ml at 100% of 10 mm Tris-HCl, pH 8.0, 1 m NaCl at a flow rate of 1 ml/min. Absorbance of oligoSia was monitored at 214 nm. The elution profiles were processed with MS Excel and Prism 4.03 (GraphPad Software Inc.). For the analysis of soluble polySia cleavage, 400 μg of substrate were incubated with 2 pmol of endoNF (control or mutants) for appropriate incubation times. Samples were processed as described above for oligoSia cleavage, but separation on Mono Q HR 5/50 column was performed using a different segmented linear gradient: 2 ml at 0%, 2 ml at 0-8%, 7 ml at 8-20%, 26 ml at 20-45%, 1 ml at 45-100%, and 5 ml at 100% of 10 mm Tris-HCl, pH 8.0, 1 m NaCl.
Determination of Endosialidase Activity Using Soluble PolySia as Substrate—The enzymatic activity of purified endoNF variants was determined by means of the thiobarbituric acid assay as described previously (11, 23). Serial dilutions of sialic acid monomer were used as a standard. Kinetic parameters Km and kcat values were determined globally in Prism 4.03 (GraphPad Software Inc.).
Determination of Endosialidase Activity Using Surface-bound PolySia as Substrate—Cellstar 96-well plates (Greiner BIO-ONE) were coated with 40 ng of polySia in PBS (10 mm sodium phosphate buffer, pH 7.4, 150 mm NaCl) per well for 1 h. Blocking was performed with 200 μl/well 1% (w/v) BSA in PBS. Coating and blocking were each followed by three washing steps with PBS to remove unbound material. Purified active endoNF variants were applied in 40 μl per well in serial dilutions in 1% BSA/PBS and incubated for 30 min at 37 °C. EndoNF and cleavage products of polySia were removed by three washing steps, each with 200 μl of PBS for 5 min with orbital shaking at 1000 rpm. Residual polySia was detected using 200 ng/well monoclonal antibody 735 (monoclonal antibody 735 (24) in 1% BSA/PBS) and subsequently goat anti-mouse antibody-conjugated to horseradish peroxidase (Southern Biotechnology Associates, 1:2000 in 1% BSA/PBS). Each antibody was incubated for 1 h at room temperature with orbital shaking at 1000 rpm and followed by three washing steps with PBS. Chromogenic reaction was performed with 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Roche Applied Science) according to the manufacturer's guidelines. Data were analyzed using Prism 4.03.
In Vitro Binding Assay—Cellstar 96-well plates (Greiner BIO-ONE) were coated as described in the previous section. Serial dilutions of inactive endoNF variants were applied in 40 μl per well to polySia-coated microplates and incubated for 1 h at room temperature with orbital shaking at 1000 rpm. After washing three times each with 200 μl of PBS for 5 min at 1000 rpm on a shaker the N-terminal Strep-tag II of EndoNF was detected by incubating Strep-Tactin conjugated to horseradish peroxidase (IBA GmbH, Göttingen, Germany; 1:5000 in 1% BSA/PBS) for 1 h at room temperature with orbital shaking at 1000 rpm followed by three PBS washing steps. Color development was performed with 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Roche Applied Science). Data were analyzed using Prism 4.03.
RESULTS
The Enzymatic Activity of Proteolytically Processed and Unprocessed endoNF—Previous work has shown that proteolytic processing of endoNF is not a prerequisite for enzymatic activity (11). However, it remains unknown whether the release of the CTD affects the kinetics of polySia digestion.
To address this question, the enzymatic activities of the mature, proteolytically processed endoNF (ΔN-endoNF*, control) and the non-cleavable mutant ΔN-endoNF-S911A (c(S), Fig. 1A) were compared. All endosialidases used throughout this study are lacking the N-terminal capsid binding domain (indicated by ΔN). As shown previously, this does not influence complex formation, activity, and thermostability (11, 14, 15). The scheme presented in Fig. 1A describes the nomenclature, and Table 1 summarizes the details of endoNF variants generated in the course of this study. For efficient purification of unprocessed (c(S)) and processed enzymes, the proteins were expressed with N-terminal Strep-tag II and C-terminal His6 tag as previously published (14).
TABLE 1.
Notation of EndoNF variants
The ΔN-endoNF variants listed below were analyzed in this study. A convenient notation is introduced to facilitate reading (left column), with a, active site (lower half), b, binding site in the β-prism domain, and c, proteolytic cleavage site at Ser-911 (according to Fig. 1). The respective amino acids exchanged to alanine (see right column) are given in parentheses. All variants contain an N-terminal Strep-tag II. The non-cleavable mutants c(S) and a(RR)/c(S) additionally contain a C-terminal His6 tag.
| Protein | Amino acid substitutions |
|---|---|
| Control | |
| b(R) | R837A |
| b(S) | S848A |
| b(Q) | Q853A |
| b(RS) | R837A/S848A |
| b(RQ) | R837A/Q853A |
| b(SQ) | S848A/Q853A |
| b(RSQ) | R837A/S848A/Q853A |
| c(S) | S911A |
| Control-a(RR) | R596A/R647A |
| a(RR)/b(R) | R596A/R647A/R837A |
| a(RR)/b(S) | R596A/R647A/S848A |
| a(RR)/b(Q) | R596A/R647A/Q853A |
| a(RR)/b(RS) | R596A/R647A/R837A/S848A |
| a(RR)/b(SQ) | R596A/R647A/S848A/Q853A |
| a(RR)/c(S) | R596A/R647A/S911A |
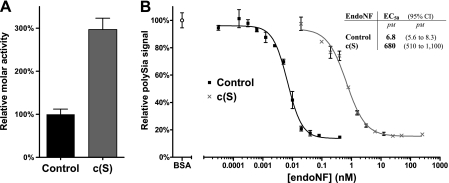
Enzymatic activities of purified proteins were determined in two assay systems. The thiobarbituric acid assay was used to monitor enzymatic activities on soluble polySia (lipid anchor-free polySia prepared according to Ref. 21) (Fig. 2A). An end-point ELISA-based assay was applied to measure enzymatic activity on surface-bound polySia (Fig. 2B). Microtiter plates were coated with polySia containing a lipid anchor (20) to allow efficient attachment of polySia to the solid phase (14). After endoNF digestion and removal of the enzyme, remaining substrate was detected with the polySia-specific monoclonal antibody 735, which binds to α2,8-linked polySia ≥8 residues (22, 24, 25).
FIGURE 2.
EndoNF activity assays. A, relative molar activities of ΔN-endoNF* (Control) and ΔN-endoNF-S911A (c(S)) as determined in the thiobarbituric acid assay. Data are means ± S.D. of two independent experiments performed in duplicates. The control was set to 100%. B, enzymatic activity of ΔN-endoNF* (Control) and ΔN-endoNF-S911A (c(S)) determined in an ELISA-based assay. In each experiment, maximal loading of microtiter plates with polySia was determined after BSA blocking and set to 100%. Samples are plotted as relative values and represent surface-bound polySia remaining after treatment with serial enzyme dilutions (for details see “Experimental Procedures”). Each value represents the mean of two independent experiments performed in duplicates. The concentration (x axis) is scaled logarithmically. Error bars represent ±S.D. Inset table: EC50 values determined for the respective endoNF variant are giving the protein concentration of half-maximal polySia removal.
Strikingly, a 3-times higher molar activity of c(S) over control was found using soluble polySia substrates (Fig. 2A), whereas the activity of c(S) using immobilized polySia was drastically diminished. As shown in Fig. 2B, 680 pm c(S) and only 6.8 pm control were required for half-maximal depolymerization of the solid-phase-bound substrate. To further interpret these apparently contradictory results, we investigated the possibility of CTD-induced conformational changes in the catalytic site of the mutant c(S) in kinetic studies. However, because endoNF possesses secondary substrate binding sites (14) (Fig. 1B), the determination of kinetic parameters required the use of the minimal substrate that is large enough to be bound and cleaved, but too short to interfere with secondary binding sites.
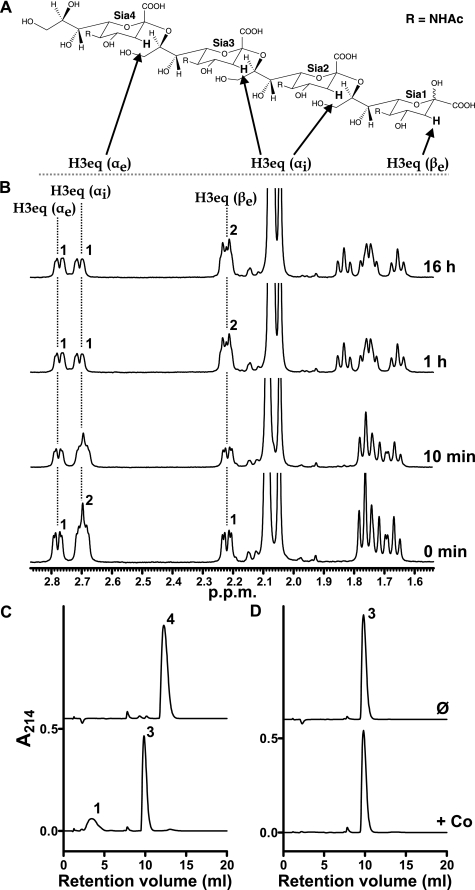
Sialic Acid Tetramer Is the Minimal endoNF Substrate—Previous studies by Troy and co-workers (26) have shown that endosialidase purified from bacteriophage K1F cleaves α2,8-linked sialic acid oligomers with a minimum degree of polymerization of five (DP5 oligoSia).
To confirm the minimal substrate for the purified recombinant ΔN-endoNF* (control) we developed a sensitive and robust 1H NMR-based assay. In this system the distinct chemical shifts of the H3 equatorial protons (H3eq) of the four sialic acid units of DP4 oligoSia were monitored (Fig. 3, A and B). In general, sialic acid moieties that constitute the reducing end of polySia chains are predominantly β-configured resulting in a chemical shift of the corresponding H3eq (βe) at 2.22 ppm. Non-reducing end residues exhibit pure α-configuration with a chemical shift for the H3eq proton at 2.78 ppm (H3eq (αe)). The H3eq protons of the two internal α-configured sialic acid residues (H3eq (αi)) that form the major part of α2,8-linked polySia are overlapped with an identical chemical shift of 2.70 ppm. The relative integrals obtained for the H3eq signals of DP4 oligoSia at t = 0 min revealed a ratio of 1:2:1 for H3eq (αe):H3eq (αi):H3eq (βe) (see Fig. 3B). Addition of ΔN-endoNF* to DP4 oligoSia results in a 1:1:2 integral ratio for the H3eq protons after 1-h incubation. This observation leads to the assumption that the sialic acid tetramer was cleaved by the endosialidase. However, an H3eq integral ratio of 1:1:2 did not correspond to a symmetrical cleavage of DP4 oligoSia into two DP2 oligoSia molecules (expected proton integral ratio 2:0:2). Instead, the observed proton integral ratio of 1:1:2 is a strong indication that DP4 oligoSia was cleaved into one DP3 oligoSia (one H3eq (αe) and one H3eq (αi) proton) and one monomeric sialic acid molecule (one H3eq (βe) proton). Moreover, the analysis of the spectral data that were collected after 1 and 16 h clearly excluded a potential cleavage of DP3 oligoSia (expected H3eq integral ratio 1:0:3). The results obtained by 1H NMR spectroscopy were corroborated in a parallel anion-exchange chromatography experiment, that showed the DP4 oligoSia was cleaved into monomer and DP3 oligoSia after incubation with the active control enzyme (+ Co, Fig. 3C), whereas no cleavage products were found when this experiment was carried out with DP2 (data not shown) and DP3 (Fig. 3D) oligoSia.
FIGURE 3.
Determination of the minimal endoNF substrate. A, structure of DP4 oligoSia. The equatorial protons at position C3 of the individual sialic acid units of DP4 are depicted. B, 1H NMR spectra of 1 mg of DP4 oligoSia incubated with 55 pmol of ΔN-endoNF* (control). 1H NMR spectra were acquired at different time intervals in deuterated 10 mm phosphate buffer, 150 mm NaCl, pH 7.4, in D2O at 298 K. Numbers above peaks indicate the calculated relative integrals. C and D, monitoring of oligoSia cleavage products by Mono Q anion-exchange chromatography. Samples of 200 μg of DP4 (C) or DP3 oligoSia (D) were incubated either with buffer (Ø) or 1 pmol of ΔN-endoNF* (+Co), respectively, for 16 h at 37 °C. Cleavage products were separated on a Mono Q HR 5/50 column. Elution profiles are separated by an appropriate y axis offset. Numbers above peaks indicate the DP of oligoSia. Experiments were carried out in duplicates with identical results.
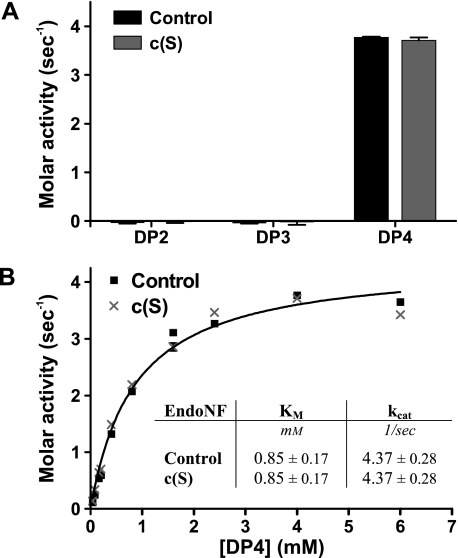
Control and Mutant endoNF Exhibit Similar Kinetics with the Minimal Substrate DP4 OligoSia—Kinetic parameters were determined using the thiobarbituric acid assay and DP4 oligoSia as the minimal substrate. As shown in Fig. 4A, DP4 oligoSia (but not DP2 or DP3 oligoSia) was efficiently hydrolyzed by both control endoNF and c(S) mutant. Importantly, identical kinetic parameters (Fig. 4B; Km = 0.85 mm and kcat = 4.37 s-1) were determined for the two enzyme variants. Although this experiment clearly supported the notion that the presence of the CTD in c(S) has no influence on the conformation and/or stability of the catalytic center, it did not explain the dramatic differences in activity observed when the enzymes were tested with polySia in either solution or immobilized form (see Fig. 2, A and B).
FIGURE 4.
Control enzyme (ΔN-endoNF*) and mutant c(S) cleave the minimal substrate DP4 with identical kinetics. A, molar activities on DP2-4 oligoSia were determined by the thiobarbituric acid assay in two independent experiments. B, kinetic parameters of endoNF variants on the minimal substrate DP4 oligoSia were determined by the thiobarbituric acid assay (in two independent experiments). The curves were best fitted globally with shared parameters. Inset table in B, kinetic parameters determined for both endoNF variants ± 95% confidence interval. Values in A and B express the amount of released reducing ends of sialic acid in mol per mol of enzyme and per second (1/s). Error bars represent ±S.D.
Proteolytic Maturation and Integrity of Site b Are Essential for Efficient PolySia Binding—One of the polySia interaction sites outside the catalytic center identified previously in the crystal structure (site b) is located in the stalk domain in close proximity to site c where the CTD is proteolytically released (see Fig. 1A). Binding studies were carried out to address the question of whether the presence of the CTD may affect polySia binding to site b, thus leading to a reduction in the stability of the enzyme-polySia complex and consequently in the observed changes in the kinetics of polymer cleavage. Essential in this study was the use of an enzymatically inactive variant of endoNF. A respective mutant termed a(RR) for active site mutant R596A/R647A (see Table 1 and inset Site a in Fig. 1B) has previously been generated based on structural information (14). Although the enzymatic activity is abolished in a(RR), the maturation of the protein into SDS-resistant trimeric complexes and binding to polySia is unaffected (14).
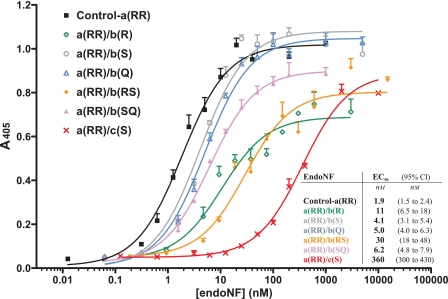
The c(S) mutation was introduced into the a(RR) background giving clone a(RR)/c(S). Purified proteins produced from both clones (a(RR) and a(RR)/c(S)) were tested for binding to immobilized polySia. The half-maximal effective concentration (EC50) of 1.9 nm demonstrated high affinity binding of a(RR) to polySia. In contrast, the mutant a(RR)/c(S) showed a drastically reduced binding with an EC50 of 360 nm (see Fig. 5, black and red curves).
FIGURE 5.
Binding of enzymatically inactive ΔN-endoNF-a(RR) variants to surface-bound polySia. The binding assay was performed as described under “Experimental Procedures” in four independent experiments. The concentration (x axis) is scaled logarithmically. Error bars above the curves represent ±S.E. Inset table: EC50 values determined in the in vitro binding assay of the respective inactive endoNF variant are giving the protein concentration of half-maximal binding. 95% CI, 95% confidence interval.
To further investigate whether the reduction in binding affinity is a consequence of interference with binding site b, this site was selectively disrupted in the a(RR) mutant. The information provided by the crystal structure (14) was used to guide the production of mutants. Amino acid residues that build up binding site b (Arg-837, Ser-848, and Gln-853) are depicted in the inset Site b in Fig. 1B. These residues were individually and in combination exchanged to alanine resulting in a total of seven b-mutants (see Table 1). In an attempt to produce the purified recombinant proteins the double b-mutant R837A/Q853A (a(RR)/b(RQ)) as well as the triple b-mutant R837A/S848A/Q853A (a(RR)/b(RSQ)) were found to be insoluble (data not shown), whereas all other variants were soluble. Moreover, analyses carried out by size exclusion chromatography (supplemental Table S1) demonstrated that all soluble b-mutants formed proteolytically matured homotrimers. However, homotrimers formed from b-mutants, did not attain SDS resistance, with the single exception of a(RR)/b(S) as determined by SDS-PAGE (supplemental Fig. S1). SDS resistance was described previously as a criterion for the kinetically stabilized ΔN-endoNF* (11, 15).
Binding of inactive b-mutants to surface-bound polySia was analyzed in the ELISA-based assay (Fig. 5). EC50 values varying between 4.1 nm for the SDS-resistant mutant a(RR)/b(S) (gray curve, cf. supplemental Fig. S1) and 30 nm for the double b-mutant a(RR)/b(RS) (orange curve) clearly demonstrated that the integrity of the binding site of the stalk domain is essential to optimize binding between endoNF and polySia chains. Interestingly, the single point mutation R837A of the mutant a(RR)/b(R) already increased the EC50 by a factor of 5 (green curve), and the defect was potentiated in the double b-mutant a(RR)/b(RS) (orange curve). Combined with the reduced protein stability found in SDS-PAGE analysis (supplemental Fig. S1) these data emphasize that Arg-837 plays an important role for both protein folding (cf. Ref. 15) and formation of the Sia binding site.
In total, mutations introduced into polySia binding site b decreased polySia binding. This effect is similar to the effect caused by lack of proteolytic processing in c(S). Therefore, the CTD preserved in c(S) masks or destabilizes site b and weakens the “lectin properties” of endoNF.
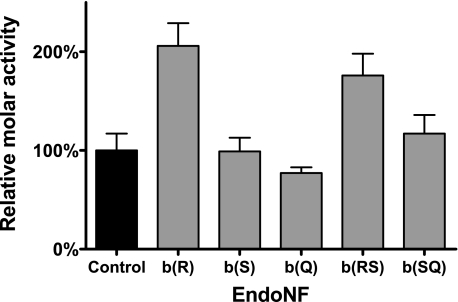
Depletion of Binding Site b Increases endoNF Activity toward Soluble Substrate—As shown for c(S), the decreased binding to polySia results in an increased enzymatic activity using soluble polySia (Fig. 2A). To test whether this is also the case for proteins carrying mutations in site b we generated enzymatically active forms of the soluble b-mutants (see Table 1). Purified proteins were analyzed in the thiobarbituric acid assay in comparison to the control (Fig. 6). In accordance with our hypothesis, protein variants with mutations that significantly affected polySia binding (cf. Fig. 5) showed significantly increased molar activities (b(R) and b(RS)), while proteins with only minor binding deficits (b(S), b(Q), and b(SQ)) were active within the control range.
FIGURE 6.
Relative molar activities of ΔN-endoNF* variants with single and double amino acid exchanges in the sialic acid binding site b. Activities were determined by the thiobarbituric acid assay using soluble polySia as substrate. Data are means ± S.D. of two independent experiments performed in duplicates. The molar activity of control was set to 100%.
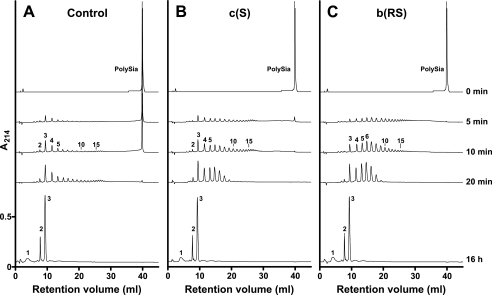
Mutations in the Stalk Domain Influence Processivity of endoNF—Based on the observation that bacteriophages infecting encapsulated bacteria create narrow paths through the thick capsules, capsule degradation could occur in a processive manner (27-29). A suggestion for the molecular mode of action has recently been provided in line with the reconstruction of the coliphage K1E (30). However, the question of whether processivity is an intrinsic feature of the tailspike or imposed upon the phage particle due to the interplay of six copies of the tailspike protein has not been investigated as yet. To address this point, soluble polySia was digested with equimolar amounts of control endoNF and the mutants c(S) and b(RS). Reaction products were monitored by anion-exchange chromatography in a time-dependent manner (Fig. 7). The control enzyme released short oligoSia species with DP3 as the main product. The long-chain starting material was still detectable after 10 min (Fig. 7A). Conversely, in digests performed with the stalk domain mutants c(S) and b(RS) long-chain polySia was cleaved more rapidly. After <5 min all starting material was digested into fragments with DPs ranging from 3 to >15. Interestingly, the production of DP3 oligoSia appeared to have similar kinetics in digests carried out with the control and c(S) protein (Fig. 7, compare A and B), whereas cleavage products generated by b(RS) were more random (Fig. 7C). After 16 h the digestions were complete with DP1, DP2, and DP3 oligoSia representing the end products of endoNF digestion (cf. Figs. 3 and 4). The results of this experiment prove processivity for ΔN-endoNF* and confirm that multipoint binding of the polySia chain is crucial to maintain processivity.
FIGURE 7.
Cleavage characteristics of control (ΔN-endoNF*) and mutant endoNF variants. Anion-exchange chromatography was used to directly monitor the cleavage products generated by control and mutant endoNF. A-C, after digest of 200 μg of polySia with 1 pmol of the respective endoNF variant for the indicated time points cleavage products as well as the polymeric substrate (PolySia, 0 min) were separated on a Mono Q HR 5/50 column. Elution profiles are drawn at the same scale and separated by an appropriate y axis offset. Numbers above peaks indicate the DP of oligoSia. The experiment was repeated twice with identical results.
DISCUSSION
Many pathogenic bacteria are decorated by a dense polysaccharide capsule, which protects the bacterium from desiccation and immune surveillance (31). The capsular polysaccharide layer can be up to 400 nm thick and provides an efficient shield against bacteriophage infections (3, 32). However, in parallel to the evolution of bacterial capsules, specialized phages emerged with tailspike proteins that carry polysaccharide hydrolase or lyase activities to allow depolymerization of the respective host capsule. Electron micrographs of phages attacking encapsulated bacteria revealed that penetration of the capsule by the phage particle mostly occurs in a processive and unidirectional manner leading to the formation of a narrow tunnel (27-29). Based on the three-dimensional cryoelectron microscopy reconstruction of the E. coli K1-specific phage K1E, processive degradation of the capsule was suggested to proceed by an intermolecular interplay of the six copies of endosialidase tailspikes that are symmetrically arranged around the tail (30).
Using endoNF, the endosialidase of coliphage K1F, we now show that the isolated tailspike protein exhibits processivity by itself, giving new insight into the molecular machinery enabling phages to infect encapsulated bacteria. We demonstrate that endoNF processivity is based on its unique multidomain structure and depends on an intact substrate binding site b located in the β-prism of the stalk domain.
Previously, we have postulated that site b plays an important role in fixing the phage particle to the outer membrane of the host by binding the oligomeric remnants of the depolymerized capsular polysaccharide (14). By abrogating site b we now show that this site is not only important for efficient binding of endoNF to polySia but also converts endoNF into a processive enzyme. The crystal structure of endoNF in complex with oligoSia (14) suggested that polySia is wrapped around the enzyme with the non-reducing end pointing toward the active site and the reducing end toward the C terminus of the stalk domain. During processive cleavage of polySia, site b acts in advance of the active site and guides endoNF along the polySia chain in a directed manner from the non-reducing to the reducing end. Because in vivo polySia is fixed by its reducing end to the outer membrane of E. coli K1, the orientation of site b toward the reducing end ensures that endoNF and thereby the whole phage is not detached from the bacterium during capsule depolymerization. Two experimental approaches have provided clear evidence for this model. Firstly, the binding studies displayed in Fig. 5 clearly show reduced binding affinity to surface-bound polySia for the mutant b(RS) lacking a functional site b. Secondly, the assay shown in Fig. 7 directly monitored processivity (19, 33, 34) and highlighted significant differences between control enzyme and the mutant b(RS). When long polySia chains (DP >130) were digested by the control enzyme the predominant initial cleavage product was found to be DP3 oligoSia and highly polymeric substrate persisted over >10 min. In contrast, when digests were performed with the mutant b(RS), long polySia chains disappeared rapidly (<5 min) paralleled by the appearance of random-sized cleavage products. The cleavage pattern observed for the control enzyme indicates a prolonged association between enzyme and substrate that allows several cleavage steps before dissociation, which results in short cleavage products. By contrast, mutant b(RS) exhibited an increased dissociation probability resulting in an increased random fragmentation of the polymeric substrate. Thus, the reduced binding affinity caused by alanine substitution of two critical amino acid residues (Arg-837 and Ser-848) within site b is directly translated in a loss of processivity.
Similarly, reduced binding to surface-bound polySia and loss of processivity were also observed for the non-cleavable endoNF variant c(S), which mimics the unprocessed precursor protein. The possibility that the presence of the CTD induces conformational changes within the active site could be excluded by the fact that identical kinetic data were obtained for control and mutant enzyme with the minimal substrate DP4 oligoSia. Consequently, the 190-fold decrease in binding affinity observed for c(S) could be interpreted due to loss of secondary polySia binding sites, i.e. binding sites that are located outside the catalytic center. Because site b is located in close proximity to the CTD, a direct interference between CTD and site b is most likely.
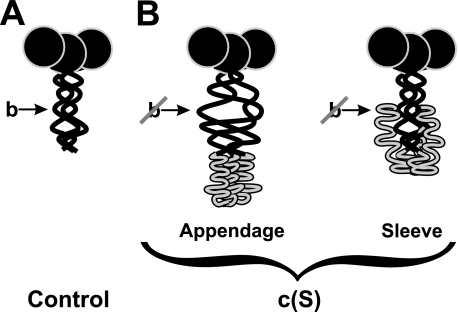
As depicted schematically in Fig. 8, we propose two models to explain this interference. In the “Appendage” model (Fig. 8B), the CTD is shown as a linear extension that keeps the stalk domain in an immature state that is incompatible with proper binding of polySia (cf. Fig. 8A schematically showing the mature protein). Only cleavage and release of the CTD provides the folding enthalpy necessary to attain the mature conformation of the stalk and to generate binding site b. In the “Sleeve” model, the CTD is wrapped around the maturely folded stalk, thereby masking site b.
FIGURE 8.
Two proposed folding models of structural influence of the CTD on site b. Schematic representations of endoNF variants with the β-propeller shown as black circle, the stalk domain as black curve, and the CTD is shown in gray (adapted from Ref. 15). A, ΔN-endoNF* (Control) with proper folded stalk domain (location of site b is indicated by an arrow). B, two possible folding models of the CTD (gray) in (c(S)) in which site b is affected: in the Appendage model the preserved CTD induces a conformational change in the stalk domain, which abolishes the binding site b. The Sleeve model suggests that the CTD interacts with regions of the stalk domain and hides site b.
Both previous and current data support the Sleeve model. Firstly, preliminary data obtained for crystals of the c(S) mutant4 revealed no significant difference in the N-terminal portion of endoNF (amino acids 246-911) compared with the previously described structure of the processed enzyme (14). Additional electron density corresponding to the CTD was found not only at the C-terminal end of the stalk but also surrounding the stalk domain. These findings argue against the Appendage model, which is based on a misfolded stalk domain.
Secondly, we have previously shown that the chaperone function of the CTD essentially depends on the integrity of a series of highly conserved amino acid residues. Single point mutations of these positions were sufficient to prevent generation of folded functional endosialidases (11, 15). It is likely that CTD-assisted folding involves the formation of protein-protein interactions between CTD and the N-terminal protein portion, which, as proposed in the Sleeve model, could take place between CTD and stalk domain. Proteolytic cleavage may be required to terminate these interactions for instance by enabling conformational changes in the CTD that facilitate dissociation from the stalk.
A third argument for the Sleeve model is the abrogation of SDS resistance observed for site b-mutants (cf. supplemental Fig. S1). Single mutations of Arg-837 and Gln-853 resulted in SDS-sensitive protein complexes. This observation was unexpected, because the targeted residues protrude out of the stalk, do not interact with other amino acids, and thus should not play a crucial role in stabilizing the rigid trimeric stalk domain (14). However, if these amino acids contribute to the interaction between stalk and CTD during folding, alanine substitutions could interfere with CTD-assisted folding, resulting in misfolded and therefore less stable enzyme complexes. In line with this hypothesis, the site b double mutant b(RQ) and triple mutant b(RSQ) were expressed exclusively as insoluble proteins.
The unprocessed variant c(S) is noteworthy, because it showed a 12-fold lower affinity toward solid-phase-bound polySia than the b(RS) mutant. One explanation for this might be that binding of polySia to the stalk is not restricted to interactions with residues Arg-837 and Ser-848 but mediated by a number of weak protein-carbohydrate interactions that were not detected in endoNF crystals soaked with DP5 oligoSia (14). In the c(S) variant, the CTD might conceal a large part of the stalk domain, as proposed by the Sleeve model, and thus prevent polySia binding more efficiently than abrogation of site b by two amino acid exchanges.
Interestingly, endoNF mutants that showed reduced binding to surface-bound polySia exhibited increased enzymatic activity toward soluble polySia, strongly arguing that neither CTD release nor integrity of site b is essential for efficient degradation of soluble substrate. However, interference with binding of polySia to the stalk seems to facilitate dissociation from the polymer, thereby allowing faster re-association with new substrate. A gain of cleavage efficiency toward soluble substrate at the expense of processivity and activity toward solid-phase bound substrate has been described for other enzymes that degrade biopolymers like cellulose (cellulases) and chitin (chitinases) (19, 33, 35-38). Processivity in these studies was measured by the fast appearance of short oligomeric cleavage products and a simultaneous slow decrease of long polymer chains (17-19). In the present study, this assay system was adapted to provide direct proof for the processive activity of endoNF.
In line with the fact that endoNF is an endo-acting sialidase oligomeric cleavage products were found for both processive and non-processive variants. By contrast, if the minimal acceptor substrate DP4 was used, a sialic acid monomer was released, opening the question whether this residue is released from the reducing end by endo-activity or from the non-reducing end by a not yet described exo-activity. However, cleavage of DP4 also produced DP3, which is the predominant cleavage product released by processive endoNF from long polySia chains. Therefore, it is quite likely that endoNF acts exclusively in an endo-manner and that a glycosidic linkage within oligo- or polySia can be hydrolyzed only if the cleavage site is followed by at least three sialic acids at the non-reducing end.
Based on the observed frequent horizontal gene transfer the concept of modular evolution of bacteriophages has been developed (39-41). In line with this concept, endosialidase genes seem to have evolved by genomic insertion of a bacterial exosialidase into a tailspike gene (12). The polySia binding site b within the tailspike-borne stalk domain might have evolved subsequently by a series of point mutations. The multifunctional structure generated in this way gave rise to an effective adhesin for attachment to the host surface and to a processive degradation machinery for unidirectional penetration of the capsule. In addition, the multidomain structure confers high protein stability by combining the sialidase β-propeller with an intertwining β-helix (14, 15), which, in summary, crucially impacts phage fitness.
Structural data at the atomic level are also available for the tailspike endoglycosidases of Salmonella phage P22, Shigella flexneri phage Sf6, and coliphage HK620 (42-44), which all infect bacterial hosts decorated with a dense layer of lipopolysaccharide. Similar to endosialidases, the tailspike proteins of these phages combine receptor-destroying activity with substrate binding subsites. Although no direct proof is available yet, the organization of binding subsites in these tailspikes implicates a contribution in sliding along the polymeric substrate. Therefore, it is conceivable that processivity is an inherent feature of tailspike proteins with a polysaccharide depolymerase activity.
Supplementary Material
Acknowledgments
Andrea Bethe is acknowledged for excellent technical assistance.
This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG) in the framework of DFG Research Unit 548 (Grants Ge801/7-1 and 7-2).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: endoN, endosialidase (endo-N-acylneuraminidase); BSA, bovine serum albumin; CTD, C-terminal domain; DP, degree of polymerization; EC50, half-maximal effective concentration; ΔN-endoNF, endo-N-acylneuraminidase or endosialidase (EC 3.2.1.129) of coliphage K1F lacking the N-terminal 245 amino acids (ΔN) of the capsid binding domain (14, 15); ΔN-endoNF*, catalytic part (amino acids 246-911) proteolytically released from ΔN-endoNF (11, 15); oligoSia, oligosialic acid; polySia, polysialic acid; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline.
K. Stummeyer, A. Dickmanns, M. Mühlenhoff, R. Gerardy-Schahn, and R. Ficner, unpublished data.
References
- 1.Robbins, J. B., McCracken, G. H., Jr., Gotschlich, E. C., Orskov, F., Orskov, I., and Hanson, L. A. (1974) N. Engl. J. Med. 290 1216-1220 [DOI] [PubMed] [Google Scholar]
- 2.Sarff, L. D., McCracken, G. H., Schiffer, M. S., Glode, M. P., Robbins, J. B., Orskov, I., and Orskov, F. (1975) Lancet 1 1099-1104 [DOI] [PubMed] [Google Scholar]
- 3.Scholl, D., Adhya, S., and Merril, C. (2005) Appl. Environ. Microbiol. 71 4872-4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutishauser, U. (2008) Nat. Rev. Neurosci. 9 26-35 [DOI] [PubMed] [Google Scholar]
- 5.Kleene, R., and Schachner, M. (2004) Nat. Rev. Neurosci. 5 195-208 [DOI] [PubMed] [Google Scholar]
- 6.Hildebrandt, H., Mühlenhoff, M., and Gerardy-Schahn, R. (2008) Neurochem. Res. 10.1007/s11064-008-9724-7 [DOI] [PubMed]
- 7.Gerardy-Schahn, R., Bethe, A., Brennecke, T., Mühlenhoff, M., Eckhardt, M., Ziesing, S., Lottspeich, F., and Frosch, M. (1995) Mol. Microbiol. 16 441-450 [DOI] [PubMed] [Google Scholar]
- 8.Long, G. S., Bryant, J. M., Taylor, P. W., and Luzio, J. P. (1995) Biochem. J. 309 543-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machida, Y., Hattori, K., Miyake, K., Kawase, Y., Kawase, M., and Iijima, S. (2000) J. Biosci. Bioeng. 90 62-68 [DOI] [PubMed] [Google Scholar]
- 10.Scholl, D., Rogers, S., Adhya, S., and Merril, C. R. (2001) J. Virol. 75 2509-2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlenhoff, M., Stummeyer, K., Grove, M., Sauerborn, M., and Gerardy-Schahn, R. (2003) J. Biol. Chem. 278 12634-12644 [DOI] [PubMed] [Google Scholar]
- 12.Stummeyer, K., Schwarzer, D., Claus, H., Vogel, U., Gerardy-Schahn, R., and Mühlenhoff, M. (2006) Mol. Microbiol. 60 1123-1135 [DOI] [PubMed] [Google Scholar]
- 13.Jakobsson, E., Jokilammi, A., Aalto, J., Ollikka, P., Lehtonen, J. V., Hirvonen, H., and Finne, J. (2007) Biochem. J. 405 465-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stummeyer, K., Dickmanns, A., Mühlenhoff, M., Gerardy-Schahn, R., and Ficner, R. (2005) Nat. Struct. Mol. Biol. 12 90-96 [DOI] [PubMed] [Google Scholar]
- 15.Schwarzer, D., Stummeyer, K., Gerardy-Schahn, R., and Mühlenhoff, M. (2007) J. Biol. Chem. 282 2821-2831 [DOI] [PubMed] [Google Scholar]
- 16.Weigele, P. R., Scanlon, E., and King, J. (2003) J. Bacteriol. 185 4022-4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Ossowski, I., Stahlberg, J., Koivula, A., Piens, K., Becker, D., Boer, H., Harle, R., Harris, M., Divne, C., Mahdi, S., Zhao, Y., Driguez, H., Claeyssens, M., Sinnott, M. L., and Teeri, T. T. (2003) J. Mol. Biol. 333 817-829 [DOI] [PubMed] [Google Scholar]
- 18.Zhou, W., Irwin, D. C., Escovar-Kousen, J., and Wilson, D. B. (2004) Biochemistry 43 9655-9663 [DOI] [PubMed] [Google Scholar]
- 19.Horn, S. J., Sikorski, P., Cederkvist, J. B., Vaaje-Kolstad, G., Sorlie, M., Synstad, B., Vriend, G., Varum, K. M., and Eijsink, V. G. H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18089-18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decher, G., Ringsdorf, H., Venzmer, J., Bitter-Suermann, D., and Weisgerber, C. (1990) Biochim. Biophys. Acta Biomembr. 1023 357-364 [DOI] [PubMed] [Google Scholar]
- 21.Rode, B., Endres, C., Ran, C., Stahl, F., Beutel, S., Kasper, C., Galuska, S., Geyer, R., Mühlenhoff, M., Gerardy-Schahn, R., and Scheper, T. (2008) J. Biotechnol. 135 202-209 [DOI] [PubMed] [Google Scholar]
- 22.Frosch, M., Roberts, I., Gorgen, I., Metzger, S., Boulnois, G. J., and Bitter-Suermann, D. (1987) Microb. Pathog. 2 319-326 [DOI] [PubMed] [Google Scholar]
- 23.Skoza, L., and Mohos, S. (1976) Biochem. J. 159 457-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frosch, M., Gorgen, I., Boulnois, G. J., Timmis, K. N., and Bitter-Suermann, D. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 1194-1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häyrinen, J., Haseley, S., Talaga, P., Mühlenhoff, M., Finne, J., and Vliegenthart, J. F. (2002) Mol. Immunol. 39 399-411 [DOI] [PubMed] [Google Scholar]
- 26.Hallenbeck, P. C., Vimr, E. R., Yu, F., Bassler, B., and Troy, F. A. (1987) J. Biol. Chem. 262 3553-3561 [PubMed] [Google Scholar]
- 27.Lindberg, A. A. (1977) in Surface Carbohydrates of the Prokaryotic Cell (Sutherland, I. W., ed) pp. 289-356, Academic Press, London
- 28.Sutherland, I. W. (1977) in Surface Carbohydrates of the Prokaryotic Cell (Sutherland, I. W., ed) pp. 209-245, Academic Press, London
- 29.Bayer, M. E., Thurow, H., and Bayer, M. H. (1979) Virology 94 95-118 [DOI] [PubMed] [Google Scholar]
- 30.Leiman, P. G., Battisti, A. J., Bowman, V. D., Stummeyer, K., Mühlenhoff, M., Gerardy-Schahn, R., Scholl, D., and Molineux, I. J. (2007) J. Mol. Biol. 371 836-849 [DOI] [PubMed] [Google Scholar]
- 31.Schnaitman, C. A. (2001) in Molecular Medical Microbiology (Sussman, M., ed) pp. 93-136, Vol. 1, Academic Press, New York [Google Scholar]
- 32.Hughes, K. A., Sutherland, I. W., and Jones, M. V. (1998) Microbiology 144 3039-3047 [DOI] [PubMed] [Google Scholar]
- 33.Horn, S. J., Sørbotten, A., Synstad, B., Sikorski, P., Sorlie, M., Varum, K. M., and Eijsink, V. G. (2006) FEBS J. 273 491-503 [DOI] [PubMed] [Google Scholar]
- 34.Sørbotten, A., Horn, S. J., Eijsink, V. G., and Varum, K. M. (2005) FEBS J. 272 538-549 [DOI] [PubMed] [Google Scholar]
- 35.Koivula, A., Kinnari, T., Harjunpaa, V., Ruohonen, L., Teleman, A., Drakenberg, T., Rouvinen, J., Jones, T. A., and Teeri, T. T. (1998) FEBS Lett. 429 341-346 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, S., Irwin, D. C., and Wilson, D. B. (2000) Eur. J. Biochem. 267 3101-3115 [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, T., Ariga, Y., Sato, U., Toratani, T., Hashimoto, M., Nikaidou, N., Kezuka, Y., Nonaka, T., and Sugiyama, J. (2003) Biochem. J. 376 237-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katouno, F., Taguchi, M., Sakurai, K., Uchiyama, T., Nikaidou, N., Nonaka, T., Sugiyama, J., and Watanabe, T. (2004) J. Biochem. 136 163-168 [DOI] [PubMed] [Google Scholar]
- 39.Hendrix, R. W., Lawrence, J. G., Hatfull, G. F., and Casjens, S. (2000) Trends Microbiol. 8 504-508 [DOI] [PubMed] [Google Scholar]
- 40.Hendrix, R. W. (2002) Theor. Popul. Biol. 61 471-480 [DOI] [PubMed] [Google Scholar]
- 41.Casjens, S. R. (2005) Curr. Opin. Microbiol. 8 451-458 [DOI] [PubMed] [Google Scholar]
- 42.Steinbacher, S., Baxa, U., Miller, S., Weintraub, A., Seckler, R., and Huber, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10584-10588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbirz, S., Müller, J. J., Uetrecht, C., Clark, A. J., Heinemann, U., and Seckler, R. (2008) Mol. Microbiol. 69 303-316 [DOI] [PubMed] [Google Scholar]
- 44.Müller, J. J., Barbirz, S., Heinle, K., Freiberg, A., Seckler, R., and Heinemann, U. (2008) Structure 16 766-775 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.