Abstract
We have developed a technique called the generation of longer cDNA fragments from serial analysis of gene expression (SAGE) tags for gene identification (GLGI), to convert SAGE tags of 10 bases into their corresponding 3′ cDNA fragments covering hundred bases. A primer containing the 10-base SAGE tag is used as the sense primer, and a single base anchored oligo(dT) primer is used as an antisense primer in PCR, together with Pfu DNA polymerase. By using this approach, a cDNA fragment extending from the SAGE tag toward the 3′ end of the corresponding sequence can be generated. Application of the GLGI technique can solve two critical issues in applying the SAGE technique: one is that a longer fragment corresponding to a SAGE tag, which has no match in databases, can be generated for further studies; the other is that the specific fragment corresponding to a SAGE tag can be identified from multiple sequences that match the same SAGE tag. The development of the GLGI method provides several potential applications. First, it provides a strategy for even wider application of the SAGE technique for quantitative analysis of global gene expression. Second, a combined application of SAGE/GLGI can be used to complete the catalogue of the expressed genes in human and in other eukaryotic species. Third, it can be used to identify the 3′ cDNA sequence from any exon within a gene. It can also be used to confirm the reality of exons predicted by bioinformatic tools in genomic sequences. Fourth, a combined application of SAGE/GLGI can be applied to define the 3′ boundary of expressed genes in the genomic sequences in human and in other eukaryotic genomes.
A particular biological event in a cell is largely controlled by the expression of multiple genes at the correct time and in a spatially appropriate manner. Monitoring the pattern of gene expression under various physiological and pathological conditions is a critical step in understanding these biological processes and for potential intervention. Because of the large number of genes expressed in higher eukaryotic genomes, powerful tools are needed to characterize the overall pattern of gene expression. The successful development of the serial analysis of gene expression (SAGE) technique is an important milestone in this regard (1). In the SAGE technique, a short sequence tag with 10-base nucleotides representing each expressed sequence is excised, and the tags from different expressed sequences are ligated for sequencing analysis. This strategy provides maximal coverage of the expressed genes for gene identification at the whole genome level while keeping the sequencing analysis at a manageable scale. Application of the SAGE technique has provided valuable information in various biological systems (2–6).
However, there are two problems when applying the SAGE tag sequence for gene identification. The first one is that many SAGE tags identified have no match to known sequences in databases (2, 3). These tags may represent previously unidentified genes. It is difficult, however, to use this tag information for further characterization of the corresponding genes because of their short length. The second problem is that certain SAGE tag sequences have multiple matches with sequences in the databases. These matched sequences have no similarity to each other except that they share the same SAGE tag sequence. This feature makes it difficult to determine the correct sequence in a particular tissue corresponding to a SAGE tag among these matched sequences.
To overcome these problems, we have developed a technique called the generation of longer cDNA fragments from SAGE tags for gene identification (GLGI). The key feature of this technique is the use of a sequence containing a SAGE tag as the sense primer, an anchored oligo(dT) as the antisense primer, and Pfu DNA polymerase for PCR amplification. By using this approach, a SAGE tag sequence can be converted immediately into a longer cDNA fragment containing up to several hundred bases from the SAGE tag to the 3′ end of the corresponding cDNA. The development of the GLGI technique overcomes the two obstacles discussed above and should have wide application in SAGE-related techniques for global analysis of gene expression.
Materials and Methods
SAGE Tags.
A group of SAGE tags with 10 bases was selected from the SAGE tag sequences generated from epithelium cells of normal colon (ref. 2; http://www.ncbi.nlm.nih.gov/SAGE/sagerec.cgi?rec=166). Each selected SAGE tag sequence was checked in the Unigene database (http://www.ncbi.nlm.nih.gov/SAGE/SAGEtag.cgi?tag) to identify it as a matched or an unmatched tag sequence. Each matched sequence was given the appropriate Unigene identification number. Both matched and unmatched tags were used in the experiments.
RNA Samples and cDNA Synthesis.
The same RNA sample from epithelium cells of normal human colon tissue was used for this experiment (2). RNA samples from 24 different human tissues were also used for the detection of multiple expression (CLONTECH). First-strand cDNAs were generated through oligo(dT) priming with a cDNA synthesis kit (Life Technologies), following the manufacturer's instructions. The remaining free oligo(dT) primers in cDNA samples were removed by using a MicroSpin S-300 column (Amersham Pharmacia).
PCR Conditions.
Pfu DNA polymerase (Stratagene) was used with 10× buffer (200 mM Tris⋅HCl, pH 8.8/100 mM KCl/100 mM (NH4)2SO4/20 mM MgSO4/1% Triton X-100/1 mg/ml BSA). MgCl2 (2 mM) was added in each reaction to increase the [Mg2+]. The PCR mixture contained 1× buffer, 2 mM MgCl2, 0.3 mM dNTP, 0.04 units/μl Pfu polymerase, 3 ng/μl sense primer, and 1.5 ng/μl anchored oligo(dT) primer (single or mixture) in a final volume of 20 or 50 μl. The PCRs were performed first at 94°C for 1 min, followed by five cycles at 94°C for 20 s, 50–53°C for 20 s, and 72°C for 20 s. The conditions were then changed to 25 cycles at 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s. The reactions were kept at 72°C for 5 min for the last cycle.
DNA Cloning and Sequencing.
PCR-amplified fragments were cloned into pCR-Blunt vector (Invitrogen). Positive clones were screened by using PCR with M13 reverse and M13 forward (−20) primers located in the vector or by using EcoRI digestion. Plasmids were prepared with a plasmid purification kit (Qiagen, Valencia, CA). Sequencing reactions were performed with PE big-dye kit (PE Applied Biosystems, Foster City, CA) with M13 reverse primer, following the manufacturer's instructions.
Database Search.
All the sequences generated from the clones were searched by using the blast program for alignment (http://www.ncbi.nlm.nih.gov/BLAST/).
Results and Discussion
General Strategy.
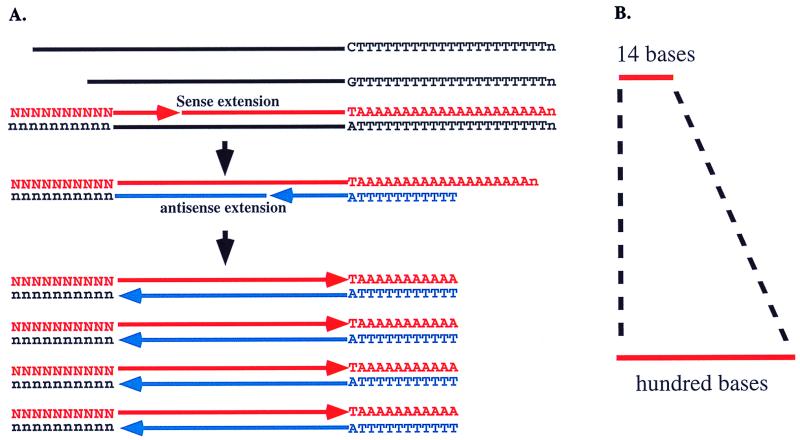
We reasoned that the amplification of a particular template corresponding to a particular SAGE tag should be possible by using a combination of a sense primer containing a SAGE tag sequence and an anchored oligo(dT) primer, (Fig. 1). In this process, only the cDNA templates containing the binding sequences for the SAGE tag will be annealed and extended in the first PCR cycle. In the second cycle, the extension will happen only from the anchored oligo(dT) primer that annealed at the 5′ end of the poly(dA) sequences with the anchored nucleotide correctly paired to the last nucleotide before the poly(dA) sequence. Extension of all other anchored primers annealed along the poly(dA) sequences will be blocked because of the presence of the anchor nucleotide. The resulting extended templates will exclude poly(dA)/(dT) sequences. Only the cDNA templates containing the SAGE tag sequence will undergo exponential amplification in the following PCR cycles. Thus, only copies of the same size will be generated.
Figure 1.
Schematic for GLGI. (A) In this process, first-strand cDNA synthesized by oligo(dT) is used for PCR. In the first cycle, the template with the SAGE tag binding site is annealed by the sense primer and extended to the end of the template. In the second cycle, extension occurs only from the anchored oligo(dT) primer annealed and paired correctly at the beginning of poly(dA) sequences. Exponential amplification occurs only for the template with the SAGE tag binding site. (B) The result of GLGI will be the conversion of 10 bases of SAGE tag to a hundred bases of 3′ cDNA fragment.
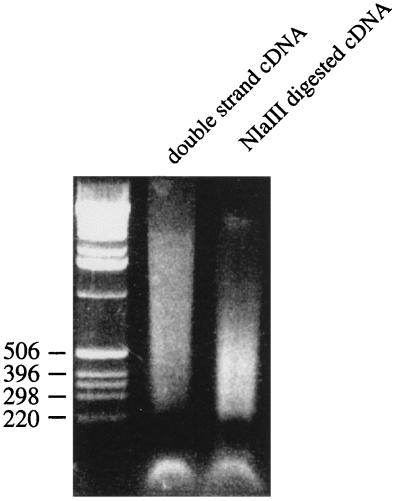
The expected size distribution of amplified sequences with this strategy should be up to several hundred bases, because of the use of NlaIII digestion in the SAGE process for SAGE tag collection (1). NlaIII is a restriction enzyme recognizing CATG. As shown in Fig. 2, the size distribution of NlaIII-digested cDNA was centered between 200 and 500 base pairs.
Figure 2.
Size distribution of NlaIII-digested cDNA. Double-strand cDNA was digested by NlaIII and electrophoresed on a 1.5% agarose gel to show the size distribution of the digested fragments.
Design of Primer.
Each SAGE tag contains only a 10-base sequence. To increase the length of the primers for efficient PCR priming, CATG, a NlaIII recognition site used for collecting SAGE tag fragments (1), was added 5′ of the SAGE tag. A BamHI recognition site, GGATCC, was added 5′ of the primer to increase the primer size and to provide a potential site for subcloning. For the anchored oligo(dT) primers, a single-base anchor dA, dG, or dC was attached to the 3′ end of the oligo(dT) primer (7–11). To determine the best length of oligo(dT) sequences, different numbers of dT nucleotides from 11 to 20 were tested, with dT11 giving the best results.
Optimizing PCR Conditions.
Various PCR conditions were tested to maximize the specificity and efficiency of amplification. In the PCR, either the anchored primers were combined separately with each sense primer, or a mixture of equal amounts of dA-, dG-, and dC-anchored primers was used with the sense primer. Pfu DNA polymerase was chosen for the PCR amplification, because it showed greater fidelity of amplification compared with regular Taq DNA polymerase (ref. 12 and data not shown). The Mg2+ concentration played an important role in determining the specificity and the yield of the PCR products. Satisfactory results were usually obtained at the final concentration of 4 mM Mg2+. The number of PCR cycles is important to maintain the specificity of the amplification. Overamplification with a high number of PCR cycles could result in nonspecific amplification (data not shown).
Amplification of Longer Sequences from SAGE Tags.
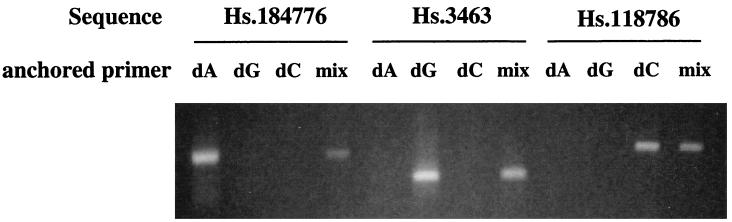
A group of SAGE tags generated from colon tissues was selected for the analysis (ref. 2; Table 1). PCR was performed with each sense primer containing the SAGE tag sequence and individual or mixed anchored oligo(dT) primers, combined with cDNAs from colon tissue generated by oligo(dT) priming. The PCR products were electrophoresed through an agarose gel and cloned into a vector for sequencing analysis. Fig. 3 shows examples of the PCR amplification with three SAGE tags that matched to known sequences. The last nucleotide before the poly(dA) sequences for those three sequences (Hs.184776, Hs.3463, and Hs.118786) is dT, dC, and dG, respectively. We obtained the expected results. The amplification occurred only in the reaction with dA-, dG-, and dC-anchored oligo(dT) for these three sequences. When the dA-, dG-, and dC-anchored oligo(dT) primers were mixed for each reaction, the same amplification products were generated, even though the amplification efficiency was lower because of the competition of binding between these three primers. These data indicate that the reaction can be simplified into a single reaction by using a combination of dA-, dG-, and dC-anchored oligo(dT) primers. Table 1 summarizes the results generated from these experiments. For the matched SAGE tag sequences, amplification occurred when the correct anchor primers were used, except for Hs.194659, which was amplified by dG-anchored oligo(dT), but the matched sequences ended with dT. The size distribution of these amplified fragments ranged from 77 to 382 base pairs. cDNA fragments were also generated from three unmatched SAGE tags, and they represent previously unidentified sequences.
Table 1.
Summary of GLGI results from SAGE tags
| SAGE tags (10 base) | Unigene identification no. | 3′ end nucleotide in matched sequences* | Amplified by anchored oligo (dT) | Length of sequence bases | Match to original sequence† |
|---|---|---|---|---|---|
| GGAAGGTTTA | Hs.105484 | dT/dG | dT | 77 | + |
| AGATCCCAAG | Hs.50813 | dC/dG | dC | 84 | + |
| CTTATGGTCC | Hs.179608 | dT | dT | 86 | + |
| AGGATGGTCC | Hs.71779 | dC | dC | 112 | + |
| GTCATCACCA | Hs.32966 | dC | dC | 119 | + |
| GACCAGTGGC | Hs.143131 | dC/dT | dC | 135 | + |
| CTGTTGGTGA | Hs.3463 | dC | dC | 148 | + |
| ACTGGGTCTA | Hs.227823 | dG | dG | 150 | + |
| TACGGTGTGG | Hs.105460 | dC | dC | 166 | + |
| CGGTGGGACC | Hs.99175 | dC/dT/dG | dC | 200 | + |
| CCTTCAAATC | Hs.23118 | dC/dT | dC | 220 | + |
| GGAGGCGCTC | Hs.33455 | dT/dG | dT | 238 | + |
| AAGAAGATAG | Hs.73848 | dT | dT | 317 | + |
| GATCCCAACT | Hs.118786 | dG/dT/dC | dG | 329 | + |
| GAACAGCTCA | Hs.194659 | dT | dG | 382 | + |
| AGGTGACTGG | — | — | dC | 156 | − |
| CACCTAGTTG | — | — | dT | 170 | − |
| CCTGTCTGCC | — | — | dT | 249 | − |
*The 3′ end nucleotides from all the sequences were included in each matched Unigene cluster.
†The amplified sequences were matched to databases again. The last three sequences have no matches and represent previously unidentified sequences.
Figure 3.
Specific amplification of 3′ sequences corresponding to a specific SAGE tag sequence by GLGI. In the PCR, each SAGE tag sequence was used as the sense primer, each single dA, dG, or dC or a mixture of three anchored oligo(dT) primers was used as the antisense primer. The 3′ end nucleotide for Hs.184776 is dT; that for Hs.3463 is dC; and that for Hs.118786 is dG.
Identifying the Correct Sequence from Multiple Sequences That Matched with the Same SAGE Tag.
When matching SAGE tag sequences in databases, a single SAGE tag may align with several sequences. For example, 9 of 40 SAGE tag sequences show matches to multiple Unigene clusters (2). Other than sharing the same SAGE tag sequence, these matched sequences have no homology and are derived from different tissues. To test this issue experimentally, 12 SAGE tags were used for amplification with cDNA samples from 24 different human tissues. Of these 12 tags, 4 generated multiple templates. For example, the SAGE tag GTCATCACCA generated five different sequences from five different tissues (fetal liver, skeletal muscle, spinal cord, trachea, and colon) and two different sequences from the same tissue (spinal cord; Table 2). All of these fragments contained the same SAGE tag sequence, but the rest of the sequences showed no homology. Among these sequences, the ones from colon tissue all matched the previous amplified sequences in the colon (Table 1). These data indicate that a SAGE tag itself may not be sufficient to serve as a unique identifier for a particular sequence when several sequences share the same SAGE tag sequences. It is important to distinguish which one of the matched sequences is the correct sequence corresponding to the particular SAGE tag. To avoid the uncertainty when different sequences are expressed from different tissues, it will be necessary to generate the fragment from the same tissue used to generate the SAGE tag. Our observations also indicate that relying only on a database search to identify the sequence corresponding to a SAGE tag may provide misleading information. Direct amplification of the specific template with our strategy will be very useful for confirmation of the validity of a particular SAGE tag.
Table 2.
Detection of heterogeneous sequences in various tissues containing the same SAGE tag
| SAGE tag | Positive tissues | Unigene identification no. | Length of sequence |
|---|---|---|---|
| CGGTGGGACC | Colon, thymus, small intestine | Hs.99175 | 200 |
| Small intestine | No match | 368 | |
| Thymus | No match | 90 | |
| AGATCCCAAG | Colon, heart, placenta, thymus | Hs.50813 | 84 |
| Placenta | No match | 53 | |
| Skeletal muscle | Hs.85937 | 282 | |
| Testis | No match | 227 | |
| Thymus, placenta | No match | 51 | |
| CTTATGGTCC | Bone marrow | Hs.237416 | 393 |
| Bone marrow | No match | 144 | |
| Colon | Hs.179608 | 86 | |
| GTCATCACCA | Fetal liver, spinal cord | Hs.222346 | 125 |
| Skeletal muscle | Hs.1288 | 399 | |
| Spinal cord | Hs.9641 | 394 | |
| Trachea | No match | 225 | |
| Colon | Hs.32966 | 136 |
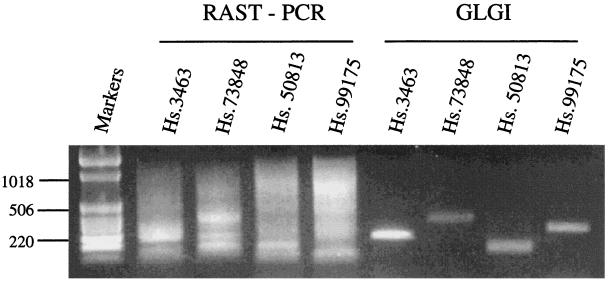
During the course of our research, we became aware of a report describing a method of rapid reverse transcription–PCR analysis of unknown SAGE tags (13). The authors used sense primers that were designed based on SAGE tags. However, the antisense primer was the M13 sequence connected to the 5′ end of oligo(dT) used for cDNA synthesis. In the process of cDNA synthesis, oligo(dT) primers anneal randomly along the poly(A) sequences in the mRNA templates. The resulting cDNAs include various lengths of poly(dA)/(dT) sequences at the 3′ end of the cDNA, even from the same mRNA template. Using the M13 sequence connected to the oligo(dT) as the antisense primer for PCR will generate multiple fragments with different sizes or a smear caused by the inclusion of different lengths of poly(dA)/(dT) sequences. Using the conditions described in that paper (13), we obtained the results we expected, namely smears (Fig. 4).
Figure 4.
Comparison between rapid reverse transcription–PCR analysis of unknown SAGE tags and GLGI. A set of four SAGE tags was chosen for the analysis. The same RNA from human colon and sense primers was used for both methods. The conditions used for rapid reverse transcription–PCR analysis of unknown SAGE tags followed the procedures described in ref. 13.
The development of the GLGI method provides several potential applications. First, it provides a strategy for even wider application of the SAGE technique for quantitative analysis of global gene expression. Second, a combined application of SAGE/GLGI can be used to complete the catalogue of the expressed genes in human and in other eukaryotic species. Third, it can be used to identify the 3′ cDNA sequence from any exon within a gene. It can also be used to confirm the reality of exons predicted by bioinformatic tools in genomic sequences. Fourth, a combined application of SAGE/GLGI can be applied to define the 3′ boundary of expressed genes in the genomic sequences in human and in other eukaryotic genomes.
Acknowledgments
We thank Dr. Lin Zhang (John Hopkins University, Baltimore) for his generous support with SAGE information and RNA samples. We also thank Dr. Bert Vogelstein and Dr. Kenneth Kinzler (John Hopkins University) for their valuable comments. We thank Mr. Xiangyang Xu for his assistance in DNA sequencing. This research was supported in part by National Cancer Institute Grants CA42557 (to J.D.R.) and CA78862-01 (to J.D.R. and S.M.W.) and a grant from the G. Harold and Lelia Y. Mathers Charitable Foundation (to S.M.W.).
Abbreviations
- SAGE
serial analysis of gene expression
- GLGI
generation of longer cDNA fragments from SAGE tags for gene identification
Footnotes
References
- 1.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 4.Madden S L, Galella E A, Zhu J, Bertelsen A H, Beaudry G A. Oncogene. 1997;15:1079–1085. doi: 10.1038/sj.onc.1201091. [DOI] [PubMed] [Google Scholar]
- 5.Hibi K, Liu Q, Beaudry G A, Madden S L, Westra W H, Wehage S L, Yang S C, Heitmiller R F, Bertelsen A H, Sidransky D, et al. Cancer Res. 1998;58:5690–5694. [PubMed] [Google Scholar]
- 6.Hashimoto S, Suzuki T, Dong H Y, Nagai S, Yamazaki N, Matsushima K. Blood. 1999;94:845–852. [PubMed] [Google Scholar]
- 7.Khan A S, Wilcox A S, Hopkins J A, Silela J M. Nucleic Acids Res. 1991;19:1715. doi: 10.1093/nar/19.7.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiriangkum J, Vainshtein I, Elliott J F. Nucleic Acids Res. 1992;20:3793–3794. doi: 10.1093/nar/20.14.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang P, Pardee A B. Science. 1992;257:967–970. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 10.Liang P, Zhu W, Zhang X, Guo Z, O'Connell R P, Averboukh L, Wang F, Pardee A B. Nucleic Acids Res. 1994;22:5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S M, Rowley J D. Proc Natl Acad Sci USA. 1998;95:11909–11914. doi: 10.1073/pnas.95.20.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundberg K S, Shoemaker D D, Adams M W, Short J M, Sorge J A, Mathur E J. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg A, van der Leij J, Poppema S. Nucleic Acids Res. 1999;27:e17. doi: 10.1093/nar/27.17.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]