Abstract
Complement receptor 2 (CR2, CD21) is a cell membrane protein, with 15 or 16 extracellular short consensus repeats (SCRs), that promotes B lymphocyte responses and bridges innate and acquired immunity. The most distally located SCRs (SCR1-2) mediate the interaction of CR2 with its four known ligands (C3d, Epstein-Barr virus gp350, interferon-α, and CD23). Inhibitory monoclonal antibodies against SCR1-2 block binding of all ligands. To develop ligand-specific inhibitors that would also assist in identifying residues unique to each receptor-ligand interaction, phage were selected from randomly generated libraries by panning with recombinant SCR1-2, followed by specific ligand-driven elution. Derived peptides were tested by competition ELISA. One peptide, C3dp1 (APQHLSSQYSRT) exhibited ligand-specific inhibition at midmicromolar IC50. C3d was titrated into 15N-labeled SCR1-2, which revealed chemical shift changes indicative of specific intermolecular interactions. With backbone assignments made, the chemical shift changes were mapped onto the crystal structure of SCR1-2. With regard to C3d, the binding surface includes regions of SCR1, SCR2, and the inter-SCR linker, specifically residues Arg13, Tyr16, Arg28, Tyr29, Ser32, Thr34, Lys48, Asp56, Lys57, Tyr68, Arg83, Gly84, Asn101, Asn105, and Ser109. SCR1 and SCR2 demonstrated distinct binding modes. The CR2 binding surface incorporating SCR1 is inconsistent with a previous x-ray CR2-C3d co-crystal analysis but consistent with mutagenesis, x-ray neutron scattering, and inhibitory monoclonal antibody epitope mapping. Titration with C3dp1 yielded chemical shift changes (Arg13, Tyr16, Thr34, Lys48, Asp56, Lys57, Tyr68, Arg83, Gly84, Asn105, and Ser109) overlapping with C3d, indicating that C3dp1 interacts at the same CR2 site as C3d.
Complement receptor 2 (CR2, CD21) is a 145-kDa Type I transmembrane protein composed of 15 or 16 short consensus repeat (SCR)2 domains, a 28-amino acid transmembrane domain, and a short 34-amino acid intracellular tail (1-5). Human CR2 is a cell surface receptor that plays an integral role in the immune system (6-9). On B cells, where CR2 is primarily expressed, CR2 is found in complex with other membrane proteins, including CD19, and plays a crucial role in normal immune responses (10, 11). CR2 is expressed primarily on mature B cells (12) but is also found on a subset of T cells (13, 14) and follicular dendritic cells (15). CR2 has four classes of ligands. The four classes include the C3 activation fragments iC3b, C3dg, and C3d (16, 17), the Epstein-Barr virus surface glycoprotein gp350/220 (18-20), the low affinity IgE receptor CD23 (21, 22), and the cytokine interferon α (IFN-α) (23, 24).
The primary function of CR2 is as a B cell co-receptor for antigen-mediated B cell activation through enhanced signal transduction. CR2 co-ligation with surface IgM using monoclonal antibodies (25-29), C3d covalently linked to antigen (30), or biotin-conjugated C3dg complexed with biotinylated anti-IgM (31) results in increased intracellular calcium release and activation of tyrosine and mitogen-activated protein kinases. Co-activation of the BCR via C3d interacting with CR2/CD19 greatly amplifies an immune response, even without adjuvant (30).
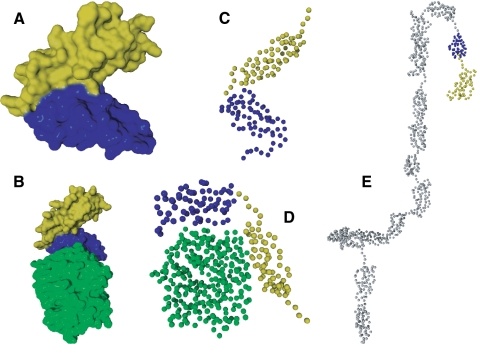
Structural studies of CR2 and its interaction with the C3d ligand have revealed inconsistent results. For example, the structures of CR2 SCR1-2 and CR2 SCR1-2 in complex with C3d derived by x-ray crystallography both show a tightly compacted V shape, where SCR1 contacts SCR2, with SCR domains forming two arms of a V (32, 33) (Fig. 1, A and B), and only SCR2 was seen to contact C3d (Fig. 1B). In contrast, using solution scattering and modeling methods, CR2 SCR1-2 was shown to be in a more extended conformation (Fig. 1, C and D), and in complex with C3d, both SCR domains appeared to be physically contacting C3d (34, 35) (Fig. 1D). Full-length CR2 has also been analyzed with solution scattering and modeling techniques, which suggested an extended conformation for the SCR1-2 domains (34) (Fig. 1E).
FIGURE 1.
Previously described structures of CR2 and ligands of CR2. A, x-ray crystal structure of CR2 SCR1-2 (32). B, x-ray co-crystal structure of CR2 SCR1-SCR2-bound C3d (33). C, x-ray and neutron scattering best fit solution model of CR2 SCR1-2 (35). D, x-ray and neutron scattering best fit co-solution model of CR2 SCR1-SCR2-bound C3d (35). E, x-ray and neutron scattering best fit solution model of full-length CR2 (34). Green, C3d; yellow, CR2 SCR1; blue, CR2 SCR2; gray, CR2 SCR3 to -15.
Site-directed mutagenesis has also been used with CR2 to elucidate binding regions of CR2 SCR1-2. The CR2-C3d interaction has been studied, and the results of site-directed mutagenesis suggest that both SCR domains are required for binding and that the interaction is in part charge-mediated (36). Residues found to be required for interaction were Arg13, Tyr16, Ser32, Arg28, Arg36, Lys41, Lys50, Lys57, Lys67, and Arg83.
Despite substantial progress in understanding CR2-ligand interactions, several key questions remain. For example, the CR2-C3d co-crystal structure suggests that only SCR2 contacts C3d and that SCR1-2 dimerize when bound to C3d. The observation that only SCR2 contacts C3d was inconsistent with the solution scattering model, monoclonal antibody mapping, and mutagenesis studies, all of which clearly suggest that both SCR1-2 are needed for the interaction with C3d to take place. In addition, dimerization has not been observed in the solution phase (35). Furthermore, there is still no consensus about which residues might be uniquely required for each CR2-ligand interaction, and importantly, there is no method available for selectively inhibiting each ligand, since anti-CR2 monoclonal antibodies directed to SCR1-2 inhibit all ligands. Such inhibitors would be useful to tease out the physiological functions of each ligand-bound state.
To further study the binding site on CR2 SCR1-2 for C3d we have employed a 2-fold approach. First, we have generated ligand-selective inhibitory peptides. Second, using isotopically labeled CR2 SCR1-2 as well as the aforementioned peptide and full-length C3d, we have evaluated in vitro binding using chemical shift analysis and NMR, a high resolution structural tool. The results from the experiments carried out further define the C3d binding site on CR2 SCR1-2 and indicate that both SCR1-2 are in direct contact with C3d in solution.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins—Human CR2 SCR1-2 for NMR studies was expressed in Pichia pastoris using a BioFlo 110 Fermenter (New Brunswick Scientific, Edison, NJ), as previously described (37). Briefly, a single colony was grown in 5 ml of Pichia basal salt medium containing 1% glycerol (BMG) overnight at 30 °C and 250 rpm, expanded to 50 ml of BMG (24 h), and finally expanded to 300 ml of BMG (24 h). The inoculation culture was centrifuged at 2500 × g 25 °C and resuspended in 30 ml of BMG. The 30-ml inoculation culture was used to inoculate 1 liter of minimal Pichia basal medium containing 40 g of glycerol. Dissolved O2 concentration was maintained at 40%, the temperature was maintained at 30 °C, and pH was maintained at 5 using 2 m KOH. Initial feeds were batch glycerol feeds; transition to methanol was eased by a methanol injection before an exponential methanol feed profile was initiated. Methanol induction lasted for 2 days, after which the culture was centrifuged to remove cellular debris. The supernatant was exchanged into 10 mm formate, pH 4, before being passed over an SP-Sepharose column (2 × 5 ml of SP HiTrap columns; GE Biosciences, Pittsburgh, PA), followed by a CR2 affinity column, generated in house by binding glutathione S-transferase-C3d to a GSTrap column (GE Biosciences). CR2 was eluted with an increasing linear NaCl gradient, 0-1.0 m in 1/3× phosphate-buffered saline (1/3× PBS; 1.6 mm MgCl2, 0.9 mm KCl, 0.5 mm KH2PO4, 45.6 mm NaCl, 2.7 mm Na2HPO4, pH 7.4). Finally, CR2 SCR1-2 was purified by size exclusion chromatography. Purity and identity of CR2 were monitored via SDS-PAGE, Western blot analysis, and mass spectrometry. Both 15N and 15N-13C isotopically labeled proteins were prepared using this strategy. For 15N isotopically labeled CR2 [15N]ammonium sulfate was used. For 15N-13C isotopically labeled CR2 [15N]ammonium sulfate, [13C]glycerol and [13C]methanol were used. Isotopically enriched chemicals were purchased from Isotec Inc. (Miamisburg, OH).
Human CR2 SCR1-2 for peptide discovery and ELISA binding studies was generated using the pMAL-P2X expression system in Escherichia coli as previously described (38, 39). Briefly, ampicillin-resistant colonies were used to start overnight cultures that were expanded to 1 liter and grown at 37 °C until an A600 of 0.3 was obtained. Cultures were induced with 0.3 mm isopropyl-β-d-thiogalactoside at 30 °C overnight before harvesting by centrifugation. Harvested pellets were resuspended in amylose column buffer (20 mm Tris-HCl, pH 7.4, 0.2 m NaCl, 1 mm EDTA) and lysed by sonication. Lysate was clarified by centrifugation and applied to an amylose resin column (New England Biosciences, Ipswich, MA). Bound MBP-CR2 SCR1-2 was eluted from the column using amylose column elution buffer (amylose column buffer plus 10 mm maltose). Finally, the MBP-CR2 SCR1-2 was purified by size exclusion chromatography. Purity and identity of MBP-CR2 was monitored via SDS-PAGE and Western blot analysis.
Human C3d for peptide discovery, ELISA binding studies, and NMR titrations was generated using the pGEX expression system in E. coli as previously described (36). Briefly, ampicillin-resistant colonies were used to start overnight cultures that were expanded to 1 liter and grown at 37 °C until an A600 of 0.3 was achieved. Cultures were induced with 0.3 mm isopropyl-β-d-thiogalactoside at 30 °C overnight before harvesting by centrifugation. Harvested pellets were resuspended in glutathione S-transferase column buffer (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 1 mm EDTA) and lysed by sonication. Lysate was clarified by centrifugation and applied to a GStrap column (GE Biosciences). C3d was cleaved from the column by digesting with 50 units of thrombin overnight at 4 °C and subsequently purified by size exclusion chromatography. Purity of C3d was monitored via SDS-PAGE.
Purification of Epstein-Barr virus gp350 was completed as previously described (37). Briefly, gp350 was produced by infecting Sf9 insect cells with the gp350-packaged baculovirus particles (pVI-Bac transfer vector, C-terminal polyhistidine tag) at a multiplicity of infection of 3. The baculoviral supernatant was concentrated, buffered with 10 mm TRIS-HCl with 10 mm imidazole, pH 7.4, and applied to a 5-ml HiTrap column (GE Biosciences) and subsequently eluted with a linear imidazole gradient. Purity and identity of gp350 were monitored via SDS-PAGE and Western blot analysis.
Human IFN-α for ELISA binding studies was cloned de novo from isolated human genomic DNA. DNA corresponding to residues 1-169 of human IFN-α-2a was PCR-amplified and ligated into the pMAL-p2x (New England Biosciences, Ipswich, MA) expression vector and subsequently transformed into E. coli BL21 cells. After DNA sequence analysis confirmed that the correct DNA sequence was isolated and ligated, protein production was optimized. Briefly, ampicillin-resistant colonies were used to start overnight cultures that were expanded to 1 liter and grown at 37 °C until an A600 of 0.3 was obtained. Cultures were induced with 0.3 mm isopropyl-β-d-thiogalactoside at 25 °C overnight before harvesting by centrifugation. Harvested pellets were resuspended in amylose column buffer (20 mm Tris-HCl, pH 7.4, 0.2 m NaCl, 1 mm EDTA) and lysed by sonication. Lysate was clarified by centrifugation and applied to an amylose resin column (New England Biosciences, Ipswich, MA). Bound MBP-IFN-α was eluted from the column using amylose column elution buffer (amylose column buffer plus 10 mm maltose). After elution, the MBP tag was cleaved overnight at 4 °C with Factor Xa (New England Biosciences). Finally, IFN-α was purified by size exclusion chromatography. Purity and identity of IFN-α were monitored via SDS-PAGE and Western blot analysis.
Identification of Selective Peptide Inhibitors—Peptides were identified using phage display libraries of 12-mer peptides from New England Biosciences. MBP-CR2, in 0.1 m NaHCO3, pH 8.6, was immobilized to 96-well plates overnight in a humidified container at 4 °C. After washing the plate with TBST (50 mm Tris-HCl pH 7.5, 150 mm NaCl, and 0.1% Tween 20), 4 × 1010 phage were diluted with 100 μl of TBST and added to each coated well for 60 min at room temperature. Bound phage were then eluted with 1 mm C3d in TBS (50 mm Tris-HCl, pH 7.5, 150 mm NaCl). Approximately 1 μl of eluate was titered to determine the concentrations of phage to use for subsequent panning stages. Each additional (up to three total) panning step utilized TBST with 0.5% Tween 20 instead of 0.1%. After the third panning step, ∼30 positive plaques from infected colonies were sequenced by automated DNA sequencing methods at the University of Colorado Denver School of Medicine Cancer Core to determine a consensus inhibitory peptide. The consensus sequences were synthesized as peptides using standard solid phase peptide synthesis protocols at the University of Colorado Denver School of Medicine Peptide Core.
Inhibitory ELISA—Inhibition of binding was tested using a modified competition ELISA. Briefly, Costar enzyme immunoassay/radioassay 96-well half-area flat bottom high binding plates (Corning Glass) were coated with 5 μg of C3d, gp350, or IFN-α overnight at 4 °C in ELISA coating buffer (15 mm Na2CO3, 35 mm NaHCO3 pH 8.8). After coating the plate with one ligand, a potential inhibitory peptide was incubated at increasing concentrations with 2 μg of MBP-CR2 in 1/3× PBS (1.6 mm MgCl2, 0.9 mm KCl, 0.5 mm KH2PO4, 45.6 mm NaCl, 2.7 mm Na2HPO4, pH 7.4). The peptide MBP-CR2 mixture was then incubated with the ligand bound plate for 1 h at 25 °C. Following washing, bound MBP-CR2 was detected using α-MBP-HRP; the percentage bound and thus the efficacy of inhibition by the peptide was calculated by the equation, percentage inhibition = (((abs(I) - abs(N))/(abs(N)) × 100), where abs(I) represents the absorbance of inhibited CR2-ligand interaction and abs(N) is the absorbance of uninhibited CR2-ligand interaction.
NMR Analysis—NMR experiments were carried out on Varian 600-, 800-, and 900-MHz magnets housed in the Rocky Mountain Regional NMR Facility at the University of Colorado Denver School of Medicine campus (600 and 900 MHz) and in the W. M. Keck High Field NMR Facility at the University of Colorado Boulder campus (800 MHz). The uniformly 15N-13C-labeled SCR1-2 domains of CR2 in 1/3× PBS were used to sequentially assign the 15N TROSY-HSQC (40) by using HNCACB (41), CBCA(CO)NH (42), and 15N-edited NOESY-HSQC (43) three-dimensional spectra. The NMR data were processed with nmrPipe (44) and analyzed with ccpNMR (45). Chemical shift changes were monitored using ccpNMR by overlaying TROSY-HSQC spectra from free CR2 SCR1-2 and CR2 SCR1-2 with increasing concentrations of either full-length C3d or inhibitory peptides. The dissociation constants (Kd) were determined by a nonlinear least-squares analysis using Prism and the equation, Δ=Δmax × ((([L] + [P] + KD) - sqrt(sqr([L] + [P] + KD) - (4 × [P] × [L])))/(2 × [P])), where [L] is concentration of C3d or the peptide, [P] is concentration of CR2 (SCR1-2), Δ is observed chemical shift change, and Δmax is the difference in chemical shifts of the free and the ligand-bound protein.
RESULTS
Identification of Ligand-selective Inhibitory Peptides—Linear consensus peptides were identified by phage display library methods. Phage were targeted to recombinant CR2 SCR1-2, and binding phage were competed off with each ligand to attempt to identify peptides that would block binding of one but not the other ligand. After three subsequent panning rounds with increasing detergent concentrations, selected phage were sequenced. A consensus peptide sequence was determined (Table 1), and the corresponding peptide was synthesized by conventional solid-phase techniques (46).
TABLE 1.
Identified inhibitory peptides against CR2-ligand interactions
Shown is the inhibitory peptide against the CR2-C3d interaction identified by phage display and its control peptide. The reported IC50 values represent an average and S.D. of three trials.
| Peptide name | Peptide sequence | IC50 |
|---|---|---|
| μm | ||
| C3dp1 | APQHLSSQYSRT | 100 ± 8 |
| Control | APAHLSSQ | >500 |
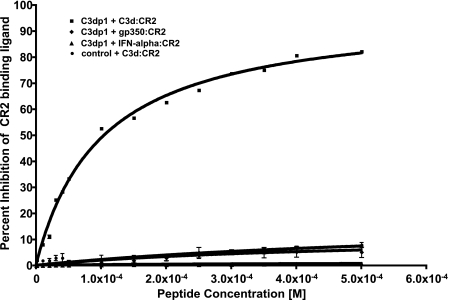
Peptide Competition ELISA—C3dp1 was tested in increasing concentration against its respective ligand (C3d), gp350, and IFN-α. Percentage inhibition was calculated and plotted against peptide concentration to develop an inhibition curve. From these data, an IC50 was calculated and listed in Table 1. The IC50 for the C3d peptide (C3dp1) was 100 ± 8 μm, and for the control peptide was greater than 500 μm. The peptide C3dp1 was found to demonstrate dose-dependent inhibition (Fig. 2). Of particular importance, the inhibitory peptide presented here is specific to the CR2-C3d interaction and does not inhibit the CR2-gp350 or the CR2-IFN-α interaction.
FIGURE 2.
CR2-ligand competition using ELISA with bound ligand and solution phase CR2 with increasing amounts of inhibitory peptides. Ligands include C3d, gp350, and IFN-α. The mean and S.D. of at least three replicates are reported. Peptide IC50 values against the CR2-C3d interaction are reported in Table 1.
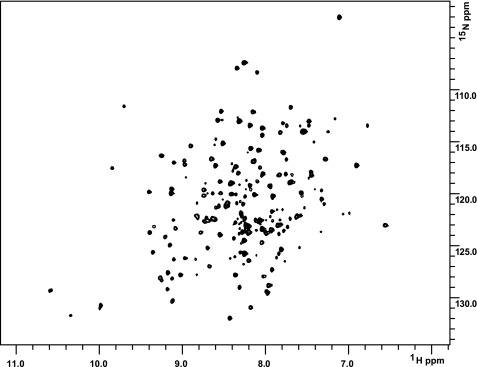
NMR CR2 Assignment and Chemical Shift Mapping—CR2 SCR1-2 expressed in P. pastoris appeared to be a correctly folded protein, as first evidenced by the purification process, whereby protein was purified by binding to a C3d-Sepharose column, and second by 1H-15N HSQC spectra (Fig. 3). As is evident in Fig. 3, the line shapes of resonances are not identical. Such differential line broadening is indicative of a highly dynamic protein and suggests that localized motions occur on the microsecond to millisecond time scale.
FIGURE 3.
1H-15N TROSY-HSQC spectra reveal that CR2 SCR1-2 is folded in solution. 1H-15N TROSY-HSQC spectra of 15N-labeled CR2 SCR1-2 (0.4 mm in 1/3× PBS) collected at 25 °C on a Varian 900 MHz spectrometer. Spectra were processed with nmrPipe and viewed with ccpNMR.
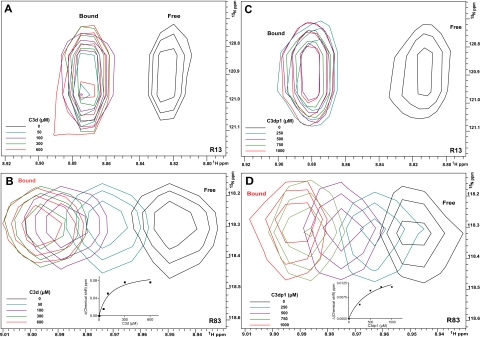
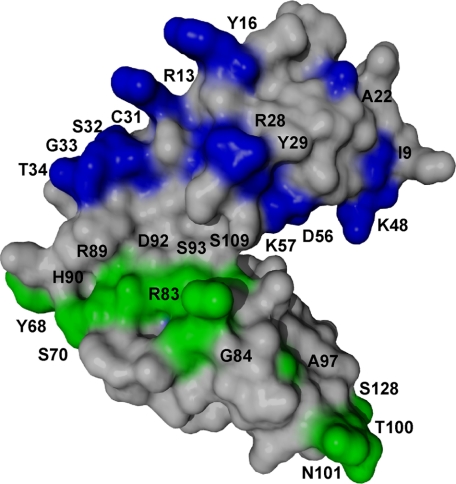
Following resonance assignment, full-length unlabeled C3d was titrated into a uniformly 15N-labeled CR2 SCR1-2 sample, and the 1H-15N chemical shifts were monitored (Fig. 4, A and B). Two modes of binding were observed. The first is characterized by the disappearance and reappearance of resonances, indicative of a tight binding interaction (Fig. 4A). The second is characterized by specific resonance chemical shift changes with no loss or addition of intensity, indicative of a weaker interaction (Fig. 4B). Those residues showing the tight interaction were residues Ile9, Arg13, Tyr16, Ala22, Arg28, Tyr29, Cys31, Ser32, Gly33, and Thr34. These residues encompass the SCR1 domain (Fig. 5). The residues showing a weaker binding interaction were residues Lys48, Asp56, Lys57, Tyr68, Ser70, Arg83, Gly84, Arg89, His90, Asp92, Ser93, Ala97, Thr100, Asn101, Asn105, Ser109, Arg122, and Ser128. These residues encompass both the SCR2 domain and the intra-SCR linker region (Fig. 5). These results suggest two distinct noncontiguous interaction faces on CR2 SCR1-2 and suggest that the linker region allows SCR1 and SCR2 to be oriented such that each intimately contacts C3d. The weaker Kd calculated from the NMR titration was 130 ± 60 μm; the associated binding curve is shown as the inset in Fig. 4B. For the tighter interaction using the minimal observed chemical shift difference between free and bound resonances (about 45 Hz) and assuming a diffusion-limited on rate of ∼10 × 8 m-1s-1, an upper limit to the binding constant was calculated as ∼45 μm (Table 2).
FIGURE 4.
NMR titration analysis reveals that SCR1 and SCR2 of CR2 are both involved in the ligation with C3d and C3d-inhibitory peptide C3dp1. A and B, five superimposed 1H-15N TROSY-HSQC spectra of 15N-labeled CR2 SCR1-2 (0.3 mm in 1/3× PBS) collected during titration with increasing amounts of C3d. Black, no C3d; blue, with 50 μm C3d; purple, with 100 μm C3d; green, with 300 μm C3d; red, with 600 μm C3d. Inset on B, the binding curve for the CR2-C3d weak interaction. C and D, five superimposed 1H-15N TROSY-HSQC spectra of 15N-labeled CR2 SCR1-2 (0.3 mm in 1/3× PBS) collected during titration with increasing amounts of C3dp1. Black, no C3dp1; blue, with 250 μm C3dp1; purple, with 500 μm C3dp1; green, with 750 μm C3dp1; red, with 1000 μm C3dp1. Inset in D, binding curve for the CR2-C3dp1 weak interaction. Residues monitored were Arg13 (A and C) and Arg83 (B and D). The numbering system used here within for CR2 is based on the reported amino acid sequence for the mature protein (5).
FIGURE 5.
Surface representation of CR2 SCR1-2 x-ray crystal structure in its ligand-bound state (C3d not shown) with NMR-determined C3d binding residues mapped out. Gray residues represent residues unaffected by C3dp titration. The blue residues on SCR1 represent the tight binding affinity interactions on CR2 between CR2 and C3d. The green residues on the linker and SCR2 represent the weaker affinity binding interactions on CR2 between CR2 and C3d.
TABLE 2.
Binding constants from NMR titrations
Shown are the weak and upper limit to the tight binding constants for the CR2-C3d and CR2-C3dp1 interactions. The binding constants were calculated using NMR titrations, monitoring chemical shift changes. The binding curves are found in Fig. 4, B and D. UL, upper limit.
| Ligand/Peptide | Binding mode | Kd |
|---|---|---|
| μm | ||
| C3d | Tight, UL | 45 |
| Weak | 130 ± 60 | |
| C3dp1 | Tight, UL | 45 |
| Weak | 500 ± 250 |
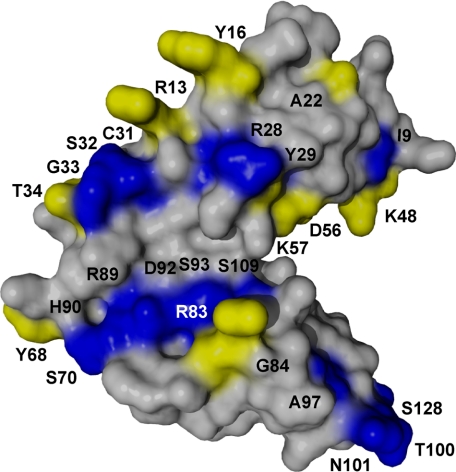
C3dp1 was also titrated into a uniformly 15N-labeled CR2 SCR1-2 sample, and the 1H-15N chemical shifts were monitored (Fig. 4, C and D). The chemical shift changes resulting from peptide binding show two binding interactions, similar to full-length C3d. The residues showing tight interactions were residues Arg13, Tyr16, Thr34, whereas the weaker interaction was observed with residues Lys48, Asp56, Lys57, Tyr68, Arg83, Gly84, Asn105, and Ser109. Again, these residues encompass both SCR domains and linker region as well as overlapping with the CR2-C3d chemical shifts. The interacting residues were mapped onto the surface of the x-ray structure (Fig. 6). These data demonstrate that this peptide is specifically interacting with the same residues as the full-length ligand. The weak Kd calculated from the NMR titration was 500 ± 250 μm (Table 2), with the associated binding curve shown as the inset in Fig. 4D. The tight CR2-C3dp1 interaction is similar to that of the CR2-C3d tight interaction (see above). Residues involved in both interactions were mapped onto the surface (Fig. 6), which shows that all residues involved in the CR2-C3dp1 interaction are involved in the CR2-C3d interaction.
FIGURE 6.
Surface representation of CR2 SCR1-2; summary of NMR binding maps. Shown is a summary of the binding maps of CR2-C3d and CR2-C3dp1. Gray residues represent residues that are not involved in either binding event. Blue residues represent residues only involved in C3d binding to CR2. Yellow residues represent residues involved in both C3d and C3dp1 binding to CR2.
DISCUSSION
Herein we have presented a two-pronged approach to study CR2-ligand interactions in the solution phase. First, we have identified a peptide that differentially inhibits C3d. The peptide and its control were tested using competition ELISA to reveal ligand-specific and -selective inhibition at a micromolar concentration against the C3d ligand. Second, we have utilized NMR with full-length C3d and C3dp1 in order to determine the residues on CR2 SCR1-2 that are involved in the CR2-C3d interaction.
To our knowledge, this is the first high resolution solution state study that is directed at the interaction between SCR1-2 and its ligands. We have also developed the first CR2 ligand-specific inhibitor that can be used to study the C3d interaction in vitro and potentially in vivo.
Previous SCR deletion mutagenesis studies have demonstrated that both SCR1 and SCR2 of CR2 are required for interaction between C3d (10, 21, 47, 48). Subsequent biochemical studies were consistent with a requirement for both SCR1 and SCR2 in the interaction with C3d (37, 49). However, contrary to suggestions in earlier studies, the x-ray co-crystal structure showed only SCR2 to be in contact with C3d (33). Subsequent amino acid mutagenesis data confirmed the SCR2 x-ray cocrystal contact site as well as providing support for the biochemical data in that multiple amino acids in SCR1 were required for a high affinity C3d interaction (36). The solution structure models of CR2-C3d complexes determined from analytical centrifugation and x-ray/neutron scattering studies also point to both SCR1 and SCR2 interacting with C3d (35). Herein we sought to extend our understanding beyond previous studies by using a new approach to map specific residues on CR2. To assist in this analysis, we also identified a specific inhibitory peptide against C3d. This peptide, along with full-length ligand, was subsequently used to map specific regions on CR2 SCR1-2 that are involved in each interaction.
Along with mutational analysis previously completed in our laboratory, which also suggests that both SCR domains are required for the interaction to occur in the solution phase, our current studies suggest a consensus model for the CR2 side of the CR2-C3d interaction in solution. Specifically, the data presented here suggest that there are two noncontinuous surfaces on the CR2 N-terminal two SCR domains that both interact with C3d (Fig. 5). In addition, our data also suggest a two-surface binding model where each SCR domain interacts with C3d in specific regions on each SCR domain and with differing binding affinities. SCR1 residues include Arg13, Tyr16, Ala22, Arg28, Tyr29, Lys48, Asp56, and Lys57 and exhibit a tighter binding interaction; SCR2 residues include Arg83, Gly84, Arg89, His90, Asp92, Ser93, Ala97, Ser109, Thr100, Asn101, and Ser128 and exhibit a weaker binding interaction. With a flexible linker, it can be imagined that the two SCR domains form a sandwich around C3d. Many resonances disappear due to the size of this large complex and to the increased tumbling time; therefore, alternative labeling techniques are necessary to observe such resonances.
Our data also suggest that the linker region plays an important role in the interaction. Although linker-derived interactions with ligand have been suggested before, they are more concretely shown here from specific chemical shift changes from residue Tyr68. In addition, with one of the longest inter-SCR linker region in SCR-containing proteins, it is reasonable to propose that the linker is flexible, and the two SCR domains could be oriented in multiple conformations, thus allowing interaction with varied ligands.
It is also relevant to note that it is not only charged residues that are suggested to interact with C3d, since there are also hydroxyl-containing amino acids that are apparently involved. Previous models of the charge dependence of the CR2-C3d interaction suggested long range contributions to binding from specific charged residues on CR2 (50, 51). Herein we have identified several of these residues as being directly involved in the CR2-C3d interaction; specifically, these residues are Arg13, Arg28, Tyr29, Arg83, Arg89, His90, and Asp92. Our data support that of the charge dependent models; however, we have also shown that hydroxyl-containing amino acids are involved in the CR2-C3d interaction, probably through short range interactions.
With regard to the Cys31 residue that was identified during the study, cysteine residues in the SCR domains are found in disulfide linkages; this residue's amide-bond being surface-exposed. Therefore, chemical shift changes from these residues are probably due to an overall conformational change in each SCR domain, although it may be subtle or possibly through a hydrogen-bonding interface.
The inhibitory peptide against C3d was also mapped onto the surface of CR2 SCR1-2 in the same way as the full-length ligand. Interestingly, the mapping of the peptide ligand overlaps with mapping of the full-length ligand (Fig. 6). The peptide, being much smaller in size than C3d, should in principle interact with a smaller number of amino acids than C3d. Interestingly, however, the amino acids that were mapped are on both SCR domains and the linker region (Fig. 6). The ability of a short peptide to interact with spatially disparate surfaces suggests that there are two binding events. This is suggested by two different modes of chemical shift tendencies that were observed on overlaid HSQC spectra (Fig. 4), which are indicative of separate or different binding events (52). In aggregate, however, these results indicate that the peptide specifically interacts in the same area with the same amino acids as the full-length ligand (Fig. 6). These results suggest that the approach of using peptide to map binding sites is valid and may have particular utility with larger ligands, such as gp350, that are likely to be too large for NMR methods.
It is also notable that the peptide CR2 interaction is very similar to full-length C3d interaction, with a tight interaction of ∼45 μm, whereas each also has weaker interactions. It is likely that the two binding affinities are not quite an order of magnitude apart; thus, both binding events can be observed simultaneously. The C3d weak interaction is tighter than C3dp1 weak interaction, which is expected. These results are consistent with previously reported Kd values for the CR2-C3d interaction in low salt conditions (23, 53), although they differ in orders of magnitude. This disparate finding is probably due to the previous Kd values being derived from a surface binding model, closer to the in vivo behavior, whereas the current study is a completely solution-based method. The two binding events that are evident in the two Kd values reported are mapped onto the crystal structure and suggest that there is one high affinity interaction and one lower affinity interaction. The data presented here suggest a two-site binding model that agrees well with two previously reported two-site binding models that were derived using a different methodology (surface plasmon resonance), and both report a low and high affinity interaction (23, 54). The high affinity interaction appears to be present with SCR1 and is represented by the immediate loss of resonances in the first titration point, where the first titration point is 50 μm C3d. The lower affinity interaction appears to be present in the linker region and SCR2 and is represented by the chemical shift change that changes in a dose-dependent manner. The higher Kd for the peptide is to be expected, since the peptide shows no similarity to sequences on C3d. Of additional interest is that the peptide has only potentially positively charged amino acids, thus not acting in a charge-dependent manner as has been shown to be required for the CR2-C3d interaction (50, 51).
Finally, the inhibitory peptide described here represents the first available ligand-selective inhibitor for CR2-ligand interactions. This peptide in principle could be used to study the specific outcomes of CR2-C3d interactions during an ongoing immune response in an in vivo animal model. In addition, it might be possible to use this peptide to direct molecules to CR2 in vivo.
In contrast to the emerging consensus regarding the CR2 side of the CR2-C3d interface, it is still uncertain where CR2 binds C3d (33, 36, 55). Our data do not directly address this question but are consistent with the co-crystal structure determination showing the SCR2-C3d interaction. However, substantial work is still required to better understand the site(s) to which SCR1 and SCR2 bind C3d.
In summary, our approach to map the binding surface of CR2 SCR1-2 to C3d has revealed a binding surface composed of both SCR domains and the inter-SCR linker region on CR2 at differing binding affinities. It is relevant to note that these surfaces agree well with previous mutagenesis and solution scattering experiments but differ from the co-crystal structure of CR2-C3d in that both SCR1 and SCR2 participate in the interaction. It is important to keep in mind that this body of research is aimed at the CR2 SCR1-2 side of the CR2-C3d interaction and does not address the C3d side of the interaction; however, this interaction is currently being addressed in our laboratory. An inhibitory peptide was also identified from a large random peptide library. This peptide has been shown to be selective against the C3d ligand, and the C3dp1 peptide maps on CR2 at a similar site as the full-length ligand. This peptide represents the first available ligand-selective inhibitor of CR2 and opens possibilities of studying the effects of specific CR2 ligands in the context of an ongoing immune response in vivo.
This work was supported, in whole or in part, by National Institutes of Health Grant R0-1 CA053615 (to V. M. H.). This work was also supported by an American Heart Association predoctoral fellowship (to J. M. K.) and assisted by the University of Colorado Denver-Rocky Mountain Regional 900-MHz NMR Facility. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SCR, short consensus repeat; IFN, interferon; PBS, phosphate-buffered saline; ELISA, enzyme-linked immunosorbent assay.
References
- 1.Fujisaku, A., Harley, J. B., Frank, M. B., Gruner, B. A., Frazier, B., and Holers, V. M. (1989) J. Biol. Chem. 264 2118-2125 [PubMed] [Google Scholar]
- 2.Moore, M. D., Cooper, N. R., Tack, B. F., and Nemerow, G. R. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 9194-9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weis, J. J., Fearon, D. T., Klickstein, L. B., Wong, W. W., Richards, S. A., de Bruyn Kops, A., Smith, J. A., and Weis, J. H. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 5639-5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weis, J. J., Toothaker, L. E., Smith, J. A., Weis, J. H., and Fearon, D. T. (1988) J. Exp. Med. 167 1047-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingeroth, J. D., Clabby, M. L., and Strominger, J. D. (1988) J. Virol. 62 1442-1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahearn, J. M., and Fearon, D. T. (1989) Adv. Immunol. 46 183-219 [DOI] [PubMed] [Google Scholar]
- 7.Cooper, N. R., Moore, M. D., and Nemerow, G. R. (1988) Annu. Rev. Immunol. 6 85-113 [DOI] [PubMed] [Google Scholar]
- 8.Holers, V. M. (1995) in Principles and Practices of Clinical Immunology (Rich, R., ed.) pp. 363-391, St. Louis, MO
- 9.Tolnay, M., and Tsokos, G. C. (1998) Clin. Immunol. Immunopathol. 88 123-132 [DOI] [PubMed] [Google Scholar]
- 10.Martin, D. R., Yuryev, A., Kalli, K. R., Fearon, D. T., and Ahearn, J. M. (1991) J. Exp. Med. 174 1299-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuveson, D. A., Ahearn, J. M., Matsumoto, A. K., and Fearon, D. T. (1991) J. Exp. Med. 173 1083-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tedder, T. F., Clement, L. T., and Cooper, M. D. (1984) J. Immunol. 133 678-683 [PubMed] [Google Scholar]
- 13.Fischer, E., Delibrias, C., and Kazatchkine, M. D. (1991) J. Immunol. 146 865-869 [PubMed] [Google Scholar]
- 14.Levy, E., Ambrus, J., Kahl, L., Molina, H., Tung, K., and Holers, V. M. (1992) Clin. and Exp. Immunol. 90 235-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynes, M., Aubert, J. P., Cohen, J. H., Audouin, J., Tricottet, V., Diebold, J., and Kazatchkine, M. D. (1985) J. Immunol. 135 2687-2694 [PubMed] [Google Scholar]
- 16.Iida, K., Nadler, L., and Nussenzweig, V. (1983) J. Exp. Med. 158 1021-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weis, J. J., Tedder, T. F., and Fearon, D. T. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 881-885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingeroth, J. D., Weis, J. J., Tedder, T. F., Strominger, J. L., Biro, P. A., and Fearon, D. T. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 4510-4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemerow, G. R., Houghten, R. A., Moore, M. D., and Cooper, N. R. (1989) Cell 56 369-377 [DOI] [PubMed] [Google Scholar]
- 20.Nemerow, G. R., Wolfert, R., McNaughton, M. E., and Cooper, N. R. (1985) J. Virol. 55 347-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aubry, J. P., Pochon, S., Gauchat, J. F., Nueda-Marin, A., Holers, V. M., Graber, P., Siegfried, C., and Bonnefoy, J. Y. (1994) J. Immunol. 152 5806-5813 [PubMed] [Google Scholar]
- 22.Aubry, J. P., Pochon, S., Graber, P., Jansen, K. U., and Bonnefoy, J. Y. (1992) Nature 358 505-507 [DOI] [PubMed] [Google Scholar]
- 23.Asokan, R., Hua, J., Young, K. A., Gould, H. J., Hannan, J. P., Kraus, D. M., Szakonyi, G., Grundy, G. J., Chen, X. S., Crow, M. K., and Holers, V. M. (2006) J. Immunol. 177 383-394 [DOI] [PubMed] [Google Scholar]
- 24.Delcayre, A. X., Salas, F., Mathur, S., Kovats, K., Lotz, M., and Lernhardt, W. (1991) EMBO J. 10 919-926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohnsack, J. F., and Cooper, N. R. (1988) J. Immunol. 141 2569-2576 [PubMed] [Google Scholar]
- 26.Carter, R. H., and Fearon, D. T. (1989) J. Immunol. 143 1755-1760 [PubMed] [Google Scholar]
- 27.Carter, R. H., and Fearon, D. T. (1992) Science 256 105-1071373518 [Google Scholar]
- 28.Carter, R. H., Spycher, M. O., Ng, Y. C., Hoffman, R., and Fearon, D. T. (1988) J. Immunol. 141 457-463 [PubMed] [Google Scholar]
- 29.Luxembourg, A. T., and Cooper, N. R. (1994) J. Immunol. 153 4448-4457 [PubMed] [Google Scholar]
- 30.Dempsey, P. W., Allison, M. E., Akkaraju, S., Goodnow, C. C., and Fearon, D. T. (1996) Science 271 348-350 [DOI] [PubMed] [Google Scholar]
- 31.Lyubchenko, T., dal Porto, J., Cambier, J. C., and Holers, V. M. (2005) J. Immunol. 174 3264-3272 [DOI] [PubMed] [Google Scholar]
- 32.Prota, A. E., Sage, D. R., Stehle, T., and Fingeroth, J. D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10641-10646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szakonyi, G., Guthridge, J. M., Li, D., Young, K., Holers, V. M., and Chen, X. S. (2001) Science 292 1725-1728 [DOI] [PubMed] [Google Scholar]
- 34.Gilbert, H. E., Asokan, R., Holers, V. M., and Perkins, S. J. (2006) J. Mol. Biol. 362 1132-1147 [DOI] [PubMed] [Google Scholar]
- 35.Gilbert, H. E., Eaton, J. T., Hannan, J. P., Holers, V. M., and Perkins, S. J. (2005) J. Mol. Biol. 346 859-873 [DOI] [PubMed] [Google Scholar]
- 36.Hannan, J. P., Young, K. A., Guthridge, J. M., Asokan, R., Szakonyi, G., Chen, X. S., and Holers, V. M. (2005) J. Mol. Biol. 346 845-858 [DOI] [PubMed] [Google Scholar]
- 37.Guthridge, J. M., Rakstang, J. K., Young, K. A., Hinshelwood, J., Aslam, M., Robertson, A., Gipson, M. G., Sarrias, M. R., Moore, W. T., Meagher, M., Karp, D., Lambris, J. D., Perkins, S. J., and Holers, V. M. (2001) Biochemistry 40 5931-5941 [DOI] [PubMed] [Google Scholar]
- 38.Young, K. A., Chen, X. S., Holers, V. M., and Hannan, J. P. (2007) J. Biol. Chem. 282 36614-36625 [DOI] [PubMed] [Google Scholar]
- 39.Young, K. A., Herbert, A. P., Barlow, P. N., Holers, V. M., and Hannan, J. P. (2008) J. Virol. 82 11217-11227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12366-12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittkekind, M., and Mueller, L. (1993) J. Magn. Reson. 101 201-205 [Google Scholar]
- 42.Grzesiek, S., and Bax, A. (1992) J. Magn. Reson. 96 432-440 [Google Scholar]
- 43.Zuiderweg, E. R., and Fesik, S. W. (1989) Biochemistry 28 2387-2391 [DOI] [PubMed] [Google Scholar]
- 44.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 45.Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, M., Ulrich, E. L., Markley, J. L., Ionides, J., and Laue, E. D. (2005) Proteins 59 687-696 [DOI] [PubMed] [Google Scholar]
- 46.Merrifield, R. B. (1969) Adv. Enzymol. Mol. Biol. 32 221-296 [DOI] [PubMed] [Google Scholar]
- 47.Carel, J. C., Myones, B. L., Frazier, B., and Holers, V. M. (1990) J. Biol. Chem. 265 12293-12299 [PubMed] [Google Scholar]
- 48.Molina, H., Brenner, C., Jacobi, S., Gorka, J., Carel, J. C., Kinoshita, T., and Holers, V. M. (1991) J. Biol. Chem. 266 12173-12179 [PubMed] [Google Scholar]
- 49.Guthridge, J. M., Young, K., Gipson, M. G., Sarrias, M. R., Szakonyi, G., Chen, X. S., Malaspina, A., Donoghue, E., James, J. A., Lambris, J. D., Moir, S. A., Perkins, S. J., and Holers, V. M. (2001) J. Immunol. 167 5758-5766 [DOI] [PubMed] [Google Scholar]
- 50.Morikis, D., and Lambris, J. D. (2004) J. Immunol. 172 7537-7547 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., Mallik, B., and Morikis, D. (2007) J. Mol. Biol. 369 567-583 [DOI] [PubMed] [Google Scholar]
- 52.Cavanagh, J. (2007) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., pp. 753-759, Academic Press, Inc., New York
- 53.Li, K., Okemefuna, A. I., Gor, J., Hannan, J. P., Asokan, R., Holers, V. M., and Perkins, S. J. (2008) J. Mol. Biol. 384 137-150 [DOI] [PubMed] [Google Scholar]
- 54.Sarrias, M. R., Franchini, S., Canziani, G., Argyropoulos, E., Moore, W. T., Sahu, A., and Lambris, J. D. (2001) J. Immunol. 167 1490-1499 [DOI] [PubMed] [Google Scholar]
- 55.Clemenza, L., and Isenman, D. E. (2000) J. Immunol. 165 3839-3848 [DOI] [PubMed] [Google Scholar]