Abstract
In G0 and G1, DNA double strand breaks are repaired by nonhomologous end joining, whereas in S and G2, they are also repaired by homologous recombination. The human CtIP protein controls double strand break (DSB) resection, an event that occurs effectively only in S/G2 and that promotes homologous recombination but not non-homologous end joining. Here, we mutate a highly conserved cyclin-dependent kinase (CDK) target motif in CtIP and reveal that mutating Thr-847 to Ala impairs resection, whereas mutating it to Glu to mimic constitutive phosphorylation does not. Moreover, we show that unlike cells expressing wild-type CtIP, cells expressing the Thr-to-Glu mutant resect DSBs even after CDK inhibition. Finally, we establish that Thr-847 mutations to either Ala or Glu affect DSB repair efficiency, cause hypersensitivity toward DSB-generating agents, and affect the frequency and nature of radiation-induced chromosomal rearrangements. These results suggest that CDK-mediated control of resection in human cells operates by mechanisms similar to those recently established in yeast.
DNA double strand breaks (DSBs)2 are highly cytotoxic lesions that can lead to mutations, chromosomal aberrations, or cell death. Defects in DSB signaling and/or repair can cause pathologies, including neurodegenerative disease and cancer predisposition. DSBs are repaired by two main mechanisms (1, 2): non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ ligates broken DNA ends without requiring extensive sequence complementarity and assumes the greatest importance in G0 and G1 (3). By contrast, HR is generally restricted to S and G2, where it can ensure accurate repair by using sister chromatid sequences as the repair template (4-6). Such cell cycle control of DSB repair is important because if HR is employed in G1, it can generate gross chromosomal rearrangements by using spurious homologous sequences as repair templates.
Although various mechanisms likely control HR, a prime site of regulation is at the level of 5′ to 3′ DSB resection. Resection is needed for HR but not for NHEJ and is governed by CDK activity in yeast and mammalian cells, occurring effectively in S/G2 but not G0/G1 (5-7). Recent work has shown that a key target for this control in yeast is the Sae2 protein, which is phosphorylated on Ser-267 by CDK to promote resection (8). Notably, Sae2 counterparts have been identified in other organisms, including vertebrates (9-12), and with the exception of Schizosaccharomyces pombe Ctp1 (9), they all share a short homologous region in their C termini containing a CDK consensus site that aligns with Ser-267 of Sae2 (10-12). We have recently shown that mutating Sae2 Ser-267 to Ala to prevent its phosphorylation impairs resection and consequently reduces HR, whereas altering Ser-267 to Glu mimics constitutive phosphorylation and allows some resection even in the absence of CDK activity (8). Here, we carry out analogous studies on the equivalent CDK consensus motif of CtIP and thus provide evidence that CDK-mediated control of DSB resection operates by conserved mechanisms in Saccharomyces cerevisiae and humans.
EXPERIMENTAL PROCEDURES
Cell Culture, siRNA Transfection, and Cell Survival—U2OS cells stably expressing GFP-CtIP variants were grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (BioSera), 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich) supplemented with 0.5 mg/ml G418 (Invitrogen). siRNA duplex against CtIP (GCUAAAACAGGAACGAAUC; MWG Biotec) was previously described (11), and siRNA transfections were performed with 50 nm final oligonucleotide concentrations and Oligofectamine (Invitrogen). Experiments were performed 48 h after transfection. Cell survival assays were performed as described previously (11).
Immunoblotting—Extracts were prepared in 4% SDS, 20% glycerol, 120 mm Tris-HCl, pH 6.8, and proteins were resolved by SDS-PAGE and transferred to nitrocellulose followed by immunoblotting. R. Baer (Columbia University) provided a mouse monoclonal CtIP antibody, and X. Yu (University of Michigan Medical School) provided the phospho-Ser-327 antibody. Other antibodies were from AbCam (Mre11, RPA32), Bethyl Laboratories (RPA32-pS4/S8), Santa Cruz Biotechnology (BRCA1, GST), and Roche Applied Science (GFP).
In Vitro CDK Assay—A GST-fused version of the CtIP C-terminal region (residues 790-897) was affinity-purified with glutathione-Sepharose 4B (Amersham Biosciences), incubated with recombinant CDK2/cyclin A (Upstate Biotechnology) in the presence of [γ-32P]ATP according to the manufacturer's instructions, separated by 10% SDS-PAGE, and transferred to a nitrocellulose membrane. Proteins were detected with an anti-GST antibody, and phosphorylation was visualized by autoradiography.
Laser Microirradiation—Generation of localized DNA damage by laser was done as previously (13). Cells were fixed with 4% paraformaldehyde (w/v) in PBS for 15 min, treated with 0.2% Triton X-100 in PBS for 10 min, washed three times with PBS, and then co-immunostained with antibodies against γH2AX (Cell Signaling Technology), cyclin A (Santa Cruz Biotechnology), and RPA32 (Lab Vision Corp.). For detection, Alexa Fluor 594- (red) and 647- (far red) conjugated secondary antibodies were used (Molecular Probes, Paisley, UK). Samples were visualized with an Olympus inverted confocal laser microscope by sequential scanning of the emission channels.
Immunofluorescence Microscopy—For RPA focus detection, U2OS cells expressing GFP-CtIP fusions were transfected with CtIP siRNA, and 2 days later, they were treated with 1 μm camptothecin or 10 Gy of ionizing radiation (IR) and collected 1 h afterward. Following pre-extraction for 5 min on ice (25 mm Hepes, pH 7.4, 50 mm NaCl, 1 mm EDTA, 3 mm MgCl2, 300 mm sucrose, and 0.5% Triton X-100), cells were fixed with 4% paraformaldehyde (w/v) in PBS for 15 min. Coverslips were washed three times with PBS and then co-immunostained as above.
Random Plasmid Integration—Assays were as described previously with minor modifications (14). One day after transfection with CtIP siRNA, cells were transfected with PvuI-linearized pCDNA4-HISMAX-LacZ (Invitrogen). The following day, cells were collected, counted, and plated on three plates, one containing 1 mg/ml Zeocin. One day after plating, cells on a plate lacking Zeocin were assessed for transfection efficiency by a β-galactosidase staining kit (Invitrogen), and the other two plates were incubated for 10-14 days at 37 °C for colony formation. Colonies were stained with 0.5% crystal violet, 20% ethanol and counted. Integration events (number of colonies on plates containing Zeocin) were normalized to transfection (number of β-galactosidase-positive cells) and plating efficiencies (colonies after 14 days of growth without Zeocin).
Chromosomal Analyses—Following CtIP depletion, cells were exposed to 2 Gy of IR and then allowed to recover at 37 °C for 8 h in fresh medium before chromosome preparation. Within these 8 h, cells were treated with caffeine (2 mm final concentration) for the last 5 h to allow cells with gross chromosomal rearrangements (GCRs) to overcome the G2/M checkpoint and enter mitosis, and for the last 3 h, they were treated with Colcemid (KaryoMAX, Invitrogen; final concentration 0.1 mg/ml) to induce chromosome condensation. Cells were then harvested and treated with 0.075 m KCl for 10 min at 37 °C, fixed in methanol/acetic acid (3/1), washed twice with methanol/acetic acid (3/1), and then spread on a glass microscope slide, air-dried, and 4′,6-diamidino-2-phenylindole-stained.
RESULTS
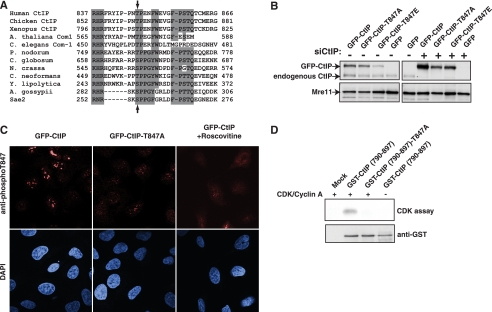
CtIP Function Is Impaired by Mutating Thr-847—A similar CDK consensus sequence to that encompassing Sae2 Ser-267 is present in nearly all Sae2/CtIP orthologues, with the analogous residue of human CtIP being Thr-847 (Fig. 1A). To investigate the potential functions of Thr-847 phosphorylation, we generated stable U2OS cell clones that expressed siRNA-resistant wild-type GFP-tagged CtIP, or siRNA-resistant CtIP derivatives in which Thr-847 was changed to an unphosphorylatable Ala (GFP-CtIP-T847A) or to a negatively charged Glu residue (GFP-CtIP-T847E) to mimic constitutive phosphorylation. We selected for clones that expressed the engineered protein at similar levels to endogenous CtIP (Fig. 1B), and a stable cell line expressing GFP alone was generated as a negative control. As shown in Fig. 1B, transfection with the CtIP siRNA oligonucleotide effectively depleted endogenous CtIP but not the siRNA-resistant GFP-CtIP fusion proteins.
FIGURE 1.
Functional effects of mutating Thr-847 of CtIP. A, alignment of the region conserved among Sae2/CtIP orthologues. Arrows show the position of the conserved CtIP Thr-847 and Sae2 Ser-267. A. thaliana, Arabidopsis thaliana; C. elegans, Caenorhabditis elegans; P. nodorum, Phaeosphaeria nodorum; C. globosum, Chaetomium globosum; N. crassa, Neurospora crassa; C. neoformans, Cryptococcus neoformans; Y. lipolytica, Yarrowia lipolytica; A. gossypii, Ashbya gossypii. B, expression levels of GFP-CtIP derivatives in stably transfected clones before (left) or after (right) siRNA depletion of endogenous CtIP (siCtIP). C, representative confocal microscope images of cells expressing wild-type or T847A CtIP variants after immunostaining with a phospho-specific antibody raised against phosphorylated Thr-847. Cells were incubated in the presence of the CDK inhibitor roscovitine where indicated. D, a GST-fused wild-type or T847A mutant CtIP C-terminal fragment (residues 750-897) was affinity-purified with glutathione-Sepharose 4B and then incubated with [γ-32P]ATP in the presence or absence of recombinant CDK2/cyclin A, separated by 10% SDS-PAGE, and transferred to nitrocellulose membrane. Proteins were detected with an anti-GST antibody (bottom), and phosphorylation was visualized by autoradiography (CDK assay; top).
As an approach to detect phosphorylation on CtIP Thr-847, we made several attempts to generate phospho-specific antisera against this site. When we assessed the resulting antisera by Western immunoblotting, however, this strategy was undermined by the antibodies recognizing additional phosphorylation sites on CtIP (data not shown). By contrast, when we used the antibodies in indirect immunofluorescence studies, we observed that a subpopulation (∼60%) of cells expressing wild-type GFP-CtIP exhibited a strongly positive signal, whereas staining was much weaker in cells expressing the CtIP variant, GFP-CtIP-T847A (we assume that the remaining, weak signal observed with the T847A mutant was due to cross-reactivity with other phosphorylation sites). Consistent with the strongly positive signal reflecting CDK-mediated modification of CtIP on Thr-847, this signal was markedly diminished when cells were pretreated with the CDK inhibitor roscovitine (Fig. 1C). To further substantiate the idea that CtIP is phosphorylated on Thr-847 by CDK, we purified bacterially expressed wild-type and Thr-847 mutated versions of a C-terminal fragment of CtIP tagged with GST. Next, we subjected these proteins to in vitro CDK phosphorylation assays with purified CDK/cyclin A and radioactive ATP (Fig. 1D). Notably, when the wild-type CtIP fragment was used, radioactive phosphate was incorporated in a CDK-dependent manner. By contrast, little or no radioactivity was incorporated into the CtIP derivative bearing the Thr-847 Thr-to-Ala mutation (Fig. 1D). Collectively, these findings thereby provided strong support for CtIP Thr-847 indeed being a CDK target.
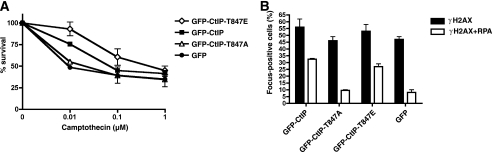
To assess the potential functions of CtIP Thr-847 phosphorylation, we siRNA-depleted endogenous CtIP from the cell clones expressing GFP-CtIP variants (Fig. 1B) and then assessed their survival following acute (1 h) treatments with various concentrations of camptothecin. In agreement with our previous data (8), cells expressing the T847A CtIP mutant were more sensitive to camptothecin than cells expressing wild-type CtIP and were essentially as sensitive as control cells expressing GFP alone (Fig. 2A). By contrast, cells expressing the T847E CtIP mutant were not hypersensitive to camptothecin and were, in fact, more resistant than cells expressing wild-type CtIP (Fig. 2A). As camptothecin cytotoxicity depends on DNA replication, it was possible that the above differences in survival reflected different cell cycle distributions among the various cell clones. However, fluorescence-activated cell sorter analyses revealed only minor differences in cell cycle distributions between the cell lines (supplemental Fig. 1A). Furthermore, all cell lines exhibited similar levels of camptothecin-induced DSB formation, as assessed by the appearance of phosphorylated histone H2AX (γH2AX) foci (Fig. 2B, black bars). However, when we analyzed the same γH2AX-positive cells for formation of RPA-coated ssDNA, significant differences were observed. RPA foci formed efficiently in cells expressing wild-type CtIP or the T847E mutant but not in cells expressing the T847A mutant or GFP alone (Fig. 2B, white bars). These results indicated that the ability of CtIP to promote camptothecin resistance is impaired when Thr-847 is mutated to an unphosphorylatable Ala residue but not when it is altered to a phospho-mimicking Glu residue. Furthermore, the camptothecin sensitivity phenotype imparted by the CtIP T847A mutation might reflect impaired processing of camptothecin-induced DSBs into ssDNA.
FIGURE 2.
Effects of CtIP mutations on cellular responses to camptothecin-induced DNA damage. A, cell survival after exposing cells expressing GFP-CtIP fusions to the indicated doses of camptothecin; averages and standard deviations (error bars) of three independent experiments are shown. B, quantification of γH2AX and RPA foci-positive cells and γH2AX focus-positive cells for the indicated CtIP variants after 1 h of treatment with 1 μm camptothecin. Averages and standard deviations (error bars) of four independent experiments are shown. At least 200 cells were counted per experiment.
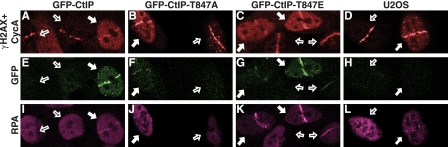
CtIP Mutations Affect Recruitment of CtIP and RPA to DNA Damage—CtIP modulates responses to DNA damage in a cell cycle-dependent manner and is only effectively recruited to sites of laser-induced damage in cyclin A-positive cells (S and G2 cells), when RPA tracts are also formed (11). As CtIP depletion impairs ssDNA formation after camptothecin treatment, this suggests that assessment of camptothecin-induced RPA focus formation would be an effective way to test the effects of mutating CtIP Thr-847. However, this approach has several limitations. First, camptothecin primarily yields DSBs only in S-phase; second, CtIP recruitment into discernible foci is difficult to observe after camptothecin treatment (11); and third, to observe RPA recruitment to sites of DNA damage, cellular preextraction is required, which impedes co-staining for soluble cell cycle markers such as cyclins. To overcome these problems, we used laser DNA-damaging microirradiation. Thus, we laser-irradiated the previously described cell clones stably expressing wild-type or mutated CtIP derivatives after they had been siRNA-depleted of endogenous CtIP. Next, we assessed the appearance of DNA damage tracts by immunofluorescence with antibodies against RPA (to detect RPA-coated ssDNA), γH2AX (to detect damaged chromatin), and cyclin A (to distinguish G1 cells from S/G2 cells. Due to limitations in the number of channels available in the microscope and in the number of non-cross-reacting secondary antibodies, γH2AX and cyclin A were analyzed in the same channel). As shown in Fig. 3, A-D, laser-induced γH2AX tracts were clearly evident in all irradiated cells, irrespective of whether or not they displayed pannuclear cyclin A staining. By contrast, and as reported previously (11), wild-type GFP-CtIP was recruited to damage sites in all S/G2 cells that stained positive for cyclin A (Fig. 3E, filled arrows) but not in cyclin A-negative G1 cells (open arrows). Notably, although a similar recruitment kinetics was observed for the CtIP-T847A mutant (supplemental Fig. 2), RPA recruitment was readily observed in S/G2 cells expressing wild-type CtIP but not in S/G2 cells expressing CtIP-T847A (Fig. 3, I and J), implying that Thr-847 phosphorylation regulates the ability of CtIP to promote resection. In line with this, whereas CtIP-T847E was effectively recruited to damage sites in S/G2 (Fig. 3G, filled arrows; supplemental Fig. 2 for kinetics), it was also recruited to some degree in G1 cells (more than 80% of G1 cells showed weak but visible GFP lines; open arrows). This suggests that mimicking constitutive phosphorylation enhances CtIP activity and raises the possibility that this might allow some CtIP function even in G1 cells. Indeed, cells expressing CtIP-T847E generally displayed more pronounced RPA recruitment than cells expressing wild-type CtIP (Fig. 3, K and I, respectively), and furthermore, weak RPA recruitment was evident in more than 90% of G1 cells expressing CtIP-T847E (Fig. 3K, open arrows), although this was less pronounced than in S/G2 cells (filled arrows). These findings thus suggest that CtIP Thr-847 controls resection during the cell cycle but imply that other CDK-mediated phosphorylations are also needed for optimal resection to occur.
FIGURE 3.
Effects of CtIP mutations on recruitment of proteins to laser-induced damage. Representative images of cells expressing GFP-CtIP variants after laser damage are shown. Cells were immunostained for RPA (magenta) and γH2AX plus cyclin A (red). Damaged cells not expressing cyclin A (G1) and cells positive for cyclin A (S/G2) are marked with empty and filled arrows, respectively.
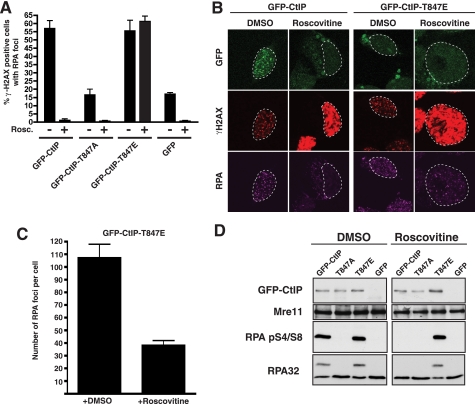
CtIP-T847E Promotes Resection Even Following CDK Inhibition—Although the above data suggest that mimicking constitutive phosphorylation of CtIP Thr-847 can partially overcome the CDK requirement for DNA DSB processing, it was formally possible that the cyclin A-negative CtIP-T847E cells displaying RPA recruitment in Fig. 3 were in early S-phase, when CDK was already active, but cyclin A levels were too weak for detection. To address this possibility, we siRNA-depleted endogenous CtIP from cells stably expressing siRNA-resistant GFP-CtIP variants and then treated them with DMSO (negative control) or with the CDK inhibitor, roscovitine. (Supplemental Fig. 1B shows that the fluorescence-activated cell sorter distributions of DMSO- and roscovitine-treated samples were similar, presumably reflecting inhibition of cell cycle transitions by roscovitine.) Next, we treated the cells with X-rays. We chose x-ray treatment because it generates DSBs in all cell cycle phases and allowed us to damage a larger number of cells than we could with laser microirradiation. Subsequently, we assessed cells for DSB formation (γH2AX foci) and ssDNA production (RPA foci). In line with our previous results, DMSO-treated cells expressing wild-type GFP-CtIP or GFP-CtIP-T847E effectively formed RPA foci, whereas cells expressing GFP-CtIP T847A or GFP alone did not (Fig. 4A). Similar results were obtained when we detected ssDNA with an anti-bromodeoxyuridine antibody labeling method (supplemental Fig. 3), indicating that CtIP Thr-847 indeed controls ssDNA formation. Taken together with our other data, these findings therefore indicate that CtIP phosphorylation on Thr-847 controls ssDNA formation and RPA recruitment to sites of damaged DNA induced by camptothecin, laser microirradiation, or X-rays. Furthermore, we found that although roscovitine severely curtailed RPA focus formation following exposure to IR in cells expressing wild-type CtIP, it did not prevent RPA focus formation in cells expressing CtIP-T847E (Fig. 4A). Nevertheless, careful comparisons revealed that roscovitine treatment did reduce the intensity and number of RPA foci in cells expressing CtIP-T847E (Fig. 4, B and C), showing that although CtIP-T847E permits resection even after CDK inhibition, this resection is less extensive than in the presence of CDK activity.
FIGURE 4.
CtIP mutations affect DSB processing. A, cells expressing CtIP variants were treated with DMSO (-) or 25 μm roscovitine (Rosc.)(+) and then irradiated with 10 Gy of IR. One h later, cells were immunostained for RPA or γH2AX. Averages and standard deviations (error bars) of three independent experiments are shown. At least 200 cells were counted per experiment. B, representative images of cells treated in A. C, the number of RPA foci per cell in cells expressing the GFP-CtIP-T847E mutant in the presence or absence of the CDK inhibitor roscovitine. Error bars, standard deviations. D, an immunoblot of protein extracts, collected 1 h after irradiation (10 Gy), of cells expressing the indicated GFP-CtIP fusions. Panels to the left and right contain samples derived from cells treated in the absence or presence of roscovitine, respectively.
Although the study of focus formation by microscopy is used commonly in the DNA damage-response field, it has some limitations. On the one hand, foci are complex structures in which several types of damage can coexist and, therefore, different DNA repair pathways can operate at the same locations. In addition, to be visible, the foci must contain thousands of protein molecules, meaning that more subtle events close to the DNA lesions might be missed. To complement our data with focus formation, we therefore prepared extracts from DNA-damaged or control cells and then analyzed them by Western immunoblotting for phosphorylation on Ser-4 and Ser-8 of the 32-kDa subunit of RPA (RPA32). These modifications are generated after different types of DNA damage (15, 16) by mechanisms that involve the DNA-dependent protein kinase (DNA-PK (17)). Although the precise roles for these RPA32 phosphorylations are not known, because they affect the affinity of RPA toward both ssDNA and double-stranded DNA (18) and increase the interaction of RPA with the recombination proteins Rad51 and Rad52 (19), it has been proposed that RPA Ser-4/8 phosphorylation facilitates RPA eviction and homologous recombination. Importantly, as RPA Ser-4/8 phosphorylation appears to only occur after DNA resection (11, 17), the detection of this modified form of RPA is a very sensitive readout of DNA end processing. Notably, we readily detected RPA32 Ser-4/8 phosphorylation, both by using phospho-specific antisera and by reduced mobility of the protein on SDS-PAGE, in irradiated cells expressing wild-type CtIP or CtIP-T847E but not in cells expressing GFP alone or GFP-CtIP T847A (Fig. 4D). Furthermore, we found that RPA32 Ser-4/8 phosphorylation was slightly higher in cells expressing CtIP-T847E than in cells expressing wild-type CtIP (Fig. 4D, left panels). Moreover, although roscovitine abolished RPA Ser-4/8 phosphorylation in cells expressing wild-type CtIP, residual RPA phosphorylation was still evident after roscovitine treatment of cells expressing CtIP-T847E (Fig. 4D, right panels). These data therefore provide strong support for CtIP Thr-847 phosphorylation playing a key role in mediating cell cycle control of DSB resection and RPA recruitment.
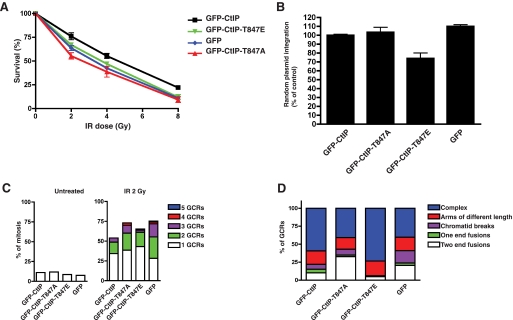
Mutation of CtIP Thr-847 Influences Genome Stability—Controlling DSB resection is thought to help ensure that the most appropriate DSB repair pathway is used at each cell cycle stage. In budding yeast, we have shown that this control depends on Sae2 Ser-267 phosphorylation by CDK and that defects in this mechanism lead to imbalances between NHEJ and HR, with the Sae2-S267A mutant favoring the former and Sae2-S267E the latter (8). Consequently, although Sae2-S267E mutant cells can repair S-phase-induced DSBs (such as are generated by camptothecin) that are mainly repaired by HR, they are mildly hypersensitive to IR due to deregulated ssDNA formation that appears to impair NHEJ in G1. To determine whether a similar phenomenon occurs in human cells, we siRNA-depleted endogenous CtIP from cells stably expressing siRNA-resistant wild-type or Thr-847 mutated GFP-CtIP derivatives and then assessed cell survival after IR. Strikingly, and in contrast to the survival data after camptothecin exposure (Fig. 2A), both CtIP T847A and CtIP-T847E mutant cells were mildly hypersensitive to IR, similar to cells lacking CtIP (Fig. 5A). To test the idea that hypersensitivity of CtIP-T847E cells might reflect impaired NHEJ, we assessed the ability of these and the other cells to randomly integrate plasmid DNA into their genome, a process that is mediated by NHEJ (14, 20). As shown in Fig. 5B, CtIP-T847E cells but not CtIP T847A or CtIP-depleted cells were mildly but significantly defective in this process, thus suggesting that the IR hypersensitivity of CtIP-T847E cells at least partly reflected impaired NHEJ.
FIGURE 5.
Effects of CtIP mutations on DNA repair and chromosome integrity. A, the survival of U2OS cells expressing GFP-CtIP fusions after treatment with IR. Averages and standard deviations (error bars) of three independent experiments are shown. B, the effects of CtIP mutations on NHEJ efficiency as measured by random plasmid integration. Frequencies of integration were normalized to the values of wild-type GFP-CtIP, set as 100%. Averages and standard deviations (error bars) of three independent experiments are shown. C, the percentage of mitoses showing 1, 2, 3, 4, or 5 GCRs in cells expressing GFP-CtIP variants either untreated (left) or treated with 2 Gy of IR (right). At least 100 mitoses were analyzed per experiment. Error bars, standard deviations. D, the percentage of each GCR type in the cells analyzed in C.
As a complementary approach to assess the effects of CtIP Thr-847 mutations, we examined the cell lines for radiation-induced GCRs. These studies revealed that 2 Gy of irradiation increased GCR formation in cells expressing wild-type CtIP, with around 50% of ensuing mitoses presenting at least one GCR event (Fig. 5C). Furthermore, somewhat more pronounced increases in GCR formation were observed in cells depleted of CtIP (GFP control) or expressing the CtIP mutants. Moreover, especially for cells expressing CtIP-T847A or the GFP control, there was an increase in the proportion of mitoses exhibiting multiple GCR events (Fig. 5C). In addition to determining overall GCR frequencies in the various cell lines, we also analyzed the data qualitatively by classifying GCRs into the following categories (Fig. 5D and supplemental Fig. 4): “two-end fusions” in which both arms of two chromosomes are fused; “one-end fusions” in which only one chromosomal arm of two chromosomes is fused (both one-end and two-end fusions probably result from NHEJ); “arms of different length” in which chromosome arms exhibit different lengths, probably due to insertions, deletions, or non-reciprocal exchanges; “chromatid breaks” in which one of the chromatids is broken but is held together by cohesion with its sister chromatid; and “complex GCRs” in which two or more chromosomes have suffered reciprocal exchanges, probably through aberrant HR. Strikingly, when compared with cells expressing wild-type CtIP, those expressing CtIP-T847E displayed an increased prevalence of complex GCRs and a reduction of fusions, which would be in agreement with increased aberrant HR and decreased NHEJ in the CtIP-T847E mutant (Fig. 5D). By contrast, cells expressing CtIP-T847A displayed the opposite pattern, exhibiting decreased complex GCRs and increased fusions, which would be consistent with increased NHEJ and reduced HR in this context (this pattern was similar to that of control cells expressing GFP alone; Fig. 5D). Collectively, these data therefore support a model in which CDK-mediated phosphorylation of CtIP Thr-847 controls DSB resection and thereby influences choice between the NHEJ and HR pathways of DSB repair.
DISCUSSION
We have investigated the mechanisms by which human cells control DNA end resection during the cell cycle. Collectively, our data provide strong support for a model in which resection is controlled by CDK-mediated modification of CtIP on Thr-847 in a manner that is analogous to CDK targeting of a related consensus site (Ser-267) on the budding yeast CtIP counterpart, Sae2 (8). In light of these findings and the fact that analogous CDK target sites are found in almost all eukaryotic Sae2/CtIP counterparts, we speculate that this mode of resection control represents a widespread mechanism to regulate DSB repair during the cell cycle.
Modification of CtIP Thr-847 Modulates DNA Resection during the Cell Cycle—Our results reveal that CtIP Thr-847 is needed for effective ssDNA generation, RPA recruitment, and RPA phosphorylation in response to camptothecin, laser-induced DNA damage, or ionizing radiation. Furthermore, we have established that mutating Thr-847 to Glu to mimic constitutive phosphorylation allows some resection in the absence of CDK activity, although this is not as effective as resection in S or G2 cells. Although it is possible that the T847E mutation does not fully mimic phosphorylation on this site, we think that this is unlikely because in S/G2 cells, the CtIP-T847E mutant promoted resection at least as well as wild-type CtIP. Instead, we favor a model (similar that to proposed for yeast Sae2 (Ref. 8)) in which CDK targeting of CtIP Thr-847 contributes to DSB resection but in which additional CDK targets, on other proteins or on CtIP itself, are needed for optimal resection. In this regard, we note that CtIP is CDK-phosphorylated on Ser-327 to promote interactions with BRCA1, a protein implicated in events occurring at resected DSBs (21). Furthermore, although we have generated phospho-specific antisera against peptides containing CtIP Thr-847 and they recognize CtIP only when CDKs are active, phospho-reactivity on Western blots was not abolished when CtIP was mutated on Thr-847 or was doubly mutated on Ser-327 and Thr-847 (data not shown). Although these results have unfortunately precluded the use of such antibodies to study Thr-847 phosphorylation by Western blot analyses, they nevertheless point to there being other CDK-dependent sites on CtIP that might potentially control its function.
Effects of Phosphorylation on CtIP Activity—How phosphorylation of CtIP modulates resection is still not well understood. The CtIP counterpart in budding yeast, Sae2, has been shown to be an endonuclease that acts cooperatively with the Mre11-Rad50-Xrs2 (MRX) complex in vitro (22). Furthermore, combining CtIP with human Mre11 and Rad50 was shown to produce an endonuclease activity that neither component exhibited alone, although it is not yet known whether CtIP stimulates Mre11 or vice versa or both (11). Notably, the in vitro results for the yeast and human proteins were obtained in the absence of any known protein phosphorylation events, suggesting that phosphorylation of Sae2/CtIP is not absolutely required for its observed biochemical functions. Nevertheless, it is established that both CDK-dependent and checkpoint-dependent phosphorylations are required for activation of Sae2/CtIP in vivo (8, 21, 23). Although it is not yet clear why there are these apparent differences between the in vivo and in vitro data, it is possible that, within the cell, Sae2/MRX and CtIP/MRN (Mre11-Rad50-Nbs1) operate in a more stringent environment and/or do so in the presence of inhibitory proteins such that their resection functions are only exhibited upon activating phosphorylations. Alternatively, or in addition, it is possible that these modifications promote interactions with additional factors that might act as positive regulators of DNA resection in S/G2. In this regard, it is noteworthy that the interaction between CtIP and BRCA1 is controlled by CDK-mediated phosphorylation of CtIP Ser-327 and that such an interaction is required for CtIP recruitment to repair centers (21). Although this suggests that CDK-mediated Thr-847 phosphorylation might prime CtIP for further phosphorylation on Ser-327, thus allowing CtIP recruitment to repair foci, several lines of evidence argue against this. First, CtIP T847A is recruited to sites of DNA damage with similar kinetics to those of wild-type CtIP or the T847E mutant (supplemental Fig. 2). Second, we have found that the T847A mutant is still phosphorylated on Ser-327 and that it is still able to interact with BRCA1 (supplemental Fig. 5). Notably, we have also found that Thr-847 phosphorylation does not control the interaction between CtIP and MRN (supplemental Fig. 5) despite the fact that this interaction occurs through interactions that involve the CtIP C-terminal region (11). Hence, we propose that CDK activity is likely to control CtIP activity in at least two ways: one operating through Thr-847 that affects CtIP function by a mechanism that also operates in S. cerevisiae (via Sae2 Ser-267) and a second, higher eukaryote-specific mechanism that operates through BRCA1 interacting with CtIP phosphorylated on Ser-327. Notably, this latter mechanism seems to play a crucial role in bringing about CtIP recruitment to DNA damage sites (21), a mechanism that does not appear to exist in budding yeast, where it has been shown that Sae2 is always chromatin-bound and can localize to sites of DNA damage in both G1 and S/G2 independently of any factors tested so far (24, 25).
Deregulated DNA End Resection Leads to DNA Damage Hypersensitivity—Irrespective of the precise mechanisms at play, what seems clear is that CDK targeting of CtIP/Sae2 is used to restrict resection in phases of the cell cycle when NHEJ is favored and activate resection in S and G2, when HR is employed. Indeed, we have shown that lack of CtIP or an inability to activate CtIP by CDK-mediated phosphorylation on Thr-847 causes hypersensitivity toward DSB-generating agents, at least partly due to defective HR in S/G2. This is especially relevant when DNA DSBs are generated by agents such as camptothecin in S-phase, when HR is required for DNA replication restart. Conversely, our data imply that artificial activation of CtIP by mimicking constitutive phosphorylation of Thr-847 (which allows some DNA resection in the absence of CDK activity) can also result in deleterious consequences and DNA damage hypersensitivity. Therefore, the fine regulation of DNA resection during the cell cycle appears to be critical to ensure that the cell uses the DSB repair pathway most appropriate to its cell cycle status. Notably, we have found that both impaired resection and constitutive activation of resection (by blocking or mimicking constitutive phosphorylation of CtIP Thr-847, respectively) lead to increased sensitivity to DNA-damaging agents and increased formation of GCRs, although by differing mechanisms. Thus, although hampering resection in S/G2 leads to increased chromosomal fusions, probably due to increased NHEJ, hyperactivation of resection results in DSBs being processed at inappropriate times. Such aberrantly processed ends can then engage in futile HR cycles, resulting in complex GCRs and causing cell mortality, probably by mitotic catastrophe.
It is noteworthy that not only CtIP mutations but also reduced CtIP levels and CtIP overexpression have been reported in various cancer cell lines (26-29) and that reduced CtIP levels lead to tumor formation in mouse models (30). Although these effects could reflect transcriptional functions for CtIP (31), it is tempting to speculate that they arise due to deregulated DSB resection, thus causing mutations and GCRs that foster tumor formation. For instance, overexpression of CtIP, especially in G1 when CtIP levels are normally low, might hyperactivate the DNA resection machinery, thus causing spurious HR or leading to large deletions at sites of NHEJ. On the other hand, CtIP-inactivating mutations or CtIP haplo-insufficiency could resemble what we have observed with CtIP T847A mutants, in which GCRs apparently arise due to reduced HR. If deregulated DSB resection is found in certain cancer cells, this raises the exciting prospect that such defects could be exploited by intelligent tailoring of existing DNA-damaging chemotherapies and/or by using compounds that selectively target specific DNA repair pathways.
Conserved Mechanisms for Regulating DSB Processing by CtIP/Sae2 Orthologues—The data we have obtained through mutating CtIP Thr-847 have a strong resonance with results derived through analyzing the effects of mutating the analogous motif of S. cerevisiae Sae2 (8). It therefore seems that this aspect of CDK-mediated control of DSB processing and repair has been highly conserved throughout evolution. As analogous CDK target sites are present in almost all eukaryotic counterparts of Sae2/CtIP (10-12), it is tempting to speculate that similar control mechanisms operate in diverse eukaryotes to modulate DSB repair pathway choice during the cell cycle. Strikingly, the only Sae2/CtIP homologue known that lacks a site analogous to Thr-847 of human CtIP is S. pombe Ctp1 (9). Nevertheless, S. pombe Ctp1 is phosphorylated on other sites by CDK, and its abundance is strongly regulated during the cell cycle at the transcriptional level, suggesting that these mechanisms serve to control its activity (9, 32). Contrary to S. cerevisiae Sae2, which is constitutively expressed in all phases of the cell cycle, human CtIP is also regulated at the transcriptional level, its amounts being highest in S and G2. It is therefore tempting to speculate that human CtIP combines characteristics exhibited by both S. cerevisiae Sae2 and S. pombe Ctp1. Furthermore, it is noteworthy that the functions of CtIP-interacting proteins such as BRCA1, which promotes CtIP recruitment to sites of DNA damage, and retinoblastoma protein 1 (RB), which controls the G1/S transition, are also cell cycle-regulated (28, 33-35). This suggests that multiple layers of regulation combine to fine-tune CtIP activity during the cell cycle, perhaps to provide even more stringent control of DSB resection than is evident in the yeast systems.
Supplementary Material
Acknowledgments
We thank Andreas Meier for providing the U2OS-GFP strain and Julia Coates, Jeanine Harrigan, Andreas Meier, Sophie Polo, Yaron Galanty, and Alessandro Sartori for technical advice. We also thank J. Harrigan and K. Dry for critical reading of the manuscript. CtIP and CtIP phospho-Ser-327 antibodies were kindly provided by R. Baer (Columbia University) and X. Yu (University of Michigan Medical School).
This work was supported by grants from Cancer Research UK and the European Community (Integrated Project DNA repair, Grant LSHG-CT-2005-512113, and Genomic Instability in Cancer and Precancer, Grant HEALTH-F2-2007-201630) (to S. P. J.) and Biotechnology and Biological Sciences Research Council Research Grant BBF0016651 (to P. H.).
The on-line version of this article (available at http://www.jbc.org) contains five supplemental figures.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: DSB, double strand break; NHEJ, non-homologous end joining; HR, homologous recombination; CDK, cyclin-dependent kinase; siRNA, small interfering RNA; GFP, green fluorescent protein; DMSO, dimethyl sulfoxide; GST, glutathione S-transferase; PBS, phosphate-buffered saline; IR, ionizing radiation; Gy, grays; GCR, gross chromosomal rearrangement; ssDNA, single-stranded DNA.
References
- 1.Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008) Cell Res. 18 134-147 [DOI] [PubMed] [Google Scholar]
- 2.Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y., and Takeda, S. (2006) DNA Repair 5 1021-1029 [DOI] [PubMed] [Google Scholar]
- 3.Lieber, M. R. (2008) J. Biol. Chem. 283 1-5 [DOI] [PubMed] [Google Scholar]
- 4.West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4 435-445 [DOI] [PubMed] [Google Scholar]
- 5.Aylon, Y., Liefshitz, B., and Kupiec, M. (2004) EMBO J. 23 4868-4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ira, G., Pellicioli, A., Balijja, A., Wang, X., Fiorani, S., Carotenuto, W., Liberi, G., Bressan, D., Wan, L., Hollingsworth, N. M., Haber, J. E., and Foiani, M. (2004) Nature 431 1011-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jazayeri, A., Falck, J., Lukas, C., Bartek, J., Smith, G. M., Lukas, J., and Jackson, S. P. (2006) Nat. Cell Biol. 8 37-45 [DOI] [PubMed] [Google Scholar]
- 8.Huertas, P., Cortés-Ledesma, F., Sartori, A. A., Aguilera, A., and Jackson, S. P. (2008) Nature 455 689-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limbo, O., Chahwan, C., Yamada, Y., de Bruin, R. A., Wittenberg, C., and Russell, P. (2007) Mol. Cell 28 134-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penkner, A., Portik-Dobos, Z., Tang, L., Schnabel, R., Novatchkova, M., Jantsch, V., and Loidl, J. (2007) EMBO J. 26 5071-5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartori, A. A., Lukas, C., Coates, J., Mistrik, M., Fu, S., Bartek, J., Baer, R., Lukas, J., and Jackson, S. P. (2007) Nature 450 509-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uanschou, C., Siwiec, T., Pedrosa-Harand, A., Kerzendorfer, C., Sanchez-Moran, E., Novatchkova, M., Akimcheva, S., Woglar, A., Klein, F., and Schlogelhofer, P. (2007) EMBO J. 26 5061-5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman, J. R., and Jackson, S. P. (2008) EMBO Rep. 9 795-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stucki, M., Clapperton, J. A., Mohammad, D., Yaffe, M. B., Smerdon, S. J., and Jackson, S. P. (2005) Cell 123 1213-1226 [DOI] [PubMed] [Google Scholar]
- 15.Shao, R. G., Cao, C. X., Zhang, H., Kohn, K. W., Wold, M. S., and Pommier, Y. (1999) EMBO J. 18 1397-1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakasai, R., Shinohe, K., Ichijima, Y., Okita, N., Shibata, A., Asahina, K., and Teraoka, H. (2006) Genes Cells 11 237-246 [DOI] [PubMed] [Google Scholar]
- 17.Cruet-Hennequart, S., Glynn, M. T., Murillo, L. S., Coyne, S., and Carty, M. P. (2008) DNA Repair 7 582-596 [DOI] [PubMed] [Google Scholar]
- 18.Binz, S. K., Sheehan, A. M., and Wold, M. S. (2004) DNA Repair 3 1015-1024 [DOI] [PubMed] [Google Scholar]
- 19.Wu, Xiaoming, Yang, Zhengguan, Liu, Yiyong, Zou, Yue (2005) Biochem. J. 391 473-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington, J., Chih-lin, H., Gerton, J., Bosma, G., and Lieber, M. R. (1992) Mol. Cell. Biol. 12 4758-4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, X., and Chen, J. (2004) Mol. Cell. Biol. 24 9478-9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengsfeld, B. M., Rattray, A. J., Bhaskara, V., Ghirlando, R., and Paull, T. T. (2007) Mol. Cell 28 638-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baroni, E., Viscardi, V., Cartagena-Lirola, H., Lucchini, G., and Longhese, M. P. (2004) Mol. Cell. Biol. 24 4151-4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H.-S., Vijayakumar, S., Reger, M., Harrison, J. C., Haber, J. E., Weil, C. F., and Petrini, J. H. J. (2008) Genetics 178 711-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004) Cell 118 699-713 [DOI] [PubMed] [Google Scholar]
- 26.Liu, F., and Lee, W.-H. (2006) Mol. Cell. Biol. 26 3124-3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, J., Lv, S., Qin, Y., Shu, F., Xu, Y., Chen, J., Xu, B., Sun, X., and Wu, J. (2007) Biochim. Biophys. Acta 1770 273-278 [DOI] [PubMed] [Google Scholar]
- 28.Wong, A. K., Ormonde, P. A., Pero, R., Chen, Y., Lian, L., Salada, G., Berry, S., Lawrence, Q., Dayananth, P., Ha, P., Tavtigian, S. V., Teng, D. H., and Bartel, P. L. (1998) Oncogene 17 2279-2285 [DOI] [PubMed] [Google Scholar]
- 29.Vilkki, S., Launonen, V., Karhu, A., Sistonen, P., Vastrik, I., and Aaltonen, L. A. (2002) J. Med. Genet. 39 785-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, P.-L., Liu, F., Cai, S., Lin, X., Li, A., Chen, Y., Gu, B., Lee, E. Y.-H. L., and Lee, W.-H. (2005) Mol. Cell. Biol. 25 3535-3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, G., and Lee, W.-H. (2006) Cell Cycle 5 1592-1596 [DOI] [PubMed] [Google Scholar]
- 32.Akamatsu, Y., Murayama, Y., Yamada, T., Nakazaki, T., Tsutsui, Y., Ohta, K., and Iwasaki, H. (2008) Mol. Cell. Biol. 28 3639-3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fusco, C., Reymond, A., and Zervos, A. S. (1998) Genomics 51 351-358 [DOI] [PubMed] [Google Scholar]
- 34.Yu, X., Wu, L. C., Bowcock, A. M., Aronheim, A., and Baer, R. (1998) J. Biol. Chem. 273 25338-25392 [DOI] [PubMed] [Google Scholar]
- 35.Chen, L., Nievera, C. J., Lee, A. Y.-L., and Wu, X. (2008) J. Biol. Chem. 283 7713-7720 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.