Abstract
The proto-oncogene Ras undergoes a series of post-translational modifications at its carboxyl-terminal CAAX motif that are essential for its proper membrane localization and function. One step in this process is the cleavage of the CAAX motif by the enzyme Ras-converting enzyme 1 (RCE1). Here we show that the deubiquitinating enzyme USP17 negatively regulates the activity of RCE1. We demonstrate that USP17 expression blocks Ras membrane localization and activation, thereby inhibiting phosphorylation of the downstream kinases MEK and ERK. Furthermore, we show that this effect is caused by the loss of RCE1 catalytic activity as a result of its deubiquitination by USP17. We also show that USP17 and RCE1 co-localize at the endoplasmic reticulum and that USP17 cannot block proliferation or Ras membrane localization in RCE1 null cells. These studies demonstrate that USP17 modulates Ras processing and activation, at least in part, by regulating RCE1 activity.
Five families of deubiquitinating enzymes consisting of ∼95 members have been identified including the ubiquitin carboxyl-terminal hydrolases, the ubiquitin-specific proteases (USPs),2 the Machado-Joseph disease protein domain proteases, the ovarian tumor proteases, and the JAMM motif proteases (1). The USPs, including the DUB/USP17 subfamily, are cysteine proteases identified by the inclusion of typical histidine and cysteine boxes within their catalytic domain (1).
In recent years there has been significant progress in our understanding of the specific functions of many deubiquitinating enzymes. These include regulation of processes such as NF-κB activation (CYLD, A20, Cezanne, and USP31) (2-8), the initiation of DNA repair (USP1) (9, 10), and control of the cell cycle (USP16) (11). In addition, several have been implicated in cancer including CYLD, the tumor suppressor gene responsible for familial cylindromatosis (12), HAUSP/USP7 for deubiquitination of mdm-2 and effects on p53 stability (13, 14), and Unp/USP4, which is overexpressed in both lung and adrenocortical carcinomas (15, 16). Also, mutations of ubiquitin carboxyl-terminal hydrolase L1 have been identified in one family with a history of Parkinson disease (17), and a particular polymorphism in ubiquitin carboxyl-terminal hydrolase L1 has been associated with reduced susceptibility to Parkinson disease (18).
The DUB/USP17 subfamily were originally identified as murine hematopoietic specific genes (DUB-1, DUB-1A, and DUB-2) induced in response to a range of cytokines (19, 20). We have identified a human orthologue DUB-3/USP17 (subsequently referred to as USP17) that is also cytokine-regulated (21, 22).
Several lines of evidence suggest that the DUB/USP17 family regulate cell growth and survival. First, a number of murine (DUB-1, DUB-1A, and DUB-2) and human (USP17) family members have been shown to be cytokine-inducible immediate early genes (19-21, 23) and DUB-1 expression resulted in cell cycle arrest prior to S phase (24). Additionally, DUB-2 expression can markedly inhibit apoptosis induced by cytokine withdrawal (25), and we have reported that constitutive expression of human USP17 can block cell proliferation (21). However, the targets of USP17 have not been identified.
The small GTPase p21Ras (Ras) has long been recognized as an important regulator of cell proliferation, and constitutively active mutants have been associated with many cancers (26, 27). Although Ras is activated by cycling from the GDP- to the GTP-bound states, membrane localization is also essential for its activation. This requires a series of processing events that involve modifications of its carboxyl-terminal CAAX motif. Briefly, the cysteine residue is isoprenylated by a farnesyltransferase (FTase) (28), and Ras is localized to the endoplasmic reticulum (ER) where the three carboxyl-terminal amino acids (i.e. the -AAX) are removed by Ras-converting enzyme 1 (RCE1), an ER integral transmembrane protease (29-31). Subsequently, the exposed isoprenylcysteine is methylated by an ER transmembrane methyltransferase called isoprenylcysteine carboxyl methyltransferase (ICMT) (32). Depending on the Ras isoform, it may then be palmitoylated (33, 34) prior to plasma membrane trafficking.
Little is known about the mode of action of RCE1. It had initially been identified as a cysteine protease, but subsequent studies would suggest that it has metalloprotease-like activity (35, 36). Nonetheless, it has been clearly shown that loss of RCE1 blocks proper Ras localization and impedes cell growth and transformation (30, 37, 38).
In this study we show that USP17 disrupts Ras plasma membrane localization and activation and markedly inhibits proliferation. Moreover USP17 also blocks RCE1 activity, and this effect is dependent on USP17 deubiquitinating activity.
EXPERIMENTAL PROCEDURES
Plasmids—The pME18S-USP17-FLAG, pME18S-USP17CS-FLAG, pMX-ires-EGFP-USP17-FLAG, and pMX-ires-EGFP-USP17CS-FLAG expression constructs have been previously described (21). Peroxisomal 3-oxoacyl-CoA thiolase A and Galectin1 cDNAs were tagged with the HA epitope at their amino termini by standard PCR-based methods. Each cDNA was amplified by PCR, using a 5′ oligonucleotide containing an XbaI site as well as a 3′ oligonucleotide containing a BglII site. The XbaI/BglII PCR fragment was subcloned between the XbaI and BglII sites of a modified pCEFL vector in frame with the HA epitope. RCE1 cDNA was tagged with the FLAG epitope at its carboxyl termini as described previously (21). The HA and FLAG epitope-tagged RCE1 was produced as above using a 5′ oligonucleotide containing an XbaI site and a 3′ oligonucleotide lacking a stop codon but containing a FLAG epitope sequence followed by a BglII site. The pEFIRES-HA-ubiquitin, pEFIRES-HA-R63K, pEFIRES-HA-R48K, GFP-H-Ras, pCMV.TAG2A.FLAG.FTaseα, pcDNA3.1-CYLD-HA, pcDNA3.1-CYLDCS-HA, and pSUPER-USP17shRNA (shRNA1 target sequence, GCAGGAAGATGCCCATGAA) constructs were obtained as gifts from various sources (see “Acknowledgments”). The pRS-USP17shRNA (shRNA2 target sequence, GATGATTTGGCTCCTGTGGCAAGACAGCT) construct was purchased from Origene Technologies (Rockville, MD).
Cell Cultures and Transfections—The IL-3-dependent pro-B cell lines Ba/F3, PlatinumE, and 293T were cultured as described previously (21). HeLa and MDA-MB-231 (kind gift from Dr. David Waugh, Queen's University Belfast) cells were grown in Dulbecco's modified Eagle's medium (PAA Laboratories, Somerset, UK) supplemented with 10% fetal calf serum, 1% penicillin/streptomycin, 1% l-glutamine, and 1% sodium pyruvate. RCE1-/- MEF and RCE1+/+ MEF cells (kind gift of Dr. S. G. Young, University of California, San Francisco) were grown as described previously (31). Transfectants were generated using FuGENE 6 transfection reagent (Roche Applied Science), as specified by the manufacturer.
Cell Lysis, Immunoprecipitations, and Immunoblotting—Whole cell lysates and immunoprecipitations were generated and separated prior to immunoblotting as previously described (21). The following antibodies were used: anti-FLAG (M2; Sigma, Dorset, UK); anti-DUB-3/USP17 (Fusion Antibodies, Belfast, UK); anti-phospho-ERK1/2, ERK2, phosho-MEK1/2, MEK1/2, HA or pan-Ras (Cell Signaling); anti-FTaseα (BD Biosciences); and anti-HA (12ca5; Roche Applied Science).
Confocal Microscopy—MEF, Cos7 or HeLa cells were seeded at 2.5 × 104 cells/1.7 cm2 well of LabTek II, CC2-treated chamber slides (Nalge Nunc, Hereford, UK). Cells, as necessary, were transfected with 0.25 μg of each plasmid using FuGENE 6 transfection reagent (Roche Applied Science). 24 h after transfection, or 48 h after seeding, the cells were fixed with 4% paraformaldehyde in CBS for 20 min at room temperature. The slides were stained as necessary with the appropriate antibodies and washed with phosphate-buffered saline. The following antibodies were used: anti-FLAG (M2, Sigma), anti-DUB-3/USP17 (Fusion Antibodies), anti-HA (12ca5; Roche Applied Science), and anti-calnexin (Jackson Laboratories). The slides were then viewed on a Zeiss LSM 510 META NLO Confocal Microscope. The images presented in the same panels were captured using standardized settings and exposure times (Zeiss, Hertfordshire, UK).
Retroviral Infection—Retroviral infections of RCE1+/+ MEF and RCE1-/- MEF cells were carried out as previously described (21).
Ras RBD Pulldown Assay—Ba/F3 cells were cultured in the presence or absence of tetracycline for 48 h before being transferred to serum-free medium for 4 h to minimize basal activation. They were then stimulated with 100 units/ml IL-3 for the indicated time points prior to lysis. MDA-MB-231 cells were transfected as previously described and rested in 0% fetal calf serum Dulbecco's modified Eagle's medium for 4 h prior to serum stimulation. The cells were immediately washed with ice-cold phosphate-buffered saline and lysed in lysis buffer containing 0.5 m Tris, pH 7.5, 0.1 m MgCl2, 0.5 m NaCl, 1% Triton, and 5% glycerol with 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride, 2.5 mm benzamidine, and 2 mm Na3VO4, and GST pulldown was carried out. Lysate was rested on ice for 10 min and spun down at 12,000 RPM for 10 min to pellet the membrane. Lysate was removed and added to RBD-GST fusion protein preassociated GST beads (Amersham Biosciences, Buckinghamshire, UK). The pulldowns were separated and immunoblotted as previously described using a pan-Ras antibody.
CAAX Cleavage Assay—The cells were lysed as previously described (39), and CAAX assays were performed using membrane fractions prepared as previously described (36), and a quenched flurogenic peptide, ABZ-KSKTKC(f)QLIM was obtained from AnaSpec. Membrane fractions (50 μl) were incubated with 50 μl of assay buffer (100 mm HEPES, pH 7.5, 5 mm MgCl2) with 40 μm of flurogenic peptide. The fluorescence generated was recorded at excitation and emission wavelengths of 320 and 420 nm, respectively, using a Cytofluor® Multi-well series 4000 spectrofluorimeter, and all reactions were performed in triplicate on a black 96-well plate.
RNA Extraction and RT-PCR—RNA was extracted and RT-PCR was carried out as previously described (21).
RESULTS
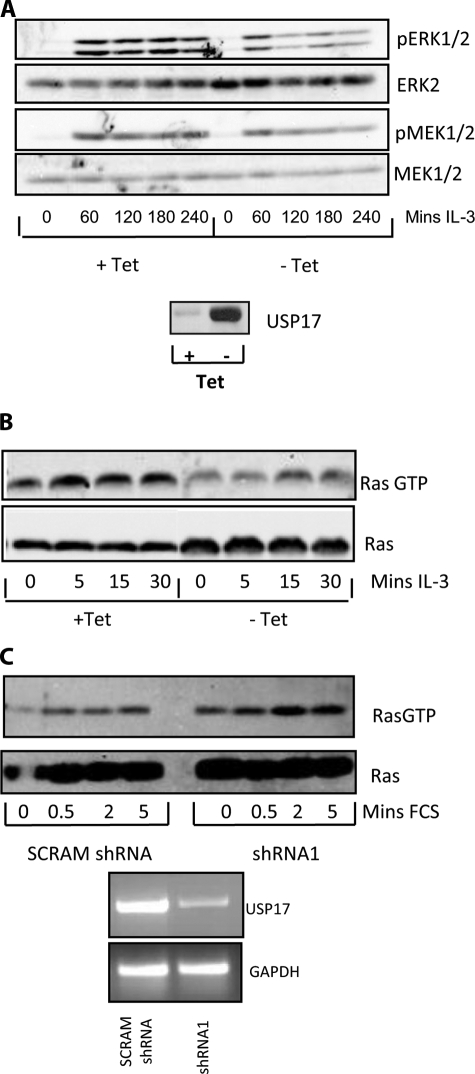
USP17 Regulates Ras/MEK/ERK Signaling—We have previously shown that constitutive expression of USP17 blocks cell growth. To determine how this occurs, we examined its effect on intracellular signaling using a previously described IL-3-dependent Ba/F3 cell line in which the expression of USP17 is blocked by tetracycline (21). Ba/F3 cells, cultured in the presence or absence of tetracycline for 48 h, were stimulated with IL-3, and activation of ERK was examined using a phospho-ERK-specific antibody. ERK1/2 activation was markedly down-regulated by USP17 expression (Fig. 1A, top panels, and supplemental Fig. S1). We also found that USP17 expression decreased the activation of upstream MEK (Fig. 1A, lower panels, and supplemental Fig. S1). These results suggested that the expression of USP17 may modulate the Ras pathway, and therefore IL-3 induced Ras activity was also assessed using a RBD pulldown assay. This assay involves using a recombinant GST-tagged Raf-RBD to pull down Ras. The Raf-RBD only interacts with active GTP-bound Ras and thus determines Ras activity. In the presence of USP17, the levels of activated Ras were found to be markedly reduced (Fig. 1B and supplemental Fig. S1).
FIGURE 1.
USP17 blocks the Ras/MEK/ERK pathway. A, Ba/F3 cells expressing tetracycline-regulated USP17 were cultured for 48 h with and without tetracycline (Tet), cultured in the absence of fetal calf serum (FCS) for 4 h, and treated with IL-3 (100 units/ml) for the time periods specified and lysates immunoblotted with phospho-specific antibodies for ERK and MEK. In addition, to show equal loading, the same immunoblots were subsequently immunoblotted with antibodies specific for ERK2 and MEK1/2. The bottom panel shows USP17 expression. B, Ba/F3 cells expressing tetracycline regulated USP17 were cultured as above and treated with IL-3 for the time periods specified. Pulldowns were performed using GST-tagged Raf-RBD fusion protein. The resultant pulldowns and protein lysates were immunoblotted with pan-Ras antibody. C, MDA-MB-231 cells transfected as indicated were cultured without serum for 4 h and stimulated with serum for the periods specified. The resultant Raf-RBD pulldowns (upper panel), and protein lysates (lower panel) were immunoblotted with a pan-Ras antibody. In addition RNA was extracted from parallel transfections, and RT-PCR was performed as indicated.
Because constitutive expression of USP17 decreased Ras activation, we also hypothesized that knockdown of endogenous USP17 would enhance Ras activation. Therefore, we transiently transfected USP17-specific shRNA (shRNA1) and observed the levels of GTP-bound Ras following serum stimulation. The shRNA efficiently knocked down USP17 expression (Fig. 1C, lower panel), resulting in a marked elevation in the level of GTP-bound Ras (Fig. 1C, upper panel, and supplemental Fig. S1). These results, which were representative of three independent experiments and subsequently have been repeated with an independent shRNA for USP17, indicated that Ras activity was regulated by USP17.
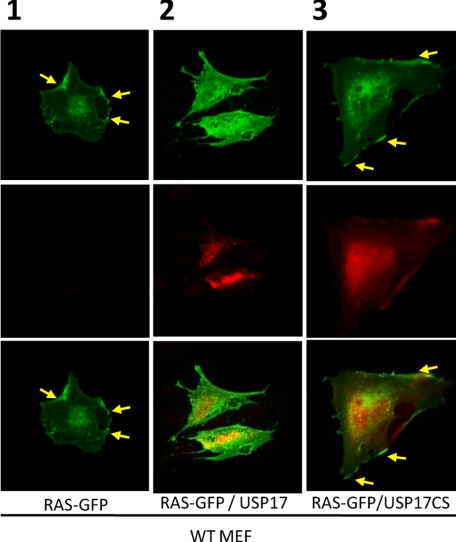
USP17 Regulates Ras Localization—These results suggested that USP17 was impeding the Ras/MEK/ERK signaling pathway. Therefore, because trafficking of Ras to the plasma membrane is required for Ras/MEK/ERK signaling, we decided to determine whether USP17 could alter Ras localization. The localization of Ras to the plasma membrane is controlled by the processing of its carboxyl terminus, and previous studies have used GFP-tagged Ras to monitor its localization. Therefore, wild type (WT) mouse embryonic fibroblasts (MEF) were transfected with constructs expressing GFP-H-Ras in conjunction with either USP17 or catalytically inactive USP17CS (Fig. 2 and supplemental Fig. S3). In more than three separate experiments, when expressed alone or in conjunction with USP17CS, the majority of GFP-H-Ras localized to the plasma membrane as expected (Fig. 2, see yellow arrows in panels 1 and 3). However, in the presence of USP17, although some GFP-H-Ras was present at the plasma membrane, in repeated experiments H-Ras showed a clear cytosolic distribution (Fig. 2, panel 2). Similar experiments using a shRNA targeting USP17 showed no effect upon GFP-H-Ras localization (supplemental Fig. S3).
FIGURE 2.
USP17 blocks Ras membrane localization. WT MEF cells transiently transfected with vectors expressing GFP-H-Ras, USP17, or USP17CS were fixed. The localization of GFP-H-Ras (top row) and USP17 or USP17CS (middle row), as well as both together (bottom row) was then assessed by confocal microscopy. In panels 1 and 3, the yellow arrows indicate membrane localization of GFP-H-Ras.
USP17 Deubiquitinates RCE1—Processing of the Ras carboxyl-terminal CAAX motif is essential for Ras membrane localization and is mediated in a multi-step process by a number of enzymes, namely FTase (28), RCE1 (29-31), and ICMT (32). On the basis of our data showing that the expression of USP17 blocked the transfer of Ras to the plasma membrane, we examined the possibility that one or more of these Ras-processing enzymes was a substrate for USP17.
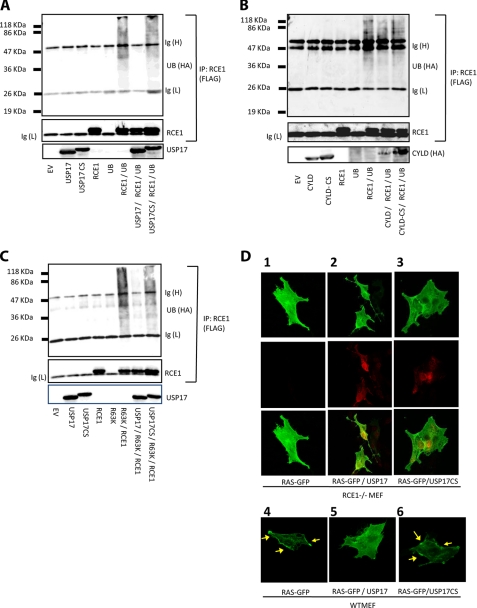
Therefore we examined whether these enzymes could be ubiquitinated and whether USP17 could reverse this modification. First we examined the Ras-processing enzymes ICMT and the FTase α-subunit but did not see any evidence of ubiquitination, even in the presence of overexpressed HA-tagged ubiquitin (results not shown). However, when we co-expressed RCE1 with ubiquitin and RCE1 immunoprecipitates were immunoblotted for the presence of ubiquitin, smearing was observed that would indicate RCE1 could be modified by this peptide (Fig. 3A, sixth lane). This smearing was removed upon expression of USP17 (Fig. 3A, seventh lane), but not USP17CS (Fig. 3A, eighth lane), indicating that catalytically competent USP17 acted to remove ubiquitin from RCE1. Moreover, this deubiquitination was specific because the deubiquitinating enzyme CYLD did not remove ubiquitin from RCE1 (Fig. 3B, seventh lane).
FIGURE 3.
USP17 deubiquitinates RCE1. 293T cells were transiently transfected with vectors expressing either RCE1-FLAG, HA-ubiquitin, USP17, or USP17CS. The lysates were extracted and immunoprecipitated where indicated with the FLAG epitope antibody and immunoblotted for: USP17, HA epitope-tagged ubiquitin, and FLAG epitope-tagged RCE1 (A); HA epitope-tagged CYLD, FLAG epitope-tagged RCE1, and HA epitope-tagged ubiquitin (B); and HA epitope-tagged K63 ubiquitin, FLAG epitope-tagged RCE1, and USP17 (C). D, RCE1-/- (panels 1-3) and WT (panels 4-6) MEF cells transiently transfected with the indicated expression vectors were fixed. The localization of GFP-H-Ras (top row) and USP17 or USP17CS (middle row), as well as both together (bottom row) was then assessed by confocal microscopy. The yellow arrows indicate membrane localization of GFP-H-Ras.
Interestingly, RCE1 protein levels were not enhanced in the presence of USP17, indicating that the ubiquitination of RCE1 was not promoting turnover of RCE1 by proteasomal degradation, and therefore we further examined the type of ubiquitin chains that form on RCE1 using mutant ubiquitin constructs (Fig. 3C). When a ubiquitin mutant that can only form K63 branched ubiquitin chains was present, the characteristic smears were once again observed, indicating that K63-branched ubiquitin chains could be attached to RCE1 and that USP17 could remove these chains (Fig. 3C). In contrast, the expression of a ubiquitin mutant that only formed K48-linked chains resulted in RCE1 degradation (supplemental Fig. S2). This indicated that K63 linked ubiquitin chains can conjugate to RCE1 and that they can be removed by USP17. Because no alteration in RCE1 protein levels were observed ± USP17, it suggested that ubiquitin was modulating RCE1 in a manner independent of protein turnover. K63 ubiquitin chains have previously been associated with modulation of protein function (39, 40), and we hypothesized that USP17 may modulate the activity of RCE1.
This hypothesis was further strengthened when we studied the localization of GFP-H-Ras in MEF cells lacking the enzyme RCE1 (Fig. 3D, panels 1-3). GFP-H-Ras expressed in RCE1 null cells exhibited cytosolic distribution, whether expressed alone or in conjunction with USP17 or USP17CS (Fig. 3D, panels 1-3). This suggested that the presence of RCE1 was required to observe the USP17-induced effect. In addition, when the localization of GFP-H-Ras in RCE1-/- MEF cells ((Fig. 3D, panels 1-3) was compared with the localization in WT MEF cells (Fig. 3D, panels 4-6), it was apparent that the pattern observed in RCE1 null cells was similar to that observed in WT MEF cells expressing USP17 (Fig. 3D, panel 5). This clearly demonstrated the dependence of RCE1 to produce the USP17-induced effect on Ras.
USP17 Regulates RCE1 Activity—To determine whether USP17 could attenuate the proteolytic activity of RCE1, we employed a CAAX cleavage assay using a farnesylated internally quenched fluorescent peptidyl substrate derived from the carboxyl terminus of K-Ras, which releases its quencher and fluoresces after RCE1-mediated cleavage (36). 293T cells were transfected with combinations of RCE1, USP17, or USP17CS, and microsomal membrane preparations were assessed for RCE1 activity. Expression of USP17, but not USP17CS, markedly reduced both the rate and total turnover of the substrate (Fig. 4A, top panel), indicating that the deubiquitinating activity of USP17 attenuated RCE1 activity. Greater than 50% reduction in RCE1 activity in the presence of USP17 was observed consistently over three independent experiments. Furthermore, equivalent RCE1 expression confirmed that the reduction in RCE1 activity was not merely due to changes in protein levels (Fig. 4A, bottom panel).
FIGURE 4.
USP17 blocks RCE1 activity. 293T cells were transiently transfected with either empty vector (EV) or vectors expressing RCE1-FLAG, USP17, USP17CS, shRNA1, or scrambled shRNA. Membrane fractions were extracted and incubated with a RCE1-specific quenched fluorescent peptidyl substrate, and activity was determined by monitoring hydrolysis of this substrate in the presence of each membrane fraction in a time course assay (A and B, top panels). Concurrently, protein lysates were extracted from a sample of the transfection, and equal amounts of protein were immunoprecipitated and analyzed as in Fig. 2 to determine that equal amounts of RCE1 were present in each sample assessed for RCE1 activity (A, lower panel, and B, lower left panel). RNA was extracted and RT-PCR was performed as indicated to check shRNA knockdown (B, lower right panel). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The ability of USP17 to block RCE1 activity was further confirmed using a shRNA specifically targeting USP17 (shRNA1) (Fig. 4B, bottom right panel). As before, 293T cells were transiently transfected with RCE1 in the presence and absence of the USP17-specific shRNA, and RCE1 activity was determined. In the presence of the shRNA, the level of RCE1 activity was consistently augmented (Fig. 4B, top panel), suggesting that removal of endogenous USP17 enhanced RCE1 activity. Collectively, these observations show that USP17 inhibits RCE1 proteolytic activity, thus attenuating Ras activation. This was further supported by the use of another USP17-specific shRNA (shRNA2), which gave similar results (supplemental Fig. S2).
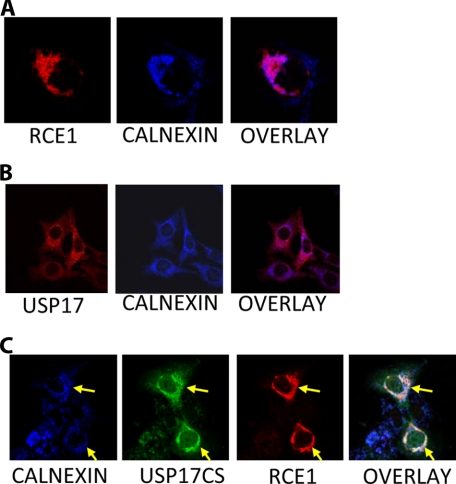
USP17 and RCE1 Co-localize at the Endoplasmic Reticulum—Previously, RCE1 had been shown localized at the ER in yeast (40), so we set out to determine its localization in mammalian cells and examine whether it co-localized with USP17. Cells transfected with RCE1 displayed a perinuclear distribution consistent with the protein being restricted to the ER (Fig. 5A, left panel). This was confirmed by simultaneous staining using an antibody to the ER marker calnexin (Fig. 5A, middle panel), which exhibited consistent and clear co-localization with RCE1 (Fig. 5A, right panel).
FIGURE 5.
USP17 and RCE1 co-localize. A, Cos7 cells were transiently transfected with HA-RCE1. RCE1 was visualized by confocal using an anti-HA antibody (first panel), and an anti-calnexin antibody (second panel) was used to visualize the ER. An overlay is also provided (third panel). B, the endogenous distribution of USP17 was examined in HeLa cells. USP17 was visualized using a USP17 specific monoclonal antibody (first panel), and an anti-calnexin antibody (second panel) was used to visualize the ER. An overlay is also provided (third panel). C, Cos7 cells were transiently transfected with USP17CS-FLAG and HA-RCE1. USP17CS was visualized using an anti-FLAG antibody (second panel) and HA-RCE1 (third panel) and the ER (first panel) were visualized as above. An overlay is also provided (fourth panel).
Localization of endogenous USP17 was examined in HeLa cells with a USP17 antibody (Fig. 5B). USP17 was distributed throughout the cell, but a more concentrated signal indicates that perinuclear distribution was observed, consistent with ER localization (Fig. 5B, left panel). This was further confirmed by simultaneous staining with calnexin (Fig. 5B, middle panel), which suggested that much of USP17 co-localized with the ER (Fig. 5B, right panel).
The localization of USP17 with RCE1 was further supported by experiments with co-transfected RCE1 and USP17CS. The localization of these proteins were examined using anti-HA (Fig. 5C, third panel), anti-FLAG (Fig. 5C, second panel), and calnexin antibodies (Fig. 5C, first panel). USP17CS exhibited marked perinuclear distribution and co-localized with RCE1 and the ER marker calnexin (Fig. 5C, first, second, and third panels, see yellow arrows), indicating that the loss of catalytic activity may trap USP17 at the ER (Fig. 5C, fourth panel, arrows).
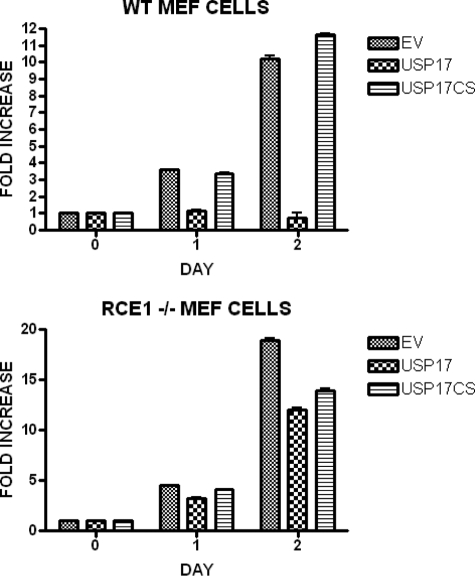
USP17 Inhibition of Cell Proliferation Is RCE1-dependent—We have previously shown that USP17 blocks cell proliferation in both NIH3T3 and Ba/F3 cells (21), and it was important to establish whether this inhibition was dependent on RCE1, as already suggested by our previous results. We therefore established WT and RCE1-/- MEF cells expressing either USP17 or USP17CS using retroviral infection with a bicistronic vector also expressing EGFP. The proportion of cells expressing USP17 was assessed using EGFP. The fraction of cells expressing USP17 in the WT MEF cells consistently dropped by more than 10-fold over a 2-week period.
Therefore, to examine this effect quantitatively, we again infected cells as above and sorted for EGFP expression 24 h after infection to obtain populations >99% EGFP positive. We measured the proliferation of these cells by trypan exclusion assay over 48 h. In three separate experiments, WT MEF cells expressing USP17 failed to proliferate, whereas those infected with empty vector (EGFP alone) or expressing the inactive USP17CS proliferated as normal (Fig. 6, upper panel). By sharp contrast, the population of RCE1-/- cells expressing USP17 proliferated at a similar rate to those expressing either USP17CS or the controls (Fig. 6, lower panel). These results demonstrated that in the absence of RCE1, USP17 was unable to block proliferation, clearly showing that the function of USP17 was dependent on RCE1.
FIGURE 6.
USP17 regulation of growth is RCE1-dependent. WT and RCE1-/- MEF cells were infected with either bicistronic retroviral vectors expressing EGFP and either USP17-FLAG or USP17CS-FLAG or with empty vector (EV). The cells were sorted upon EGFP expression to produce populations of >99% positive cells, and then the proliferation of each population was monitored by trypan blue exclusion assay. The results are illustrated as fold increase of cell numbers over time.
DISCUSSION
The enzyme RCE1 regulates Ras processing, membrane localization, and activation, but whether RCE1 activity is regulated has not been reported to date. In this study we have shown that RCE1 is ubiquitinated and that USP17 acts to remove this post-translational modification. The presence of USP17 markedly down-regulates RCE1 activity, demonstrated by the loss of Ras trafficking to the plasma membrane as well as the down-regulation of the Ras/MEK/ERK pathway. These observations represent the first evidence that this deubiquitinating enzyme regulates Ras signaling and that ubiquitination may control Ras processing.
It has become apparent that ubiquitination is a key regulatory mechanism for intracellular signaling, and several recent reports have identified crucial roles for deubiquitinating enzymes in the regulation of a number of signaling events such as NF-κB activation and DNA repair. In particular, the deubiquitinating enzyme CYLD, originally identified as a tumor suppressor involved in cylindromatosis (43), has been shown to regulate NF-κB activation by removing K63-linked polyubiquitin chains from TRAF2, TRAF6, and IKKγ (2, 3, 4). In addition, the enzyme USP1 is thought to regulate the DNA damage response and has been shown to remove monoubiquitin from both FANCD2 (9) and the sliding clamp proliferating cell nuclear antigen (10), until DNA damage triggers its auto-cleavage, allowing the accumulation of monoubiquitinated FANCD2 and proliferating cell nuclear antigen and the initiation of DNA repair.
Our observations suggest that the activity of the proto-oncogene Ras is similarly regulated by a deubiquitinating enzyme. Ras is localized to the plasma membrane through a series of processing events involving modifications of its carboxyl-terminal CAAX motif resulting in relocalization of Ras to the plasma membrane. These modifications are facilitated by a series of enzymes including FTase (28), RCE1 (29-31), and ICMT (32). Previously, modulation of FTase activity has been shown to occur through phosphorylation of the α- and β-subunits (44-46), and this phosphorylation is induced in response to growth factor stimulation (46). However, no reports have indicated that either RCE1 or ICMT can be regulated, and to date it has been assumed that these enzymes are constitutively active.
RCE1 knock-out mice die at embryonic day 15, suggesting that this enzyme plays a critical role in development, although the exact cause of death was unclear (30). Heart defects have been suggested as a possible cause of mortality and indeed have also been observed in RCE1fl/fl α-myosin heavy chainCre mice, which die between 3 and 5 months as a result of cardiomyopathy (47). More interestingly, RCE1-/- MEF cells have been shown to proliferate more slowly and be less susceptible to transformation and failed to localize farnesylated Ras to the plasma membrane (30, 38).
Our findings suggest that the expression of USP17 has similar effects to the loss of RCE1 and support the theory that USP17 regulates RCE1 activity. In particular, our initial observation that USP17 expression causes down-regulation of the Ras/MEK/ERK pathway would fit with a previous study showing that in cells lacking RCE1, ERK phosphorylation was not eliminated but was clearly down-regulated (47). In addition, the ability of USP17 to relocalize GFP-H-Ras from the plasma membrane to the cytosol is consistent with a previous paper showing that the absence of RCE1 results in similarly altered localization (38).
The hypothesis that USP17 could act on RCE1 was confirmed by the observation that RCE1 was ubiquitinated and that USP17 could specifically deubiquitinate RCE1. In addition, we were able to demonstrate that USP17 could markedly block RCE1 activity and that knockdown of endogenous USP17 using a shRNA consistently boosted this activity.
Compellingly, we showed that K63 ubiquitin chains could be conjugated to RCE1 and that these chains can be removed by USP17. Ubiquitination, normally associated with protein stability, can also affect protein function, and the addition of K63-linked polyubiquitin chains has previously been shown to allow the activation of TRAF2, TRAF6, and IKKγ (48). This is required for NF-κB activation, and the removal of these chains by CYLD has been shown to negatively regulate the NF-κB signaling pathway (2, 3, 5).
It is as yet unclear how the addition of K63-linked ubiquitin chains plays a role in the regulation of RCE1 activity, but their addition could result in a conformational change that results in the activation of this enzyme. Alternatively, they could allow the recruitment of interacting proteins necessary for RCE1 activity or the dislodgement of inhibitors. Whatever the case, further studies will be required to determine exactly how this ubiquitination contributes to the regulation of RCE1.
The observation that endogenous USP17 and RCE1 are co-localized at the ER and that overexpressed inactive USP17CS appears trapped at the ER indicated that these enzymes may interact. This is interesting because other catalytically inactive proteases, such as matrix metalloproteases, have been used to trap their target substrates (49). The importance of the relationship between USP17 and RCE1 was further strengthened by the observation that USP17 could not block proliferation of cells lacking RCE1. These observations strongly support a direct link between the two enzymes and suggest that RCE1 is downstream of USP17.
We have previously demonstrated that USP17 is transiently induced in response to cytokines, including IL-4 and IL-6 (21). Our current findings would indicate that its role may be to regulate signaling through the Ras/MEK/ERK pathway, possibly in a feedback loop. Previously members of the SOCS family, induced by the JAK-STAT signaling pathway, have been shown to act in this manner to regulate the STAT pathway (50). USP17 would appear to have a similar function in the Ras/MEK/ERK pathway by blocking Ras processing, and it could be hypothesized that this rapid and transient USP17 expression could limit signaling in this pathway.
Many other CAAX box-containing proteins such as RhoA, Rac1, and Cdc42 have been implicated in cell cycle control (51), and a block of RCE1 activity should influence their processing and signaling. This may be the case, but there is conflicting evidence whether or not RCE1 and ICMT are important in the processing of other GTPases, clearly showing that further study of CAAX box processing is required (38, 51). Whatever the case, there are many prenylated proteins important for proliferation such as those already mentioned and the likes of CENP-E, CENP-F, RhoB, and Rheb, and studies of farnesyltransferase inhibitors have suggested that Ras is not the only prenylated protein that contributes to proliferation (52). Interestingly, USP17 transfected WT MEF cells showed altered morphology. It is possible that the constitutive expression of USP17 may, as mentioned above, affect the processing of other CAAX box-containing proteins, such as Rac1, Cdc42, and Rap1, which have been linked to the regulation of the cytoskeleton. A previous study (38) had suggested that the loss of RCE1 affected the localization and function of Ras proteins but had no effect on the localization and function of Rho proteins including Rac1 and Cdc42. However, a more recent study (51) has now suggested that GTPases other than Ras are affected by the loss of RCE1, and this may explain the changes in morphology. It is also interesting to note that the morphology of the RCE1-/- cells is similar to that of the WT MEF cells transfected with USP17. Therefore this could suggest that, although we see a specific effect on Ras localization and signaling, other substrates for RCE1 could contribute to the observed effects on cell growth.
In summary, we have shown that USP17 can regulate Ras localization and Ras/MEK/ERK signaling, at least in part, through regulation of the ubiquitination and activity of RCE1. These results implicate USP17 as a novel regulator of the Ras processing pathway, and because Ras is a key proto-oncogene in a number of cancer types, this processing pathway has become a focus for therapeutic intervention. Indeed, inhibitors of FTase have already been used in the clinic, and inhibitors of RCE1 and ICMT are under development (53). Thus the knowledge that Ras processing and activation are regulated by deubiquitination not only advances our understanding of this pathway but may be of relevance for future cancer therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Prof. Yossi Yarden (Weizmann Institute, Rehovot, Israel) (pEFIRES-HA-ubiquitin, pEFIRES-HA-R63K, and pEFIRES-HA-R48K), Prof. Rene Bernards (Netherlands Cancer Institute, Amsterdam, The Netherlands) (pcDNA3.1-CYLD-HA and pcDNA3.1-CYLDCS-HA), Dr. Rene Medema (University Medical Center, Utrecht) (pSUPER-USP17shRNA), Dr. S. G. Young (University of California, San Francisco) (GFP-H-Ras, RCE1+/+ and RCE1-/- MEF cells), Dr. David Waugh (Queen's University Belfast) (MDA-MB-231) and Dr. Marc Goalstone (University of Colorado, Denver) (pCMV.TAG2A.FLAG.FTaseα) for the kind gifts. We also acknowledge Dr. Irwin Hollander (Wyeth Research) for help with regards to the CAAX cleavage assay, James Jonkman (Princess Margaret Hospital, University Health Network, Toronto, Canada) for help regarding confocal microscopy, and Dr. Alfonso Blanco Fernández (University College Dublin, Dublin, Ireland) for help with cell sorting.
This work was supported in part by the Biotechnology and Biological Sciences Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1-S3.
Footnotes
The abbreviations used are: USP, ubiquitin-specific protease; RCE, Ras-converting enzyme; ER, endoplasmic reticulum; FTase, farnesyltransferase; HA, hemagglutinin; IL, interleukin; ERK, extracellular signal-regulated kinase; MEK, mitogen-activated protein kinase/ERK kinase; GST, glutathione S-transferase; RT, reverse transcription; RBD, Raf-Ras-binding domain; shRNA, small hairpin RNA; GFP, green fluorescent protein; WT, wild type; MEF, mouse embryonic fibroblast(s); EGFP, enhanced GFP; ICMT, isoprenylcysteine carboxyl methyltransferase.
References
- 1.Nijman, S. M., Luna-Vargas, M., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., and Bernards, R. (2005) Cell 123 773-786 [DOI] [PubMed] [Google Scholar]
- 2.Trompouki, E., Hatzivassiliou, E., Tsichritzis, T., Farmer, H., Ashworth, A., and Mosialos, G. (2003) Nature 424 793-796 [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp, T. R., Nijman, S. M., Dirac, A. M., and Bernards, R. (2003) Nature 424 797-801 [DOI] [PubMed] [Google Scholar]
- 4.Kovalenko, A., Chable-Bessia, C., Cantarella, G., Israel, A., Wallach, D., and Courtois, G. (2003) Nature 424 801-805 [DOI] [PubMed] [Google Scholar]
- 5.Wertz, I. E., O'Rourke, K. M., Zhou, H., Eby, M., Aravind, L., Seshagiri, S., Wu, P., Wiesmann, C., Baker, R., Boone, D. L., Ma, A., Koonin, E. V., and Dixit, V. M. (2004) Nature 430 694-699 [DOI] [PubMed] [Google Scholar]
- 6.Boone, D. L., Turer, E. E., Lee, E. G., Ahmad, R. C., Wheeler, M. T., Tsui, C., Hurley, P., Chien, M., Chai, S., Hitotsumatsu, O., McNally, E., Pickart, C., and Ma, A. (2004) Nat. Immunol. 5 1052-1060 [DOI] [PubMed] [Google Scholar]
- 7.Evans, P. C., Smith, T. S., Lai, M. J., Williams, M. G., Burke, D. F., Heyninck, K., Kreike, M. M., Beyaert, R., Blundell, T. L., and Kilshaw P. J. (2003) J. Biol. Chem. 278 23180-23186 [DOI] [PubMed] [Google Scholar]
- 8.Tzimas, C., Michailidou, G., Arsenakis, M., Kieff, E., Mosialos, G., and Hatzivassiliou, E. G. (2006) Cell Signal. 18 83-92 [DOI] [PubMed] [Google Scholar]
- 9.Nijman, S. M., Huang, T. T., Dirac, A. M., Brummelkamp, T. R., Kerkhoven, R. M., D'Andrea, A. D., and Bernards, R. (2005) Mol. Cell 17 331-339 [DOI] [PubMed] [Google Scholar]
- 10.Huang, T. T., Nijman, S. M., Mirchandani, K. D., Galardy, P. J., Cohn, M. A., Haas, W., Gygi, S. P., Ploegh, H. L., Bernards, R., and D'Andrea, A. D. (2006) Nat. Cell Biol. 8 339-347 [DOI] [PubMed] [Google Scholar]
- 11.Joo, H. Y., Zhai, L., Yang, C., Nie, S., Erdjument-Bromage, H., Tempst, P., Chang, C., and Wang, H. (2007) Nature 449 1068-1072 [DOI] [PubMed] [Google Scholar]
- 12.Bowen, S., Gill, M., Lee, D. A., Fisher, G., Geronemus, R. G., Vazquez, M. E., and Celebi, J. T. (2005) J. Investig. Dermatol. 124 919-920 [DOI] [PubMed] [Google Scholar]
- 13.Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J., and Gu, W. (2002) Nature 416 648-653 [DOI] [PubMed] [Google Scholar]
- 14.Sheng, Y., Saridakis, V., Sarkari, F., Duan, S., Wu, T., Arrowsmith, C. H., and Frappier, L. (2006) Nat. Struct. Mol. Biol. 13 285-291 [DOI] [PubMed] [Google Scholar]
- 15.Gray, D. A., Inazawa, J., Gupta, K., Wong, A., Ueda, R., and Takahashi, T. (1995) Oncogene 10 2179-2183 [PubMed] [Google Scholar]
- 16.Velazquez-Fernandez, D., Laurell, C., Geli, J., Hoog, A., Odeberg, J., Kjellman, M., Lundeberg, J., Hamberger, B., Nilsson, P., and Backdahl, M. (2005) Surgery 138 1087-1094 [DOI] [PubMed] [Google Scholar]
- 17.Lincoln, S., Vaughan, J., Wood, N., Baker, M., Adamson, J., Gwinn-Hardy, K., Lynch, T., Hardy, J., and Farrer, M. (1999) Neuroreport 10 427-429 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, J., Hattori, N., Leroy, E., Morris, H. R., Kubo, S., Kobayashi, T., Wood, N. W., Polymeropoulos, M. H., and Y. Mizuno, Y. (2000) Parkinsonism Relat. Disord. 6 195-197 [DOI] [PubMed] [Google Scholar]
- 19.Zhu, Y., Os Pless, M., Inhorn, R., Mathey-Prevot, B., and D'Andrea, A. D. (1996) Mol. Cell Biol. 16 4808-4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, Y., Lambert, K., Corless, C., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., and D'Andrea, A. D. (1997) J. Biol. Chem. 272 51-57 [DOI] [PubMed] [Google Scholar]
- 21.Burrows, J. F., McGrattan, M. J., Rascle, A., Humbert, M., Baek, K. H., and Johnston, J. A. (2004) J. Biol. Chem. 279 13993-14000 [DOI] [PubMed] [Google Scholar]
- 22.Burrows, J. F., McGrattan, M. J., and Johnston, J. A. (2005) Genomics 85 524-529 [DOI] [PubMed] [Google Scholar]
- 23.Baek, K.-H., Kim, M. S., Kim, Y. S., Shin, J. M., and Choi, H. K. (2004) J. Biol. Chem. 279 2368-2376 [DOI] [PubMed] [Google Scholar]
- 24.Zhu, Y., Carroll, M., Papa, F. R., HochStrasser, M., and D'Andrea, A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3275-3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Migone, T., Humbert, M., Rascle, A., Sanden, D., D'Andrea, A., and Johnston, J. A. (2001) Blood 98 1935-1941 [DOI] [PubMed] [Google Scholar]
- 26.Shields, J. M., Pruitt, K., McFall, A., Shaub, A., and Der, C. J. (2000) Trends Cell Biol. 10 147-154 [DOI] [PubMed] [Google Scholar]
- 27.Malumbres, M., and Barbacid, M. (2003) Nat. Rev. Cancer 3 459-465 [DOI] [PubMed] [Google Scholar]
- 28.Reiss, Y., Goldstein, J. L., Seabra, M. C., Casey, P. J., and Brown, M. S. (1990) Cell 62 81-88 [DOI] [PubMed] [Google Scholar]
- 29.Boyartchuk, V. L., Ashby, M. N., and Rine, J. (1997) Science 275 1796-1800 [DOI] [PubMed] [Google Scholar]
- 30.Kim, E., Ambroziak, P., Otto, J. C., Taylor, B., Ashby, M., Shannon, K., Casey, P. J., and Young, S. G. (1999) J. Biol. Chem. 274 8383-8390 [DOI] [PubMed] [Google Scholar]
- 31.Otto, J. C., Kim, E., Young, S. G., and Casey, P. J. (1999) J. Biol. Chem. 274 8379-8382 [DOI] [PubMed] [Google Scholar]
- 32.Hrycyna, C. A., Sapperstein, S. K., Clarke, S., and Michaelis, S. (1991) EMBO J. 10 1699-1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choy, E., Chiu, V. K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I. E., and Philips, M. R. (1999) Cell 98 69-80 [DOI] [PubMed] [Google Scholar]
- 34.Apolloni, A., Prior, I. A., Lindsay, M., Parton, R. G., and Hancock, J. F. (2000) Mol. Cell Biol. 20 2475-2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolence, J. M., Steward, L. E., Dolence, E. K., Wong, D. H., and Poulter, C. D. (2000) Biochemistry 39 4096-4104 [DOI] [PubMed] [Google Scholar]
- 36.Plummer, L. J., Hildebrandt, E. R., Porter, S. B., Rogers, V. A., McCracken, J., and Schmidt, W. K. (2006) J. Biol. Chem. 281 4596-4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergo, M. O., Ambroziak, P., Gregory, C., George, A., Otto, J. C., Kim, E., Nagase, H., Casey, P. J., Balmain, A., and Young, S. G. (2002) Mol. Cell Biol. 22 171-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelson, D., Ali, W., Chiu, V. K., Bergo, M., Silletti, J., Wright, L., Young, S. G., and Philips, M. (2005) Mol. Biol. Cell 16 1606-1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickhart, C. M. (2000) Trends Biochem. Sci., 25 544-548 [DOI] [PubMed] [Google Scholar]
- 40.Ikeda, F., and Dikic, I. (2008) EMBO Rep. 9 536-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deleted in proof
- 42.Deleted in proof
- 43.Bignell, G. R., Warren, W., Seal, S., Takahashi, M., Rapley, E., Barfoot, R., Green, H., Brown, C., Biggs, P. J., Lakhani, S. R., Jones, C., Hansen, J., Blair, E., Hofmann, B., Siebert, R., Turner, G., Evans, D. G., Schrander-Stumpel, C., Beemer, F. A., van Den Ouweland, A., Halley, D., Delpech, B., Cleveland, M. G., Leigh, I., Leisti, J., and Rasmussen, S. (2000) Nat. Genet. 25 160-165 [DOI] [PubMed] [Google Scholar]
- 44.Kumar, A., and Mehta, K. D. (1997) Neurosci. Lett. 231 143-146 [DOI] [PubMed] [Google Scholar]
- 45.Kumar, A., Beresini, M. H., Dhawan, P., and Mehta, K. D. (1996) Biochem. Biophys. Res. Commun. 222 445-452 [DOI] [PubMed] [Google Scholar]
- 46.Goalstone, M., Carel, K., Leitner, J. W., and Draznin, B. (1997) Endocrinology 138 5119-5124 [DOI] [PubMed] [Google Scholar]
- 47.Bergo, M. O., Lieu, H. D., Gavino, B. J., Ambroziak, P., Otto, J. C., Casey, P. J., Walker, Q. M., and Young, S. G. (2004) J. Biol. Chem. 279 4729-4736 [DOI] [PubMed] [Google Scholar]
- 48.Terzic. J., Marinovic-Terzic, I., Ikeda, F., and Dikic, I. (2007) Biochem. Soc. Trans. 35 942-945 [DOI] [PubMed] [Google Scholar]
- 49.Overall, C. M., Tam, E. M., Kappelhoff, R., Connor, A., Ewart, T., Morrison, C. J., Puente, X., López-Otín, C., and Seth, A. (2004) Biol. Chem. 385 493-504 [DOI] [PubMed] [Google Scholar]
- 50.Elliott, J., and Johnston, J. A. (2004) Trends Immunol. 25 434-440 [DOI] [PubMed] [Google Scholar]
- 51.Coleman, M. L., Marshall, C. J., and Olson, M. F. (2004) Nat. Rev. Mol. Cell Biol. 5 355-366 [DOI] [PubMed] [Google Scholar]
- 52.Roberts, P. J., Mitin, N., Keller, P. J., Chenette, E. J., Madigan, J. P., Currin, R. O., Cox, A. D., Wilson, O., Kirschmeier, P., and Der, C. J. (2008) J. Biol. Chem. 283 25150-25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan, S., and Yeung, S. J. (2005) Cancer Res. 65 9109-9112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.