Abstract
Helicases play critical roles in all aspects of nucleic acid metabolism by catalyzing the remodeling of DNA and RNA structures. UvrD is an abundant helicase in Escherichia coli with well characterized functions in mismatch and nucleotide excision repair and a possible role in displacement of proteins such as RecA from single-stranded DNA. The mismatch repair protein MutL is known to stimulate UvrD. Here we show that the nucleotide excision repair proteins UvrA and UvrB can together stimulate UvrD-catalyzed unwinding of a range of DNA substrates containing strand discontinuities, including forked DNA substrates. The stimulation is specific for UvrD, as UvrAB failed to stimulate Rep helicase, a UvrD homologue. Moreover, although UvrAB can promote limited strand displacement, stimulation of UvrD did not require the strand displacement function of UvrAB. We conclude that UvrAB, like MutL, modulate UvrD helicase activity. This stimulation likely plays a role in DNA strand and protein displacement by UvrD in nucleotide excision repair. Promotion of UvrD-catalyzed unwinding of nicked duplexes by UvrAB may also explain the need for UvrAB and UvrD in Okazaki fragment processing in cells lacking DNA polymerase I. More generally, these data support the idea that helicase activity is regulated in vivo, with helicases acting as part of multisubunit complexes rather than in isolation.
Helicases and translocases use the energy derived from NTP hydrolysis to translocate along single-stranded or double-stranded nucleic acids to remodel nucleic acid structures. Many of these enzymes have RecA-like motor domains, reflecting close similarities in translocation mechanisms (1-3). Specificity is often, therefore, conferred by additional domains within these motor enzymes (4). It is also becoming apparent that helicases and translocases often function as part of larger multisubunit complexes rather than in isolation and that such interactions impact on motor function and specificity. The complex interactions between replicative helicases and other proteins acting at the replication fork have long been known to be critical for replisome function (5-7). Many helicases also interact with, and their activities modulated by, ssDNA2-binding proteins (8-13), while helicase/translocase interactions with RNA polymerases are emerging (14, 15).
The Escherichia coli 3′-5′ helicase UvrD (16) has roles in mismatch (17) and nucleotide excision repair (18) and may also act to displace proteins such as RecA at replication forks or ssDNA gaps in duplex DNA (19-21). UvrD is likely the most abundant helicase in E. coli (22). There is also a second helicase in E. coli, Rep, that shares 40% identity with UvrD but has no known role in mismatch or nucleotide excision repair or in protein displacement in vivo. Employment of UvrD, but not Rep, in a diversity of roles in vivo might, therefore, demand specific physical or functional interactions between UvrD and other proteins in each system. However, specific interaction of UvrD has only been documented with a component of the mismatch repair system, MutL (23). This interaction appears to be essential in allowing a motor with limited dsDNA processivity to unwind the large tracts of DNA necessary during mismatch repair (23-26).
Little is known concerning the protein displacement function of UvrD in vivo. Lack of UvrD leads to a hyperrecombination phenotype (27), whereas UvrD can both promote (20, 21) and inhibit (20) RecA-catalyzed strand exchange in vitro, depending on reaction conditions. UvrD might, therefore, promote turnover of RecA-ssDNA complexes at blocked forks and gaps in duplex DNA. Inhibition of RecA function at blocked forks might facilitate other pathways of fork repair that do not rely on strand exchange, possibly minimizing the risks to genome stability that blocked forks present (28). However, whether abortion of strand exchange by UvrD in vivo requires other proteins is unknown. In contrast, the role of UvrD in nucleotide excision repair is well characterized. Nucleotide excision repair is needed to remove and replace bulky lesions within the genome with a complex of UvrA and -B, executing the initial damage recognition (for review, see Refs. 29 and 30). Secondary DNA damage recognition by UvrB is thought to be achieved by ATP-driven translocation of UvrB (31, 32), with damaged nucleotides sterically hindering this translocation process (33-37). However, the UvrAB complex has very limited strand displacement activity (32) which probably reflects a strand opening function rather than processive translocation in the manner of a typical helicase. The end result of this two-step damage recognition mechanism is the dissociation of UvrA and recruitment of the endonuclease UvrC via a UvrB-UvrC interaction (38, 39). Cleavage on the 3′ and then the 5′ side of the DNA damage by UvrC (40, 41) is then followed by displacement of the cleaved strand and turnover of both UvrB and C by UvrD (42), probably via an interaction between UvrB and UvrD (43).
Nucleotide excision repair proteins also have a function in DNA replication. Strains with mutations in the DNA polymerase I gene, polA, have major defects in joining of Okazaki fragments (44, 45). However, under restricted growth conditions ΔpolA strains can survive (46). Alternative means therefore exist for joining of Okazaki fragments as long as the rate of DNA replication and, therefore, the rate of Okazaki fragment production is reduced. This alternative pathway(s) of Okazaki fragment processing is poorly defined. However, viability of ΔpolA strains is dependent on the presence of UvrA, -B, and -D (47-51), suggesting that these three nucleotide excision repair proteins play key roles in this alternative pathway. Moreover, deletion of uvrC enhances the viability of ΔpolA cells implying that UvrC activity inhibits this alternative pathway (51). These in vivo data were explained by a model in which UvrAB binds to nicks in newly synthesized lagging strand DNA even in the absence of DNA damage (51). Promotion of Okazaki fragment processing was proposed to involve conferral of a specific direction on UvrD-catalyzed unwinding by binding of UvrAB, promoting removal of the RNA primers by a presumed nuclease, and subsequent sealing of the gap by a DNA polymerase and ligase.
To probe the possible function of UvrAB in modulation of UvrD helicase activity, we have analyzed their activities on a range of DNA substrates in vitro. UvrAB stimulated UvrD helicase activity on nicked duplexes. However, UvrAB had little effect on UvrD polarity; UvrD with or without UvrAB translocated preferentially on the continuous DNA strand on nicked substrates. Stimulation was also apparent on other DNA substrates containing strand discontinuities, including forked DNA substrates. UvrAB binding of any DNA structures containing strand discontinuities might, therefore, promote UvrD-catalyzed unwinding of such structures. The strand displacement function of UvrAB was not essential for stimulation of UvrD, suggesting that stimulation did not simply reflect a reduction in the UvrD processivity barrier by limited unwinding of the substrate by UvrAB. Our findings imply that stimulation of UvrD helicase activity is required in both nucleotide excision and mismatch repair. These data also explain the need for UvrA, -B, and -D to maintain viability in the absence of DNA polymerase I, but they also raise questions regarding the consequences of such an activity in normal wild type cells.
MATERIALS AND METHODS
DNA Substrates—DNA substrates were constructed by annealing complementary oligonucleotides, one of which in each structure was labeled with [γ-32P]ATP at the 5′ end and purified by gel electrophoresis (52). Sequences of the oligonucleotides are shown in Table 1, whereas the oligonucleotide composition of DNA structures is shown in Table 2. All oligonucleotides were supplied by Eurogentec.
TABLE 1.
Oligonucleotide sequences used in construction of DNA substrates
All oligonucleotides are written 5′ to 3′.
| Oligonucleotide | Sequence |
|---|---|
| 1 | AACGTCATAGACGATTACATTGGACTATCTACGTCCGAGGCTCGCGCCGCAGACTCATTTAGCCCTTATCCGTATTG CGGTCTCGAGTCGCCATGGACGGTCGACCTGTAGAAGGCTTGC |
| 2 | GCAAGCCTTCTACAGGTCGACCGTCCATGGCGACTCGAGACCGCAATACGGATAAGGGCT |
| 3 | AAATGAGTCTGCGGCGCGAGCCTCGGACGTAGATAGTCCAATGTAATCGTCTATGACGTT |
| 4 | AGCCCTTATCCGTATTGCGGTCTCGAGTCGCCATGGACGGTCGACCTGTAGAAGGCTTGC |
| 5 | GACTATCTACGTCCGAGGCTCGCGCCGCAGACTCATTTAGCCCTTATCCGTATTGCGGTCTCGAGTCGCCATGGAC GGTCGACCTGTAGAAGGCTTGC |
| 6 | GCAAGCCTTCTACAGGTCGACCGTCCATGGCGACTCGAGACCGCAATACGGATAAGGGCTGAGCACGCCGACGAACA TTCACCACGCCAGACCACGTA |
| 7 | TACGTGGTCTGGCGTGGTGAATGTTCGTCGGCGTGCTC |
| 8 | AAATGAGTCTGCGGCGCGAGCCTCGGACGTAGATAGTC |
| 9 | AACGTCATAGACGATTACATTGCTACATGGAGCTGTCTAGAGGATCCGAC |
| 10 | GTCGGATCCTCTAGACAGCTCCATG |
| 11 | TAGCAATGTAATCGTCTATGACGTT |
| 12 | GTCGGATCCTCTAGACAGCTCCATGATCACTGGCACTGGTAGAATTCGGC |
| 13 | GCCGAATTCTACCAGTGCCAGTGAT |
TABLE 2.
Oligonucleotide composition of DNA structures used in this study
Oligonucleotide numbers refer to Table 1. Asterisks indicate the 5′-labeled oligonucleotide in each substrate.
| Substrate number | Oligonucleotide numbers |
|---|---|
| 1 | 1 + 2* + 3 |
| 2 | 1 + 2 + 3* |
| 3 | 1 + 3* |
| 4 | 1 + 2* |
| 5 | 2* + 4 |
| 6 | 5* + 6 + 7 + 8 |
| 7 | 9* + 10 + 11 |
| 8 | 9* + 11 + 12 + 13 |
Proteins—All protein concentrations are stated in terms of monomers. UvrA, wild type UvrB, UvrBK45A, and SSB were overexpressed and purified as described (33, 53-55).
UvrD was overexpressed from a pETDuet vector (Merck) kindly supplied by Dr. Nigel Savery (University of Bristol). pETDuetuvrD was transformed into BL21-AI (Merck) and a 100-ml culture in LB plus 100 μg/ml ampicillin grown at 37 °C until the A650 reached 0.6, at which point the culture was stored at +4 °C overnight. The next morning the cells were pelleted by centrifugation and resuspended in 10 ml of fresh LB. This resuspension was used to inoculate 8 liters of LB plus ampicillin in a stirred vessel fermenter. The cells were grown to an A650 of 0.6 at 37 °C, then isopropyl 1-thio-β-d-galactopyranoside and arabinose were added to final concentrations of 1 mm and 0.2%, respectively. Growth was continued at 37 °C for a further 3 h before harvesting the cells by centrifugation. The cells were then resuspended (at 50% (w/w)) in 50 mm Tris-HCl, pH 7.5, at 4 °C, and 10% sucrose before flash-freezing in liquid nitrogen. The cell paste was thawed and Tris, pH 8.3, EDTA, EGTA, DTT, NaCl, and lysozyme were added to final concentrations of 50, 5, 0.5, 1, and 200 mm and 0.2 mg/ml, respectively. The mixture was left on ice for 30 min before the addition of sodium deoxycholate and deoxyribonuclease I to final concentrations of 0.05% and 5 μg/ml, respectively. Incubation was continued on ice for 30 min. NaCl was then added to a final concentration of 500 mm, and the mixture was sonicated on ice and then centrifuged at 16,000 rpm for 20 min at 4 °C in a Sorvall SS34 rotor. The supernatant was collected and then diluted by the addition of buffer A (20 mm Tris-HCl, pH 8.3, 1 mm EDTA, 0.5 mm EGTA, 1 mm DTT) to a conductivity equivalent to buffer A plus 100 mm NaCl. This diluted supernatant was then applied to a 22-ml heparin-agarose column (Sigma Aldrich) equilibrated in buffer A plus 100 mm NaCl. This and subsequent chromatography was performed at 4 °C. The column was washed with six column volumes of buffer A plus 100 mm NaCl before eluting proteins with a 32-column-volume gradient of buffer A plus 100-650 mm NaCl. UvrD eluted at ∼500 mm NaCl. Buffer A was added to the pooled fractions until the conductivity was reduced to the equivalent of buffer A plus 200 mm NaCl. This diluted sample was then loaded on to a 5-ml Hi-Trap Q-Sepharose column (GE Healthcare) equilibrated in buffer A plus 100 mm NaCl. The column was washed with six column volumes of buffer A plus 100 mm NaCl and then a 55-column-volume gradient of buffer A plus 100-600 mm NaCl. Fractions containing UvrD were then loaded onto a Sephacryl S200 26/60 gel filtration column (GE Healthcare) equilibrated in buffer A plus 200 mm NaCl. Fractions containing UvrD were pooled and diluted with buffer A until a conductivity equivalent to buffer A plus 200 mm NaCl was achieved. The diluted sample was then loaded on to a 1-ml Q-Sepharose column equilibrated in buffer A plus 200 mm NaCl and washed with 6 column volumes of buffer A plus 200 mm NaCl, and then protein was eluted with buffer A plus 700 mm NaCl. Fractions containing UvrD were pooled and then dialyzed overnight at 4 °C in 20 mm Tris-HCl, pH 8.3, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1 mm DTT, and 50% glycerol (v/v) before freezing in liquid nitrogen and subsequent storage at -80 °C.
Rep was overexpressed from a pET21arep clone kindly supplied by Dr. Ken Marians (Memorial Sloan-Kettering Cancer Center, New York). This clone was introduced into BL21 cells (Merck) using LB plus 50 μg/ml carbenicillin. 80 ml of F-medium (56) containing 50 μg/ml carbenicillin was inoculated with a single colony and incubated overnight at 37 °C. Cells were pelleted, resuspended in fresh F-medium, and inoculated into 8l of F-medium containing 100 μg/ml ampicillin in a stirred vessel fermenter. The culture was incubated at 37 °C until an A650 of 0.6 was reached, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm, and incubation was continued for a further 3 h at 37 °C. Cells were pelleted and then resuspended in Tris-HCl and sucrose and frozen as above. Cells were thawed on ice and lysed using Brij 58 and lysozyme (57). Nucleic acids were precipitated by the addition of Polymin P to a final concentration of 0.075% and centrifuged in a Sorvall SS34 at 4 °C for 20 min at 16,000 rpm. Proteins were precipitated from the supernatant by the addition of solid ammonium sulfate to 50%. The ammonium sulfate pellet was resuspended in a sufficient volume of buffer B (50 mm imidazole, pH 6.5, 1 mm EDTA, 1 mm DTT) such that the conductivity was equivalent to buffer B plus 50 mm NaCl. Proteins were then loaded onto a 50-ml SP-Sepharose column (GE Healthcare), washed with 3 column volumes of buffer B plus 50 mm NaCl, and then eluted using a 10-column-volume gradient of buffer B from 50 to 800 mm NaCl. Fractions containing Rep (centered on 370 mm) were pooled, and protein was precipitated with 50% ammonium sulfate followed by resuspension in buffer C (50 mm Tris, pH 7.5, 1 mm EDTA, 1 mm DTT) plus 50 mm NaCl. The resuspension was loaded onto a Hi-load Superdex 200 26/60 column (GE Healthcare) run in buffer C plus 50 mm NaCl. Fractions containing Rep were pooled and loaded directly onto a 3.5-ml heparin-agarose column equilibrated in buffer C plus 50 mm NaCl. The column was washed with 3 column volumes of buffer C plus 50 mm NaCl followed by a 10-column-volume gradient of buffer C plus 50-800 mm NaCl. Rep eluted at ∼530 mm NaCl. Positive fractions were pooled, diluted with buffer C to a conductivity equivalent to buffer C plus 50 mm NaCl, and then loaded on to a 1.4-ml dsDNA cellulose column (Sigma Aldrich) equilibrated in buffer C plus 50 mm NaCl. The column was washed with 3 column volumes of buffer C plus 50 mm NaCl, and then proteins were eluted using five column volumes of buffer C plus 50-1000 mm NaCl. Rep eluted at ∼700 mm NaCl. Positive fractions were pooled and diluted with buffer C to a conductivity equivalent to buffer C plus 50 mm NaCl before loading onto an ssDNA cellulose column (Sigma Aldrich) equilibrated in buffer C plus 50 mm NaCl. The column was washed with 3 column volumes of buffer C plus 50 mm NaCl before development with 20 column volumes of buffer C plus 50-1000 mm NaCl. Rep eluted at ∼650 mm NaCl. Positive fractions were pooled and dialyzed against buffer B plus 50 mm NaCl, then loaded on to a Mono S 5/5 HR column (GE Healthcare) equilibrated in buffer B plus 50 mm NaCl. Proteins were eluted with 30 column volumes of buffer B plus 50-800 mm NaCl. Fractions containing Rep were pooled and dialyzed into 50 mm Tris, pH 7.5, 1 mm DTT, 1 mm EDTA, 200 mm NaCl, and 40% glycerol (v/v) before freezing as above.
DNA Unwinding Assays—Assays were performed in 10-μl volumes using 1 nm DNA substrate in 50 mm HEPES, pH 8, 4 mm MgCl2, 2 mm DTT, 2 mm ATP, and 0.04 mg/ml bovine serum albumin. All reaction components except proteins were assembled and incubated for 2 min at 37 °C. When included, SSB was then added and incubation continued for 2 min. UvrA and/or UvrB were then added, and incubation was continued for 2 min. UvrD or Rep was then added, and incubation was continued at 37 °C for 10 min. Reactions were terminated by the addition of 2.5 μl of 100 mm Tris-HCl, pH 7.5, 5% SDS, 200 mm EDTA, and 10 mg/ml proteinase K. Termination was carried out at 37 °C for 20 min. 2.5 μl of 0.25% bromphenol blue, and 30% glycerol was then added. Samples were loaded onto 16 × 10-cm 10% polyacrylamide gels in 1× Tris-buffered EDTA and electrophoresed at 190 V for 90-150 min depending on substrate structure. Gels were dried and analyzed using phosphorimaging and autoradiography.
Time courses were performed in a similar manner except that reaction volumes were increased, with 10-μl aliquots removed at the indicated times. Time 0 was the point at which UvrD was added to the reaction.
RESULTS
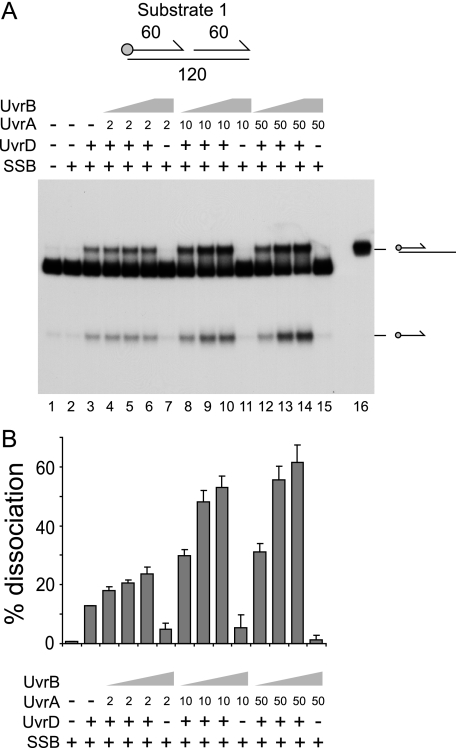
UvrA and -B Promote Unwinding of Nicked Duplex Substrates by UvrD—In the absence of DNA polymerase I, UvrAB and UvrD may be needed to unwind DNA at Okazaki fragment junctions (51). These junctions can be viewed as nicked duplexes. We tested the effects of UvrAB on UvrD-catalyzed unwinding of a model nicked duplex having two 60-bp duplex arms (Fig. 1). SSB was also included, given that the product of any unwinding, ssDNA, would be bound by SSB in vivo. The addition of UvrD generated two products, one of which migrated above the substrate, whereas the other migrated below (Fig. 1A, lane 3). The upper product proved to be a partial duplex product generated by unwinding of the downstream 60-mer oligonucleotide (Fig. 1A, lane 16). The slower migration of this product as compared with the initial substrate was likely due to secondary structure formation within the exposed 60 bases of ssDNA upon deproteinization of the reactions before loading on the gel. The faster-migrating product was the labeled upstream 60-mer oligonucleotide. UvrD could therefore unwind both 60-mer strands, although whether displacement of the labeled oligonucleotide was a secondary product of unwinding the partial duplex product cannot be ascertained from this experiment.
FIGURE 1.
UvrA and -B stimulate UvrD-catalyzed unwinding of a nicked duplex. A, unwinding of substrate 1 (numbers indicate the length in base pairs of each duplex) in the presence of SSB (125 nm), UvrD (10 nm), UvrA (nm concentrations shown), and UvrB (10 nm in lanes 4, 8, and 12, 100 nm in lanes 5, 9, and 13, and 500 nm in lanes 6, 7, 10, 11, 14, and 15). Lane 16 contained a partial duplex marker. In the substrate diagram, the circle represents the position of the 5′ 32P label, whereas arrows represent the 3′ ends of oligonucleotides. B, degree of unwinding of substrate 1 in lanes 2-15. Data represent the means of two experiments.
A range of concentrations of UvrA and UvrB were tested for effects on UvrD-catalyzed unwinding of substrate 1. Stimulation of the total levels of unwinding was observed at all concentrations of UvrA and UvrB (Fig. 1). Maximal stimulation was achieved with 10 nm UvrA or above and 100 nm UvrB or above (Fig. 1B). In the absence of UvrD, UvrAB failed to promote significant displacement of any of the DNA strands within the substrate (Fig. 1A, lanes 7, 11, and 15). Although UvrAB does have limited strand displacement activity, displacement is restricted to duplexes of less than 30 bp (32), which correlates with the lack of strand displacement of the 60-bp duplexes present in substrate 1. The presence of UvrAB enhanced production of both the partial duplex and the single-stranded oligonucleotide products (Fig. 1A). Thus, UvrAB had no major effect on the polarity of UvrD-catalyzed unwinding of substrate 1.
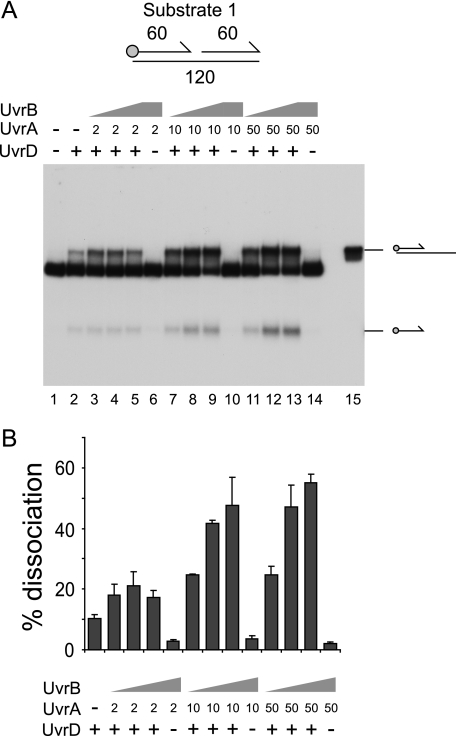
The contribution of SSB to stimulation of UvrD by UvrAB was also analyzed using substrate 1. In the absence of SSB, stimulation of UvrD-catalyzed unwinding of substrate 1 by UvrAB was again observed (Fig. 2). Therefore, SSB plays no essential role in stimulation of UvrD by UvrAB. Production of both the partial duplex and the single-stranded oligonucleotide was enhanced (Fig. 2A, compare lane 2 with lanes 9 and 13). Thus, as seen in the presence of SSB, UvrAB had no major effect on the polarity of unwinding by UvrD.
FIGURE 2.
Stimulation of UvrD by UvrAB is not dependent on SSB. A, unwinding of substrate 1 in the presence of UvrD (10 nm), UvrA (nm concentrations shown), and UvrB (10 nm in lanes 3, 7, and 11, 100 nm in lanes 4, 8, and 12 and 500 nm in lanes 5, 6, 9, 10, 13, and 14). B, degree of unwinding of substrate 1 in lanes 2-14. Data represent the means of two experiments.
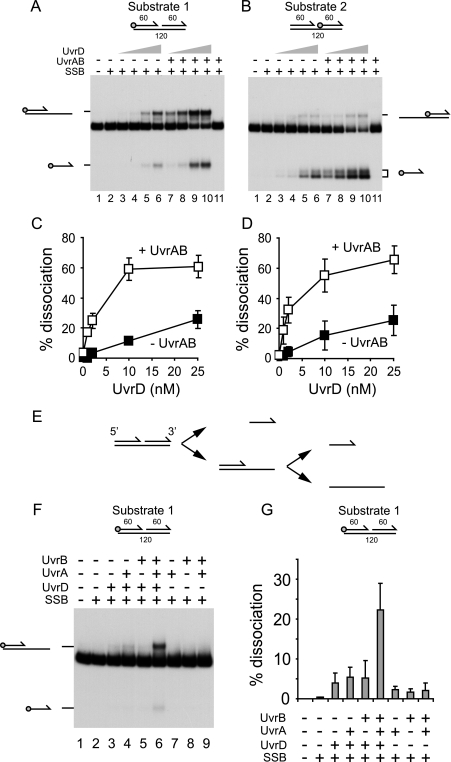
UvrD was then titrated into reactions containing 10 nm UvrA and 100 nm UvrB, concentrations which gave near-maximal stimulation of UvrD, and SSB (Fig. 1). Alternately labeled versions of the same nicked duplex employed in Fig. 1 were also used to investigate the directionality of unwinding. On substrate 1, stimulation by UvrAB was observed at all concentrations of UvrD tested (Fig. 3, A and C). The presence of UvrAB resulted in a dramatic reduction in the concentration of UvrD required for strand displacement; 2 nm UvrD in the presence of UvrAB gave similar levels of unwinding as 25 nm UvrD in the absence of UvrAB (Fig. 3C). As in Fig. 1, UvrAB had little impact on the directionality of UvrD-catalyzed unwinding of substrate 1. UvrAB also stimulated unwinding of substrate 2 at all concentrations of UvrD tested (Fig. 3, B and D). UvrAB-dependent stimulation of unwinding of substrate 2 by UvrD was also observed in the absence of SSB (supplemental Fig. 1).
FIGURE 3.
Effects of UvrAB on strand displacement and direction of translocation by UvrD. A, unwinding of substrate 1 by UvrD (1, 2, 10, and 25 nm) in the absence and presence of UvrA (10 nm) and UvrB (100 nm). SSB was also present (125 nm). B, unwinding of substrate 2 using the same protein concentrations as described in A. Note that the labeled 60-mer oligonucleotide product migrated as a doublet probably due to the formation of secondary structures within the oligonucleotide after deproteinization before loading on the gel. C, quantification of UvrD-catalyzed unwinding of substrate 1 in the absence and presence of UvrAB. D, quantification of UvrD-catalyzed unwinding of substrate 2 in the absence and presence of UvrAB. E, model of unwinding of the nicked duplex used in A and B. F, unwinding of substrate 1 by the indicated combinations of UvrD (2 nm), UvrA (10 nm), UvrB (100 nm), and SSB (125 nm). G, quantification of the unwinding of substrate 1 in F.
The only striking difference between the unwinding of substrates 1 and 2 was the distribution of partial duplex and single-stranded oligonucleotide products. Both types of product were generated from substrate 1 (Fig. 3A), whereas primarily single-stranded oligonucleotide was generated from substrate 2 (Fig. 3B). Note the single-stranded oligonucleotide of substrate 2 migrated as two bands, most probably due to formation of secondary structures after deproteinization.
This pattern implies that unwinding of the nicked duplex occurred initially by unwinding of the downstream 60-mer oligonucleotide, with the upstream 60-mer being a secondary product of unwinding (Fig. 3E). This orientation of unwinding by UvrD was observed both in the absence and the presence of UvrAB (Fig. 3, A and B). This pattern of unwinding was also maintained regardless of the presence or absence of SSB (compare Fig. 3, A and B, with supplemental Fig. 1A). Therefore, UvrAB appears to stimulate strand displacement by UvrD but not to affect the directionality of UvrD-catalyzed unwinding of nicked duplexes.
The above data demonstrate that a combination of UvrA and UvrB promote strand displacement by UvrD on DNA substrates containing strand discontinuities regardless of the presence or absence of SSB. However, UvrA or UvrB on their own elicited no significant stimulation of UvrD helicase activity in either the presence (Fig. 3, F and G) or absence of SSB (supplemental Fig. 2). Thus, stimulation of UvrD requires both UvrA and UvrB, presumably as a UvrAB complex.
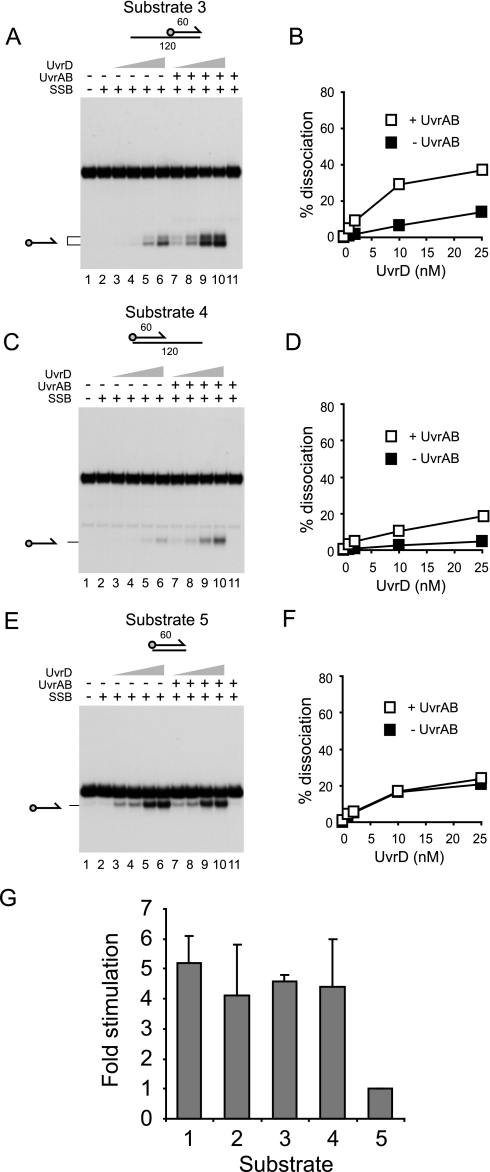
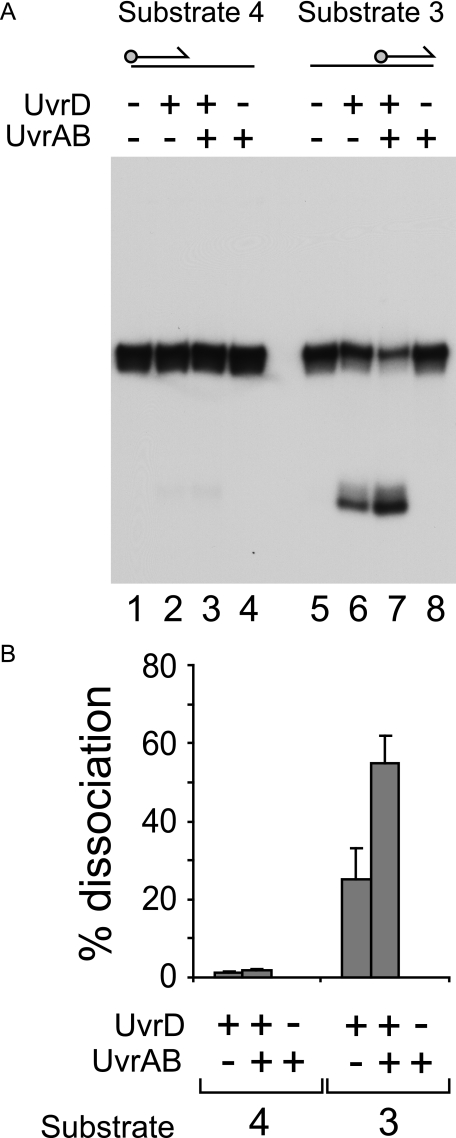
UvrAB Stimulates UvrD Helicase at DNA Strand Discontinuities—We analyzed which features of the nicked duplex substrates were responsible for the observed stimulation of UvrD by UvrAB. In the presence of SSB, stimulation was observed on partial duplexes having either a 3′ (substrate 3) or a 5′ ssDNA tail (substrate 4) (Fig. 4, A and C, respectively). However, unwinding of substrates 3 and 4 was reduced as compared with substrates 1 and 2 both in the presence and the absence of UvrAB (compare Fig. 3, C and D, with Fig. 4, B and D). For substrate three this was probably due to binding of the 3′ ssDNA tail by SSB, thereby inhibiting access of UvrD to the DNA substrate (58). For substrate 4, unwinding of the duplex by the 3′-5′ helicase UvrD (16) could only initiate either by translocation along the 120-mer strand from the blunt duplex end or by translocation along the 60-mer from the 3′ end of that strand. In either situation, a 3′ ssDNA tail would be lacking. Although UvrD can initiate unwinding from DNA ends lacking any ssDNA, this reaction requires higher concentrations of UvrD than those needed for unwinding of substrates with 3′ ssDNA tails (59).
FIGURE 4.
Stimulation of UvrD by UvrAB requires the presence of strand discontinuities within the DNA substrate. A, C, and E, unwinding of substrates 3, 4, and 5 by UvrD (1, 2, 10, and 25 nm) in the absence and presence of UvrA (10 nm) and UvrB (100 nm). SSB was also present (125 nm). Note that, as in Fig. 3, the labeled 60-mer oligonucleotide product generated from substrate 3 migrated as a doublet. B, D, and F, quantification of UvrD-catalyzed unwinding of substrates 3, 4, and 5 in the absence and presence of UvrAB. G, comparison of the extent of stimulation of UvrD-catalyzed unwinding of substrates 1-5 by 10 nm UvrA and 100 nm UvrB. UvrD and SSB were present at concentrations of 10 and 125 nm, respectively.
Substrates 1-4 all possessed strand discontinuities. We tested whether such discontinuities were a prerequisite for stimulation of UvrD by analyzing unwinding of a 60-bp duplex. No stimulation of UvrD by UvrAB was detected at any concentration of UvrD tested (Fig. 4, E and F). Furthermore, no stimulation could be detected using duplexes of 38 or 25 bp (data not shown). We conclude that stimulation of UvrD by UvrAB requires the presence of a strand discontinuity within the DNA structure to be unwound.
Given the different levels of unwinding of substrates 1-5, the degree of stimulation of UvrD was compared on these substrates in the presence of SSB. With 10 nm UvrD, a concentration which gave measurable levels of unwinding of these substrates both with and without UvrAB, there was no significant difference in the degree of stimulation with substrates 1-4 (Fig. 4G). Thus, the presence of a strand discontinuity within the DNA substrate, regardless of the presence of ssDNA, is essential for UvrAB-dependent stimulation of UvrD.
Partial duplex substrates 3 and 4 each contained 60 bases of ssDNA. Most, if not all, ssDNA would be bound by SSB in vivo, and so it was of interest to analyze any possible role of SSB in UvrD function on these substrates. For substrate 3, omission of SSB resulted in increased unwinding by UvrD in the absence of UvrAB (compare Fig. 4, A (lane 5) and B, with Fig. 5A (lane 6) and B). This difference is consistent with SSB binding to the 3′ ssDNA tail in substrate 3 and inhibiting access by UvrD, as suggested above. However, stimulation of UvrD by UvrAB was still observed in the absence of SSB (Fig. 5). Thus, SSB is not essential for UvrAB-dependent stimulation of UvrD.
FIGURE 5.
Effect of SSB on unwinding of substrates containing ssDNA. A, unwinding of substrates 3 and 4 by UvrD (10 nm) in the absence and presence of UvrA (10 nm) and UvrB (100 nm). Note, as in Fig. 3, the labeled 60-mer oligonucleotide product generated from substrate 3 migrated as a doublet. B, quantification of UvrD-catalyzed unwinding of substrates 3 and 4 in the absence and presence of UvrAB. Protein concentrations are as in A.
In contrast to substrate 3, unwinding of substrate 4 by UvrD required SSB regardless of the presence or absence of UvrAB (compare Fig. 4D with Fig. 5B). Thus, binding of SSB to the ssDNA in substrate 4 promoted unwinding. This promotion might have occurred either by helix destabilization by SSB at the ssDNA-dsDNA junction of substrate 4 or by prevention of secondary structure formation in the 60-base ssDNA tail. This requirement likely reflects the 5′ ssDNA tail of substrate 4 being of the wrong polarity to promote the 3′-5′ helicase activity of UvrD.
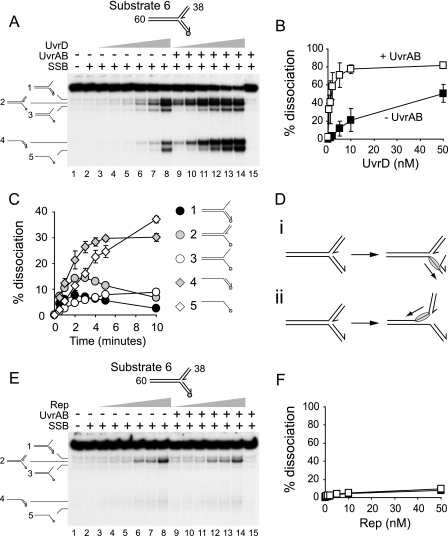
UvrAB Promote Unwinding of Branched DNA Substrates by UvrD—Strand discontinuities are also present at replication forks and recombination intermediates, possible targets of UvrD action in vivo (21). We therefore assessed whether UvrA and UvrB stimulated unwinding of branched DNA structures by UvrD. Initially, we attempted to employ forked DNA structures in which all three duplex arms were 60 bp, but secondary structure formation in long ssDNA regions, formed by unwinding and subsequent deproteinization, complicated interpretation of the experiments (data not shown). We therefore used a forked structure having a 60-bp parental duplex and two 38-bp arms equivalent to the leading and lagging strand arms at a replication fork (Fig. 6). Unwinding of this fork by UvrD in the presence of SSB was promoted by the addition of both UvrA and UvrB (Fig. 6, A and B). With 10 nm UvrD, the presence of UvrAB increased levels of unwinding ∼4-fold (Fig. 6B). This degree of stimulation is similar to that seen on substrates 1-4 (Fig. 4G), supporting the conclusion that the only essential requirement for stimulation of UvrD by UvrAB is the presence of a strand discontinuity within the DNA substrate. No strand displacement was observed by UvrAB in the absence of UvrD (Fig. 6A, lane 15), and as before, stimulation required both UvrA and UvrB (data not shown). Stimulation was also observed in the absence of SSB (supplemental Fig. 3), supporting the conclusion that SSB plays no essential role in this stimulation.
FIGURE 6.
UvrAB stimulate UvrD- but not Rep-catalyzed unwinding of forked DNA. A, unwinding of substrate 6 by 0.5, 1, 2, 5, 10, and 50 nm UvrD. UvrA (10 nm), UvrB (100 nm), and SSB (125 nm) were present as indicated. B, quantification of total levels of unwinding in A. C, time dependence of product formation in the presence of 5 nm UvrD, 125 nm SSB, 10 nm UvrA, and 100 nm UvrD. D, model of unwinding of substrate 6 by UvrD. E, unwinding of substrate 6 by 0.5, 1, 2, 5, 10, and 50 nm Rep. UvrA and -B and SSB were present as in A. F, quantification of total levels of unwinding in E. Black and white squares indicate unwinding by Rep in the absence and the presence of UvrAB, respectively.
The products of unwinding forked DNA were complex. In the absence of UvrAB the main product of UvrD-catalyzed unwinding was a fork lacking the “lagging” strand (Fig. 6A, lane 8, product 2). In the presence of UvrAB, levels of all five detectable products of unwinding were elevated (Fig. 6A, compare lanes 3-8 with 9-14). To establish the primary products of unwinding in the presence of UvrAB, DNA product formation was analyzed as a function of time (Fig. 6C). The main products of unwinding at earlier times were the partial duplex and the fork lacking the lagging strand (products 4 and 2, respectively, in Fig. 6C). At later times single-stranded oligonucleotide pre-dominated, presumably by secondary strand displacement reactions. Thus, UvrD, in the absence of UvrAB, displaced primarily the lagging strand (Fig. 6, A, lane 8, and Di). However, in the presence of UvrAB, both the lagging strand and the 60-bp parental duplex were unwound (Fig. 6, C and D, i and ii). This modulation of UvrD directionality on the fork by UvrAB is in apparent contrast to the observed lack of any directionality effects using the nicked duplex substrates (Fig. 3, A and B). However, the effective processivity of UvrD in isolation is 40-50 bp (60). Generation of product 2 required the displacement of a 38-bp duplex, whereas product 4 required unwinding of a 60-bp duplex. The dominance of product 2 formation in the absence of UvrAB is likely to be due to this discrepancy in duplex sizes.
Specificity of UvrD Stimulation by UvrAB—The high degree of sequence and structural homology between E. coli Rep and UvrD helicases suggests that both helicases unwind DNA via similar mechanisms (2, 3). Although Rep has no known role in nucleotide excision repair (18), this helicase may act at blocked replication forks (61). We, therefore, tested the specificity of stimulation by comparing the effects of UvrAB on UvrD- and Rep-catalyzed unwinding of a forked DNA substrate. No stimulation of Rep-catalyzed strand displacement was observed either in the presence of SSB (Fig. 6, E and F) or in the absence of SSB (supplemental Fig. 3). Similar results were obtained with a nicked duplex substrate (data not shown). We conclude that UvrAB specifically stimulates UvrD helicase.
The Strand Displacement Activity of UvrAB Is Not Essential for Stimulation of UvrD—Given the UvrD processivity of 40-50 bp (60), stimulation of unwinding of 60-bp (Fig. 1) and even 38-bp (Fig. 6A) duplexes might simply reflect the lowering of the processivity barrier by localized UvrAB-catalyzed strand displacement of 30 bp or less (32). Stimulation of UvrD but not Rep (Fig. 6, A and E) argues against such a model, as any nonspecific UvrAB-dependent reduction in the number of base pairs to be unwound would be predicted to stimulate both helicases. However, it remained possible that limited strand opening by UvrAB generated suitable DNA substrates for UvrD but not for Rep.
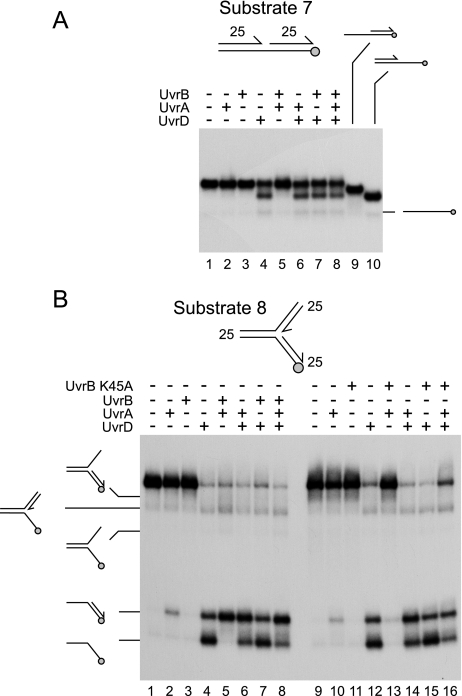
Initially, strand displacement by UvrAB was tested on a small nicked duplex with 25-bp arms, as wild type UvrAB is known to be able to displace duplexes of such lengths (32). SSB was omitted from these reactions due to its ability to destabilize short duplex regions of DNA. UvrD unwound this substrate with the same directionality seen on larger nicked duplexes, preferentially displacing the downstream 25mer oligonucleotide (Fig. 7A, compare lane 4 with 9 and 10). Thus, UvrD translocated away from the nick in the 3′-5′ direction along the continuous DNA strand even in the absence of UvrAB. However, no strand displacement by UvrAB in the absence of UvrD could be detected (Fig. 7A, lane 5). Strand displacement was also not observed with any combination of UvrA and -B on partial, as opposed to nicked, duplex substrates (data not shown). Moreover, no stimulation of UvrD-catalyzed unwinding of substrate 7 could be detected with UvrA, UvrB, or UvrAB (Fig. 7A compare lane 4 with lanes 6-8). This contrasted with the UvrAB-dependent stimulation observed on the nicked duplex substrates having 60-bp arms (Figs. 1, 2 and 3).
FIGURE 7.
Strand displacement catalyzed by UvrA and/or UvrB on nicked and forked DNA substrates. A, strand displacement on a nicked duplex substrate with 25-bp arms by UvrD, UvrA, and/or wild type UvrB present at 1, 10, and 100 nm, respectively. Lanes 9 and 10 contained partial duplex markers. B, strand displacement on a forked DNA substrate with 25-bp arms in the presence of UvrD, UvrA, and/or wild type UvrB (lanes 1-8) and UvrB K45A (lanes 9-16). Protein concentrations were as in A. Products of unwinding are indicated on the left.
The inability to detect strand displacement by UvrAB on small nicked duplexes was a surprise in the light of previous studies (32, 62). One possible explanation is that substrate 7 was of insufficient length to permit formation of a UvrAB-DNA complex in an appropriate conformation to allow strand separation. This possibility led us to test a forked DNA substrate with 25-bp arms. This substrate was unwound by UvrD (Fig. 7B, lane 4). A small amount of strand displacement was also detected with UvrA only, but none was detected with UvrB only (Fig. 7B, lanes 2 and 3, respectively). However, a combination of UvrA and UvrB promoted dissociation of the fork (Fig. 7B, lane 5). These data indicate that duplex arms of 25 bp in length are sufficient to allow UvrAB-catalyzed strand displacement but that such strand displacement requires branched rather than linear DNA substrates. The presence of UvrA with or without UvrB also altered the distribution of products generated by UvrD (Fig. 7B, compare lanes 4, 6, 7, and 8), implying that binding of UvrA could affect access of UvrD to DNA.
Given the rapid unwinding of substrate 8 by UvrD in isolation and by UvrAB, it was not possible to establish whether UvrA/B stimulated unwinding by UvrD. However, the ability of wild type UvrA and UvrB to promote dissociation of substrate 8 allowed testing of the strand displacement function of UvrB mutant K45A. This mutant is deficient in ATP hydrolysis and unable to promote displacement of DNA strands within partial duplex substrates in the presence of UvrA (63). UvrA plus UvrB K45A was also deficient in strand displacement on the forked substrate 8, in contrast to the wild type combination of proteins (Fig. 7B compare lanes 13 and 5, respectively). The presence of UvrB K45A also inhibited the small amount of strand displacement promoted by UvrA alone (Fig. 7B, compare lanes 10 and 13).
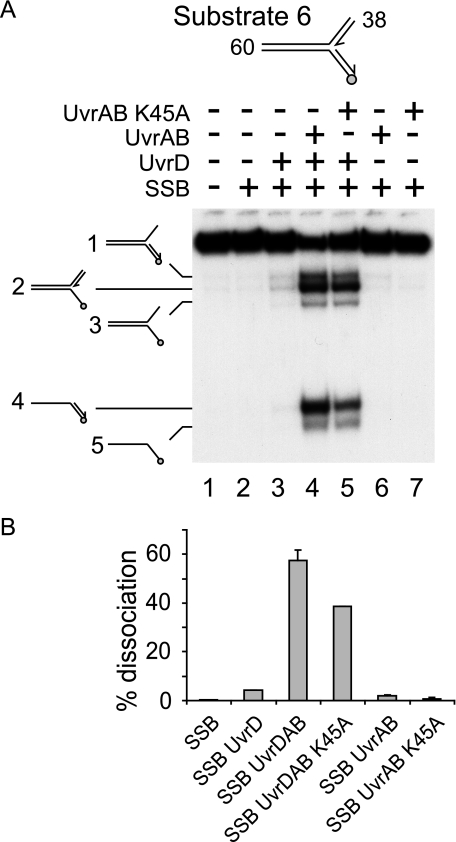
We exploited the lack of strand displacement on forked DNA by UvrA plus UvrB K45A to test whether such a displacement reaction was essential for the observed stimulation of UvrD helicase. Because the difference in strand destabilization by wild type UvrB and UvrB K45A was only apparent on a branched DNA substrate (Fig. 7B), the effects of UvrB K45A on stimulation of UvrD-catalyzed unwinding of the large forked substrate 6 was analyzed. Comparison of wild type UvrAB with UvrA plus UvrB K45A demonstrated that both types of complex stimulated unwinding by UvrD of this fork in the presence of SSB (Fig. 8A compare lane 3 with lanes 4 and 5). Stimulation by both types of UvrAB complex was also observed in the absence of SSB (supplemental Fig. 4). However, stimulation was reduced when comparing wild type with mutant UvrB in the presence of SSB (Fig. 8B), suggesting that ATP hydrolysis by UvrB may play some role in stimulation of UvrD. Such a partial requirement for ATP hydrolysis might reflect UvrAB-catalyzed strand opening contributing to some extent to stimulation of UvrD. However, other explanations are possible such as translocation of UvrAB along the DNA being required to achieve optimal positioning for stimulation of UvrD.
FIGURE 8.
Stimulation of UvrD helicase is effected in the absence of UvrAB helicase activity. A, unwinding of substrate 6 by UvrD and/or UvrA plus either wild type UvrB or UvrB K45A. UvrD, UvrA, UvrB (both wild type and mutant), and SSB were present at 2, 10, 100, and 125 nm, respectively. B, quantification of extent of unwinding.
Regardless of any role of ATP hydrolysis by UvrB in stimulation of UvrD, the lack of correlation between stimulation of UvrD and strand displacement by UvrAB (Figs. 7 and 8 and supplemental Fig. 4) together with the inability of UvrAB to stimulate Rep (Fig. 6, E and F) indicate that stimulation of UvrD cannot be explained solely by a UvrAB-catalyzed reduction in the number of base pairs of DNA to be unwound by UvrD.
DISCUSSION
This study demonstrates that a combination of UvrA and UvrB can promote UvrD-catalyzed unwinding of both branched and unbranched DNA structures containing strand discontinuities. Although a strand discontinuity within the DNA substrate is essential for this stimulation, the presence of ssDNA is not required (compare Figs. 3 and 4). Furthermore, although SSB can modulate UvrD helicase activity on substrates containing ssDNA, SSB is not required for stimulation (compare Figs. 4 and 5). These data indicate that the activity of the most abundant helicase in E. coli is regulated not just by a component of the mismatch repair apparatus (64) but also by nucleotide excision repair proteins. Thus, the two well defined processes that employ UvrD both involve modulation of its helicase activity. UvrD may also promote displacement of proteins, especially RecA, from DNA (19-21). Whether this protein displacement function requires specific recruitment of UvrD or merely reflects the abundance of UvrD in vivo remains unknown.
Facilitation by UvrAB of nicked duplex unwinding by UvrD provides an explanation as to why UvrA, -B, and -D are all required to maintain viability in the absence of DNA polymerase I (51). Without the 5′ to 3′ exonuclease and DNA synthesis functions of this polymerase, UvrA, -B, and -D are needed for efficient displacement of strands at junctions between Okazaki fragments. Other (unidentified) exonucleases and DNA polymerases might then be able to remove the displaced RNA primer and extend the 3′ end of the upstream Okazaki fragment to fill the gap before sealing by DNA ligase (51). A requirement for UvrAB in addition to UvrD could reflect the need for UvrAB to impose a specific directionality of UvrD catalysis at Okazaki fragment junctions, facilitating displacement of the strand containing the RNA primer at the junction rather than the upstream strand (51). However, our data demonstrate that UvrAB have little impact on UvrD directionality at nicked duplex substrates in vitro; regardless of the presence of UvrAB, UvrD preferentially unwound the downstream fragment equivalent to the 5′ end, rather than the 3′ end, of an Okazaki fragment (Figs. 3, A and B, and 7A; supplemental Fig. 1) (65). Instead, UvrAB stimulated UvrD-catalyzed strand displacement rather than re-orientating this displacement reaction (Fig. 3; supplemental Fig. 1). Note that our observation of preferential translocation of UvrD along the continuous DNA strand at a nick contrasts with the bidirectional movement of UvrD from such a structure reported previously (66). This discrepancy might reflect the use of a large excess of UvrD in relation to DNA substrate used in the previous report.
The lack of stimulation of Rep by UvrAB (Fig. 6, E and F; supplemental Fig. 3) suggests that stimulation of UvrD by UvrAB was not due only to the additive effects of UvrD- and UvrAB-catalyzed strand displacement. Such a mechanism would be expected to lead to nonspecific stimulation of both Rep and UvrD helicase activity. The retention of UvrD stimulation by a mutant UvrB incapable of strand displacement supports this conclusion (Figs. 7B and 8; supplemental Fig. 4). However, wild type UvrB ATPase activity is needed in vivo to allow colony formation in the absence of DNA polymerase I (51). This in vivo requirement might reflect the observed decrease in stimulation of UvrD helicase by UvrA plus UvrB K45A (Fig. 8). However, whether the need for ATP hydrolysis by UvrB to obtain maximal stimulation of UvrD reflects a contribution by UvrAB strand displacement to this stimulation is unclear. Alternative explanations are possible. For example, turnover of UvrA plus UvrB K45A on undamaged DNA may be reduced as compared with the wild type proteins (67), providing a kinetic barrier to binding of gaps between Okazaki fragments in ΔpolA cells.
Enhancement of UvrD-catalyzed unwinding by UvrAB could reflect either increased processivity of UvrD-catalyzed unwinding from nicks or increased loading of UvrD molecules at nicks. Note that enhanced loading (that is, increased association) of UvrD molecules would also lead to an increase in apparent processivity (68-70). Indeed, it is possible that stimulation may be effected by a combination of both these mechanisms. Regardless of the exact mechanism of stimulation, it is tempting to speculate that an interaction between UvrB and UvrD (43) is responsible for enhancement of UvrD helicase. However, testing the importance of any UvrB-UvrD interaction will require isolation of mutations in UvrB and/or UvrD that abrogate such an interaction. UvrD also plays a key role in mismatch repair, initiating unwinding from a MutH-catalyzed nick and displacing up to 1 kilobase of the DNA strand containing the mismatch (24). Interaction of UvrD with MutL bound at the single strand incision is essential in allowing a motor with limited dsDNA processivity to unwind such large tracts of DNA (23-26). Stimulation of UvrD by MutL is likely to occur via increased association, rather than decreased dissociation, of UvrD with the DNA substrate (26, 64). However, it should not be assumed that the mechanism of UvrD stimulation is the same for MutL and UvrAB, given the disparities between the structure and function of these proteins.
The observed stimulation of UvrD at a variety of DNA structures containing strand discontinuities, including branched DNA structures, raises the question of whether this reaction has any physiological role in wild type cells. Although UvrAB stimulation of UvrD explains the need for these three proteins in the absence of DNA polymerase I (51), wild type cells would presumably not need UvrABD for efficient Okazaki fragment processing. It remains possible that stimulation of UvrD by UvrAB does have an as yet unidentified physiological role in wild type cells. However, this stimulation might simply be a reflection of the need for UvrD to displace the 12/13-mer damaged oligonucleotide and to promote UvrB/C turnover during nucleotide excision repair (18, 42). Thus, targeting by UvrABD of strand discontinuities in DNA might be an unavoidable and possibly deleterious consequence of the need to recruit a DNA helicase for nucleotide excision repair. One way that gratuitous unwinding from strand discontinuities by UvrABD might be reduced is the maintenance of UvrA at 20 copies per cell in the absence of high levels of DNA damage, in contrast to UvrB and UvrD (250 and 5000-8000 molecules per cell, respectively) (for review, see Ref. 71). Stimulation of UvrD is dependent on UvrA as well as UvrB (Fig. 3, F and G; supplemental Fig. 2), and so limiting concentrations of UvrA would restrict stimulation of UvrD helicase activity. However, any such control must be relaxed in the presence of high levels of DNA damage via SOS induction of uvrA, uvrB, and uvrD to facilitate nucleotide excision repair. Any deleterious consequences of stimulating UvrD helicase to initiate unwinding at strand discontinuities might, therefore, be a price that must be paid for enhanced removal of DNA damage.
Supplementary Material
Acknowledgments
We thank Steve Matson for supplying purified UvrD used in preliminary experiments, Nigel Savery for providing cloned uvrD and for communicating results before publication, and Ken Marians for providing a rep overexpression clone.
This work was funded by the Medical Research Council and Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom (to P. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-4.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; SSB, single strand binding protein; DTT, dithiothreitol.
References
- 1.Subramanya, H. S., Bird, L. E., Brannigan, J. A., and Wigley, D. B. (1996) Nature 384 379-383 [DOI] [PubMed] [Google Scholar]
- 2.Korolev, S., Hsieh, J., Gauss, G. H., Lohman, T. M., and Waksman, G. (1997) Cell 90 635-647 [DOI] [PubMed] [Google Scholar]
- 3.Lee, J. Y., and Yang, W. (2006) Cell 127 1349-1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007) Annu. Rev. Biochem. 76 23-50 [DOI] [PubMed] [Google Scholar]
- 5.Wahle, E., Lasken, R. S., and Kornberg, A. (1989) J. Biol. Chem. 264 2469-2475 [PubMed] [Google Scholar]
- 6.LeBowitz, J. H., and McMacken, R. (1986) J. Biol. Chem. 261 4738-4748 [PubMed] [Google Scholar]
- 7.Kim, S., Dallmann, H. G., McHenry, C. S., and Marians, K. J. (1996) Cell 84 643-650 [DOI] [PubMed] [Google Scholar]
- 8.Tanaka, T., and Nasmyth, K. (1998) EMBO J. 17 5182-5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosh, R. M., Jr., Orren, D. K., Nehlin, J. O., Ravn, P. H., Kenny, M. K., Machwe, A., and Bohr, V. A. (1999) J. Biol. Chem. 274 18341-18350 [DOI] [PubMed] [Google Scholar]
- 10.Ma, Y., Wang, T., Villemain, J. L., Giedroc, D. P., and Morrical, S. W. (2004) J. Biol. Chem. 279 19035-19045 [DOI] [PubMed] [Google Scholar]
- 11.Cadman, C. J., Lopper, M., Moon, P. B., Keck, J. L., and McGlynn, P. (2005) J. Biol. Chem. 280 39693-39700 [DOI] [PubMed] [Google Scholar]
- 12.Shereda, R. D., Bernstein, D. A., and Keck, J. L. (2007) J. Biol. Chem. 282 19247-19258 [DOI] [PubMed] [Google Scholar]
- 13.Slocum, S. L., Buss, J. A., Kimura, Y., and Bianco, P. R. (2007) J. Mol. Biol. 367 647-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, J. S., Marr, M. T., and Roberts, J. W. (2002) Cell 109 757-767 [DOI] [PubMed] [Google Scholar]
- 15.Aygun, O., Svejstrup, J., and Liu, Y. L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8580-8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson, S. W. (1986) J. Biol. Chem. 261 10169-10175 [PubMed] [Google Scholar]
- 17.Lahue, R. S., Au, K. G., and Modrich, P. (1989) Science 245 160-164 [DOI] [PubMed] [Google Scholar]
- 18.Husain, I., Van Houten, B., Thomas, D. C., Abdel-Monem, M., and Sancar, A. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 6774-6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yancey-Wrona, J. E., and Matson, S. W. (1992) Nucleic Acids Res. 20 6713-6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morel, P., Hejna, J. A., Ehrlich, S. D., and Cassuto, E. (1993) Nucleic Acids Res. 21 3205-3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veaute, X., Delmas, S., Selva, M., Jeusset, J., Le Cam, E., Matic, I., Fabre, F., and Petit, M. A. (2005) EMBO J. 24 180-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arthur, H. M., and Eastlake, P. B. (1983) Gene (Amst.) 25 309-316 [DOI] [PubMed] [Google Scholar]
- 23.Hall, M. C., Jordan, J. R., and Matson, S. W. (1998) EMBO J. 17 1535-1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dao, V., and Modrich, P. (1998) J. Biol. Chem. 273 9202-9207 [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi, M., Dao, V., and Modrich, P. (1998) J. Biol. Chem. 273 9197-9201 [DOI] [PubMed] [Google Scholar]
- 26.Mechanic, L. E., Frankel, B. A., and Matson, S. W. (2000) J. Biol. Chem. 275 38337-38346 [DOI] [PubMed] [Google Scholar]
- 27.Arthur, H. M., and Lloyd, R. G. (1980) Mol. Gen. Genet. 180 185-191 [DOI] [PubMed] [Google Scholar]
- 28.Mahdi, A. A., Buckman, C., Harris, L., and Lloyd, R. G. (2006) Genes Dev. 20 2135-2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goosen, N., and Moolenaar, G. F. (2001) Res. Microbiol. 152 401-409 [DOI] [PubMed] [Google Scholar]
- 30.Van Houten, B., Croteau, D. L., DellaVecchia, M. J., Wang, H., and Kisker, C. (2005) Mutat. Res. 577 92-117 [DOI] [PubMed] [Google Scholar]
- 31.Seeley, T. W., and Grossman, L. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 6577-6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordienko, I., and Rupp, W. D. (1997) EMBO J. 16 889-895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moolenaar, G. F., Hoglund, L., and Goosen, N. (2001) EMBO J. 20 6140-6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skorvaga, M., Theis, K., Mandavilli, B. S., Kisker, C., and Van Houten, B. (2002) J. Biol. Chem. 277 1553-1559 [DOI] [PubMed] [Google Scholar]
- 35.Skorvaga, M., DellaVecchia, M. J., Croteau, D. L., Theis, K., Truglio, J. J., Mandavilli, B. S., Kisker, C., and Van Houten, B. (2004) J. Biol. Chem. 279 51574-51580 [DOI] [PubMed] [Google Scholar]
- 36.Moolenaar, G. F., Schut, M., and Goosen, N. (2005) DNA Repair (Amst) 4 699-713 [DOI] [PubMed] [Google Scholar]
- 37.Truglio, J. J., Karakas, E., Rhau, B., Wang, H., DellaVecchia, M. J., Van Houten, B., and Kisker, C. (2006) Nat. Struct. Mol. Biol. 13 360-364 [DOI] [PubMed] [Google Scholar]
- 38.Orren, D. K., and Sancar, A. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 5237-5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moolenaar, G. F., Franken, K. L., van de Putte, P., and Goosen, N. (1997) Mutat. Res. 385 195-203 [DOI] [PubMed] [Google Scholar]
- 40.Sancar, A., and Rupp, W. D. (1983) Cell 33 249-260 [DOI] [PubMed] [Google Scholar]
- 41.Verhoeven, E. E., van Kesteren, M., Moolenaar, G. F., Visse, R., and Goosen, N. (2000) J. Biol. Chem. 275 5120-5123 [DOI] [PubMed] [Google Scholar]
- 42.Orren, D. K., Selby, C. P., Hearst, J. E., and Sancar, A. (1992) J. Biol. Chem. 267 780-788 [PubMed] [Google Scholar]
- 43.Ahn, B. (2000) Mol. Cells 10 592-597 [DOI] [PubMed] [Google Scholar]
- 44.Okazaki, R., Arisawa, M., and Sugino, A. (1971) Proc. Natl. Acad. Sci. U. S. A. 68 2954-2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uyemura, D., Eichler, D. C., and Lehman, I. R. (1976) J. Biol. Chem. 251 4085-4089 [PubMed] [Google Scholar]
- 46.Joyce, C. M., and Grindley, N. D. (1984) J. Bacteriol. 158 636-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horiuchi, T., and Nagata, T. (1974) Mol. Gen. Genet. 128 105-115 [DOI] [PubMed] [Google Scholar]
- 48.Morimyo, M., and Shimazu, Y. (1976) Mol. Gen. Genet. 147 243-250 [DOI] [PubMed] [Google Scholar]
- 49.Shizuya, H., and Dykhuizen, D. (1972) J. Bacteriol. 112 676-681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegel, E. C. (1973) J. Bacteriol. 113 161-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moolenaar, G. F., Moorman, C., and Goosen, N. (2000) J. Bacteriol. 182 5706-5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons, C. A., Kemper, B., and West, S. C. (1990) J. Biol. Chem. 265 9285-9289 [PubMed] [Google Scholar]
- 53.Visse, R., de Ruijter, M., Moolenaar, G. F., and van de Putte, P. (1992) J. Biol. Chem. 267 6736-6742 [PubMed] [Google Scholar]
- 54.Payne, B. T., van Knippenberg, I. C., Bell, H., Filipe, S. R., Sherratt, D. J., and McGlynn, P. (2006) Nucleic Acids Res. 34 5194-5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malta, E., Moolenaar, G. F., and Goosen, N. (2006) J. Biol. Chem. 281 2184-2194 [DOI] [PubMed] [Google Scholar]
- 56.Dallmann, H. G., Thimmig, R. L., and McHenry, C. S. (1995) J. Biol. Chem. 270 29555-29562 [PubMed] [Google Scholar]
- 57.Marians, K. J. (1995) Methods Enzymol. 262 507-521 [DOI] [PubMed] [Google Scholar]
- 58.Matson, S. W., and George, J. W. (1987) J. Biol. Chem. 262 2066-2076 [PubMed] [Google Scholar]
- 59.Runyon, G. T., and Lohman, T. M. (1989) J. Biol. Chem. 264 17502-17512 [PubMed] [Google Scholar]
- 60.Ali, J. A., and Lohman, T. M. (1997) Science 275 377-380 [DOI] [PubMed] [Google Scholar]
- 61.Heller, R. C., and Marians, K. J. (2005) J. Biol. Chem. 280 34143-34151 [DOI] [PubMed] [Google Scholar]
- 62.Oh, E. Y., and Grossman, L. (1989) J. Biol. Chem. 264 1336-1343 [PubMed] [Google Scholar]
- 63.Seeley, T. W., and Grossman, L. (1990) J. Biol. Chem. 265 7158-7165 [PubMed] [Google Scholar]
- 64.Matson, S. W., and Robertson, A. B. (2006) Nucleic Acids Res. 34 4089-4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cadman, C. J., Matson, S. W., and McGlynn, P. (2006) J. Mol. Biol. 362 18-25 [DOI] [PubMed] [Google Scholar]
- 66.Runyon, G. T., Bear, D. G., and Lohman, T. M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6383-6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malta, E., Moolenaar, G. F., and Goosen, N. (2007) Biochemistry 46 9080-9088 [DOI] [PubMed] [Google Scholar]
- 68.Maluf, N. K., Fischer, C. J., and Lohman, T. M. (2003) J. Mol. Biol. 325 913-935 [DOI] [PubMed] [Google Scholar]
- 69.Levin, M. K., Wang, Y. H., and Patel, S. S. (2004) J. Biol. Chem. 279 26005-26012 [DOI] [PubMed] [Google Scholar]
- 70.Byrd, A. K., and Raney, K. D. (2004) Nat. Struct. Mol. Biol. 11 531-538 [DOI] [PubMed] [Google Scholar]
- 71.Janion, C. (2001) Acta Biochim. Pol. 48 599-610 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.