Abstract
Background
Microbes must sense environmental stresses, transduce these signals and mount protective responses to survive in hostile environments. In this study we have tested the hypothesis that fungal stress signalling pathways have evolved rapidly in a niche-specific fashion that is independent of phylogeny. To test this hypothesis we have compared the conservation of stress signalling molecules in diverse fungal species with their stress resistance. These fungi, which include ascomycetes, basidiomycetes and microsporidia, occupy highly divergent niches from saline environments to plant or mammalian hosts.
Results
The fungi displayed significant variation in their resistance to osmotic (NaCl and sorbitol), oxidative (H2O2 and menadione) and cell wall stresses (Calcofluor White and Congo Red). There was no strict correlation between fungal phylogeny and stress resistance. Rather, the human pathogens tended to be more resistant to all three types of stress, an exception being the sensitivity of Candida albicans to the cell wall stress, Calcofluor White. In contrast, the plant pathogens were relatively sensitive to oxidative stress. The degree of conservation of osmotic, oxidative and cell wall stress signalling pathways amongst the eighteen fungal species was examined. Putative orthologues of functionally defined signalling components in Saccharomyces cerevisiae were identified by performing reciprocal BLASTP searches, and the percent amino acid identities of these orthologues recorded. This revealed that in general, central components of the osmotic, oxidative and cell wall stress signalling pathways are relatively well conserved, whereas the sensors lying upstream and transcriptional regulators lying downstream of these modules have diverged significantly. There was no obvious correlation between the degree of conservation of stress signalling pathways and the resistance of a particular fungus to the corresponding stress.
Conclusion
Our data are consistent with the hypothesis that fungal stress signalling components have undergone rapid recent evolution to tune the stress responses in a niche-specific fashion.
Background
Microbes require robust stress responses to survive changing environments, and in particular, pathogenic microbes must mount effective responses to counter the defences of their host. The cellular and molecular responses to stress involve both acute and adaptive phases. Acute responses generally attempt to minimise the damage caused by harmful effects of a stress, such as the immediate physico-mechanical forces imposed by an osmotic stress [1]. In contrast, adaptive responses generally promote the restoration of cellular homeostasis with a view to allowing the growth of the microbe under the new conditions [1-3]. Cells that fail to adapt adequately to a relatively severe stress may die [4,5].
To mount appropriate acute and adaptive responses, cells must sense the change in their environment and activate the cognate signal transduction pathways that induce these responses [4]. In this study we focused on hyperosmotic, oxidative and cell wall stress signalling pathways because these have been shown to contribute to fungal virulence and their sensitivity to antifungal drugs [6-11]. However, these stress signalling pathways have been characterized to the greatest extent in the relatively benign model yeast, Saccharomyces cerevisiae.
In S. cerevisiae, the HOG (High Osmolarity Glycerol) MAPK (Mitogen Activated Protein Kinase) pathway is required for responses to osmotic stress [4,12]. The Stress Activated MAP Kinase (SAPK) Hog1 is central to this pathway. Hog1 activity is regulated by the MAP kinase kinase, Pbs2 [13]. In turn Pbs2 activity is controlled by two independent osmosensing branches involving Sho1 and Sln1, respectively [14,15]. Sho1 is a putative osmosensor that regulates the Pbs2-Hog1 MAP kinase module directly [16], whereas Sln1 controls a phosphorelay system that down-regulates the MAP kinase module in the absence of hyperosmotic stress [4,17]. In response to hyperosmotic stress Pbs2 becomes activated, leading to the phosphorylation and nuclear accumulation of Hog1, and the subsequent activation of osmo-protective mechanisms such as the accumulation of the osmolyte, glycerol. A well-characterized model of the osmotic stress pathway in S. cerevisiae was recently described by Krantz and coworkers (2006) [18].
Oxidative stress signalling in S. cerevisiae has been reviewed by Moye-Rowley (2003) [19], and Ikner and Shiozaki (2005) [20]. The transcription factor Yap1p plays a key role in the activation of oxidative stress genes [21,22]. Yap1 activity is regulated by the upstream regulators Gpx3, Ybp1 and Tsa1. The heat shock transcription factor Hsf1p contributes to the activation of protective functions during the oxidative stress response [23,24]. In addition, HOG signalling is thought to regulate Yap1 synthesis. Also, the cAMP-protein kinase A signalling pathway down-regulates the activity of the partially redundant transcription factors, Msn2 and Msn4, which contribute to the core stress response that helps to protect S. cerevisiae against oxidative stress.
Genetic or chemical insults to the S. cerevisiae cell wall lead to the activation of the cell wall stress (or cell integrity) pathway, which mediates compensatory changes in cell wall architecture [25]. Cell wall stresses are detected by specific sensors in the plasma membrane, such as Wsc1, Wsc2, Wsc3, Mid2 and Mtl1 [26,27]. These sensors, together with phosphatidylinositol-4,5-biphosphate (PI4,5P2), stimulate nucleotide exchange on the GTP-binding protein Rho1 [28]. PI4,5P2 activates the guanosine nucleotide exchange factors (GEFs) Rom1/2 [29] at the plasma membrane [30]. Then, Rho1 activates the protein kinase C (Pck1) MAP kinase cascade. This cascade involves sequential activation of the MAPKKK Bck1, the MAPKK's Mkk1 and Mkk2, and the MAPK Mpk1/Slt2 [31-34].
Arguably these stress signalling pathways have been best characterised in S. cerevisiae. However, it is becoming clear that there are differences in stress signalling and stress sensitivities amongst fungal species, for example amongst S. cerevisiae, Schizosaccharomyces pombe and Candida albicans [35-38]. This presumably reflects their evolution in dissimilar environments where they have been exposed to different types and intensity of stress [39]. Approximately 1.5 million fungal species are thought to exist, and their great diversity reflects the heterogeneity of the niches they occupy [40]. For example, free-living ascomycetes are frequently found in the soil, tree products, plant roots and on fruit, and are often transported between substrates via insect vectors [41]. Numerous ascomycetes and basidiomycetes are important plant pathogens. Now that genome sequences are becoming available for an increasing number of diverse fungal species, it is becoming increasingly possible to perform broad bioinformatic comparisons of stress regulators across fungal species and thereby to examine the evolution of fungal stress signalling pathways. The first step in such a comparison is the identification of putative orthologues of stress signalling molecules in these fungal genomes. This approach has been used effectively to assign provisional functional annotations to protein coding genes identified by genome sequencing [42-44], to measure the effects of functional genomic variables on protein evolution rates [45-47], and applied to other areas of evolutionary genomics [48], thereby increasing our understanding of eukaryotic evolution [49,50].
The available data suggest that fungal stress signalling pathways are evolving rapidly and in a niche-specific fashion to protect different species against the contrasting environmental stresses they encounter in their diverse niches. This hypothesis implies that fungal stress resistance is evolving in a manner that is independent of fungal phylogeny. To test this hypothesis we have explored the degree of conservation of fungal stress regulators relative to their relatively well-characterised orthologues in S. cerevisiae, focusing on the osmotic, oxidative and cell wall stress pathways. We selected eighteen fungal species for this analysis, all of which have had their genomes sequenced and annotated. These species have evolved in divergent niches, and they show a wide variety of virulence phenotypes. The data have highlighted the strong conservation of particular fungal G-proteins and protein kinases involved in stress signalling, and the rapid evolution of upstream sensors and downstream transcription factors on these pathways. In addition we have performed the first direct comparison of the sensitivities of these fungi to osmotic, oxidative and cell wall stresses, thereby confirming the diversity of stress phenotypes amongst the species examined. Our data confirm the lack of correlation between stress sensitivity and the degree of conservation of stress regulators. Our data are consistent with the rapid polyphyletic evolution of fungal stress responses.
Results
Phylogenetic relationships
A preliminary objective was to reconfirm the phylogenetic relationships of the fungi under analysis (Methods). Encephalitozoon cuniculi was selected as outgroup for the reconstruction of the phylogenetic tree (Figure 1). As expected, the fifteen ascomycetes examined were separated into three well-resolved groups: the Saccharomycotina (7 genera, 99% bootstrap support), the Pezizomycotina (7 genera, 100% bootstrap support) and the Archiascomycetes (1 genus: S. pombe). The ascomycetes formed a well-supported clade, which was the sister group of the basidiomycetes. Lastly, the microsporidial species E. cuniculi is very different from the other two taxa, based on its genetic distance. This was entirely consistent with the recent work of Fitzpatrick and co-workers (2006) [51] who described similar phylogenies when they created a supertree based on 4,805 gene families from 42 complete fungal genomes. Therefore, our phylogenetic tree for the 18 fungal species of interest is robust and consistent with accepted views.
Figure 1.
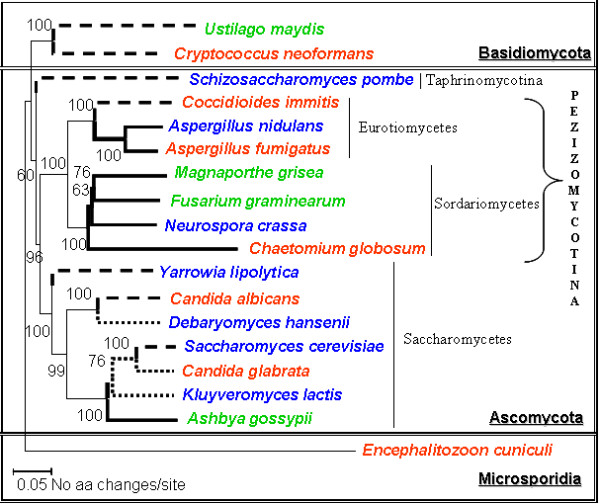
Neighbour-joining phylogeny constructed using a concatenated alignment of 8 genes for each of the 18 fungal species. Bootstrap scores for all the nodes are displayed. Encephalitozoon cuniculi was selected as an outgroup. The basidiomycetes and ascomycetes form distinct clades. Dotted lines indicate yeast-like fungi, thick straight lines indicate filamentous fungi, whereas dashed lines indicate dimorphic fungi [information adapted [144]]: red, human pathogens; green, plant pathogens; blue, benign fungi. Scale bar corresponds to 0.05 amino acid changes/site.
Phenotypic analyses
Our first main objective was to compare the sensitivities of the various fungal species to osmotic, oxidative and cell wall stresses. Fourteen of the eighteen fungal species were subjected to these tests. Cryptococcus neoformans, Coccidioides immitis, Chaetomium globosum and E. cuniculi are classified as category 3 pathogens by the Advisory Committee on Dangerous Pathogens (ACDP) and hence were excluded from this part of analysis. The sensitivities of yeast-like species were examined during their exponential growth phase (OD600 = 0.8 – 1.0: data not shown). The stress sensitivities of the filamentous fungi were examined using established approaches. Hence we examined the impact of various stresses conditions upon the germination of non-vegetative A. gossypii and M. grisea spores, Aspergillus conidia, and F. graminearum and N. crassa macroconidia [52,53]. The data are presented in Figure 2 and Tables 1, 2, 3, 4, 5 and 6 (also see additional file 1).
Figure 2.
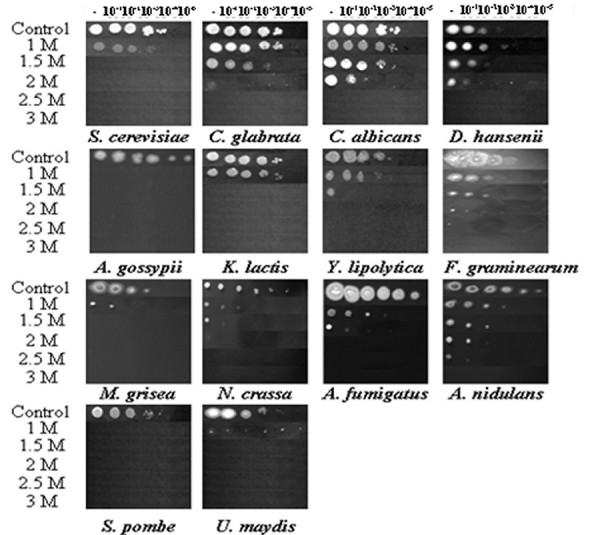
Comparison of fungal NaCl sensitivities. Growth of fungi on media containing various NaCl concentrations, the control plates lacking NaCl. Serial dilutions were plated as described in Materials and Methods.
Table 1.
Relative sensitivity of fungal species to NaCl
| *Relative growth (%) | NaCl [M] | |||||
| Species | Control | 1.0 | 1.5 | 2.0 | 2.5 | 3.0 |
| S. cerevisiae | 100 | 75 | 0 | 0 | 0 | 0 |
| C. glabrata | 100 | 100 | 70 | 15 | 0 | 0 |
| C. albicans | 100 | 100 | 95 | 50 | 0 | 0 |
| D. hansenii | 100 | 100 | 71 | 43 | 0 | 0 |
| A. gossypii | 100 | 0 | 0 | 0 | 0 | 0 |
| K. lactis | 100 | 100 | 0 | 0 | 0 | 0 |
| Y. lipolytica | 100 | 72 | 22 | 0 | 0 | 0 |
| F. graminearum | 100 | 76 | 71 | 41 | 18 | 0 |
| M. grisea | 100 | 50 | 0 | 0 | 0 | 0 |
| N. crassa | 100 | 48 | 22 | 4 | 0 | 0 |
| A. fumigatus | 100 | 54 | 29 | 0 | 0 | 0 |
| A. nidulans | 100 | 65 | 52 | 39 | 22 | 0 |
| S. pombe | 100 | 0 | 0 | 0 | 0 | 0 |
| U. maydis | 100 | 47 | 0 | 0 | 0 | 0 |
* NaCl stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the NaCl plates are shown in Fig. 2.
Table 2.
Relative sensitivity of fungal species to sorbitol
| *Relative growth (%) | Sorbitol [M] | |||
| Species | Control | 1.0 | 2.0 | 3.0 |
| S. cerevisiae | 100 | 87 | 83 | 0 |
| C. glabrata | 100 | 100 | 96 | 0 |
| C. albicans | 100 | 100 | 100 | 0 |
| D. hansenii | 100 | 79 | 50 | 0 |
| A. gossypii | 100 | 50 | 0 | 0 |
| K. lactis | 100 | 96 | 75 | 0 |
| Y. lipolytica | 100 | 94 | 69 | 0 |
| F. graminearum | 100 | 75 | 63 | 0 |
| M. grisea | 100 | 75 | 19 | 0 |
| N. crassa | 100 | 65 | 22 | 0 |
| A. fumigatus | 100 | 100 | 71 | 0 |
| A. nidulans | 100 | 79 | 63 | 13 |
| S. pombe | 100 | 78 | 33 | 0 |
| U. maydis | 100 | 46 | 0 | 0 |
* Sorbitol stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the sorbitol plates are shown in Additional File 1.
Table 3.
Relative sensitivity of fungal species to H2O2
| *Relative growth (%) | H2O2 [mM] | |||||||||||||
| Species | Control | 0.2 | 0.5 | 1.0 | 1.5 | 2.0 | 2.5 | 3.0 | 5.0 | 10 | 15 | 20 | 25 | 30 |
| S. cerevisiae | 100 | 83 | 83 | 83 | 75 | 67 | 58 | 25 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. glabrata | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 60 | 40 | 40 | 20 |
| C. albicans | 100 | 100 | 100 | 90 | 90 | 80 | 80 | 80 | 70 | 0 | 0 | 0 | 0 | 0 |
| D. hansenii | 100 | 82 | 82 | 82 | 76 | 53 | 35 | 24 | 6 | 0 | 0 | 0 | 0 | 0 |
| A. gossypii | 100 | 67 | 50 | 33 | 33 | 33 | 33 | 33 | 33 | 17 | 17 | 17 | 0 | 0 |
| K. lactis | 100 | 100 | 100 | 95 | 95 | 70 | 50 | 25 | 0 | 0 | 0 | 0 | 0 | 0 |
| Y. lipolytica | 100 | 83 | 74 | 83 | 83 | 70 | 70 | 70 | 48 | 0 | 0 | 0 | 0 | 0 |
| F. graminearum | 100 | 56 | 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. grisea | 100 | 81 | 75 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N. crassa | 100 | 96 | 71 | 71 | 54 | 38 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. fumigatus | 100 | 100 | 100 | 100 | 83 | 83 | 83 | 83 | 38 | 0 | 0 | 0 | 0 | 0 |
| A. nidulans | 100 | 83 | 83 | 83 | 67 | 67 | 67 | 50 | 17 | 0 | 0 | 0 | 0 | 0 |
| S. pombe | 100 | 100 | 100 | 100 | 94 | 81 | 75 | 50 | 0 | 0 | 0 | 0 | 0 | 0 |
| U. maydis | 100 | 65 | 65 | 65 | 60 | 60 | 55 | 35 | 0 | 0 | 0 | 0 | 0 | 0 |
* H2O2 stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the H2O2 plates are shown in Additional File 1.
Table 4.
Relative sensitivity of fungal species to menadione
| *Relative growth (%) | Menadione [mM] | ||||||||||||||
| Species | Control | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.35 | 0.4 | 0.45 | 0.5 |
| S. cerevisiae | 100 | 83 | 83 | 83 | 83 | 83 | 83 | 75 | 33 | 17 | 17 | 17 | 17 | 17 | 17 |
| C. glabrata | 100 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 79 | 63 | 46 | 33 |
| C. albicans | 100 | 100 | 100 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 78 | 70 | 70 | 70 |
| D. hansenii | 100 | 93 | 93 | 79 | 57 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. gossypii | 100 | 83 | 67 | 67 | 50 | 25 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| K. lactis | 100 | 83 | 83 | 83 | 83 | 83 | 67 | 67 | 67 | 67 | 50 | 50 | 50 | 50 | 17 |
| Y. lipolytica | 100 | 83 | 83 | 67 | 67 | 67 | 58 | 54 | 46 | 17 | 0 | 0 | 0 | 0 | 0 |
| F. graminearum | 100 | 69 | 69 | 56 | 56 | 56 | 50 | 50 | 50 | 31 | 31 | 31 | 31 | 31 | 31 |
| M. grisea | 100 | 75 | 50 | 50 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N. crassa | 100 | 71 | 63 | 42 | 33 | 33 | 33 | 33 | 17 | 17 | 17 | 4 | 4 | 4 | 4 |
| A. fumigatus | 100 | 100 | 100 | 83 | 83 | 83 | 67 | 54 | 38 | 33 | 33 | 25 | 25 | 0 | 0 |
| A. nidulans | 100 | 100 | 100 | 100 | 100 | 94 | 88 | 88 | 75 | 75 | 75 | 75 | 50 | 50 | 50 |
| S. pombe | 100 | 100 | 100 | 100 | 87 | 80 | 60 | 47 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| U. maydis | 100 | 100 | 81 | 75 | 56 | 25 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
* Menadione stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the menadione plates are shown in Additional File 1.
Table 5.
Relative sensitivity of fungal species to Calcofluor White
| *Relative growth (%) | CFW [μg/ml] | |||||||||
| Species | Control | 20 | 30 | 50 | 75 | 100 | 150 | 200 | 250 | 300 |
| S. cerevisiae | 100 | 71 | 58 | 50 | 50 | 50 | 50 | 50 | 42 | 42 |
| C. glabrata | 100 | 100 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 |
| C. albicans | 100 | 83 | 50 | 17 | 17 | 0 | 17 | 17 | 17 | 17 |
| D. hansenii | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. gossypii | 100 | 83 | 67 | 67 | 67 | 67 | 67 | 67 | 67 | 67 |
| K. lactis | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Y. lipolytica | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F. graminearum | 100 | 87 | 80 | 80 | 80 | 80 | 80 | 80 | 80 | 73 |
| M. grisea | 100 | 100 | 75 | 75 | 75 | 50 | 50 | 50 | 50 | 50 |
| N. crassa | 100 | 75 | 75 | 75 | 75 | 71 | 71 | 71 | 71 | 71 |
| A. fumigatus | 100 | 100 | 83 | 83 | 83 | 83 | 50 | 50 | 50 | 50 |
| A. nidulans | 100 | 83 | 83 | 83 | 83 | 83 | 83 | 54 | 54 | 38 |
| S. pombe | 100 | 94 | 94 | 94 | 94 | 81 | 81 | 81 | 81 | 81 |
| U. maydis | 100 | 46 | 46 | 38 | 38 | 23 | 8 | 8 | 8 | 8 |
* CFW stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the Calcofluor White plates are shown in Additional File 1.
Table 6.
Relative sensitivity of fungal species to Congo Red
| *Relative growth (%) | CR [μg/ml] | |||||||||
| Species | Control | 20 | 50 | 100 | 150 | 200 | 250 | 300 | 400 | 500 |
| S. cerevisiae | 100 | 95 | 85 | 75 | 55 | 40 | 40 | 25 | 20 | 20 |
| C. glabrata | 100 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 67 |
| C. albicans | 100 | 83 | 67 | 67 | 67 | 67 | 67 | 67 | 63 | 17 |
| D. hansenii | 100 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. gossypii | 100 | 67 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| K. lactis | 100 | 100 | 100 | 95 | 95 | 95 | 80 | 80 | 80 | 80 |
| Y. lipolytica | 100 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F. graminearum | 100 | 94 | 69 | 56 | 44 | 44 | 44 | 44 | 44 | 44 |
| M. grisea | 100 | 94 | 75 | 75 | 75 | 75 | 75 | 75 | 75 | 50 |
| N. crassa | 100 | 75 | 75 | 71 | 71 | 71 | 71 | 71 | 71 | 71 |
| A. fumigatus | 100 | 100 | 100 | 100 | 100 | 100 | 83 | 50 | 33 | 33 |
| A. nidulans | 100 | 79 | 71 | 71 | 71 | 71 | 71 | 71 | 71 | 71 |
| S. pombe | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| U. maydis | 100 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
* CR stress sensitivities were quantified by calculating the percentage growth under each condition relative to the corresponding non-stress control for that species. Examples of the Congo Red plates are shown in Additional File 1.
Osmotic stress sensitivity
The sensitivity of each fungal species to osmotic stress was examined using a range of NaCl and sorbitol concentrations, which impose ionic and non-ionic osmotic stresses, respectively [54]. S. pombe and A. gossypii exhibited the greatest sensitivity to NaCl, their growth being completely inhibited by 1 M NaCl. In contrast, C. albicans, C. glabrata and D. hansenii were the most resistant to NaCl (Figure 2, Table 1). While C. albicans and D. hansenii are members of the CTG clade (i.e. the clade of organisms in which the CTG codon is decoded as serine instead of leucine), C. glabrata is not (Figure 1). The growth of these species was only slightly inhibited by 1.5 M NaCl, and no growth was observed at concentrations above 2 M NaCl. The rest of ascomycetes showed relatively low resistance to NaCl. Of the filamentous fungi, F. graminearum and A. nidulans showed the highest resistance to NaCl. The human pathogen, A. fumigatus was more sensitive to NaCl, and the plant pathogen, U. maydis was most sensitive to this stress.
Similar observations were made when sorbitol was used to impose osmotic stress (Table 2, see also additional file 1). Once again S. pombe and A. gossypii were the most sensitive to sorbitol, and C. albicans and C. glabrata were amongst the most resistant species. Interestingly, compared with other species, the halotolerant yeast D. hansenii was relatively sensitive to sorbitol but resistant to NaCl (Tables 1 and 2). Also the human pathogen, A. fumigatus was more sensitive to NaCl than sorbitol, by comparison with the other species. Indeed all of the human pathogens tested were resistant to sorbitol. With the exception of A. gossypii, M. grisae and N. crassa, the ascomycetes tested were relatively resistant to sorbitol (Table 2). The basidiomycetes species tested (U. maydis) was sensitive to both osmotic stresses.
Oxidative stress sensitivity
The sensitivities of the fungi to oxidative stresses were tested by plating on media containing hydrogen peroxide (H2O2) and menadione (Tables 3 and 4; see also additional file 1). F. graminearum and M. grisae were particularly sensitive to H2O2 whereas the human pathogen C. glabrata was exceptionally resistant to this oxidative stress. M. grisea was also sensitive to menadione as well as H2O2, suggesting that this plant pathogen is sensitive to oxidative stresses in general. In contrast F. graminearum, C. albicans and K. lactis were relatively resistant to menadione. It is interesting to note that, in general, the human pathogens were relatively resistant to the oxidative stresses tested, whereas the plant pathogens tested were relatively sensitive.
Cell wall stress sensitivity
Finally, we tested the sensitivity of the cell wall stresses using Calcofluor White and Congo Red. These inhibitors disturb cell wall biosynthesis in S. cerevisiae and C. albicans, activating compensatory changes in cell wall architecture via the cell wall stress signalling (or cell integrity) pathway. The majority of fungi investigated were resistant to both Calcofluor White and Congo Red (Tables 5 and 6). S. pombe and C. glabrata stood out as the most resistant species to these cell wall stresses, whereas D. hansenii and Y. lipolytica were the most sensitive to these stresses. In contrast, K. lactis and C. albicans were sensitive to Calcofluor White, but relatively resistant to Congo Red. The basidiomycete U. maydis was sensitive to both inhibitors. Regarding the other plant pathogens, F. graminearum was relatively resistant to Calcofluor White, but sensitive to Congo Red. The reverse was true for M. grisae. In general the human pathogens were relatively resistant to both stresses, the exception being the sensitivity of C. albicans to Calcofluor White. The non-pathogenic Pezizomycotina were resistant to Calcofluor White and Congo Red.
Conservation of stress signalling modules
Having tested the sensitivity of the fungal species to osmotic, oxidative and cell wall stresses, we examined the degree of conservation of regulatory proteins on the corresponding stress signalling pathways. C. neoformans, C. immitis, C. globosum and E. cuniculi were included in these analyses. We assumed for the purposes of this study that the functions of orthologues are conserved across the fungal species examined. This allowed us to map putative regulators to the corresponding stress signalling pathways that have been characterized in S. cerevisiae. However, this assumption does not always hold, and the possibility that these regulators might execute alternative functions in a particular species should be borne in mind.
First, putative orthologues were identified for each S. cerevisiae protein in the other seventeen fungal species by screening for reciprocal best hits, as described in Materials and Methods. Lists of fungal orthologues are presented as additional file 2. Proteins involved in osmotic, oxidative and cell wall stress signalling were then selected, based on recent models of these pathways in S. cerevisiae. We used the model of the osmotic stress signalling pathway reported by Krantz and co-workers (2006) [18], the cell wall stress pathway described by Levin (2005) [25], and a model of the oxidative stress signalling pathway based on the reviews of Moye-Rowley (2003) [19] and Ikner and Shiozaki (2005) [20]. We then examined the conservation of each signalling pathway in each fungus by collating the percent identities for the relevant regulators, relative to their S. cerevisiae orthologues, as reported by BLASTP (Figures 3, 4 and 5). The orthologues are listed in the additional file 2.
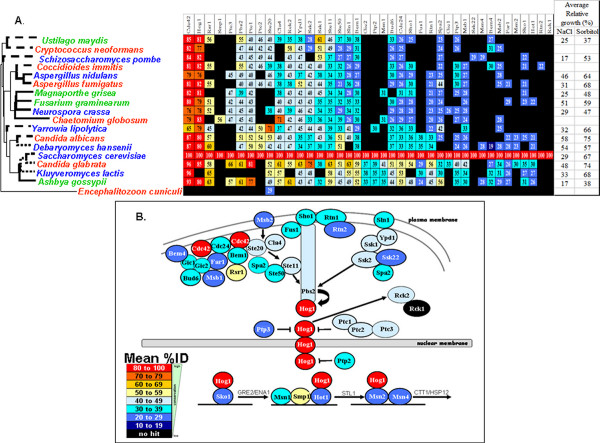
Figure 3.
Degree of conservation of fungal osmotic stress regulators. (A) Orthologues of S. cerevisiae osmotic stress regulators in the fungi analysed. The organisms are ordered according to their position in the phylogeny, and the regulators ordered according to their mean %ID across all of the fungal species examined. Closed indicates no orthologue identified. (B) Mean conservation (%ID) of osmotic stress regulators across the fungal species examined based on the model of the osmotic stress pathway in S. cerevisiae described by Krantz and co-workers (2006) [18].
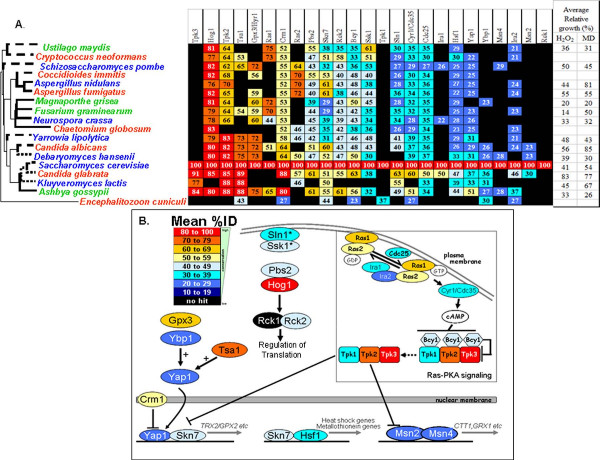
Figure 4.
Degree of conservation of fungal oxidative stress regulators. (A) Orthologues of S. cerevisiae oxidative stress regulators in the fungi analysed. As before, the fungi are ordered according to their position in the phylogeny, and the regulators ordered according to their mean %ID. Closed indicates no orthologue identified. (B) Mean conservation (%ID) of oxidative stress regulators across the fungal species examined based on the S. cerevisiae oxidative stress pathway adapted from reviews by Moye-Rowley (2003) [19] and Ikner and Shiozaki (2005) [20].
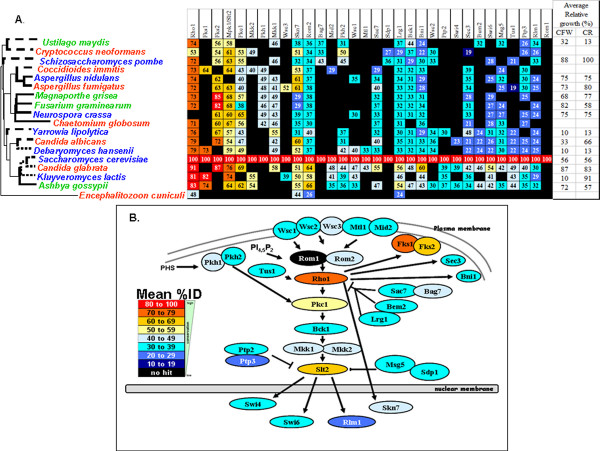
Figure 5.
Degree of conservation of fungal cell wall stress regulators. (A) Orthologues of S. cerevisiae cell wall stress regulators in the fungi analysed. As before, the fungi are ordered according to their position in the phylogeny, and the regulators ordered according to their mean %ID. Closed indicates no orthologue identified. (B) Mean conservation (%ID) of cell wall stress regulators across the fungal species examined based on the S. cerevisiae cell wall stress pathway adapted from Levin (2005) [25].
Mean percent identities were then calculated for each regulator. This allowed the comparison of osmotic, oxidative and cell wall stress signalling components across the eighteen fungal species under investigation (Figures 3, 4 and 5). The Hog1 SAPK and the G-protein Cdc42 stand out as being the most highly conserved signalling molecules in the osmotic stress pathway (Figure 3). Other central components of the MAP kinase module (Ste11, Pbs2) and phosphorelay module (Ypd1, Ssk1, Ssk2) are reasonably well conserved across the diverse species analysed. However, the sensors and transcriptional regulators that lie upstream and downstream of these models are generally poorly conserved.
With regard to oxidative stress signalling (Figure 4), members of the glutaredoxin and thioredoxin systems are well conserved (Tsa1 and Gpx3). Also, Ras (Ras1/2) and protein kinase A (Tpk1/2/3) are well conserved. (The strong conservation of Hog1 has already been mentioned.) Once again, downstream transcription factors involved in oxidative stress signalling are poorly conserved. This is the case for Yap1 orthologues, even though they are known to play key roles in the oxidative stress response in S. cerevisiae, S. pombe, C. albicans and C. glabrata [7,21,22,55]. This also holds for Msn2/4 orthologues. However in this case, while Msn2/4 orthologues contribute to oxidative stress responses in S. cerevisiae and C. glabrata, they do not do so in C. albicans [36,55,56].
Signalling components on the cell wall stress pathway also show great diversity with respect to their degree of evolutionary conservation (Figure 5). Once again some core components are strongly conserved most notably a G-protein (Rho1), protein kinase C (Pkc1), and a MAP kinase (Slt2). Also, the sensors of cell wall stresses (Wsc1/2/3, Mtl1, Mid2) are less well conserved than these central signalling components. Also, the transcription factors that are downstream targets of these signalling modules are less well conserved (Swi4/6, Rlm1). However, subunits of the glucan synthase are highly conserved (Fks1/2). These lie downstream of Rho1 signalling, and are essential for cell wall biosynthesis [25,57,58].
Individual diagrams have been created to illustrate the degree of conservation of each regulator in each fungus relative to S. cerevisiae (see additional file 3). Generally, the above observations hold across the individual fungal species we examined. In general core signalling molecules are more highly conserved than upstream and downstream components.
Discussion
In this study we have compared directly the stress sensitivities of a diverse group of fungal species for the first time (Figure 1). These species were selected on the basis that their genomes had been sequenced and annotated (Table 7). This allowed us to examine the evolutionary conservation of stress signalling components amongst ascomycete, basidiomycete and microsporidial species. Our study has focused on osmotic, oxidative and cell wall stress signalling. A number of significant observations have been made.
Table 7.
Strains and data sources
| Organisms | Strains | Ecologic Niche | References |
| A. gossypii | ATCC10895 | Cotton | [145] |
| A. fumigatus | Af293 | Decaying organic & plant material | [146] |
| A. nidulans | FGSCA4 | Tropical & subtropical regions | [147] |
| C. albicans | SC5314 | Skin, mucosa | [148] |
| C. glabrata | CBS138 | Mouth, gastrointestinal tracks | [149] |
| C. globosum* | CBS148.51 | Soil, air and plant debris | Unpublished |
| C. immitis* | RS | Soil (dessert like areas of southwest USA) | Unpublished |
| C. neoformans* | JEC21 | Soil contaminated by pigeon droppings | [150] |
| D. hansenii | CBS767 | All types of cheese, dairies, brines | [149] |
| E. cuniculi* | GB-M1 | Urine, blood, kidney | [151] |
| F. graminearum | PH-1 | Cotton, wheat, barley, bean, soybean | Unpublished |
| K. lactis | NRRLY-1140 | Milk and milk products | [149] |
| M. grisea | Guy-11 | Rice | [152] |
| N. crassa | OR74A | Dead plant matter after fibres | [153] |
| S. cerevisiae | S288C | Oak tress (oils), surface of fruits | [154] |
| S. pombe | 927C | Grapes (wine fermentation) | [155] |
| U. maydis | 521 | Soil plant material, maize (corn), grasses | Unpublished |
| Y. lipolytica | CLIB122 | Oil fields, cheese, sausages | [149] |
* C. globosum, C. immitis, C. neoformans and E. cuniculi are classified as category 3 pathogens by ACDP (Advisory Committee on Dangerous Pathogens) and hence were used only for bioinformatics analysis in current study. NCBI GI numbers are provided for E. cuniculi.
Genome sequences sources; A. gossypii: http://agd.vital-it.ch/index.html; A. fumigatus and C. neoformans: http://www.tigr.org/tdb/fungal/index.shtml; A. nidulans, C. globosum, C. immitis, F. graminearum, M. grisea, N. crassa and U. maydis http://www.broad.mit.edu/annotation/fgi/; C. albicans http://www.candidagenome.org/; C. glabrata, D. hansenii, K. lactis and Y. lipolytica http://cbi.labri.fr/Genolevures/; S. cerevisiae http://www.yeastgenome.org; S. pombe http://www.sanger.ac.uk/Projects/S_pombe/; E. cuniculi http://www.ncbi.nlm.nih.gov/Genomes/
The first main observation was that the fungal species examined displayed wide variation in their resistance to osmotic, oxidative and cell wall stresses (Tables 1, 2, 3, 4, 5 and 6). For example, while some fungi showed acute sensitivity to osmotic stress (S. pombe, A. gossypii), others were relatively resistant (C. albicans, F. graminearum). Similarly, some fungi were relatively sensitive to oxidative stress (D. hansenii, M. grisea), whereas others were extremely resistant to this type of environmental insult (C. glabrata).
Interestingly, some species showed differential sensitivities to the alternative osmotic, oxidative and cell wall stresses. For example, D. hansenii was resistant to NaCl, but less resistant to sorbitol (Tables 1 and 2). NaCl imposes a salt (ionic) stress in addition to an osmotic stress, whereas sorbitol imposes a non-ionic stress [54,59]. D. hansenii has been isolated from saline environments such as sea water [60] and concentrated brines [61]. This species is known to be more osmotolerant than S. cerevisiae [60], accumulating glycerol and to a lesser extent arabitol as compatible solutes [62,63]. This difference is due in part to more effective sodium extrusion by D. hansenii [64].
A. nidulans, F. graminearum, C. albicans, K. lactis and S. cerevisiae showed differential responses to the oxidative stresses tested (Tables 3 and 4). These fungi were more resistant to menadione than H2O2. This was consistent with the findings of Mutoh and co-authors (2005) [65] who previously reported that S. pombe is more sensitive to H2O2 than menadione. This would appear to suggest that these fungi are better able to detoxify the superoxide generated by menadione, than the peroxide anions generated by H2O2. However in S. cerevisiae, peroxide is generated from superoxide by superoxide dismutases, this peroxide subsequently being detoxified by catalases [66,67]. Therefore it might seem counterintuitive for some fungi to be more resistant to menadione. However when transition metals are present, H2O2 can also be converted to the hydroxyl radical, which is more potent than the superoxide radical [67,68]. Therefore the relative sensitivity of some fungi to H2O2 might reflect a reduced capacity to detoxify hydroxyl radicals.
Differential sensitivities to the cell wall stress were also observed (Tables 5 and 6). Calcofluor White and Congo Red responses interact with different components in the fungal cell wall. Calcofluor White binds to nascent chitin, inhibiting the assembly of chitin chains in the wall [69-72]. In contrast, Congo Red is generally thought to inhibit β-1,3-glucan assembly in the cell wall [73-76]. S. cerevisiae mutants with an increased chitin content in the cell wall are more sensitive to Calcofluor White, whereas mutants with reduced chitin are more resistant to Calcofluor White [76-78]. It follows that the differential fungal sensitivities to Calcofluor White and Congo Red might be explained, at least in part, by the different chitin and β-glucan contents of their cell walls. In S. cerevisiae, C. albicans and S. pombe, β-1,3-glucan accounts for 50 – 55% of the cell wall dry weight and is responsible for much of the mechanical strength of the cell wall [58,79,80]. Chitin is a relatively minor constituent of the yeast cell wall comprising 1 to 2% of the cell wall dry weight in S. cerevisiae, C. albicans [58,81,82]. The S. pombe cell wall was reported to contain no chitin [83-85], but more recently a small amount of chitin was detected [0.3% of dry weight: [86]]. This low chitin content probably accounts for the Calcofuor White resistance of S. pombe (Table 5). However the filamentous fungi were relatively resistant to Calcofuor White and Congo Red, and yet chitin is a major component of their cell walls [82,87]. The Neurospora cell wall contains 10–20% chitin [87-89], whereas in A. nidulans chitin constitutes up to 40% of the cell wall [90]. Therefore additional mechanisms must account for the relative Calcofuor White resistance of the filamentous fungi.
Our second main observation was that there was no clear correlation between fungal phylogeny and stress resistance (Figures 3, 4 and 5). In some cases closely related fungi displayed similar stress sensitivities. For example, the Aspergillus species examined (Eurotiomycetes) were relatively resistant to the cell wall stresses (Calcofluor White and Congo Red: Figure 5) and displayed similar responses to osmotic stresses (NaCl and sorbitol: Figure 3). The Saccharomycetes, C. albicans and D. hansenii, which also belong to the CTG clade (where the CTG codon is translated as serine, rather than leucine), were both highly resistant to NaCl (Figure 3). However in other cases, closely related fungal species displayed contrasting stress sensitivities. For example, C. glabrata was much more resistant to osmotic, oxidative and cell wall stresses than S. cerevisiae (Figures 3, 4 and 5). F. graminearum was more resistant to NaCl and menadione than M. grisae. Also, C. albicans was more resistant to H2O2, menadione and Congo Red than D. hansenii.
This was extended by more systematic analysis of individual stress response pathways. The mean percent identity was calculated for all components of a given stress response pathway in each species. These data were then plotted against the differential impact of that same stress upon S. cerevisiae and the comparator species (by calculating the difference between their mean growth inhibitions for the stress in question). If pathway sequence divergence is indicative of increasing differences in stress response, then a negative correlation would be expected of this analysis (i.e. a lower mean pathway percentage identity would correlate with greater differences in stress response). In fact in every case, the regression R2 coefficients were less than 0.09 (completely non-significant), and for all but one stress, correlations were weakly positive (see also additional file 4). Therefore, this analysis confirmed that there is no correlation between the degree of conservation stress regulators and the similarity of stress phenotypes. We conclude that fungal stress phenotypes have evolved rapidly and in a polyphyletic manner.
Presumably this rapid evolution of fungal stress phenotypes has been driven by local and niche-specific environmental pressures. If this was the case, one might expect to observe a correlation between the stress phenotype of a particular fungus and the nature of the environmental niche that it occupies. Our data are consistent with this view. For example as described above, D. hansenii, which was has been isolated from saline environments, is resistant to salt stress but less resistant to a non-ionic osmotic stress (Figure 3). Also, the human pathogens we examined (A. fumigatus, C. glabrata and C. albicans) were highly resistant to oxidative stress, whereas the plant pathogens (M. grisea, F. graminearum, A. gossypii and U. maydis) were sensitive to this type of stress (Figure 4).
Phagocytic cells are a first line of defence against fungal infections, generating superoxide, H2O2, and hydroxyl radicals in an attempt to destroy the phagocytosed pathogen [91-93]. The major fungal pathogen C. albicans activates oxidative stress responses when exposed to human blood, macrophages or neutrophils [11,94-97]. Indeed the virulence of C. albicans is dependent upon its ability to counteract oxidative stress [6,8,10,11]. Therefore it is hardly surprising that C. albicans has evolved to become relatively resistant to oxidative stress [98]. The same is true for another opportunistic fungal pathogen. C. glabrata is highly resistant to oxidative stress [55,92,99-101] even though phylogenetically it is more closely related to S. cerevisiae than to C. albicans [102].
Our third main observation was that, despite the critical importance of stress responses for the environmental robustness of the fungi, the upstream sensors and downstream transcriptional regulators on three stress signalling pathways show a low degree of sequence conservation (Figures 3, 4 and 5). Core components of the stress pathways are relatively strongly conserved. In some cases these core components play multiple cellular roles. For example, core components of the cell wall stress (cell integrity) pathway and in Ras-cAMP-protein kinase A signalling play key roles in the regulation of growth and cell polarity [30,103,104]. Therefore their strong sequence conservation is to be expected. In contrast, the upstream sensors and downstream transcriptional regulators generally play more specific roles in the detection of environmental stress and the activation of stress-specific responses. Clearly the evolutionary divergence of specific sensors or transcriptional regulators could contribute to the differential stress phenotypes of these fungal species by modulating the sensitivity of each species to a particular type of stress and tuning the strength of the molecular response to that stress. Therefore, the low degree of conservation of the upstream and downstream signalling components is entirely consistent with the rapid polyphyletic evolution of fungal stress resistance in response to niche-specific selection pressures.
Our evolutionary comparison of stress signalling components was based on the identification of the fungal orthologues of S. cerevisiae. Orthologues were defined on a genome-wide basis by identifying the best reciprocal hits. In some cases it was not possible to identify orthologues in all of the species examined. In many of these cases this probably reflects the lack of a genuine orthologue. However in some cases genuine orthologues might have fallen below the BLAST cut-off due to their low level of sequence similarity. In other cases the presence of an orthologue was not detected because of the existence of closely related paralogous gene pairs in S. cerevisiae that arose through the ancient genome duplication [105]. In these cases, the BLAST search for S. cerevisiae 'paralogue A' identified a particular fungal 'gene X', but the reciprocal BLAST search for 'gene X' identified S. cerevisiae 'paralogue B', thereby yielding no reciprocal best hit. Despite these health warnings, most commonly used orthology resources are based on reciprocal best hits. These include Clusters of Orthologues Genes [C/KOGs: [106,107]], INPARANOID [108,109] and the NCBI resource, HomoloGENE [110]. Also, it should be noted that, as our comparisons were based on S. cerevisiae, fungal proteins that exist in other species but not in S. cerevisiae will not have been identified in this study. Nevertheless, our study has provided a comprehensive list of fungal orthologues to S. cerevisiae proteins in seventeen divergent fungal species (see additional file 2). We have used this resource to study fungal stress signalling, but it is freely available for the analysis of other aspects of fungal molecular and cell biology.
Conclusion
Our comparison of the stress resistance of diverse fungal species has revealed a high degree of variation in their resistance to osmotic, oxidative and cell wall stresses. Fungal species that are closely related in phylogenetic terms did not necessarily display similar levels of stress resistance. Human pathogens tended to be more resistant to stress, with the exception of Candida albicans which was relatively sensitive to the cell wall stress, Calcofluor White. Plant pathogens tended to be sensitive to oxidative stress.
We have examined the degree of conservation of osmotic, oxidative and cell wall stress signal transduction pathways in the eighteen diverse fungal species. Central components of these signalling pathways are generally well conserved, whereas upstream sensors and downstream transcriptional regulators have diverged to a greater extent. No correlation between the degree of conservation of stress signalling pathways and the resistance of a particular fungus to the corresponding stress was observed. The data reinforce the view that stress signalling components have evolved rapidly to protect fungal species against the environmental insults they experience in their specialized niches.
Methods
Strains and growth media
The strains used in this study are summarized in Table 7. C. albicans, Candida glabrata, Debaryomyces hansenii, Kluyveromyces lactis, S. cerevisiae, S. pombe, Ustilago maydis and Yarrowia lipolytica were grown in YPD [111,112]. Ashbya gossypii mycelia or spores were grown on AFM [Ashbya full medium: [113,114]]. Aspergillus fumigatus mycelia or conidia were grown on PDA [potato dextrose agar: [115]]. Aspergillus nidulans mycelia or conidia were grown on MNVUU (minimal medium: [116,117]). Fusarium graminearum mycelia or macroconidia were grown on SNA [synthetic nutrient poor agar: [118,119]]. Magnaporthe grisea mycelia or spores were grown on CM [complete medium: [120]]. Neurospora crassa mycelia were grown on solid Vogel's medium using D-glucose instead of sucrose as described by Vogel (1956) [121] and Selitrennikoff and Sachs (1991) [122]. All strains were maintained as frozen stocks and then cultured on the appropriate media.
Using the above methods we were able to standardize the growth conditions under which the stress phenotypes for yeast and filamentous species were examined as far as was practically possible. However, these growth conditions were not optimal for some species, and this might have affected their stress resistance.
Growth conditions
C. albicans, C. glabrata, D. hansenii, K. lactis, S. cerevisiae, S. pombe, U. maydis and Y. lipolytica colonies were picked from YPD plates, inoculated into 5 ml of YPD, and incubated overnight at 30°C at 200 rpm [112].
Fragments of A. gossypii mycelia were placed on AFM agar plates and incubated for 7 days at 30°C [adapted from [123]]. Mycelial mats were then removed, resuspended in 800 μl H2O and 200 μl zymolyase-100T, and incubated at 37°C for 4 hours. 1 ml of 0.03% Triton X-100 were added and the spores were collected by centrifugation at 5000 rpm for 5 min. The spores were washed twice with 0.03% Triton X-100.
Disks of A. fumigatus mycelia (2.5 cm diameter) were inoculated on PDA plate and incubated upside down at 37°C for 5 days [115]. The conidia harvested by gentle scraping three times into 3 ml 0.1% Tween 20, and the conidial suspension filtered through 4 layers of Miracloth (Calbiochem, Merck Biosciences, Nottingham, UK) to remove hyphae.
Disks of A. nidulans mycelia (2.5 cm diameter) were inoculated on MNVUU plates and incubated upside down at 28°C for 7 days [117]. Conidia were then harvested using the same procedure as for A. fumigatus but water was used instead of Tween solution to scrape the plate.
Small fragments of agar containing F. graminearum mycelia were placed on fresh SNA plates [119]. Plates were sealed with Parafilm and incubated under blue/white light for 8–10 days at 25°C. Macroconidia were harvested using the same procedure as for A. nidulans.
Filter papers containing M. grisea mycelia were placed on CM plates and incubated upside down at 25°C for 14 days [adapted from [120]]. Spores were harvested using the same procedure as for A. nidulans.
N. crassa mycelia were grown in 250 ml conical flasks containing 40 ml solid VgS medium [adapted from [124]]. Flasks were incubated in the dark at 28°C for 3 days, and then in the light for 2 more days. Macroconidia were harvested into 50 ml dH2O, and the suspension transferred to a 15 ml Falcon tube. This was repeated three times to maximize the yield of macroconidia.
Stress sensitivity assays
Osmotic stress was applied using NaCl (0–3 M range) and Dsorbitol (0–3 M range). Oxidative stress was imposed using H2O2 (0–30 mM range) and menadione sodium bisulfate (0–0.5 mM range). Cell wall stress was applied using Calcofluor White (0–300 μg/ml range) and Congo Red (0–500 μg/ml range).
Overnight cultures of C. albicans, C. glabrata, D. hansenii, K. lactis, S. cerevisiae, S. pombe, U. maydis and Y. lipolytica grown in YPD at 30°C, were used to inoculate 10 ml of YPD to a starting OD600 of 0.1. The cells were grown at 30°C at 200 rpm to an OD600 of 0.8 – 1.0. These exponential cells were then serially diluted and 3 μl drops of each dilution (100–10-5) were spotted onto YPD plates containing the appropriate stress treatment. Growth was assessed after 2 days incubation at 30°C.
In general, the stress assays for the filamentous fungi were performed under the growth conditions described above. Exceptions were the media used for A. fumigatus and N. crassa conidial stress assays. A. fumigatus conidia were grown on YG [yeast extract agar: [125]]. For N. crassa L-sorbose was added to Vogel's growth medium to promote colonial growth [126]. Fresh spore or conidial suspensions were serially diluted and 3 μl drops of each dilution (100–10-5) were spotted onto plates containing the appropriate stress treatment. Growth of the filamentous fungi was examined after 2 days incubation, except for M. grisea, which was examined after 5 days incubation. Experiments were repeated at least three times.
Quantitative analysis of stress resistance
To semi-quantitatively compare the stress resistances of the fungal species analysed under the conditions tested, the percentage of growth of each species was calculated relative to their non-stress control for each stress condition. For each species, the total number of spots observed across the dilutions (100–10-5) for each stress condition tested was counted and expressed as percentages of those on the corresponding control plates (Tables 1, 2, 3, 4, 5 and 6). To obtain a global view of the response of each fungal species to each osmotic (NaCl and sorbitol), oxidative (H2O2 and MD) and cell wall (CFW and CR) stress the mean relative growth (%) was calculated for each species under analysis for each condition tested. To measure relative growth, the amount of growth in the presence of stress was divided by the amount of growth observed for unstressed cells of the same species and expressed as a percentage.
Phylogenetic analyses
Eight S. cerevisiae proteins were used as queries for our phylogenetic analyses: actin [127], 3- phosphoglycerate kinase [128], translation elongation factor EF-1 alpha [129], the cyclin-dependent protein kinase, Cdc28 [130], adenylate cyclase [131,132], and the transcription factors Gcn4, Mig1 and Fap1 [133-135], which belong to different gene families. These S. cerevisiae protein sequences were retrieved from the Saccharomyces Genome Database (SGD: http://www.yeastgenome.org). The sequences of the orthologues of these proteins in the other fungal species under analysis were retrieved by BLASTP [136] using the SGD protein sequences as queries (see additional file 5). Manual searches were undertaken at the National Center for Biotechnology Information using NCBI databases, including the non-redundant protein sequence database (nr database) currently containing approximately 900,000 sequences http://www.ncbi.nih.gov/BLAST/. BLASTP search parameters were set to default. Phylogenetic analyses were performed using MEGA3.1 [137] available at http://www.megasoftware.net/. Sequences were aligned using ClustalW [138]. A concatenated phylogenetic tree was then produced by neighbour-joining (NJ) clustering [139]. The phylogeny was drawn using the p-distances method to correct for multiple amino acid substitutions per site and rate heterogeneity amongst sites. The substitution of amino sites per site was 0.05 (scale bar underneath tree). Clade stability was assessed using 1000 bootstrap replicates. Phylogenetic trees were presented using TreeView [140].
Identification of putative orthologues
The complete annotated fungal genome sequences were retrieved from the databases in Table 7[144-155]. Putative orthologues were identified for each S. cerevisiae protein in each fungal species under analysis using the reciprocal best hit (rbh) method [46,141]. We define 'putative orthologues' as two proteins, one from each fungal genome, that are each other's reciprocal best hit [142]. Perl scripts were used to reformat the amino acid sequence data (formatdb), to perform reciprocal BLASTP searches, and to generate output files that provide the accession number for each orthologue, its percentage identity to the corresponding S. cerevisiae protein and the match score. Automated reciprocal BLASTP searches were performed [143] using the default parameters except that scoring parameter compositional adjustments were set to "no adjustment" and the filter parameter was set automatically to "low complexity regions". Putative orthologues were not identified for all S. cerevisiae proteins in every species above the default BLASTP cut-off, which was set to 10 by default. Only the reciprocal best hits identified in this way were considered for further analysis. Where no significant reciprocal hit was identified, the score was left blank.
Authors' contributions
EN carried out the phenotypic and bioinformatic analyses and drafted the manuscript. IA created the PERL program for the automated BLASTP searches under the supervision of MS. JQ participated in the oxidative stress analysis. IS participated in the bioinformatic analysis and helped to draft the manuscript. AJPB conceived of the study, participated in its design and coordination, and prepared the last version of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sensitivity of fungi to different stresses. Fungal stress sensitivity data: (A) sorbitol; (B) H2O2; (C) enadione sodium bisulfite; (D) Calcofluor White; (E) Congo Red.
List of fungal orthologues. Lists of reciprocal best hits: (A) complete list of all fungal orthologues; (B) osmotic stress signalling orthologues; (C) oxidative stress signalling orthologues; (D) cell wall cell stress signalling orthologues.
Conservation of fungal osmotic, oxidative and cell wall stress pathways. Figures illustrating the degree of conservation of signalling molecules on stress pathways in each of the fungal species examined: (A) osmotic stress signalling pathway; (B) oxidative stress signalling pathway; (C) cell wall cell stress signalling pathway.
No correlation exists between fungal stress phenotypes and the degree of conservation of fungal stress regulators. Plot showing no significant correlation between the degree of conservation of oxidative stress regulators and the resistance of the fungal species to oxidative stress.
Proteins used for phylogenetic analysis. Details of fungal orthologues used to construc the phylogenetic tree.
Acknowledgments
Acknowledgements
We thank CBS, LGC-Promochem, FGSC, David Denning (University of Manchester), Ken Haynes (Imperial College London), Kim Hammond-Kosack and Martin Urban (Rothamsted Research), Nick Talbot and Mick Kershaw (University of Exeter), Nick Read (University of Edinburgh), Joerg Kaemper (MPI), and Naomie Jacques (de Levures d'Interet Biotech) for providing strains and protocols. EN was supported by a studentship from the University of Aberdeen. AB was supported by the Wellcome Trust (080088) and the BBSRC (BB/C510391/1, BB/D009308/1, BB/F00513X/1).
Contributor Information
Elissavet Nikolaou, Email: e.nikolaou@abdn.ac.uk.
Ino Agrafioti, Email: ino.agrafioti@imperial.ac.uk.
Michael Stumpf, Email: m.stumpf@imperial.ac.uk.
Janet Quinn, Email: janet.quinn@newcastle.ac.uk.
Ian Stansfield, Email: i.stansfield@abdn.ac.uk.
Alistair JP Brown, Email: al.brown@abdn.ac.uk.
References
- Mager WH, Siderius M. Novel insights into the osmotic stress response of yeast. FEMS Yeast Res. 2002;2:251–257. doi: 10.1016/S1567-1356(02)00116-2. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradas-Ferreira P, Costa V. Adaptive response of the yeast Saccharomyces cerevisiae to reactive oxygen species: defenses, damage and death. Redox Rep. 2000;5:277–285. doi: 10.1179/135100000101535816. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signalling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wilkinson CRM, Watt S, Penkett CJ, Toone WM, Jones N, Bahler J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol Biol Cell. 2008;19:308–317. doi: 10.1091/mbc.E07-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol. 1999;181:700–708. doi: 10.1128/jb.181.3.700-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148:3705–3713. doi: 10.1099/00221287-148-11-3705. [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R, Navarro-Garcia F, Roman E, Negredo AI, Eisman B, Nombela C, Pla J. The HOG1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryotic Cell. 2003;2:351–361. doi: 10.1128/EC.2.2.351-361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: Transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol Biol Cell. 2004;15:456–467. doi: 10.1091/mbc.E03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 "two-component" osmosensor. Cell. 1996;86:865–75. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Sprague GF. Control of MAP kinase signalling specificity or how not to go HOG wild. Genes Dev. 1998;12:2817–20. doi: 10.1101/gad.12.18.2817. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–5. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Reiser V, Raitt DC, Saito H. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol. 2003;161:1035–40. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz M, Becit E, Hohmann S. Comparative genomics of the HOG-signalling system in fungi. Curr Genet. 2006;49:137–151. doi: 10.1007/s00294-005-0038-x. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley SW. Regulation of the transcriptional response to oxidative stress in fungi: Similarities and differences. Eukaryot Cell. 2003;2:381–389. doi: 10.1128/EC.2.3.381-389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner A, Shiozaki K. Yeast signalling pathways in the oxidative stress response. Mutat Res. 2005;569:13–27. doi: 10.1016/j.mrfmmm.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley WS, Harshman KD, Parker CS. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989;3:283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- Toone WM, Jones N. AP-1 transcription factors in yeast. Curr Opin Genet Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- Sewell AK, Yokoya F, Yu W, Miyagawa T, Murayama T, Winge DR. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J Biol Chem. 1995;270:25079–25086. doi: 10.1074/jbc.270.42.25079. [DOI] [PubMed] [Google Scholar]
- Liu XD, Thiele DJ. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- Levin D. Cell wall integrity signalling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder AL, Lee TK, Ballester R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics. 1999;152:1487–1499. doi: 10.1093/genetics/152.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavel M, Philip B, Buehrer BM, Errede B, Levin DE. Mid2 is a putative sensor for cell integrity signalling in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Valdés-Hevia MD, de la Guerra R, Gancedo C. Isolation and characterization of the gene encoding phosphoenolpyruvate carboxykinase from Saccharomyces cerevisiae. FEBS Lett. 1989;258:313–316. doi: 10.1016/0014-5793(89)81682-5. [DOI] [PubMed] [Google Scholar]
- Irie K, Takase M, Lee KS, Levin DE, Araki H, Matsumoto K, Oshima Y. Mkk1 and Mkk2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Irie K, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin DE. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–75. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Bardes ES, Ohya Y, Lew DJ. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat Cell Biol. 2001;3:417–20. doi: 10.1038/35070104. [DOI] [PubMed] [Google Scholar]
- Quinn J, Findlay VJ, Dawson K, Millar JB, Jones N, Morgan BA, Toone WM. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, Straffon M, Enjalbert B, Nantel A, Macaskill S, Whiteway M, Brown AJP. Msn2/4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen, Candida albicans. Eukaryot Cell. 2004;3:1111–1123. doi: 10.1128/EC.3.5.1111-1123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJP, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham J, Smith DA, da Silva Dantas A, Doris KS, Patterson MJ, Bruce CR, Quinn J. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol Biol Cell. 2007;18:4603–4614. doi: 10.1091/mbc.E07-06-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Ainsworth and Bisby's Dictionary of the Fungi. United Kingdom: CAB International; 1995. [Google Scholar]
- Alexopoulos CJ, Mims CW, Blackwell M. Introductory Mycology. New York: John Wiley and Sons; 1996. [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- Turchin A, Kohane IS. Gene homology resources on the World Wide Web. Physiol Genomics. 2002;11:165–177. doi: 10.1152/physiolgenomics.00112.2002. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Smith NG. Do essential genes evolve slowly? Curr Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Wall DP, Hirsh AE. A simple dependence between protein evolution rate and the number of protein-protein interactions. BMC Evol Biol. 2003;3:11. doi: 10.1186/1471-2148-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L, Kondrashov A. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- Lang BF, Seif E, Gray MW, O'Kelly CJ, Burger G. A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J Eukaryot Microbiol. 1999;46:320–326. doi: 10.1111/j.1550-7408.1999.tb04611.x. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJ, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O'Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D, Logue M, Stajich J, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Wendland J. Initial molecular characterisation of a novel Rho-type GTPase RhoH in the filamentous ascomycete Ashbya gossypii. Curr Genet. 2005;48:247–255. doi: 10.1007/s00294-005-0017-2. [DOI] [PubMed] [Google Scholar]
- Zhao W, Panepinto JC, Fortwendel JR, Fox L, Oliver BG, Askew DS, Rhodes JC. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage and virulence. Infect Immun. 2006;74:4865–4874. doi: 10.1128/IAI.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa T, Ashitani K, Yoshikawa K, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S. Comparison of transcriptional responses to osmotic stresses induced by NaCl and sorbitol additions in Saccharomyces cerevisiae using DNA microarray. J Biosci Bioeng. 2006;102:568–71. doi: 10.1263/jbb.102.568. [DOI] [PubMed] [Google Scholar]
- Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, Castano I, De las Penas A. High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell. 2008;7:814–25. doi: 10.1128/EC.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SB, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–54. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- Klis FM, Mol P, Hellingwerf K, Stanley B. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2002;26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Serrano R. Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol. 1996;165:1–52. doi: 10.1016/s0074-7696(08)62219-6. [DOI] [PubMed] [Google Scholar]
- Norkrans B. Studies on marine occurring yeasts: Growth related to pH, NaCl concentration and temperature. Arch Microbiol. 1966;54:374–392. [Google Scholar]
- Onishi H. Osmophilic yeasts. Adv Food Res. 1963;12:53–94. [PubMed] [Google Scholar]
- Gustafsson L, Norkans B. On the mechanism of salt tolerance. Production of glycerol and heat during growth of Debaryomyces hansenii. Arch Microbiol. 1976;110:177–183. doi: 10.1007/BF00690226. [DOI] [PubMed] [Google Scholar]
- Nobre MF, da Costa MS. The accumulation of polyols by the yeast Debaryomyces hansenii in response to water stress. Canadian J Microbiol. 1985;31:1061–1064. [Google Scholar]
- Norkrans B, Kylin A. Regulation of the potassium to sodium ratio and of the osmotic potential in relation to salt tolerance in yeasts. J Bacteriol. 1969;100:836–845. doi: 10.1128/jb.100.2.836-845.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh N, Kawabata M, Kitajima S. Effects of four oxidants, menadione, 1-chloro-2, 4-dinitrobenzene, hydrogen peroxide and cumene hydroperoxide, on fission yeast Schizosaccharomyces pombe. J Biochem. 2005;138:797–804. doi: 10.1093/jb/mvi179. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Veal E, Day A, Morgan B. Hydrogen peroxide sensing and signalling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Gille G, Sigler K. Oxidative stress and living cells. Folia Microbiol (Praha) 1995;40:131–152. doi: 10.1007/BF02815413. [DOI] [PubMed] [Google Scholar]
- Elorza MV, Rico H, Sentandreu R. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–82. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- Selitrennikoff CP. Calcofluor white inhibits Neurospora chitin synthetase activity [bleaching agent for fabrics] Plant Physiol Bioch. 1984;8:269–272. [Google Scholar]
- Roncero C, Duran A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol. 1988;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Tsurugi J, Watanabe T. The dependence of the conformation of a (1→ 3)-β-D-glucan on chain-length in alkaline solution. Carbohydr Res. 1973;29:397–403. [Google Scholar]
- Ogawa K, Hatano M. Circular dichroism of the complex of a (1–3)-bD-glucan with Congo Red. Carbohydr Res. 1978;67:527–535. [Google Scholar]
- Kopecka M, Gabriel M. The influence of Congo red on the cell wall and (1 → 3)-β-d-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol. 1992;158:115–126. doi: 10.1007/BF00245214. [DOI] [PubMed] [Google Scholar]
- Imai K, Noda Y, Adachi H, Yoda K. A novel endoplasmic reticulum membrane protein Rcr1 regulates chitin deposition in the cell wall of Saccharomyces cerevisiae. J Biol Chem. 2005;280:8275–8284. doi: 10.1074/jbc.M409428200. [DOI] [PubMed] [Google Scholar]
- Ram AFJ, Wolters A, Hoopen RT, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Ram AFJ, Brekelmans SSC, Oehlen LJWM, Klis FM. Identification of two cell cycle regulated genes affecting the β1, 3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- Manners DJ, Masson AJ, Patterson JC. The structure of a P-(1-*3)-D-glucan from yeast cell walls. Biochem J. 1973;135:19–30. doi: 10.1042/bj1350019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet GH. Cell walls. In: Rose AH, Harrison JS, editor. The Yeasts. New York: Academic Press; 1991. pp. 199–277. [Google Scholar]
- Kang MS, Elango N, Mattia E, Au-Young J, Robbins PW, Cabib E. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J Biol Chem. 1984;259:14966–14972. [PubMed] [Google Scholar]
- Borgia PT, Dodge CL. Characterization of Aspergillus nidulans mutants deficient in cell wall chitin or glucan. J Bacteriol. 1992;174:377–83. doi: 10.1128/jb.174.2.377-383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DA, Horisberger M, Horman I, Wursch P. The wall structure of Schizosaccharomyces pombe. J Gen Microbiol. 1974;81:199–206. doi: 10.1099/00221287-81-1-199. [DOI] [PubMed] [Google Scholar]
- Horisberger M, Vonlanthen M, Rosset J. Localization of α-galactomannan and of wheat germ agglutinin receptors in Schizosaccharomyces pombe. Arch Microbiol. 1978;119:107–111. doi: 10.1007/BF00964260. [DOI] [PubMed] [Google Scholar]
- Bulawa CE, Slater M, Cabib E, Au-Young J, Sburlati A, Adair WLJr, Robbins PW. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–25. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Sietsma JH, Wessels JG. The occurrence of glucosaminoglycan in the wall of Schizosaccharomyces pombe. J Gen Microbiol. 1990;136:2261–5. doi: 10.1099/00221287-136-11-2261. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia S. Cell Wall Chemistry, Morphogenesis, and Taxonomy of Fungi. Ann Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J. Fungal cell wall: Structure, synthesis and assembly. USA: CRC Press; 1992. [Google Scholar]
- De Nobel H, Ruiz C, Martin H, Morris W, Brul S, Molina M, Klis FM. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology. 2000;146:2121–2132. doi: 10.1099/00221287-146-9-2121. [DOI] [PubMed] [Google Scholar]
- Specht CA, Liu Y, Robbins PW, Bulawa CE, Iartchouk N, Winter KR, Riggle PJ, Rhodes JC, Dodge CL, Culp DW, Borgia PT. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet Biol. 1996;20:153–167. doi: 10.1006/fgbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Mansour MK, Levitz SM. Interactions of fungi with phagocytes. Curr Opin Microbiol. 2002;5:359–365. doi: 10.1016/s1369-5274(02)00342-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Parraga P, Hernandez JA, Arguelles JC. Role of antioxidant enzymatic defences against oxidative stress (H2O2) and the acquisition of oxidative tolerance in Candida albicans. Yeast. 2003;20:1161–1169. doi: 10.1002/yea.1029. [DOI] [PubMed] [Google Scholar]
- Thorpe GW, Fong CS, AliC N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: Oxidative-stress-response genes. PNAS. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci USA. 2003;100:11007–11012. doi: 10.1073/pnas.1834481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, MacCallum DM, Odds FC, Brown AJP. Niche-specific activation of the oxidative stress responses by the pathogenic fungus Candida albicans. Infect Immun. 2007;75:2143–2151. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J, Brown AJP. Stress Responses in Candida albicans. In: d'Enfert C, Hube B, editor. Candida: comparative and functional genomics. UK: Caister Academic Press; 2007. pp. 217–261. [Google Scholar]
- Jamieson DJ, Stephen DW, Terriere EC. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol Lett. 1996;138:83–88. doi: 10.1111/j.1574-6968.1996.tb08139.x. [DOI] [PubMed] [Google Scholar]
- Avery AM, Avery SV. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J Biol Chem. 2001;276:33730–33735. doi: 10.1074/jbc.M105672200. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Inglis D, Roman E, Pla J, Li D, Calera JA, Calderone R. Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot Cell. 2003;2:1018–1024. doi: 10.1128/EC.2.5.1018-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. "Sleeping beauty": Quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan BA, Brown AJP, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CEV, Sonnhammer ELL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer ELL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pontius JU, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. New York: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Ashby SF, Nowell W. The fungi of stigmatomycosis. Annual Botany. 1926;40:69–84. [Google Scholar]
- Altmann-Johl R, Philippsen P. AgTHR4, a new selection marker for the transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol Gen Genet. 1996;250:69–80. doi: 10.1007/BF02191826. [DOI] [PubMed] [Google Scholar]
- Vicentefranqueira R, Moreno MA, Leal F, Calera JA. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, inc-depleted environment. Eukaryot Cell. 2005;4:837–848. doi: 10.1128/EC.4.5.837-848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer E. Meiotic and mitotic recombination in Aspergillus, and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- Virag A, Harris SD. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot Cell. 2006;5:881–895. doi: 10.1128/EC.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg HI. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium -Sektion Liseola. Mitt Biol Bundesanst. 1976;169:1–117. [Google Scholar]
- Urban M, Daniels S, Mott E, Hammond-Kosack K. Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant. 2002;32:961–973. doi: 10.1046/j.1365-313x.2002.01480.x. [DOI] [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, Hamer JE. Identification and Characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ. Vogel, A convenient medium for Neurospora (medium N) Microb Genet Bull. 1956;13:42–43. [Google Scholar]
- Selitrennikoff C, Sachs MS. Lipofectin increases transformation of Neurospora crassa. Fungal Genet Newsl. 1991;38:90–91. [Google Scholar]
- Park EY, Zhang JH, Tajima S, Dwiarti L. Isolation of Ashbya gossypii mutant for an improved riboflavin production targeting for biorefinery technology. J Appl Microbiol. 2007;103:468–476. doi: 10.1111/j.1365-2672.2006.03264.x. [DOI] [PubMed] [Google Scholar]
- Davis RH. Genetic, biochemical and molecular techniques. In: Davis RH, editor. Neurospora: contributions of a model organism. USA: Oxford University Press; 2000. p. 293. [Google Scholar]
- Kontoyiannis DP, Lewis RE, May GS, Osherov N, Rinaldi MG. Aspergillus nidulans is frequently resistant to amphotericin B. Mycoses. 2002;45:406–407. doi: 10.1046/j.1439-0507.2002.00797.x. [DOI] [PubMed] [Google Scholar]
- Perkins DD. How to convert wild-type spreading growth to colonial. 2006. http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm
- Ng R, Abelson J. Isolation and sequence of the gene actin in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1980;77:3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman RA, Clarke L, Carbon J. Isolation and characterization of the yeast 3-phosphoglycerokinase gene (PGK) by an immunological screening technique. J Biol Chem. 1980;255:12073–12080. [PubMed] [Google Scholar]
- Schirmaier F, Philippsen P. Identification of the two genes coding for the translation elongation factor EF-1a of S. cerevisiae. EMBO. 1984;3:3311–3315. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. Cell cycle control in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editor. The molecular and cellular biology of the yeast Saccharomyces: Cell cycle and cell biology. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- Kataoka T, Broek D, Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985;43:493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Casperson GF, Walker N, Bourne HR. Isolation of the gene encoding adenylate cyclase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:5060–3. doi: 10.1073/pnas.82.15.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Fink GR. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfiyya LL, Iyer VR, DeRisi J, DeVit J, O'Brown P, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Loeschmann A, Deuter-Reinhard M, Hall MN. FAP1, a homologue of human transcription factor NF-X1, competes with rapamycin for binding to FKBP12 in yeast. Mol Microbiol. 2000;37:1480–93. doi: 10.1046/j.1365-2958.2000.02105.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologues. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Wall DP, Hirsh AE, Fraser HB, Kumm J, Giaever G, Eisen MB, Feldman MW. Functional genomic analysis of the rates of protein evolution. PNAS. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, van Noort V, Heijden RTJM van der, Boekhout T, Snel B, Huynen MA. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23:815–824. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pöhlmann R, Luedi P, Choi S, Wing RA, Flavier A, Gaffney TD, Philippsen P. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Peñalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Córdoba S, Rodríguez-Peña JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sánchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with the A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, de Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne B, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, de Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, We'solowski-Louvel M, Westhofl E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D'Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJM, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, MathewsoN CE, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. The Genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivarès CP. Genome sequence and gene compaction of the eukaryotic parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma L, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CPC, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O'Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schaefre M, Mueller-Auer S, Gabel S, Fuchs M, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, DreÂano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, SaÂnchez M, del Rey F, Benito J, DomõÂnguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerrutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity of fungi to different stresses. Fungal stress sensitivity data: (A) sorbitol; (B) H2O2; (C) enadione sodium bisulfite; (D) Calcofluor White; (E) Congo Red.
List of fungal orthologues. Lists of reciprocal best hits: (A) complete list of all fungal orthologues; (B) osmotic stress signalling orthologues; (C) oxidative stress signalling orthologues; (D) cell wall cell stress signalling orthologues.
Conservation of fungal osmotic, oxidative and cell wall stress pathways. Figures illustrating the degree of conservation of signalling molecules on stress pathways in each of the fungal species examined: (A) osmotic stress signalling pathway; (B) oxidative stress signalling pathway; (C) cell wall cell stress signalling pathway.
No correlation exists between fungal stress phenotypes and the degree of conservation of fungal stress regulators. Plot showing no significant correlation between the degree of conservation of oxidative stress regulators and the resistance of the fungal species to oxidative stress.
Proteins used for phylogenetic analysis. Details of fungal orthologues used to construc the phylogenetic tree.