Abstract
Immune function is likely to be a critical determinant of an organism's fitness, yet most natural animal and plant populations exhibit tremendous genetic variation for immune traits. Accumulating evidence suggests that environmental heterogeneity may retard the long-term efficiency of natural selection and even maintain polymorphism, provided alternative host genotypes are favoured under different environmental conditions. ‘Environment’ in this context refers to abiotic factors such as ambient temperature or availability of nutrient resources, genetic diversity of pathogens or competing physiological demands on the host. These factors are generally controlled in laboratory experiments measuring immune performance, but variation in them is likely to be very important in the evolution of resistance to infection. Here, we review some of the literature emphasizing the complexity of natural selection on immunity. Our aim is to describe how environmental and genetic heterogeneities, often excluded from experimentation as ‘noise’, may determine the evolutionary potential of populations or the potential for interacting species to coevolve.
Keywords: host–pathogen interactions, genotype by environment interactions, genetic variation, insect immunity, natural selection, evolution
1. Introduction
Natural plant and animal populations frequently harbour genetic variation for immune capacity, which may seem paradoxical given the likely importance of immune performance to fitness. Why is functional variation in immune capacity not purged by natural selection? Why are susceptibility alleles not eliminated? Mounting evidence suggests that environmental heterogeneity and pathogen diversity can maintain host genetic variation in immune function by favouring alternative host genotypes over time and/or space. We review here some of the literature that demonstrates how environmental variation can complicate natural selection on immunity. Much of this work has been done in invertebrate model systems owing to their amenability to experimental manipulation. The principles that emerge, however, should be broadly applicable.
The arena of natural infection is highly variable: parasite epidemics come and go, abiotic factors such as temperature fluctuate and food availability (and hence nutrition levels) may be volatile. The world in which host and parasites interact is a noisy one and disease is often context dependent. Mechanistic study of immune systems rarely accounts for such contexts. Immunological study has revealed the structural and biochemical bases for complex systems of defence, often relying on experimental designs that are greatly simplified relative to the natural world. Many experiments are conducted using non-infectious immune elicitors, experiments are generally performed under carefully controlled laboratory conditions and test subjects are typically genetically homogeneous and even highly inbred. These do not reflect the conditions that host and parasite populations experience in natural settings. Moreover, mechanistic studies of immunity tend to focus on genes involved in canonical defence responses, but resistance to infection is a whole-organism phenotype that involves much broader aspects of host physiology. A host's ability to sustain an effective immune response is profoundly affected by its overall condition, which is one key reason why immune performance is influenced by a variety of environmental and ecological factors.
Experiments conducted by evolutionary ecologists typically measure whole-organism traits related to fitness. While these are generally poor for elucidating immunological mechanism, they are much better at establishing environmental context. These contexts are essential for understanding selective pressure on the immune system. In this review, we will summarize a range of important contexts impacting host immunity: host genetic variation; fluctuating abiotic environments; genotype by environment interactions; host genotype by pathogen genotype interactions; and pleiotropic constraints. The central premise of this paper is that contextual heterogeneity generates complex natural selective pressures, acting to retard direct selection on immunity and potentially maintaining genetic polymorphism in immunocompetence. A major challenge here is not just documenting the context-specific aspects of immunity, but inferring the relative degree to which each of these factors influences the evolution and expression of disease.
2. What is environment?
Before we can proceed, we must define what we mean by ‘environment’. Most obviously, environment includes abiotic variables such as ambient temperature and the nutritional value of available resources. Environment may also include biotic variables such as the genetic diversity of the pathogens infecting the host. This may encompass genetic polymorphism within a pathogen species or taxonomic diversity among pathogens a host species is exposed to. The environment in which an immune response manifests itself may also be defined by competing physiological demands on the host at the time of infection. This suborganismal environment may of course be influenced by abiotic external environmental conditions, but it need not be. Examples of relevant physiological conditions that influence immune performance include reproductive status and metabolic state. Abiotic environmental conditions and pathogen diversity vary both temporally and geographically, and competing physiological demands vary over the lifetime of the host.
Environmental factors can have absolute effects on host resistance to infection. For example, the ambient temperature under which infections occur can quantitatively affect resistance to microbial infection in invertebrates (Elliot et al. 2003; Mitchell et al. 2005; Lazzaro et al. 2008; Linder et al. 2008). Absolute effects of environment, however, do not dramatically impact the efficacy of selection. In order for a variable environment to maintain genetic variation, for instance, alternative host genotypes must have the highest fitness in distinct environmental states (figure 1; Gillespie & Turelli 1989). Statistically, there must be a significant genotype by environment interaction (G×E) determining fitness. As will be discussed below, G×E appears to be common in natural systems, lending plausibility to the idea that environmental heterogeneity may result in the maintenance of polymorphism in natural populations. Considerable genotype by temperature interaction, for instance, has been shown for resistance of the aquatic crustacean Daphnia magna and the fruit fly Drosophila melanogaster to bacterial infection (Mitchell et al. 2005; Lazzaro et al. 2008). Analogously, host genetic variation could be maintained by pathogen diversity if distinct host genotypes are best able to combat infection by each of the pathogens a host is likely to encounter (Hamilton 1993; Frank 1994). We may, in this case, consider that fitness is determined by an interaction between host and pathogen genotypes (GH×GP). GH×GP interactions have been shown in exposure of D. magna to diverse strains of its bacterial pathogen Pasteuria ramosa (Carius et al. 2001) and to different parasite species (Decaestecker et al. 2003). To further complicate matters, the abiotic environment may alter the interaction between host and pathogen genotypes, such that resistance is determined by a three-way GH×GP×E interaction (e.g. Tétard-Jones et al. 2007).
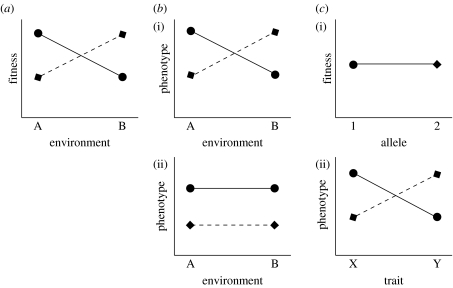
Figure 1.
G×E and local adaptation can be illustrated by imagining two environments (A and B) and one gene with two alleles (symbolized by circles and diamonds), each affecting the same trait. (a) G×E interactions maintain polymorphism when one allele gives the highest fitness in environment A, but the other allele is fitter in environment B. G×E refers to the general case where environments may vary in time or space, while local adaptation refers to the specific case where the environments are different locales. (b) The maintenance of polymorphism by G×E can occur in two ways. (i) In the first example, the phenotype produced by each allele has environment-specific values and the phenotype is positively correlated with fitness. In this case, alternative genotypes yield higher fitness in the two environments. (ii) The phenotype produced by an allele may be constant across environments, but different phenotypes are favoured in different environments, such that the correlation between phenotype and fitness changes from negative to positive depending on the environment. In this case, there is no G×E for the direct phenotype measured, but there is G×E for fitness. This latter scenario is reasonable, as many traits will have varied relationships with fitness. For example, small size might be favoured in some environments, but large size in others. (c) Pleiotropy occurs when one gene with two alleles codes for two traits (X and Y). (ii) Both traits may be under selection for larger values, but one of the two alleles codes for high phenotypic values of trait X and low values of trait Y, while the other allelic form of the gene codes for high values of trait Y. (i) Under this scenario, the overall fitness conferred by each allele is identical due to the balanced effects on both traits, so natural selection does not eliminate either of them.
Resistance to infection could also be impacted by competing physiological demands on the host. For instance, increased reproductive activity may decrease immune capacity in insects (Siva-Jothy et al. 1998; Adamo et al. 2001; McKean & Nunney 2001; Rolff & Siva-Jothy 2002). This phenotypic correlation implies a physiological connection between immune performance and reproduction wherein each process limits the other. Such physiological linkages could in principle be genetically variable, with some genotypes having larger immune responses at the expense of fecundity, but others having higher fecundity in place of strong resistance to infection. Genetically determined balances across competing fitness demands are termed life-history trade-offs. Polymorphism for relative investment in two fitness traits may be maintained by selection in heterogeneous environments (figure 1), as would be the case if the risk of infection (environment) were spatially or temporally variable, making elevated immune responses adaptive in some environments but maladaptive in others. As will be discussed below, life-history trade-offs may themselves be modulated by abiotic environment (Reznick et al. 2000).
3. The abiotic environment
In many host–pathogen or immunological studies, hosts are regarded as having ‘resistant’ or ‘susceptible’ genotypes with the assumption that the relative relationships among genotypes are fixed across environments. However, relative resistance/susceptibility is complex: it includes genetic variation plus environmentally mediated variation and, often, genotype by environment interactions (e.g. de Jong 1990; Gomulkiewicz & Kirkpatrick 1992). Thus the label susceptible may depend entirely on when and where the resistance phenotype is measured. Indeed, very small and realistic alterations in the abiotic environment may dramatically modify infection outcomes and exaggerate or diminish differences between genotypes (reviewed in Thomas & Blanford 2003). Such G×E means that while selection in one environment may predictably drive genetic change in the host population, the same selective force may have no predictable effect or may even drive allele frequencies in the opposite direction if applied under alternative environmental conditions (Gomulkiewicz & Kirkpatrick 1992; Via 1994). In a world of variable environments, G×E interactions decouple genotype and phenotype, slowing adaptive evolution and potentially resulting in the maintenance of polymorphism (Gillespie & Turelli 1989).
G×E is common. It has been most widely studied by evaluating host genotypes (GH) under environments varying in temperature, food availability and parasite dose, a proxy for the encounter rate with parasites. Temperature is the most commonly studied environmental variable, as it impacts a great range of physiological processes (e.g. Hochachka & Somero 1984; Johnston & Bennett 1996) and it greatly modifies the amount of virulence hosts suffer. GH by temperature interactions for infection-related traits are evident in a considerable range of invertebrates including orthopterans (Thomas & Blanford 2003), dipterans (Lazzaro et al. 2008), crustaceans (Mitchell et al. 2005) and paramecia (Fels & Kaltz 2006). The aquatic crustacean Daphnia provides a dramatic case where genotypes switch their rank order of susceptibility (measured either as the proportion of host becoming infected, or host fitness losses) as temperature rises from 15 to 25 degrees (Mitchell et al. 2005), a range hosts could reasonably expect to experience over short time frames or even over depth changes. Food quality (Ferguson & Read 2002; Bedhomme et al. 2004; Lambrechts et al. 2006) and parasite dose (e.g. Ben-Ami et al. 2008) also interact with genotype, although these G×E effects appear to be smaller in magnitude than those involving temperature. The range of environments that have been experimentally manipulated in G×E studies is far from comprehensive. Instead, studied environments generally reflect variables that are easy to manipulate in the laboratory, although the aspiration is clearly to make reasonable estimations of what are assumed to be key environmental fluctuations in natural habitats. Similarly, the range of traits measured may not always reflect the most important ones. However, in most cases, measured traits are arguably key parasite fitness components (e.g. infection load or parasite growth rates), host fitness components (e.g. fecundity) or traits that strongly influence both interactors (e.g. host mortality or the proportion of hosts becoming infected).
Although most examples in the literature come from invertebrates, similar phenomena have been demonstrated in vertebrates. One notable example concerns mouse infection with the nematode Heligmosomoides polygyrus, under laboratory conditions where infection success is dependent on mouse strain. These differences disappear if infection takes place in a more natural arena (Scott 1991). The prevailing explanation is that transmission rate differs between the two environments, and that susceptible strains cannot control parasite numbers during the high transmission rates achieved in controlled experiments (Scott 2006). Important G×E effects on immunity are probably the rule across all higher eukaryotes. Genotype by environment interactions in vertebrates have simply been less well studied than those in invertebrates, in part because it can be unethical to subject vertebrates to a large range of stresses.
If G×E interactions are pervasive, how should we determine their ultimate impact upon the efficacy of selection? We emphasize, first, that a number of interesting traits have not yet been sufficiently explored. Maternal effects in particular have well-established effects on immunity in vertebrates (Gershwin et al. 1985; Klasing 1998; Brinkhof et al. 1999; Tella et al. 2000), and strong (and sometimes counter-intuitive) effects on susceptibility in some invertebrates (Mitchell & Read 2005), but we know of no cases where genotype by (maternal) environment interactions have been explored (but see Tseng 2006 and Little et al. 2007 for examples of how the maternal environment may affect parasite growth traits). Second, and also relatively unexplored, is how environmental variation affects the interaction between host and parasite fitness components. It will be important to test this relationship as it is a key component of common models of pathogen evolution (Anderson & May 1982; Bremermann & Pickering 1983; Frank 1996). It is important to note that the study of G×E interactions is heavily weighted by data on whole-organism resistance phenotypes. Immune responses in the strict sense, although almost certainly contributing to the fitness cost of parasitism, have themselves rarely been characterized under varied environments. This presumably reflects the general lack of studies of environmental variation on vertebrates who have the best characterized immune responses. As immunity in invertebrates is increasingly studied, this is likely to change.
In truth, we will never achieve exhaustive study of the full range of possible environmental effects on resistance to infection and related traits. Granted, in some cases a reasonably complete picture may be obtainable simply because certain environmental variables are so clearly crucial, for example if a small shift in temperature completely arrests parasite development (e.g. Laine 2007). Similarly, it may occasionally be possible to identify infection-related traits that explain essentially all variation in host and parasite fitness, as in cases where failure to infect is massively more important than growth parameters following infection (Vale et al. 2008). Even with extreme cases, however, it may be difficult to grasp the full evolutionary consequences of G×E. While observations of G×E provide evidence that environmental variation could maintain genetic polymorphism in natural populations, it also directly raises the possibility that measured fitness parameters will differ between the field and laboratory. Thus our interpretation of ‘fitness’ obtained via laboratory work may be misleading. Ultimately, it may be necessary to put more emphasis on field studies. One approach is to directly relate the levels of environmental variation and levels of genetic diversity. Studies taking this tack have been inconclusive (Maynard Smith & Hoekstra 1980; Bell & Reboud 1997; Byers 2005), in part because such a cause–effect relationship is bound to be uncertain (Byers 2005) in the absence of prior knowledge regarding the selective history or past levels of genetic variation. Moreover, many of the quantitative models developed to examine the conditions that would favour such a link have reached mixed conclusions (Levene 1953; Gillespie & Turelli 1989; Sasaki & de Jong 1999) and tend to require strict conditions, in particular crossing reaction norms (Maynard Smith & Hoekstra 1980). As an alternative, it may be desirable to directly study natural selection and fitness within populations. This, however, also presents considerable challenges, as it requires direct observation of changing genotype frequencies (and thus the availability of effective genetic markers) coupled with knowledge of the proximal agent causing the change. In this regard, the possibility to systematically resurrect diapausing hosts and pathogens offers considerable promise (Decaestecker et al. 2007).
4. Host genotype by pathogen genotype interactions
Perhaps the most important environmental consideration of all is the biotic environment, specifically the genetically determined environment that hosts present to parasites and the genetic characteristics of parasite strains hosts find themselves infected with. When the outcome of infection depends on the combination of both host and parasite genotypes, these are called genotype by genotype interactions (GH×GP) (Schmid-Hempel & Ebert 2003; Little et al. 2005). Host genotype by parasite genotype interactions may explain as much as, or more of, the variation in infection outcomes than conventional G×E does, but immunological studies typically cannot simultaneously consider multiple genetic backgrounds of both hosts and parasites, so a gap remains in our mechanistic understanding of GH×GP effects.
Most examples of GH×GP come, again, from whole-organism studies of immunity, especially from fully factored laboratory experiments that include a variety of host and parasite genotypes. Examples span a range of taxonomic groupings, including (on the host side) crustaceans, insects and nematodes, and their interaction with infecting bacteria, protozoa or parasitoids (Carius et al. 2001; Decaestecker et al. 2003; Schulenburg & Ewbank 2004; Lambrechts et al. 2005; Dubuffet et al. 2007). Plant–pathogen systems (e.g. Burdon & Jarosz 1991; Thompson & Burdon 1992; Burdon 1994; Burdon & Thrall 1999; Salvaudon et al. 2005) are more deeply studied than animal systems, while examples involving vertebrates are the most scarce (but see Grech et al. 2006 for an effort to understand the genetic context of mouse-malaria infections). Less direct evidence for GH×GP, but with the important advantage of having been conducted in the field, is a set of temporal studies of snail–trematode interactions (Dybdahl & Lively 1995, 1998). For these interactors, there is good evidence that the parasite adapts to host genotypes and that the host genotype frequencies change over time in response, which clearly points to a GH×GP scenario. An interesting and related variant on GH×GP interactions driving temporal changes in allele frequency is suggested by viral epidemiological studies. New, and hence initially rare, viral mutants frequently sweep through hosts (or populations) whose immune systems are adapted (through somatic diversification and immune memory) to common viral genotypes. In the case of influenza, such a scenario underlies large-scale human epidemics (Sharp 2002), while in the HIV case, there is good evidence that adaptation to the immune system explains viral turnover within individual patients (e.g. Poon et al. 2007). Although these human–virus interactions are not conventional coevolutionary scenarios, and are due instead to the capacity for vertebrate immunity to ‘evolve’ within a single generation, there is also compelling evidence that virus-mediated natural selection has moulded human genomes throughout evolutionary time (Worobey et al. 2007). Whether talking about acquired immunity or real coevolution, virus–human interactions do at least highlight the importance of parasite genetic background and the biotic environment offered by hosts.
GH×GP is a special kind of G×E in that there is potential for reciprocal adaptation that is absent in genotype interactions with the abiotic environment. Pathogen genetic diversity may drive selection on the host immune system, but changes in host allele frequencies may in turn place selective pressure on the parasites. Such a coevolutionary process of ongoing adaptation and counter-adaptation could result in the persistence of phenotypic variation in susceptibility (even if the underlying genotypes are in constant flux). Presently, we lack understanding of the coevolutionary process or how important it is relevant to other diversity generation mechanisms, such as G×E. Some experimental evidence that GH×GP interactions can result in coevolutionary maintenance of genetic variation comes from studies of geographical patterns in pathogenic relationships, specifically laboratory experiments that compare infectivity of sympatric and allopatric host–pathogen combinations. These have been carried out for animal–microbe associations, as well as for insect–herbivory interactions and a variety of plant–fungal interactions (reviewed in Jaenike 1990; Thompson 1993; Kaltz & Shykoff 1998). The emerging observation that pathogenesis is stronger in sympatric combinations suggests that parasites tend to win local coevolutionary contests, presumably by the virtue of having shorter generation times and larger effective population sizes (Bremermann 1985). Globally, i.e averaging across a species' range, parasites appear to have scored no such victory. This may be due to the presence of extrinsic barriers to gene flow, which subdivide populations such that they experience multiple, independent realizations of coevolution, and following divergence, pathogens are unable to spread over the entire species range. Alternatively, if there is host genetic substructure for other reasons (perhaps due to adaptation to local abiotic environments) and parasites are then forced to adapt to whatever host genotypes they are exposed to, the global spread of parasite genotypes could also be limited.
The latter scenario raises the possibility that GH×GP interactions might themselves depend on the abiotic environment, leading to GH×GP×E interactions. These have been little studied, perhaps because fully cross-factored experiments can be unfeasibly large. Still, they are possible, as exemplified by Tétard-Jones et al. (2007) who studied the interaction between aphids (representing GP), barley (representing GH) and the presence or absence of rhizobacteria (approximating variation in E). In this study, the three-way interaction was found to be significant, explaining as much as 42 and 32 per cent of the variation in barley or aphid performance, respectively. It will be important for other studies to similarly quantify the importance (not just test for the occurrence) of higher level interactions in order to understand what such interactions mean for the evolution of complex systems. It seems obvious that GH×GP×E can promote local or global levels of genetic variation in both host and pathogen, but, presently, it is difficult to assess what higher level interactions mean for direct coevolutionary interactions.
5. Pleiotropy and competing physiologies
Resistance to infection is frequently considered to be a function of the immune system, but in actuality, resistance involves the entire physiology of the host. Indeed, it is through the effects on global host physiology that external abiotic environment generally impacts resistance. Immune performance is also strongly influenced by non-immunological physiological demands. In insects, for example, resistance to infection may be compromised after strenuous physical activity such as foraging in bumble-bees (Bombus terrestrris; König & Schmid-Hempel 1995; Doums & Schmid-Hempel 2000; Moret & Schmid-Hempel 2000), courting and mating in damselflies and fruit flies (Siva-Jothy et al. 1998; McKean & Nunney 2001; Fedorka et al. 2007) or general stresses in crickets (Adamo & Parsons 2006). Similar phenomena are observed in humans, where both intense athletic activity and sustained sleep deprivation correlate with enhanced risk of infection (Nieman 1998; Castell 2002; Bryant et al. 2004; Opp 2005). Conversely, activation of the immune system can reduce performance in other physiologies. Immune challenged D. melanogaster transiently suppress expression of non-essential metabolic genes (De Gregorio et al. 2001) and D. melanogaster larvae that survive parasitoid attacks grow to be smaller and less robust adults than unparasitized matched controls (Hoang 2001). Chronic infection can lead to major metabolic disruption in both insects (Dionne et al. 2006; Schilder & Marden 2006) and humans (Powanda & Beisel 2003), including elevation of haemolymph (blood) sugar levels and depletion of oxygen and triglyceride stores.
The interconnections between immunity and other aspects of physiology, notably metabolism, suggest that natural selection may not operate directly on immune function without exerting indirect pressure on other genetically correlated fitness traits. There thus arises a potential ‘cost’ of immunity that provides the conceptual basis for life-history trade-offs. The simple observation that immune defences are induced by infection, as opposed to being constitutively active, suggests that immune activity is in some way costly. In some cases, the costs of immunity may be direct and obvious, such as autoimmunity and autoreactivity by immunologically generated cytotoxins (Sell & Max 2001; Kumar et al. 2003; Graham et al. 2005; Sadd & Siva-Jothy 2006).
In many instances, the costs may be less direct and the mechanisms much more obscured. For example, Fedorka et al. (2007) have noted transiently reduced resistance to infection in mated D. melanogaster females. The mechanistic basis for this immunosuppression is unclear, especially since mating triggers expression of antimicrobial peptide genes, at least in reproductive tissues (Lawniczak & Begun 2004; McGraw et al. 2004; Peng et al. 2005; Domanitskaya et al. 2007). One hypothesis, however, is that the transient susceptibility is linked to altered hormonal signalling and metabolism after mating. Male seminal proteins stimulate female production of juvenile hormone (JH; Moshitzky et al. 1996) and egg maturation. JH is itself a strong repressor of antimicrobial peptide gene expression (Flatt et al. 2008) that could contribute to the observed transient susceptibility to systemic infection. The scenario is complicated by the fact that JH is also responsive to insulin-like signalling (Tatar et al. 2001). Insulin-like signalling drives catabolism of fat stores (Broughton et al. 2005; Giannakou & Partridge 2007) and is probably involved in egg production (e.g. Djawdan et al. 1998). Blocking insulin-like signalling results in increased resistance to infection (Dionne et al. 2006; Libert et al. 2008) and increased lifespan but decreased fecundity (Clancy et al. 2001). Thus, immunity, longevity, reproduction and metabolism are linked in a complex network by shared hormonal regulation of all four processes (Flatt et al. 2005). In principle, natural selection could act on genetic variation in hormone signalling, altering the balance between immunity and reproduction and resulting in modified resistance to infection even though neither insulin, JH nor other hormonal signals involved in metabolism or reproduction are considered to be components of the canonical immune system.
A common and fruitful approach to experimentally identifying genetic trade-offs has been to breed laboratory populations with enhanced immunity through artificial selection, and then to test whether selected populations differ in other measures of fitness from control unselected populations. For instance, Boots & Begon (1993) found that Indian meal moths, Plodia interpunctella, bred for increased resistance to granulosis virus developed more slowly and had lower egg viability than unselected controls. Kraaijeveld & Godfray (1997) selected D. melanogaster for resistance to the parasitoid Asobara tabida, but found that resistant larvae could be outcompeted by susceptible larvae for limited nutritional resources. Luong & Polak (2007a) showed that Drosophila nigrospiracula bred to avoid ectoparasitic mites have reduced fecundity. These experiments demonstrate the potential for life-history trade-offs in field settings, and suggest that natural selection for increased resistance might be limited by concomitant decreases in other traits related to fitness.
A caveat to trade-off studies based on selection lines is that the mutations captured during strong artificial selection may not be those that make the most important contributions to standing variation in natural populations. The array of phenotypic variation observed in natural populations may be caused by a multitude of common polymorphisms with individually small effects, punctuated by a handful of rare mutations of large effect (Orr & Irving 1997). Artificial selection will preferentially capture the mutations of large effect, regardless of their initial frequencies in the population. If these large effect mutations are more likely to have negative pleiotropic consequences, fitness costs of immunity and associated life-history trade-offs are likely to be overestimated by selection experiments. This concern is clearly illustrated by work on the malaria vector mosquito Anopheles gambiae. Anopheles gambiae selected for increased melanotic encapsulation of malaria parasites can become completely resistant (Collins et al. 1986), but suffer autoimmune damage through increased activity of oxidative free radicals (Kumar et al. 2003), suggesting a possible trade-off limiting the evolution of resistance. However, although phenotypic variation for resistance to malaria is abundant in natural A. gambiae populations, the common resistance mechanism in the wild does not appear to be melanization (Niaré et al. 2002; Riehle et al. 2006), thus suggesting that the trade-off uncovered in the laboratory may have little relevance in the field. An additional potential pitfall with selection experiments is that artificial selection can result in the fixation of large sections of chromosome flanking the selected mutation (Maynard Smith & Haigh 1974). It can therefore be difficult to determine whether experimentally observed trade-offs stem from actual pleiotropic effects of the selected mutations or from deleterious effects of mutations in the linked genes.
An alternative approach is to use quantitative genetic mapping to test the hypothesis that the same genomic regions control both resistance to parasitic infection and other fitness traits. Zhong et al. (2005) successfully mapped three quantitative trait loci (QTL) for resistance to tapeworm infection in the red flour beetle, Tribolium castaneum. These QTL co-localized with mapped QTL determining reproductive success. Genotypes with high resistance to tapeworm had lower measures of reproductive fitness, demonstrating a genetic trade-off. Wilfert et al. (2007) also mapped co-localizing QTL resistance and reproductive traits, but in this case higher resistance measures were positively correlated with reproductive fitness, suggesting that a polymorphic locus controlling general vigour affected both traits but providing no evidence of a trade-off. A disadvantage of QTL mapping, however, is that the mating structure used in constructing the mapping populations results in genetic linkages that span longer physical distances (in base pairs) than would be observed in outbred populations. It can therefore be difficult to distinguish true pleiotropy from exaggerated linkage disequilibrium, so it is unclear whether apparent trade-offs truly indicate long-term constraints on adaptation.
Perhaps the most attractive experimental approach to detecting fitness trade-offs that may operate in the field is to measure a large number of outbred individuals for both the immunity phenotype and for other fitness traits. If immunity is negatively correlated with other fitness determinants, a life-history trade-off may be operating, although genetic correlations collected in this manner lend little insight into the genetic mechanism underlying the trade-off. Using this approach, McKean et al. (2008) documented a negative genetic correlation between resistance to bacterial infection and fecundity in D. melanogaster derived from a natural population. A negative genetic correlation was also found between growth rate and parasite resistance in stickleback fish (Gasterosteus aculeatus; Barber et al. 2001). Overall, however, there seems to be less tendency to detect trade-offs when studying host genotypes that represent standing genetic variation in wild populations (Little et al. 2002; Altermatt & Ebert 2007). This may be because artificial selection experiments (Yan et al. 1997; Langand et al. 1998; Webster & Woolhouse 1999) create extreme phenotypes that perhaps exaggerate the strength of trade-offs. Whether selection experiments deceive us by focusing attention on non-natural extremes, or whether they service understanding by highlighting trade-offs too subtle to be measured in the study of wild isolates remains an open question.
Negative genetic correlations between life-history traits can be enhanced, eliminated or even rendered positive in distinct abiotic environments (Sgrò & Hoffmann 2004), with many trade-offs observed only under stressful conditions. Moret & Schmid-Hempel (2000) were able to detect a reduction in lifespan of immune-induced bumble-bees, but only when the bees were starved prior to challenge. Similarly, the trade-off between resistance to bacterial infection and fecundity in D. melanogaster is erased when the flies are provided with a protein-enriched diet (McKean et al. 2008), and was not observed in a similar experiment performed in a different laboratory (Lazzaro et al. 2008). Drosophila melanogaster larvae selected for resistance to Asobara parasitization and D. nigrospiracula selected for resistance to mites are outcompeted by susceptible larvae, but only under high density conditions (Kraaijeveld & Godfray 1997; Luong & Polak 2007b). In very rich environments, genetic correlations between physiologically linked traits may even become positive, as genetic variation for acquisition of resources from the environment becomes more prominent than variation for allocation of resources to competing traits (e.g. Reznick et al. 2000; Barber et al. 2001; Coltman et al. 2001; Wilfert et al. 2007). This is presumably the explanation for the above-mentioned positive correlation between antibacterial activity and sperm number observed by Wilfert et al. (2007). These observations underscore the importance of environmental conditions in determining the evolutionary trajectory of immune performance and should be taken into account when considering the relative importance of life-history trade-offs in the population-level maintenance of genetic variation.
6. Trade-offs within the immune system
The trade-offs discussed so far have involved disparate physiological functions, but the immune system itself is multifaceted and may exhibit trade-offs between its branches. The best-studied example of this is the antagonism between different types of T helper cell responses in the vertebrate immune system (reviewed in Fenton et al. 2008). Specifically, the T helper cell response type 1 (Th1) is directed towards microparasites such as viruses, bacteria and protozoa, while the T helper cell type 2 (Th2) response is linked to the defence against macroparasites such as helminths. Although the mechanism of antagonism between Th1 and Th2 responses is not established, it is clear that hosts cannot mount strong responses in both systems in one location. Hosts have a limited pool of T-cells, and probably must compromise one response to enhance the other. The Th1/Th2 relationship is linked to the effectors, nitric oxide synthase (NOS) and arginase: in macrophages Th1-type cytokines induce NOS while they inhibit arginase, whereas the reverse is the case for Th2-type cytokines. Thus a balance between NOS and arginase, which compete for the same substrate (l-arginine) is regulated by Th1 versus Th2 immune reactions (Munder et al. 1998). This pattern appears to be conserved out to teleosts (Joerink et al. 2006), but its origins may run even deeper, as both NOS and arginase occur in invertebrates. A role of NOS in invertebrate immunity is established (Luckhart et al. 1998), and while arginase is substantially less studied, it is characterized in Drosophila where it is non-essential in the laboratory, possibly because its function is stress or infection dependent, such is the case for many immune system genes (Samson 2000). Studies of NOS/arginase antagonism during infection in an invertebrate would be intriguing, as they might shed light on a truly ancient immunological trade-off.
Additional studies have suggested trade-offs between arms of the immune system in invertebrates. Humoral antimicrobial peptides are used to combat microbial infection (Imler & Bulet 2005), whereas phenoloxidase is a component of the oxidative and melanization defences employed against eukaryotic pathogens (Cerenius & Söderhäll 2004). In the Egyptian cotton leafworm Spodoptera littoralis, humoral antibiotic activity is weakly negatively correlated with the number of circulating haemocytes. The number of haemocytes is in turn positively correlated with the haemolymph phenoloxidase activity, a predictor of capacity for defensive melanization (Cotter et al. 2004). Negative correlations between phenoloxidase activity and haemolymph antibacterial activity have also been observed in bumble-bees (Moret & Schmid-Hempel 2001; Wilfert et al. 2007) and the cabbage looper Trichoplusia ni (Freitak et al. 2007). Optimization models suggest that hosts ought to invest in defence based on the risk of infection, or allocate defence in response to the most fitness-threatening pathogen (e.g. Graham 2001; Moret 2003). Genetic variation for allocation to either phenoloxidase or humoral antimicrobial activity may be maintained if the risk of pathogenesis by either microbes or eukaryotic parasites varies over time or space.
Heterogeneous pathogen risk could also result in the maintenance of polymorphism through direct antagonistic pleiotropy of specific mutations, as would be the case with allelic variants of a pathogen recognition molecule where alternative alleles best recognize distinct pathogens. Such models have considerable theoretical appeal (Hamilton 1993; Frank 1994) but have not been well tested empirically (but see Lazzaro et al. 2006).
7. Conclusions
It is well established in both vertebrates (Murphy 1993; Hurst & Smith 1999) and insects (Schlenke & Begun 2003; Sackton et al. 2007) that immune system genes evolve faster than other parts of the genome, suggesting the widespread occurrence of host–parasite coevolution. Generally, it seems inarguable that immune systems are finely tuned for their purpose, and such a complex adaptation can only be ascribed to long-term, powerful natural selection. At any moment in time, however, genetic variation is subject to the myriad of factors we have focused on in this review, resulting in considerable noise around the long-term evolutionary trajectory.
Why strive to understand this noise? The environmental heterogeneities we have discussed are critically important to contemporary populations. Environmental fluctuations can obscure the relationship between genotype and phenotype, and spatially or temporally heterogeneous selective pressures can result in the short-term maintenance of polymorphism. An increased abundance of standing polymorphism leaves populations with greater capacity to evolve in response to environmental shifts (Barrett & Schluter 2008). Short-term evolutionary processes are also biomedically relevant. Emerging diseases, vaccine escape mutants, genetic polymorphisms that underlie important human disease, the evolution of pathogens that cause increased virulence and the fate of introduced transgenic organisms all represent challenges for global health that are also evolutionary phenomena. As such, they require the insights of evolutionary biology, and understanding of the factors, such as those we have highlighted, to fully understand and to aid control. Plasmodium-carrying mosquitoes provide a salient example: resistance polymorphism is common, as is parasite strain variation. There is a tremendous public health motivation for understanding the short-term evolutionary dynamics of the mosquito–malaria system. Can human intervention facilitate an increase in the frequency of resistant mosquito genotypes? Can we prevent the evolution of virulent, more transmissible, parasite strains? The answers to these questions are likely to lie in the effects of environmental manipulation, G×E, GH×GP and pleiotropy.
Ultimately, any complete understanding of the evolutionary dynamics of resistance to infection in any system must include understanding the complications that environmental variation poses. Gaining such understanding should be a priority of research programmes centred on disease and evolution.
Acknowledgments
We thank Pedro Vale and Simon Fellous for discussion and comments on the manuscript. Research in the Lazzaro laboratory is supported by grants from the United States National Institutes of Health and National Science Foundation. Research in the Little laboratory is supported by the Wellcome Trust, UK.
Footnotes
One contribution of 11 to a Theme Issue ‘Ecological immunology’.
References
- Adamo S.A., Parsons N.M. The emergency life-history stage and immunity in the cricket, Gryllus texensis. Anim. Behav. 2006;72:235–244. doi:10.1016/j.anbehav.2006.01.011 [Google Scholar]
- Adamo S.A., Jensen M., Younger M. Changes in lifetime immunocompetence in male and female Gryllus texenis (formerly G. integer): trade-offs between immunity and reproduction. Anim. Behav. 2001;62:417–425. doi:10.1006/anbe.2001.1786 [Google Scholar]
- Altermatt F., Ebert D. The genotype specific competitive ability does not correlate with infection in natural Daphnia magna populations. PLoS ONE. 2007;2:e1280. doi: 10.1371/journal.pone.0001280. doi:10.1371/journal.pone.0001280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.M., May R.M. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Barber I., Arnott S.A., Braithwaite V., Andrew J., Huntingford F.A. Indirect fitness consequences of mate choice in sticklebacks: offspring of brighter males grow slowly but resist parasite infections. Proc. R. Soc. B. 2001;268:71–76. doi: 10.1098/rspb.2000.1331. doi:10.1098/rspb.2000.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R.D., Schluter D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. doi:10.1016/j.tree.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Bedhomme S., Agnew P., Sidobre C., Michalakis Y. Virulence reaction norms across a food gradient. Proc. R. Soc. B. 2004;271:739–744. doi: 10.1098/rspb.2003.2657. doi:10.1098/rspb.2003.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., Reboud X. Experimental evolution in Chlamydomonas. 2. Genetic variation in strongly contrasted environments. Heredity. 1997;78:498–506. doi:10.1038/hdy.1997.78 [Google Scholar]
- Ben-Ami F., Regoes R.R., Ebert D. A quantitative test of the relationship between parasite dose and infection probability across different host–parasite combinations. Proc. R. Soc. B. 2008;275:853–859. doi: 10.1098/rspb.2007.1544. doi:10.1098/rspb.2007.1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots M., Begon M. Trade-offs with resistance to a graulosis-virus in the Indian meal moth, examined by a laboratory evolution experiment. Funct. Ecol. 1993;7:528–534. doi:10.2307/2390128 [Google Scholar]
- Bremermann H.J. The adaptive significance of sexuality. Experientia. 1985;41:1245–1255. doi: 10.1007/BF01952067. doi:10.1007/BF01952067 [DOI] [PubMed] [Google Scholar]
- Bremermann H.J., Pickering J. A game-theoretical model of parasite virulence. J. Theor. Biol. 1983;100:411–426. doi: 10.1016/0022-5193(83)90438-1. doi:10.1016/0022-5193(83)90438-1 [DOI] [PubMed] [Google Scholar]
- Brinkhof M.W.G., Heeb P., Kolliker M., Richner H. Immunocompetence of nestling great tits in relation to rearing environment and parentage. Proc. R. Soc. B. 1999;266:2315–2322. doi:10.1098/rspb.1999.0925 [Google Scholar]
- Broughton S.J., et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl Acad. Sci. USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. doi:10.1073/pnas.0405775102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant P.A., Trinder J., Curtis N. Sick and tired: does sleep have a vital role in the immune system? Nat. Rev. Immunol. 2004;4:457–467. doi: 10.1038/nri1369. doi:10.1038/nri1369 [DOI] [PubMed] [Google Scholar]
- Burdon J.J. The distribution and origin of genes for race-specific resistance to Melamspora lini in Linum marginale. Evolution. 1994;48:1564–1575. doi: 10.1111/j.1558-5646.1994.tb02196.x. doi:10.2307/2410248 [DOI] [PubMed] [Google Scholar]
- Burdon J.J., Jarosz A.M. Host–pathogen interactions in natural populations of Linum marginale and Melampsora lini. I. Patterns of resistance and racial variation in a large host population. Evolution. 1991;45:205–217. doi: 10.1111/j.1558-5646.1991.tb05278.x. doi:10.2307/2409494 [DOI] [PubMed] [Google Scholar]
- Burdon J.J., Thrall P.H. Spatial and temporal patterns in coevolving plant and pathogen associations. Am. Nat. 1999;153(Suppl.):s15–s33. doi: 10.1086/303209. doi:10.1086/303209 [DOI] [PubMed] [Google Scholar]
- Byers D.L. Evolution in heterogeneous environments and the potential of maintenance of genetic variation in traits of adaptive significance. Genetica. 2005;123:107–124. doi: 10.1007/s10709-003-2721-5. doi:10.1007/s10709-003-2721-5 [DOI] [PubMed] [Google Scholar]
- Carius H.J., Little T.J., Ebert D. Genetic variation in a host–parasite association: potential for coevolution and frequency-dependant selection. Evolution. 2001;55:1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. doi:10.1554/0014-3820(2001)055[1136:GVIAHP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Castell L.M. Can glutamine modify the apparent immunodepression observed after prolonged, exhaustive exercise? Nutrition. 2002;18:371–375. doi: 10.1016/s0899-9007(02)00754-2. doi:10.1016/S0899-9007(02)00754-2 [DOI] [PubMed] [Google Scholar]
- Cerenius L., Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. doi:10.1111/j.0105-2896.2004.00116.x [DOI] [PubMed] [Google Scholar]
- Clancy D.J., Gems D., Harshman L.G., Oldham S., Stocker H., Hafen E., Leevers S.J., Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. doi:10.1126/science.1057991 [DOI] [PubMed] [Google Scholar]
- Collins F.H., Sakai R.K., Vernick K.D., Paskewitz S., Seeley D.C., Miller L.H., Collins W.E., Campbell C.C., Gwadz R.W. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234:607–610. doi: 10.1126/science.3532325. doi:10.1126/science.3532325 [DOI] [PubMed] [Google Scholar]
- Coltman D.W., Pilkington J., Kruuk L.E., Wilson K., Pemberton J.M. Positive genetic correlation between parasite resistance and body size in a free-living ungulate population. Evolution. 2001;55:2116–2125. doi: 10.1111/j.0014-3820.2001.tb01326.x. doi:10.1554/0014-3820(2001)055[2116:PGCBPR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cotter S.C., Kruuk L.E., Wilson K. Costs of resistance: genetic correlations and potential trade-offs in an insect immune system. J. Evol. Biol. 2004;17:421–429. doi: 10.1046/j.1420-9101.2003.00655.x. doi:10.1046/j.1420-9101.2003.00655.x [DOI] [PubMed] [Google Scholar]
- Decaestecker E., Vergote A., Ebert D., De Meester L. Evidence for strong host–clone–parasite species interactions in the Daphnia microparasite system. Evolution. 2003;57:784–792. doi: 10.1111/j.0014-3820.2003.tb00290.x. doi:10.1554/0014-3820(2003)057[0784:EFSHCS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Decaestecker E., Gaba S., Raeymaekers J.A.M., Stoks R., Van Kerckhoven L., Ebert D., De Meester L. Host–parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. doi:10.1038/nature06291 [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P.T., Rubin G.M., Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl Acad. Sci. USA. 2001;98:12 590–12 595. doi: 10.1073/pnas.221458698. doi:10.1073/pnas.221458698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G. Quantitative genetics of reaction norms. J. Evol. Biol. 1990;3:447–468. doi:10.1046/j.1420-9101.1990.3050447.x [Google Scholar]
- Dionne M.S., Pham L.N., Shirasu-Hiza M., Schneider D.S. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. doi:10.1016/j.cub.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Djawdan M., Chippindale A.K., Rose M.R., Bradley T.J. Metabolic reserves and evolved stress resistance in Drosophila melanogaster. Physiol. Zool. 1998;71:584–594. doi: 10.1086/515963. [DOI] [PubMed] [Google Scholar]
- Domanitskaya E.V., Liu H., Chen S., Kubli E. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 2007;274:5659–5668. doi: 10.1111/j.1742-4658.2007.06088.x. doi:10.1111/j.1742-4658.2007.06088.x [DOI] [PubMed] [Google Scholar]
- Doums C., Schmid-Hempel P. Immunocompetence in workers of a social insect, Bombus terrestris L., in relation to foraging activity and parasitic infection. Can. J. Zool. 2000;78:1060–1066. doi:10.1139/cjz-78-6-1060 [Google Scholar]
- Dubuffet A., Dupas S., Frey F., Drezen J.-M., Poirié M., Carton Y. Genetic interactions between the parasitoid wasp Leptopilina boulardi and its Drosophila hosts. Heredity. 2007;98:21–27. doi: 10.1038/sj.hdy.6800893. doi:10.1038/sj.hdy.6800893 [DOI] [PubMed] [Google Scholar]
- Dybdahl M.F., Lively C.M. Host–parasite interactions: infection of common clones in natural populations of a freshwater snail (Potamopyrgus antipodarum) Proc. R. Soc. B. 1995;260:99–103. doi:10.1098/rspb.1995.0065 [Google Scholar]
- Dybdahl M.F., Lively C.M. Host–parasite coevolution: evidence for rare advantage and time-lagged selection in a natural population. Evolution. 1998;52:1057–1066. doi: 10.1111/j.1558-5646.1998.tb01833.x. doi:10.2307/2411236 [DOI] [PubMed] [Google Scholar]
- Elliot S.L., Blanford S., Horton C.M., Thomas M.B. Fever and phenotype: transgenerational effect of disease on desert locust phase state. Ecol. Lett. 2003;6:830–836. doi:10.1046/j.1461-0248.2003.00487.x [Google Scholar]
- Fedorka K.M., Linder J.E., Winterhalter W., Promislow D. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc. R. Soc. B. 2007;274:1211–1217. doi: 10.1098/rspb.2006.0394. doi:10.1098/rspb.2006.0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels D., Kaltz O. Temperature-dependent transmission and latency of Holospora undulata, a micronucleus-specific parasite of the ciliate Paramecium caudatum. Proc. R. Soc. B. 2006;273:1031–1038. doi: 10.1098/rspb.2005.3404. doi:10.1098/rspb.2005.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A., Lamb T., Graham A.L. Optimality analysis of Th1/Th2 immune responses during microparasite–macroparasite co-infection, with epidemiological feedbacks. Parasitology. 2008;135:841–853. doi: 10.1017/S0031182008000310. doi:10.1017/S0031182008000310 [DOI] [PubMed] [Google Scholar]
- Ferguson H.M., Read A.F. Genetic and environmental determinants of malaria parasite virulence in mosquitoes. Proc. R. Soc. B. 2002;269:1217–1224. doi: 10.1098/rspb.2002.2023. doi:10.1098/rspb.2002.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Tu M.P., Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27:999–1010. doi: 10.1002/bies.20290. doi:10.1002/bies.20290 [DOI] [PubMed] [Google Scholar]
- Flatt T., et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J. Exp. Biol. 2008;211:2712–2724. doi: 10.1242/jeb.014878. doi:10.1242/jeb.014878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.A. Recognition polymorphism in host–parasite genetics. Phil. Trans. R. Soc. B. 1994;346:283–293. doi: 10.1098/rstb.1994.0145. doi:10.1098/rstb.1994.0145 [DOI] [PubMed] [Google Scholar]
- Frank S.A. Models of parasite virulence. Quart. Rev. Biol. 1996;71:37–78. doi: 10.1086/419267. doi:10.1086/419267 [DOI] [PubMed] [Google Scholar]
- Freitak D., Wheat C.W., Heckel D.G., Vogel H. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusia ni. BMC Biol. 2007;5:56. doi: 10.1186/1741-7007-5-56. doi:10.1186/1741-7007-5-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershwin M.E., Beach R.S., Hurley L.S. Academic Press; Orlando, FL: 1985. Nutrition and immunity. [Google Scholar]
- Giannakou M.E., Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem. Sci. 2007;32:180–188. doi: 10.1016/j.tibs.2007.02.007. doi:10.1016/j.tibs.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Gillespie J.H., Turelli M. Genotype–environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R., Kirkpatrick M. Quantitative genetics and the evolution of reaction norms. Evolution. 1992;46:390–411. doi: 10.1111/j.1558-5646.1992.tb02047.x. doi:10.2307/2409860 [DOI] [PubMed] [Google Scholar]
- Graham A.L. Use of an optimality model to solve the immunological puzzle of concomitant infection. Parasitology. 2001;122:S61–S64. doi:10.1017/S0031182000017650 [PubMed] [Google Scholar]
- Graham A.L., Allen J.E., Read A.F. Evolutionary causes and consequences of immunopathology. Annu. Rev. Ecol. Evol. Syst. 2005;36:373–397. doi:10.1146/annurev.ecolsys.36.102003.152622 [Google Scholar]
- Grech K., Watt K., Read A.F. Host–parasite interactions for virulence and resistance in a malaria model system. J. Evol. Biol. 2006;19:1620–1630. doi: 10.1111/j.1420-9101.2006.01116.x. doi:10.1111/j.1420-9101.2006.01116.x [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. Haploid dynamic polymorphism in a host with matching parasites: effects of mutations/subdivision, linkage, and patterns of selection. J. Hered. 1993;84:328–338. [Google Scholar]
- Hoang A. Immune response to parasitism reduces resistance of Drosophila melanogaster to desiccation and starvation. Evolution. 2001;55:2353–2358. doi: 10.1111/j.0014-3820.2001.tb00748.x. doi:10.1554/0014-3820(2001)055[2353:IRTPRR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hochachka P.W., Somero G.N. Princeton University Press; Princeton, NJ: 1984. Biochemical adaptation. [Google Scholar]
- Hurst L.D., Smith N.G. Do essential genes evolve slowly? Curr. Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. doi:10.1016/S0960-9822(99)80334-0 [DOI] [PubMed] [Google Scholar]
- Imler J.L., Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem. Immunol. Allergy. 2005;86:1–21. doi: 10.1159/000086648. doi:10.1159/000086648 [DOI] [PubMed] [Google Scholar]
- Jaenike J. Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 1990;21:243–273. doi:10.1146/annurev.es.21.110190.001331 [Google Scholar]
- Joerink M., Savelkoul H.F.J., Wiegertjes G.F. Evolutionary conservation of alternative activation of macrophages: structural and functional characterization of arginase 1 and 2 in carp (Cyprinus carpio L.) Mol. Immunol. 2006;43:116–1128. doi: 10.1016/j.molimm.2005.07.022. doi:10.1016/j.molimm.2005.07.022 [DOI] [PubMed] [Google Scholar]
- Johnston I.A., Bennett A.F., editors. Animals and temperature. Phenotypic and evolutionary adaptation. Society for Experimental Biology Seminar Series. vol. 59. Cambridge University Press; Cambridge, UK: 1996. [Google Scholar]
- Kaltz O., Shykoff J.A. Local adaptation in host–parasite systems. Heredity. 1998;81:361–370. doi:10.1046/j.1365-2540.1998.00435.x [Google Scholar]
- Klasing K.C. Nutritional modulation of resistance to infectious diseases. Poultry Sci. 1998;77:1119–1125. doi: 10.1093/ps/77.8.1119. [DOI] [PubMed] [Google Scholar]
- König C., Schmid-Hempel P. Foraging activity and immunocompetence in workers of the bumble bee, Bombus terrestris L. Proc. R. Soc. B. 1995;260:225–227. doi:10.1098/rspb.1995.0084 [Google Scholar]
- Kraaijeveld A.R., Godfray H.C. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature. 1997;389:278–280. doi: 10.1038/38483. doi:10.1038/38483 [DOI] [PubMed] [Google Scholar]
- Kumar S., Christophides G.K., Cantera R., Charles B., Han Y.S., Meister S., Dimopoulos G., Kafatos F.C., Barillas-Mury C. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl Acad. Sci. USA. 2003;100:14 139–14 144. doi: 10.1073/pnas.2036262100. doi:10.1073/pnas.2036262100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A.L. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant–pathogen association. J. Evol. Biol. 2007;20:2371–2378. doi: 10.1111/j.1420-9101.2007.01406.x. doi:10.1111/j.1420-9101.2007.01406.x [DOI] [PubMed] [Google Scholar]
- Lambrechts L., Halbert J., Durand P., Gouagna L.C., Koella J.C. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malar. J. 2005;4:3. doi: 10.1186/1475-2875-4-3. doi:10.1186/1475-2875-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L., Chavatte J.-M., Snounou G., Koella J.C. Environmental influence on the genetic basis of mosquito resistance to malaria parasites. Proc. R. Soc. B. 2006;273:1501–1506. doi: 10.1098/rspb.2006.3483. doi:10.1098/rspb.2006.3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langand J., Jourdane J., Coustau C., Delay B., Morand S. Cost of resistance, expressed as a delayed maturity, detected in the host–parasite system Biomphalaria glabrata–Echinostoma caproni. Heredity. 1998;80:320–325. doi:10.1046/j.1365-2540.1998.00291.x [Google Scholar]
- Lawniczak M.K., Begun D.J. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. doi:10.1139/g04-050 [DOI] [PubMed] [Google Scholar]
- Lazzaro B.P., Sackton T.B., Clark A.G. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. doi:10.1534/genetics.105.054593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B.P., Flores H.A., Lorigan J.G., Yourth C.P. Genotype by environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 2008;4:e1000025. doi: 10.1371/journal.ppat.1000025. doi:10.1371/journal.ppat.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 1953;87:331–333. doi:10.1086/281792 [Google Scholar]
- Libert S., Chao Y., Zwiener J., Pletcher S.D. Realized immune response is enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol. Immunol. 2008;45:810–817. doi: 10.1016/j.molimm.2007.06.353. doi:10.1016/j.molimm.2007.06.353 [DOI] [PubMed] [Google Scholar]
- Linder J.E., Owers K.A., Promislow D.E. The effects of temperature on host–pathogen interactions in D. melanogaster: who benefits? J. Insect Physiol. 2008;54:297–308. doi: 10.1016/j.jinsphys.2007.10.001. doi:10.1016/j.jinsphys.2007.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little T.J., Carius H.-J., Sakwinska O., Ebert D. Competitiveness and life-history characteristics of Daphnia with respect to susceptibility to a parasite. J. Evol. Biol. 2002;15:796–802. doi:10.1046/j.1420-9101.2002.00436.x [Google Scholar]
- Little T.J., Hultmark D., Read A.F. Invertebrate immunity and the limits of mechanistic immunology. Nat. Immunol. 2005;6:651–654. doi: 10.1038/ni1219. doi:10.1038/ni1219 [DOI] [PubMed] [Google Scholar]
- Little T.J., Birch J., Vale P., Tseng M. Parasite transgenerational effects on infection. Evol. Ecol. Res. 2007;9:459–469. [Google Scholar]
- Luckhart S., Vodovotz Y., Cui L., Rosenberg R. The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc. Natl Acad. Sci. USA. 1998;95:5700–5705. doi: 10.1073/pnas.95.10.5700. doi:10.1073/pnas.95.10.5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong L.T., Polak M. Costs of resistance in the Drosophila–Macrocheles system: a negative genetic correlation between ectoparasite resistance and reproduction. Evolution. 2007a;61:1391–1402. doi: 10.1111/j.1558-5646.2007.00116.x. doi:10.1111/j.1558-5646.2007.00116.x [DOI] [PubMed] [Google Scholar]
- Luong L.T., Polak M. Environment-dependent trade-offs between ectoparasite resistance and larval competitive ability in the Drosophila–Macrocheles system. Heredity. 2007b;99:632–640. doi: 10.1038/sj.hdy.6801040. doi:10.1038/sj.hdy.6801040 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Maynard Smith J., Hoekstra R. Polymorphism in a varied environment: how robust are the models? Genet. Res. 1980;35:45–57. doi: 10.1017/s0016672300013926. [DOI] [PubMed] [Google Scholar]
- McGraw L.A., Gibson G., Clark A.G., Wolfner M.F. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. doi:10.1016/j.cub.2004.08.028 [DOI] [PubMed] [Google Scholar]
- McKean K.A., Nunney L. Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2001;98:7904–7909. doi: 10.1073/pnas.131216398. doi:10.1073/pnas.131216398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean K.A., Yourth C.P., Lazzaro B.P., Clark A.G. The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 2008;8:76. doi: 10.1186/1471-2148-8-76. doi:10.1186/1471-2148-8-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.E., Read A.F. Poor maternal environment enhances offspring disease resistance in an invertebrate. Proc. R. Soc. B. 2005;272:2601–2607. doi: 10.1098/rspb.2005.3253. doi:10.1098/rspb.2005.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.E., Rogers E.S., Little T.J., Read A.F. Host–parasite and genotype-by-environment interactions: temperature modifies potential for selection by a sterilizing pathogen. Evolution. 2005;59:70–80. doi:10.1554/04-526 [PubMed] [Google Scholar]
- Moret Y. Explaining variable costs of the immune response: selection for specific versus non-specific immunity and facultative life history change. Oikos. 2003;102:213–216. doi:10.1034/j.1600-0706.2003.12496.x [Google Scholar]
- Moret Y., Schmid-Hempel P. Survival for immunity: the price of immune system activation for bumblebee workers. Science. 2000;290:1166–1168. doi: 10.1126/science.290.5494.1166. doi:10.1126/science.290.5494.1166 [DOI] [PubMed] [Google Scholar]
- Moret Y., Schmid-Hempel P. Immune defence in bumble-bee offspring. Nature. 2001;414:506. doi: 10.1038/35107138. doi:10.1038/35107138 [DOI] [PubMed] [Google Scholar]
- Moshitzky P., Fleischmann I., Chaimov N., Saudan P., Klauser S., Kubli E., Applebaum S.W. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch. Insect Biochem. Physiol. 1996;32:363–374. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<363::AID-ARCH9>3.0.CO;2-T. doi:10.1002/(SICI)1520-6327(1996)32:3/4<363::AID-ARCH9>3.0.CO;2-T [DOI] [PubMed] [Google Scholar]
- Munder M., Eichmann K., Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+T cells correlates with Th1/Th2 phenotype. J. Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- Murphy P.M. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. doi:10.1016/0092-8674(93)90571-7 [DOI] [PubMed] [Google Scholar]
- Niaré O., et al. Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science. 2002;298:213–216. doi: 10.1126/science.1073420. doi:10.1126/science.1073420 [DOI] [PubMed] [Google Scholar]
- Nieman D.C. Exercise and resistance to infection. Can. J. Physiol. Pharmacol. 1998;76:573–580. doi: 10.1139/cjpp-76-5-573. doi:10.1139/cjpp-76-5-573 [DOI] [PubMed] [Google Scholar]
- Opp M.R. Cytokines and sleep. Sleep Med. Rev. 2005;9:355–364. doi: 10.1016/j.smrv.2005.01.002. doi:10.1016/j.smrv.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Orr H.A., Irving S. The genetics of adaptation: the genetic basis of resistance to wasp parasitism in Drosophila melanogaster. Evolution. 1997;51:1877–1885. doi: 10.1111/j.1558-5646.1997.tb05110.x. doi:10.2307/2411009 [DOI] [PubMed] [Google Scholar]
- Peng J., Zipperlen P., Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr. Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. doi:10.1016/j.cub.2005.08.048 [DOI] [PubMed] [Google Scholar]
- Poon A.F.Y., Kosakovsky Pond S.L., Bennett P., Richman D.D., Leigh Brown A.J., Frost S.D.W. Adaptation to human populations is revealed by within-host polymorphisms in HIV-1 and hepatitis C virus. PLoS Pathog. 2007;3:e45. doi: 10.1371/journal.ppat.0030045. doi:10.1371/journal.ppat.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powanda M.C., Beisel W.R. Metabolic effects of infection on protein and energy status. J. Nutr. 2003;133:322S–327S. doi: 10.1093/jn/133.1.322S. [DOI] [PubMed] [Google Scholar]
- Reznick D., Nunney L., Tessier A. Big houses, big cars, superfleas and the costs of reproduction. Trends Ecol. Evol. 2000;15:421–425. doi: 10.1016/s0169-5347(00)01941-8. doi:10.1016/S0169-5347(00)01941-8 [DOI] [PubMed] [Google Scholar]
- Riehle M.M., et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. doi:10.1126/science.1124153 [DOI] [PubMed] [Google Scholar]
- Rolff J., Siva-Jothy M.T. Copulation corrupts immunity: a mechanism for a cost of mating in insects. Proc. Natl Acad. Sci. USA. 2002;99:9916–9918. doi: 10.1073/pnas.152271999. doi:10.1073/pnas.152271999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T.B., Lazzaro B.P., Schlenke T.A., Evans J.D., Hultmark D., Clark A.G. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. doi:10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- Sadd B.M., Siva-Jothy M.T. Self-harm caused by an insect's innate immunity. Proc. R. Soc. B. 2006;273:2571–2574. doi: 10.1098/rspb.2006.3574. doi:10.1098/rspb.2006.3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaudon L., Héraudet V., Shykoff J.A. Parasite–host fitness trade-offs change with parasite identity: genotype-specific interactions in a plant–pathogen system. Evolution. 2005;59:2518–2524. doi:10.1554/05-299.1 [PubMed] [Google Scholar]
- Samson M.-L. Drosophila arginase is produced from a nonvital gene that contains the elav locus within its third intron. J. Biol. Chem. 2000;275:31 107–31 114. doi: 10.1074/jbc.M001346200. doi:10.1074/jbc.M001346200 [DOI] [PubMed] [Google Scholar]
- Sasaki A., de Jong G. Density dependence and unpredictable selection in a heterogeneous environment: compromise and polymorphism in the ESS reaction norm. Evolution. 1999;53:1329–1342. doi: 10.1111/j.1558-5646.1999.tb05398.x. doi:10.2307/2640880 [DOI] [PubMed] [Google Scholar]
- Schilder R.J., Marden J.H. Metabolic syndrome and obesity in an insect. Proc. Natl Acad. Sci. USA. 2006;103:18 805–18 809. doi: 10.1073/pnas.0603156103. doi:10.1073/pnas.0603156103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T.A., Begun D.J. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P., Ebert D. On the evolutionary ecology of specific immune defence. Trends Ecol. Evol. 2003;18:27–32. doi:10.1016/S0169-5347(02)00013-7 [Google Scholar]
- Schulenburg H., Ewbank J.J. Diversity and specificity in the interactions between the nematode Caenorhabditis elegans and the pathogenic bacterium Serratia marcescens. BMC Evol. Biol. 2004;4:49. doi: 10.1186/1471-2148-4-49. doi:10.1186/1471-2148-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M.E. Heligmosomoides polygrus (Nematoda): susceptible and resistant strains are indistinguishable following natural infection. Parasitology. 1991;103:429–438. doi: 10.1017/s0031182000059953. [DOI] [PubMed] [Google Scholar]
- Scott M.E. High transmission rates restore expression of genetically determined susceptibility of mice to nematode infections. Parasitology. 2006;132:669–679. doi: 10.1017/S0031182005009583. doi:10.1017/S0031182005009583 [DOI] [PubMed] [Google Scholar]
- Sell S., Max E.E. 6th edn. ASM Press; Washington, DC: 2001. Immunology, immunopathology and immunity. [Google Scholar]
- Sgrò C.M., Hoffmann A.A. Genetic correlations, tradeoffs and environmental variation. Heredity. 2004;93:241–248. doi: 10.1038/sj.hdy.6800532. doi:10.1038/sj.hdy.6800532 [DOI] [PubMed] [Google Scholar]
- Sharp P.M. Origins of human virus diversity. Cell. 2002;108:305–312. doi: 10.1016/s0092-8674(02)00639-6. doi:10.1016/S0092-8674(02)00639-6 [DOI] [PubMed] [Google Scholar]
- Siva-Jothy M.T., Tsubaki Y., Hooper R.E. Decreased immune response as a proximate cost of copulation and oviposition in a damselfly. Physiol. Entomol. 1998;23:274–277. doi:10.1046/j.1365-3032.1998.233090.x [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M.P., Yin C.M., Garofalo R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. doi:10.1126/science.1057987 [DOI] [PubMed] [Google Scholar]
- Tella J.L., Bortolotti G.R., Dawson R.D., Forero M.G. The T-cell-mediated immune response and return rate of fledgling American kestrels are positively correlated with parental clutch size. Proc. R. Soc. B. 2000;267:891–895. doi: 10.1098/rspb.2000.1086. doi:10.1098/rspb.2000.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétard-Jones C., Kertesz M.A., Gallois P., Preziosi R.F. Genotype by genotype interactions modified by a third species in a plant insect system. Am. Nat. 2007;170:492–499. doi: 10.1086/520115. doi:10.1086/520115 [DOI] [PubMed] [Google Scholar]
- Thomas M.B., Blanford S. Thermal biology in insect–parasite interactions. Trends Ecol. Evol. 2003;18:344–350. doi:10.1016/S0169-5347(03)00069-7 [Google Scholar]
- Thompson J.N. Preference heirarchies and the origin of geographic specialization in host use in swallowtail butterflies. Evolution. 1993;47:1585–1594. doi: 10.1111/j.1558-5646.1993.tb02177.x. doi:10.2307/2410169 [DOI] [PubMed] [Google Scholar]
- Thompson J.N., Burdon J.J. Gene-for-gene coevolution between plants and parasites. Nature. 1992;360:121–125. doi:10.1038/360121a0 [Google Scholar]
- Tseng M. Interactions between the parasite's previous and current environment mediate the outcome of parasite infection. Am. Nat. 2006;168:565–571. doi: 10.1086/507997. doi:10.1086/507997 [DOI] [PubMed] [Google Scholar]
- Vale P., Stjernman M., Little T.J. Temperature dependent costs of parasitism and the maintenance of polymorphism under genotype-by-environment interactions. J. Evol. Biol. 2008;21:1418–1427. doi: 10.1111/j.1420-9101.2008.01555.x. doi:10.1111/j.1420-9101.2008.01555.x [DOI] [PubMed] [Google Scholar]
- Via S. The evolution of phenotypic plasticity: what do we really know? In: Real L.A., editor. Ecological genetics. Princeton University Press; Princeton, NJ: 1994. pp. 35–57. [Google Scholar]
- Webster J.P., Woolhouse M.E.J. Cost of resistance: relationship between reduced fertility and increased resistance in a snail–schistosome host–parasite system. Proc. R. Soc. B. 1999;266:391–396. doi:10.1098/rspb.1999.0650 [Google Scholar]
- Wilfert L., Gadau J., Schmid-Hempel P. The genetic architecture of immune defense and reproduction in male Bombus terrestris bumblebees. Evolution. 2007;61:804–815. doi: 10.1111/j.1558-5646.2007.00079.x. doi:10.1111/j.1558-5646.2007.00079.x [DOI] [PubMed] [Google Scholar]
- Worobey M., Bjork A., Wertheim J.O. Point, couterpoint: the evolution of pathogenic viruses and their human hosts. Annu. Rev. Ecol. Syst. 2007;38:515–540. doi:10.1146/annurev.ecolsys.38.091206.095722 [Google Scholar]
- Yan G., Severson D.W., Christensen B.M. Costs and benefits of mosquito refractoriness to malaria parasites: implications for genetic variability of mosquitoes and genetic control of malaria. Evolution. 1997;51:441–450. doi: 10.1111/j.1558-5646.1997.tb02431.x. doi:10.2307/2411116 [DOI] [PubMed] [Google Scholar]
- Zhong D., Pai A., Yan G. Costly resistance to parasitism: evidence from simultaneous quantitative trait loci mapping for resistance and fitness in Tribolium castaneum. Genetics. 2005;169:2127–2135. doi: 10.1534/genetics.104.038794. doi:10.1534/genetics.104.038794 [DOI] [PMC free article] [PubMed] [Google Scholar]