Abstract
An adaptive immune response is usually initiated only if a major histocompatibility complex (MHC) molecule presents pathogen-derived peptides to T-cells. Every MHC molecule can present only peptides that match its peptide-binding groove. Thus, it seems advantageous for an individual to express many different MHC molecules to be able to resist many different pathogens. However, although MHC genes are the most polymorphic genes of vertebrates, each individual has only a very small subset of the diversity at the population level. This is an evolutionary paradox. We provide an overview of the current data on infection studies and mate-choice experiments and conclude that overall evidence suggests that intermediate intra-individual MHC diversity is optimal. Selective forces that may set an upper limit to intra-individual MHC diversity are discussed. An updated mathematical model based on recent findings on T-cell selection can predict the natural range of intra-individual MHC diversity. Thus, the aim of our review is to evaluate whether the number of MHC alleles usually present in individuals may be optimal to balance the advantages of presenting an increased range of peptides versus the disadvantages of an increased loss of T-cells.
Keywords: polymorphism, major histocompatibility complex, heterozygote advantage, optimum, allele counting, T-cell receptor repertoire
1. Introduction
The enormous polymorphism of major histocompatibility complex (MHC) genes struck researchers in the mid-1960s, when the function of MHC molecules was still unknown (e.g. Snell 1968). No other set of genes in jawed vertebrates is more polymorphic than the MHC (Janeway et al. 2005). More recently, a picture of how natural selection acts on the MHC has emerged, based on advances in immunology, molecular evolution, evolutionary ecology and behavioural ecology, as we review in the following. Immunological research has revealed that MHC molecules play a crucial role in the adaptive immune system of jawed vertebrates. In a nutshell, MHC molecules bind peptides from pathogens and present them on the cell surface: the so-called ‘antigen presentation’. A response of the adaptive immune system is initiated if T-cells recognize such peptide–MHC complexes (see box 1 for a more detailed description). Any given MHC molecule type can present only peptides that match its peptide-binding groove (Falk et al. 1991). Different MHC molecules may present different peptides of a given pathogen, while some MHC molecules may be incapable of presenting any peptide of the focal pathogen. Therefore, MHC alleles are expected to differ in their potential to mediate resistance to a given pathogen. Indeed, it has been shown repeatedly that resistance to a specific pathogen or parasite is associated with the presence of certain MHC alleles in the host (e.g. Hill et al. 1991; Paterson et al. 1998; Langefors et al. 2001; Grimholt et al. 2003; Bonneaud et al. 2005; Harf & Sommer 2005; Schad et al. 2005; reviews by Martin & Carrington 2005; Milinski 2006). While hosts benefit from expressing MHC molecules that can efficiently present antigen, selection on pathogens should favour strains that evade MHC presentation and T-cell recognition. There is evidence that pathogens benefit from mutations changing the amino acid composition of those peptides that are efficiently presented by MHC molecules common in the host population or the infected individual (Decamposlima et al. 1993; Allen et al. 2000).
Box 1. Major histocompatibility complex.
The major histocompatibility complex (MHC) is the most polymorphic gene cluster in vertebrate genomes. Among other proteins, it encodes a set of membrane glycoproteins called MHC molecules. The physiological function of classical MHC molecules is to present peptides derived from proteins in the body to T-cells. Peptide presentation is a short term denoting the process of a peptide binding to a groove-shaped structure at the top of the MHC molecule and the resulting peptide–MHC complex being shuttled to the cell surface, where it can interact with T-cell receptors (TCRs). Based on the structure of the MHC molecules, the origin of the presented peptides and the type of T-cells the MHC molecules interact with, two classes of classical MHC molecules can be distinguished. Generally speaking, while MHC I molecules present peptides generated from proteins in the cytosol to CD8+ T-cells, MHC II molecules present peptides derived from endocytosed proteins to CD4+ T-cells. Sequencing of numerous peptides eluted from MHC molecules has enabled researchers to identify shared residues of the peptides bound to specific MHC molecules. A typical MHC I molecule presents peptides that are 8–11 amino acids long and have specific residues at two main anchor positions and one of a set of amino acids at auxiliary anchor positions (Rammensee et al. 1997). An adaptive immune response is initiated if a T-cell recognizes—that is binds strongly to—a peptide–MHC complex. Note that MHC molecules present peptides derived from pathogens as well as peptides derived from self-proteins. Self-tolerance of the adaptive immune system is ensured mainly by eliminating T-cell precursors with receptors specific to self-peptide–MHC complexes (see also box 2). Apart from the classical class I and II MHC molecules, the mammalian MHC region comprises genes (‘class III’) that encode a heterogeneous group of proteins with immunological or non-immunological functions. While these proteins are structurally different from classical MHC molecules, non-classical class I and II MHC molecules (also encoded in the MHC region) are structurally closely related to their classical counterparts. Non-classical class I MHC molecules present conserved microbial epitopes (e.g. lipids and sugars) to γδ T-cells or interact with receptors on natural killer cells, while non-classical class II MHC molecules are involved in the loading of peptides onto classical MHC II molecules. Non-classical MHC molecules are not particularly polymorphic and not further considered in this review.
Based on the fact that every individual MHC allele mediates resistance only to a subset of pathogens, the following characteristic population genetic features of classical MHC class I and II genes appear to make sense: (i) MHC genes are the most polymorphic genes identified in vertebrates, (ii) polymorphism is highest in those regions of MHC genes that encode the peptide-binding groove, (iii) the polymorphism is not a result of neutral evolution but is maintained by Darwinian selection, and (iv) long-diverged species have been shown to share allelic lineages of the MHC (reviews by Edwards & Hedrick 1998; Piertney & Oliver 2006). If resistance to specific pathogens is linked to the presence of certain MHC alleles, balancing selection, for example negative frequency-dependent selection, can maintain a large number of different MHC alleles in a population for many generations (Borghans et al. 2004).
2. The paradox: why is intra-individual MHC diversity relatively low?
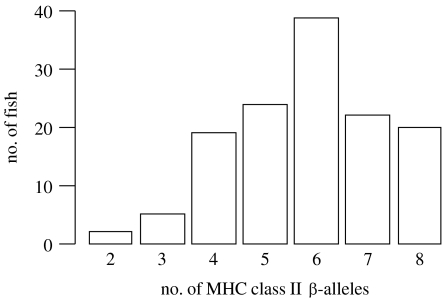
However, the paradox remains as to why the mechanism that leads to high polymorphism at the population level does not lead to high intra-individual MHC diversity. Given that any MHC molecule can mediate resistance only to a few pathogens, one would expect the expression of many different MHC alleles and loci per individual. In reality, however, the intra-individual MHC diversity (that is the overall number of different classical MHC alleles at all loci) is surprisingly low compared with the large diversity of MHC alleles at the population level. Given the potential for evolutionary duplication and diversification of individual loci (Lawlor et al. 1990), this is even more puzzling. In other large gene families (e.g. the olfactory receptor genes with more than 1000 functional loci in the mouse genome; Zhang & Firestein 2002), a substantial part of the population diversity is represented in every single individual. By contrast, only a minute fraction of the MHC diversity at the population level is present in any one individual. For example, in humans, there are in total approximately 2000 known alleles at only three classical MHC I loci (IMGT database described by Robinson et al. 2003). Small individual MHC diversity seems to be adaptive: polyploid clawed frogs (Xenopus sp.) have silenced ‘surplus’ MHC loci over evolutionary time (Kobel & Dupasquier 1986), while the duplicated loci of many other genes have been shown to remain active (Flajnik 1996). This suggests that selection may disfavour individuals with high MHC diversity. In humans, genes encoding the α- and β-chains of antigen-presenting MHC II molecules are clustered in three subregions of the MHC. In the so-called DR subregion of the human MHC, nine loci that encode for MHC II β-chains have been identified. Functional genes occur at four of these loci; however, no DR haplotype is known in which more than two DRB genes (that is genes in the DR region encoding for MHC II β-chains) are expressed (Rammensee et al. 1997). Obviously, no strong selection pressure maximizes the number of functional genes in DR haplotypes. The number of different MHC II β-alleles varies markedly among individuals of three-spined sticklebacks (Gasterosteus aculeatus). In natural populations, individuals with an intermediate number of different MHC II β-alleles are the most frequent genotype (figure 1; Reusch et al. 2001) and females with a higher number of alleles prefer males with fewer alleles. Follow-up studies on MHC-based mate choice in sticklebacks found evidence that females aim at an intermediate, rather than a maximal, level of MHC diversity in their offspring (Aeschlimann et al. 2003; Milinski et al. 2005). Similar results have been obtained in mate-choice experiments with brown trout (Salmo trutta). Females prefer males of intermediate MHC dissimilarity (Forsberg et al. 2007). Studies on human mate choice have found female preferences for males with dissimilar MHC genotypes, no preference or even a preference for mates of similar MHC genotypes. Milinski (2006) pointed out that these seemingly contradictory results may be compatible if human females aimed at an optimal number of combined MHC alleles: accordingly, females should prefer dissimilar mates in less outbred populations (as found by Wedekind et al. 1995), whereas they should prefer similar mates in more outbred populations (as found by Jacob et al. 2002). Moreover, Ihara et al. (2000) showed suggestive evidence that an overlap of two MHC alleles was more frequent than a smaller or larger overlap in Japanese couples. Taken together, these observations suggest that there is a selective force acting against high MHC diversity in individuals. Identifying the nature of this selective force may give us important insights into the workings of the immune system and is crucial for understanding the dynamics of MHC evolution. We review studies that assess the impact of intra-individual MHC diversity on immunocompetence (i.e. the ability of an organism to protect itself from pathogens and parasites; Viney et al. 2005) and discuss why maximal MHC diversity may not be optimal for the functionality of an individual's immune system.
Figure 1.
Frequency distribution of the number of MHC II β-alleles per individual in 144 stickleback fish from a lake population. Adapted from Reusch et al. (2001).
3. Which intra-individual MHC diversity is optimal? Population surveys and experimental studies
(a) Results of surveys and experimental studies
Doherty & Zinkernagel (1975) suggested that MHC heterozygotes should have the highest immunocompetence (the ‘heterozygote advantage’ hypothesis), because such individuals can present a broader range of pathogen-derived peptides. Since then, many studies have tried to assess the effect of MHC diversity on host immunocompetence and fitness. The results are mixed and ambiguous. Some studies support the heterozygote advantage hypothesis (e.g. Thursz et al. 1997; Carrington et al. 1999; Jeffery et al. 2000). Other researchers find no evidence for a correlation between MHC diversity and resistance (e.g. Paterson et al. 1998; Langefors et al. 2001; Lohm et al. 2002; Wedekind et al. 2005). Many studies contradict the heterozygote advantage hypothesis. They show that haplotype heterozygotes can be more susceptible than haplotype homozygotes (e.g. Hill et al. 1991; Pitcher & Neff 2006; Ilmonen et al. 2007). In a meta-analysis on experimental infection studies with mice, Penn (2002) found that 10 out of 17 studies indicated that MHC-dependent resistance was dominant or over-dominant whereas 5 out of 17 studies suggested that resistance was under-dominant or recessive. However, some studies found that individuals with an intermediate level of MHC heterozygosity fared the best (e.g. Wegner et al. 2003a,b; Bonneaud et al. 2004; Madsen & Ujvari 2006).
(b) Factors that may explain why results are mixed
All studies of this type face complex challenges. First, it is difficult to disentangle the effect of MHC I and II diversities from effects of other loci and from effects caused by the presence of specific MHC alleles. Conclusions as to which MHC diversity is optimal can be biased if MHC alleles associated with resistance are over-represented in one of the diversity classes (Penn 2002; Lipsitch et al. 2003). This can confound population surveys that compare disease outcomes in heterozygotes as a group to homozygotes as a group (e.g. Thursz et al. 1997). Since evolving pathogens mainly evade presentation by the most abundant MHC molecules, rare MHC alleles are more likely to confer resistance than common MHC alleles. These protective, rare alleles are likely to be concentrated in heterozygotes. Findings that heterozygotes are more resistant than homozygotes therefore need not be evidence that maximal MHC diversity is beneficial, but may be due to the fact that rare alleles are advantageous (Lipsitch et al. 2003).
Second, a recent study by Ilmonen et al. (2007) has provided evidence that an inbred genetic background changes the effect of MHC diversity on immunocompetence. While these researchers found some evidence for heterozygote advantage in inbred strains (Penn et al. 2002), their results obtained with mice of diverse genetic backgrounds indicated that resistance to different strains of Salmonella enterica was mostly recessive (Ilmonen et al. 2007). This finding highlights the issue of whether results from inbred mouse strains are relevant for understanding genetically diverse natural populations. However, many researchers studying individuals with a genetically diverse background or natural populations focus on immunological ‘non-model’ species, for which MHC typing is often indirect and thus less accurate. Surveys of natural populations have the inherent drawback that evidence is only correlational (e.g. Thursz et al. 1997; Carrington et al. 1999; Wegner et al. 2003b; Madsen & Ujvari 2006).
Third, it is often unclear how the parameters assessed as a function of MHC diversity relate to host fitness. The first study designed to test the heterozygote advantage hypothesis (Doherty & Zinkernagel 1975) illustrates that this is not straightforward. Although MHC heterozygous mice mounted a more vigorous immune response against lymphocytic choriomeningitis virus than MHC homozygous mice, MHC homozygotes survived whereas all MHC heterozygotes died. The reason for this outcome was that the pathology (death) was due to the elevated immune response, not the infection per se.
Animals in the wild are likely to be challenged with multiple parasites or pathogens during their lifetime (Potts & Slev 1995). A (hypothetical) example shows how this important natural context affects conclusions drawn from single infection studies. Assume that individuals homozygous for the A allele at a certain MHC locus (AA individuals) survive a challenge with pathogen 1 with a likelihood of 10 per cent, while their survival probability when infected with pathogen 2 is 90 per cent. Individuals homozygous for allele B (BB individuals) have survival probabilities 90 per cent for pathogen 1 and 10 per cent for pathogen 2, while heterozygotes (AB individuals) have survival probabilities of 50 per cent when challenged with either of the two pathogens. Single infection studies would indicate that MHC diversity does not affect the average resistance (on average 50% survival among both homozygotes and heterozygotes). By contrast, infections of individuals with both pathogens would reveal a clear advantage for heterozygotes (survival probability for AB individuals is 0.5×0.5=0.25, while AA and BB individuals survive with a much lower probability: 0.1×0.9=0.09). In general, heterozygosity would be advantageous if the survival probability of the heterozygote is higher than the geometric mean of the survival probabilities of the homozygotes. Therefore, single infection experiments may lead to qualitatively different results than those in which individuals are challenged with multiple parasites or pathogens. Since individuals in natural populations face multiple parasites and pathogens, the latter studies may reflect the natural situation better.
(c) Conclusion
Owing to experimental difficulties, the question of which level of intra-individual MHC diversity is optimal remains unanswered. Many studies that have tested the heterozygote advantage hypothesis do not support it. Some carefully designed studies involving the experimental infections of hosts of a genetically diverse background with multiple parasites or pathogen strains (e.g. Wegner et al. 2003a; Ilmonen et al. 2007) clearly show that high intra-individual MHC diversities can reduce immunocompetence. So far, all studies have been limited by the number of MHC I and II loci naturally occurring in studied species. Moreover, many experimental studies have compared only those individuals that are completely homozygous at all MHC loci with individuals completely heterozygous at all MHC loci. As a result of their design, these studies cannot find an optimum at intermediate MHC diversities. Irrespective of their results, they can never refute the hypothesis that there is a selective force against high MHC diversities. It is tempting to conclude that evidence from infection experiments and surveys supports the hypothesis that intermediate individual MHC diversity is optimal. The results of mate-choice experiments discussed in §2 provide a second, independent line of evidence that points to an optimum at intermediate intra-individual MHC diversity.
4. Hypotheses for explaining why high intra-individual MHC diversity is not selected for
(a) T-cell repertoire depletion hypothesis
(i) Introduction to the hypothesis
It has been suggested that high intra-individual MHC diversity may result in a depletion of the mature T-cell repertoire and thereby reduce immunocompetence (Vidovic & Matzinger 1988; Lawlor et al. 1990). The proportion of precursor T-cells that are eliminated in two selection events during T-cell maturation (see box 2) may critically depend on intra-individual MHC diversity. Only precursor T-cells with TCRs that can interact with at least one of the individual's MHC molecules survive positive selection. During negative selection, T-cells with TCRs that strongly bind to MHC molecules in complex with antigen derived from the individual's own proteins are eliminated: this prevents autoimmune reactions. Increasing intra-individual MHC diversity is therefore expected to result in greater survival of T-cells during positive selection, but reduced survival during negative selection. If negative selection increases over proportionally with MHC diversity, the TCR repertoire of individuals with high MHC diversity will be severely depleted.
Box 2. T-cells and T-cell maturation.
αβ T-cells (referred to as T-cells in this text) interact with peptide–MHC complexes via their αβ TCR. In each precursor T-cell, the genes encoding the α- and β-chains of the TCR are generated in an essentially random process, which involves the rearrangement of subgenic fragments at the TCR α- and β-loci, leading to the expression of an αβ heterodimer at the cell surface. Each individual T-cell usually expresses a single receptor with a unique specificity. Although a single T-cell thus recognizes only a small set of peptide–MHC complexes, the structural diversity of TCRs expressed on all T-cells generated in an individual ensures that the organism can respond to a great variety of antigens. During their maturation in the thymus, T-cells (called thymocytes during their maturation phase) have to pass two quality control steps, in which their randomly generated TCRs are checked. In a process called positive selection, TCRs are tested for general reactivity to the individual's MHC molecules. Only T-cells with receptors that are functional in the context of the individual's MHC molecules pass this checkpoint. After having undergone positive selection, T-cells downregulate expression of one of the two coreceptors, CD4 and CD8, and develop into either CD4, single positive, or CD8, single positive, T-cells. During negative selection, T-cells, whose receptors react strongly with self-peptide–MHC complexes and which might cause autoimmune diseases if they matured, are eliminated. However, induction of self-tolerance is not limited to the process of deleting self-reactive T-cells in the thymus but additionally encompasses apoptosis, induction of anergy and silencing of effector T-cells by regulatory T-cells in the periphery.
The pathology of AIDS, which is characterized by a profound decrease in the number of CD4 T-cells (a CD4 T-cell count of less than 200 cells μl−1 blood is used as a definition for AIDS by the US Center for Disease Control (1992); healthy human adults have approx. 1000 CD4 T-cells μl−1 of blood), indicates how much the adaptive immune system depends on a large pool of T-cells. Patients die from opportunistic infections that would normally pose no problem. Experimental studies in mice (reviewed by Nikolich-Zugich et al. 2004) indicate that reductions in TCR diversity (by more than 50% compared with wild-type diversity) can lead to reduced immunity to specific pathogens. It is essentially undisputed that large MHC diversity entails extensive negative selection and that a severe depletion of the TCR repertoire reduces immunocompetence. However, it is unclear whether the quantitative effect of MHC diversity on the efficiency of negative selection is strong enough to explain why individuals typically do not express more than 10–20 MHC molecules.
(ii) Modelling approaches to assess the effect of T-cell repertoire depletion
The verbal argument, that the observed intermediate level of intra-individual MHC diversity may be the result of a trade-off between a high presentation probability for pathogen-derived peptides and a high probability that resulting peptide–MHC complexes are recognized by T-cells, has been formalized by Nowak et al. (1992). Borghans et al. (2003) extended this model in order to account for the fact that T-cells can only be negatively selected by MHC molecules that also mediate their positive selection (this is because weak binding between the TCR and self-peptide–MHC complex is sufficient for positive selection, whereas strong binding causes negative selection of the T-cell). Consider infection of a host with M different MHC molecules by a pathogen from which e different peptides are generated. To calculate the probability Pi that the host initiates an immune response against the pathogen, the following reasoning is used:
Let q denote the probability that a given peptide is presented by a given MHC molecule type, r denote the probability that a given TCR recognizes a given peptide–MHC complex and R(M) be the size of the mature TCR repertoire of an individual with M different MHC molecules. Then, Pi(M) is given by Nowak et al. (1992)
| (4.1) |
In their extension of the model by Nowak et al. (1992), Borghans et al. (2003) assumed that the size of the mature TCR repertoire is the size of the repertoire that survived positive selection minus the number of T-cells that are negatively selected. Here, R0 denotes the size of the pre-selection TCR repertoire.
This approach makes use of the fact that all thymocytes that get negatively selected interact strongly with self-peptide–MHC complexes and are thus also positively selected (since weak interactions with self-peptide–MHC complexes are sufficient for positive selection). Let p (n) denote the probabilities that a pre-selection T-cell is positively (negatively) selected by a single MHC molecule. R(M) can then be expressed as (Borghans et al. 2003)
| (4.2) |
As a first approximation, positive and negative selection is evaluated on all MHC molecules, because thymocytes are committed to the CD4 or CD8 lineage at least partly depending on the class of the MHC molecule that mediates their positive selection (Germain 2002; Singer 2002). Based on experimental studies, Borghans et al. (2003) obtained the following parameter estimates: q=0.02, r=10−5, R0=1010, e=25, p=0.01, n=0.005 and predicted that the size of the mature TCR repertoire increased until the MHC diversity exceeded 140 different molecules. The probability Pi of mounting an immune response against a pathogen reaches its maximum at even higher MHC diversities. Therefore, the authors argue that the advantage of low intra-individual MHC diversity cannot result from a large TCR repertoire.
However, a rejection of the T-cell repertoire depletion hypothesis may be premature. Equation (4.2) models selection (positive and negative) of T-cells by the M different MHC molecules as subsequent, independent events. As p is small (p=0.01), different MHC molecules mediate the positive selection of almost non-overlapping parts of the pre-selection TCR repertoire. If M is in the range of intra-individual MHC diversities commonly observed in nature, the product pM is small. Since n<p, we can approximate R(M) by
Thus, the size of the mature TCR repertoire increases linearly with the MHC diversity. However, as outlined below, recent advances in immunology suggest that the impact of MHC diversity on positive selection efficiency may be small to negligible. This leads to slightly modified models that predict a much smaller optimum for MHC diversity.
(iii) Immunological evidence
4.2.1.1 Limited reliability of activation markers
Indications that positive selection efficiency increases with MHC diversity come from studies using activation markers such as CD69. These markers are known to be transiently upregulated by pre-selection thymocytes upon interaction with positively selecting peptide–MHC complexes. Thus, positively selected thymocytes can, in principle, be discriminated on the basis of activation marker expression from those that die by neglect. It is unclear whether studies employing this technique can accurately measure the proportion of pre-selection thymocytes that are positively selected in vivo. In cell culture, negatively selecting ligands seem to be more potent inducers of CD69 upregulation than positively selecting ligands (Daniels et al. 2006). This could be due to the fact that (i) negatively selecting ligands induce stronger upregulation of CD69, so that more thymocytes are grouped as CD69+, or (ii) negatively selecting ligands are capable of positively selecting more thymocytes in culture. While option (i) implies that the percentage of CD69+ cells may increase although positive selection efficiency does not, option (ii) alludes to the problem of whether positive selection efficiency in vitro accurately reflects that in vivo.
Studies of freshly extracted thymi that determine only the percentage of double positive thymocytes that are CD69+ and not absolute thymocyte numbers (Monteiro et al. 2005) cannot demonstrate that the analysed subsample represents the composition of the thymocyte populations in vivo. This is because thymocytes that strongly interact with stromal cells are more difficult to extract: they may therefore be under-represented in the analysis. Proportions of CD69+ thymocytes and thymocytes positive for other activation markers (e.g. CD5) can deviate considerably in the same experimental set-up (e.g. 10.1±2.5 CD69+; N=15 and 18.5±2.9 CD5+; N=6 in cultures of MHC naive thymocytes with H-2b stroma; Merkenschlager et al. 1997). This illustrates the point that the proportion of positively selected cells cannot be directly inferred from the proportion of activation-marker-positive cells.
4.2.1.2 Conserved features of MHC molecules play a key role in the process of positive selection
At present, inferring the impact of MHC diversity on positive selection efficiency based on the following considerations may be more reliable. Interactions between the TCR and a peptide–MHC complex can formally be classified into (i) interactions between the TCR and the specific peptide, (ii) interactions between the TCR and amino acid residues unique to a particular MHC molecule type, and (iii) contributions by interactions between the TCR and the conserved features of MHC molecules. If interactions with conserved features of MHC molecules play a key role in the process of positive selection, repertoires positively selected by different MHC molecules will be largely overlapping. Non-overlapping repertoires would result if unique features of MHC molecules and bound peptides were decisive.
Studies analysing the TCR repertoire of mice expressing a single peptide–MHC II complex found considerable numbers (20–50% of wild-type) of polyclonal CD4 single positive thymocytes (that express the full range of Vβ-segments) despite the extreme reduction in the diversity of positively selecting ligands (Fung-Leung et al. 1996; Ignatowicz et al. 1996; Martin et al. 1996). This suggests that, although positive selection of some T-cells is known to depend on the presence of specific peptide–MHC complexes (e.g. Santori et al. 2002), interaction with conserved features shared by most MHC molecules is sufficient to induce positive selection in a large proportion of thymocytes.
A study by Huseby et al. (2005) strongly supports this view. The authors assessed the TCR repertoire prior to negative selection using a system in which thymocytes were exposed to less stringent negative selection (I Ab - SP mice). TCRs from many mature T-cells in these mice were very cross-reactive and some TCRs even interacted with peptide–MHC complexes of both class I and class II MHC molecules. These findings contradict the traditional view that nearly all pre-selection TCRs are highly specific and that positive selection picks out the few TCRs that are specific to the individual's MHC molecules. Instead, the gene segments for the TCR α- and β-chains, which are encoded in the germ line, may give rise to an inherently high affinity of TCRs to MHC molecules. This high default affinity is thought to be based on conserved interactions, e.g. with the backbone atoms of the MHC α-helices flanking the peptide-binding groove. The fundamental contribution of evolutionarily selected interactions is expected to allow positive selection of a broad spectrum of T-cells by a single peptide–MHC complex (as observed, Huseby et al. 2005). Since positive selection is mainly mediated by conserved interactions, many of these positively selected TCRs are expected to be very cross-reactive (as observed, Huseby et al. 2005). In this scenario, it is extensive negative selection (Huseby et al. 2003) that ensures peptide and MHC specificity of the mature TCR repertoire by deleting cross-reactive T-cells.
As interactions with conserved features shared by most MHC molecules play a key role in positive selection, a substantial increase in positive selection efficiency with MHC diversity is not expected. Because negative selection prevents autoimmune reactions, it must be highly peptide- and MHC-specific. In contrast to positive selection, the effect of negative selection on the TCR repertoire size is therefore expected to increase with MHC diversity.
(iv) A model in which positive selection efficiency is independent of MHC diversity
Here, we modify the model described above, assuming that MHC diversity has no impact on positive selection efficiency. We assume that the fraction of T-cells that are positively selected is p*. Let n* denote the probability of a positively selected T-cell to be negatively selected by a given MHC molecule. It follows that:
| (4.3) |
The probability of an immune response, Pi(M), is again given by equation (4.1). Experimental studies suggest that n* is in the range of 0.25–0.9 (van Meerwijk et al. 1997; Zerrahn et al. 1997; Huseby et al. 2003) and p* is approximately 0.2 (Merkenschlager et al. 1997; Monteiro et al. 2005). Previous studies with mice suggest R0=109 (derived from Casrouge et al. 2000), q=0.02 (Kast et al. 1994) and r=10−5 (Blattman et al. 2002).
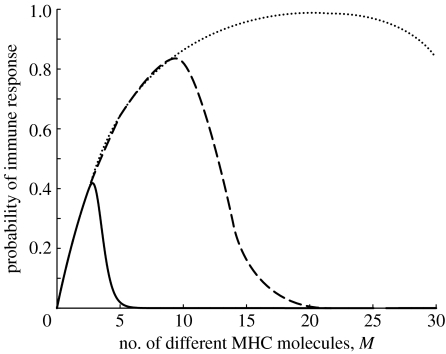
This leads to an exponential decrease of the size of the mature TCR repertoire R(M) with increasing MHC diversity (figure 2). For n* in the range compatible with experimental evidence, the probability Pi of mounting an immune response against a given pathogen reaches a maximum at an MHC diversity Mopt<30 (figure 3). The increase in Pi at low MHC diversity is due to the increase in the probability that peptides of a given pathogen get presented by at least one MHC molecule. TCR repertoire depletion by extensive negative selection accounts for the rapid decrease of Pi at MHC diversities exceeding Mopt. Individuals with an MHC diversity of Mopt are predicted to have the highest immunocompetence. Note that the values of Mopt are in the range 3–25 MHC molecules, which is the same order of magnitude as that typically observed in individuals. The model thus suggests that the observed intermediate MHC diversity in individuals can be explained as the result of a trade-off between the benefit of a high antigen presentation probability (which requires high MHC diversities) and the necessity to avoid extensive negative selection (which requires low MHC diversities).
Figure 2.
The exponential decrease of the size of the mature TCR repertoire R(M) (given by (equation (4.3)) with R0=109; p*=0.2) with increasing intra-individual MHC diversity M depicted for three scenarios with different stringencies of negative selection (dotted line, n*=0.25; dashed line, n*=0.5; solid line, n*=0.9).
Figure 3.
The probability Pi (given by equation (4.1)) that a response of the adaptive immune system against a pathogen is initiated plotted as a function of the intra-individual MHC diversity M for three scenarios with different stringencies of negative selection (dotted curve, n*=0.25; dashed curve, n*=0.5; solid curve, n*=0.9). Parameter values in equation (4.1) are q=0.02; r=10−5; e=10; R0=109.
Our assumption that positive selection efficiency is entirely independent of MHC diversity is also an approximation. Studies have shown that specific TCRs cannot be positively selected by every peptide–MHC complex (Santori et al. 2002). However, our assumption seems to reflect the situation found in nature more realistically than the alternative that different MHC molecules select positively almost non-overlapping parts of the TCR repertoire. Note that a small increase in positive selection efficiency with MHC diversity does not refute our conclusion that the depletion hypothesis may explain why intermediate MHC diversity is optimal, because (i) this may only cause a small shift of the predicted optimum and (ii) other factors may contribute to a reduction of the mature TCR repertoire diversity with increasing MHC diversity. These include the removal of TCRs from the functional repertoire by induction of non-responsiveness to antigen (anergy) in the thymus and the periphery as well as commitment of T-cells to the regulatory lineage. Regulatory T-cells can silence conventional T-cells in the periphery. The extensive overlap in the TCR repertoires of regulatory T-cells and conventional T-cells (Pacholczyk et al. 2007) suggests that regulatory T-cells can have a strong impact on the functional mature TCR repertoire.
(v) Saturation of Pi for genetically complex pathogens and the phenomenon of immunodominance
For genetically simple pathogens that encode only a ‘handful’ of potential epitopes the model predicts that, depending on host MHC genotype, an infection can either result in a powerful immune response or no response of the adaptive immune system at all. This prediction complies with the experimental finding that H-2k mice infected with vesicular stomatitis virus did not mount a detectable cytotoxic response, whereas the virus was highly immunogenic in H-2b and H-2d mice (Rosenthal & Zinkernagel 1981). For pathogens with more complex genomes that encode hundreds of potential epitopes, the model predicts that the response chance Pi saturates for a broad range of intra-individual MHC diversities. This result seems to suggest that intra-individual MHC diversity simply does not matter above a certain threshold of epitope diversity, because hosts will be able to mount an immune response against any pathogen they encounter (Borghans et al. 2003).
This view is at variance with experimental findings indicating that resistance to genetically complex pathogens and parasites is often associated with the presence of certain MHC alleles. Moreover, it raises the question of how the outstanding population genetic features of the MHC can be explained if pathogen-mediated balancing selection does not account for them (see §1). In accordance with the model prediction, experimentalists tend to find that hosts are often capable of responding to multiple epitopes if the pathogen is genetically complex. For example, T-cell responses are directed to 27 distinct gene products in C57BL/6 mice infected with murine cytomegalovirus encoding in total approximately 70 000 amino acids (Munks et al. 2006). Humans who are seropositive for human cytomegalovirus recognize, on average, epitopes from eight open reading frames (ORFs), although some individuals responded only to a single ORF (Sylwester et al. 2005). However, even if an individual is infected with highly complex pathogens, usually not more than a handful of epitopes mediate activation and proliferation of an overwhelming fraction of responding T-cells. This phenomenon is termed immunodominance (Sercarz et al. 1993; Yewdell 2006). Peptide–MHC complexes may differ in their capabilities to initiate an effective T-cell response due to differences in the stability of the peptide–MHC complexes, the amount of MHC molecules loaded with the specific peptide, the timing of expression and presentation (e.g. early versus late during the infectious phase of viruses, presentation of viral peptides derived from genes that are also expressed during the latent phase) and affinity of TCRs to the peptide–MHC complexes. Simple competition among T-cells for peptide–MHC binding and stimulation could be the mechanistic basis of immunodominance (Nowak et al. 1995). There is evidence that deletion of an immunodominant epitope can result in an attenuated T-cell response (Wallace et al. 1999), although an increase in the response to ‘subdominant’ epitopes can often at least partly compensate the loss of the response to the immunodominant epitope (Stock et al. 2006). It is likely that the necessity to efficiently present immunodominant peptides is the immunological basis for the findings that resistance to complex (even eukaryotic) pathogens and parasites is associated with the presence of specific MHC alleles in the host. This association can give rise to pathogen-mediated balancing selection (see §1).
Given that presentation of immunodominant epitopes as well as recognition of the resulting peptide–MHC complexes by T-cells is needed for mounting a potent immune response, an intermediate intra-individual MHC diversity could be optimal. The TCR depletion hypothesis can explain the finding by Ilmonen et al. (2007) that resistance in mice of diverse genetic backgrounds was mostly recessive, although these researchers had previously shown with inbred mice that resistance was dominant (Penn et al. 2002). The combination of diverse genetic backgrounds and high MHC diversity in MHC heterozygotes in the study by Ilmonen et al. (2007) may have led to severe T-cell repertoire depletion due to a high number of self-epitopes.
(vi) Deviations from the calculated optimum
The models discussed above can predict which level of MHC diversity maximizes the probability of an immune response in an individual with a certain pre-selection T-cell repertoire size. Selection, by contrast, will favour individuals that maximize the probability of an immune response under the condition that overall investment in the adaptive immune system does not exceed a certain limit. If increasing the pre-selection T-cell repertoire size is relatively ‘cheap’, whereas increasing MHC diversity is costly (MHC I molecules are constantly expressed in nearly all body cells in substantial numbers), selection is expected to favour individuals with an MHC diversity below the calculated optimum. By contrast, if increasing MHC diversity is cheap, an intra-individual MHC diversity above the calculated optimum should be advantageous.
(b) Level of antigen presentation
(i) The basics of T-cell activation
The activation of mature T-cells by antigen-presenting cells is a complex and poorly understood process. It involves the formation of a highly organized and stable contact zone—the immunological synapse—which consists of an outer ring of adhesion molecules and an inner cluster of TCRs with their coreceptors (Janeway et al. 2005). Ultimately, activation has to depend on a TCR recognizing—that is binding strongly and for a prolonged period—to its epitope. Structural analyses have found no differences in the conformation of a TCR that is bound to either agonistic or antagonistic peptide–MHC complexes, whereas correlations between the half-life of the TCR–peptide–MHC complex and T-cell activation have been reported (reviewed in Krogsgaard & Davis 2005). Therefore, structural rearrangements are thought to have only a minor influence on T-cell activation, while the main signal the T-cell receives seems to be the dissociation rates of its TCRs (Krogsgaard & Davis 2005). However, dissociation of the TCR from peptide–MHC complexes is inherently a stochastic process, because even low-affinity ligands can bind to TCRs for prolonged time periods albeit with reduced likelihood (George et al. 2005). A T-cell thus faces the challenge to convert the analogue input of all the interactions of its TCRs (with stochastically distributed binding times) into a digital output (activation or ignorance to the encountered antigen-presenting cell).
(ii) Is MHC diversity low because foreign peptides have to stand out against the self-background?
Van den Berg & Rand (2003) developed a model which is based on the assumption that each TCR is triggered to produce some intracellular signal if it binds to a peptide–MHC complex for a period longer than a certain threshold time. It is supposed that a T-cell is activated if the sum of all the single triggering events per unit time exceeds a certain threshold. Van den Berg & Rand (2003) argued that a T-cell can only discriminate between the noisy background of self-signals (caused by the stochastic distribution of dissociation times and the fact that the composition of self-peptide–MHC complexes varies between antigen-presenting cells) and interaction with foreign peptides (presented by MHC molecules), if the number of TCRs that are triggered by the foreign peptide (presented by an MHC molecule) stands out against the self-background. The authors show that this is the case if MHC diversity is low, so that a particular foreign peptide in complex with an MHC molecule can dominate the profile of peptide–MHC molecules of an antigen-presenting cell.
Nevertheless, the question remains whether the model captures the essential features of T-cell activation. In contrast to the model predictions, as few as 1–10 agonist peptide–MHC complexes against a background of 105–106 irrelevant peptide–MHC complexes were recognized by mature T-cells (reviewed in George et al. 2005; Krogsgaard & Davis 2005). Selection outcome in the thymus was shown to depend entirely on the affinity between a TCR and peptide–MHC complex (Daniels et al. 2006). Increasing the concentration of a positively selecting ligand could not convert it into a negatively selecting ligand (Daniels et al. 2006).
However, the finding that hosts upregulate MHC expression upon infection (e.g. Wegner et al. 2006; Matsuyama et al. 2007) suggests that increasing the number of MHC molecules (without increasing their diversity) can enhance the effect of an immune response. This could be due to the fact that a foreign peptide that can bind to a certain MHC molecule type of the host still can be out-competed by self-peptides when the MHC molecule is loaded. A sufficient number of MHC molecules may be needed to present the whole range of peptides capable of binding to the MHC molecule type. Moreover, it can be assumed that—although a single agonist peptide–MHC may be able to activate a T-cell—the probability that a T-cell encounters and reacts to a given peptide–MHC complex type is increased if the complex is present in higher numbers.
(c) Risk of autoimmune diseases
(i) Autoimmune diseases are associated with specific MHC alleles
In humans, correlations between susceptibility to certain autoimmune diseases and presence of certain MHC alleles in an individual's genotype are well established (Jones et al. 2006). For example, type 1 diabetes is associated with HLA-DQB1*0302 (Todd et al. 1987), more than 90 per cent of patients with severe rheumatoid arthritis have the HLA-DRB1*0401, HLA-DRB1*0404 allele and/or HLA-DRB1*0101 allele (Wordsworth et al. 1989) and HLA-DRB1*1501 predisposes carriers to multiple sclerosis (Oksenberg et al. 2004). Owing to the extensive linkage disequilibrium in the MHC, it has only recently become clear that in most cases the MHC alleles themselves (and not linked genes) are functionally involved in the increased susceptibility to autoimmune disorders (Jones et al. 2006).
(ii) The effect of increased MHC diversity on susceptibility to autoimmune diseases
The fact that autoimmune diseases are associated with specific MHC alleles suggests that individuals with a diverse set of MHC alleles should have a greater risk of autoimmune disorders. The initiation or exacerbation of autoimmune diseases often occurs in association with infectious diseases. It has been shown that stimulation of autoreactive T-cells by viral epitopes is one of the mechanisms underlying this association (Oldstone 1998; Zhao et al. 1998). The danger that T-cells triggered by a pathogen-derived peptide are cross-reactive to self-epitopes may increase with MHC diversity (Borghans & De Boer 2001). However, an increase in MHC diversity may also increase the likelihood that self-reactive T-cells are eliminated or inactivated during their maturation. Indeed, MHC alleles mediating dominant protection have been identified (Jones et al. 2006). For example, humans carrying the MHC II molecule encoded by HLA-DQA1*0102/HLA-DQB1*0602 are almost completely protected against diabetes even in the presence of the predisposing allele HLA-DQB1*0302 (Siebold et al. 2004). Structural studies comparing the predisposing and protecting MHC molecules suggest that differences in the P6 and P9 pockets may lead to the presentation of an expanded peptide repertoire by the protecting MHC molecule. Siebold et al. (2004) give suggestive evidence that the protecting MHC molecule causes selection of regulatory T-cells, which prevent autoimmune-mediated destruction of pancreatic β-cells.
In natural populations, selection against alleles mediating autoimmune diseases that manifest themselves at an early age may be strong, so that the frequencies of these alleles are low and possibly do not constitute a selection pressure strong enough to lead to an MHC optimum by themselves. Alleles of autoimmune diseases that become manifest only after the main phase of reproduction are not strongly selected against, and thus have no power to select for an MHC optimum either.
5. Future prospects
Species differ in the number of classical MHC class I and II loci. Salmonids possess only one expressed classical MHC class I locus (Aoyagi et al. 2002) and one MHC class II locus (Stet et al. 2002). Humans possess three classical MHC class I and three to four classical MHC class II β-loci (Rammensee et al. 1997). Cod have over 17 MHC class I and an unknown—but probably small—number of MHC class II loci (Miller et al. 2002). Given the potential for duplications in the MHC and the strong selective pressures exerted by pathogens and parasites, such differences in MHC diversity are unlikely to be simply due to phylogenetic constraints or chance. They may rather reflect adaptations to different evolutionary forces. We speculate that the host's overall investment into the adaptive immune system and adaptations to the local parasite fauna and its diversity are important factors. If intra-individual MHC diversity is the result of a trade-off between ensuring efficient presentation of pathogen-derived peptides and some selective force acting against high MHC diversity (e.g. T-cell repertoire depletion, necessity to ensure a high level of antigen presentation or risk of autoimmune diseases), individuals with an MHC diversity just high enough to present peptides of locally abundant parasites and pathogens efficiently will be selected. Low intra-individual MHC diversities may therefore be stable in populations whose individuals are predominantly challenged by a small pool of pathogens or parasites, which is relatively stable over time. River sticklebacks are challenged by a less diverse parasite fauna than lake sticklebacks (Wegner et al. 2003b). This may explain the finding that the average number of MHC II β-alleles in river sticklebacks (4.98±0.15 s.e.) is lower than that of lake sticklebacks (6.34±0.11 s.e.; Wegner 2004).
Unfortunately, crucial experimental evidence is lacking. Although many studies have found associations between resistance to a specific pathogen and presence of a certain MHC allele, the immunological basis of these correlations has only been demonstrated in a few cases involving infection with viruses. It is likely that specific MHC alleles enable the host to mount a potent response because they present certain immunodominant peptides. However, evidence that this mechanism accounts for the associations is wanting. The T-cell repertoire depletion hypothesis predicts that mature T-cell repertoire diversity decreases with increasing MHC diversity. This can be tested experimentally. In order to evaluate the mechanisms that may explain why intra-individual MHC diversity is low, the lifetime reproductive success (LRS) of genetically diverse individuals in three treatment groups ((i) pathogen-free conditions, (ii) challenged with multiple but non-virulent pathogens and (iii) challenged with multiple, virulent pathogens) could be assessed. The T-cell depletion hypothesis predicts that MHC diversity should have no influence on LRS under pathogen-free conditions (if there is no MHC-dependent mate choice). By contrast, when challenged with virulent pathogens, individuals with an intermediate MHC diversity are expected to have maximal LRS. If the risk of autoimmune diseases increases with MHC diversity, this should result in a lower LRS of MHC-diverse individuals in treatment (i) and especially (ii), if triggering of cross-reactive T-cells by foreign antigens is an important mechanism for provoking autoimmune diseases.
Acknowledgments
We thank Martin A. Nowak for discussions and contributions to the model. The reviewers' and editors' efforts to help improve the manuscript are acknowledged.
Footnotes
One contribution of 11 to a Theme Issue ‘Ecological immunology’.
References
- Aeschlimann P.B., Häberli M.A., Reusch T.B.H., Boehm T., Milinski M. Female sticklebacks Gasterosteus aculeatus use self-reference to optimize MHC allele number during mate selection. Behav. Ecol. Sociobiol. 2003;54:119–126. doi:10.1007/s00265-003-0611-6 [Google Scholar]
- Allen T.M., et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. doi:10.1038/35036559 [DOI] [PubMed] [Google Scholar]
- Aoyagi K., Dijkstra J.M., Xia C., Denda I., Ototake M., Hashimoto K., Nakanishi T. Classical MHC class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss) J. Immunol. 2002;168:260–273. doi: 10.4049/jimmunol.168.1.260. [DOI] [PubMed] [Google Scholar]
- Blattman J.N., Antia R., Sourdive D.J.D., Wang X.C., Kaech S.M., Murali-Krishna K., Altman J.D., Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 2002;195:657–664. doi: 10.1084/jem.20001021. doi:10.1084/jem.20001021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud C., Mazuc J., Chastel O., Westerdahl H., Sorci G. Terminal investment induced by immune challenge and fitness traits associated with major histocompatibility complex in the house sparrow. Evolution. 2004;58:2823–2830. doi: 10.1111/j.0014-3820.2004.tb01633.x. doi:10.1554/04-279 [DOI] [PubMed] [Google Scholar]
- Bonneaud C., Richard M., Faivre B., Westerdahl H., Sorci G. An MHC class I allele associated to the expression of T-dependent immune response in the house sparrow. Immunogenetics. 2005;57:782–789. doi: 10.1007/s00251-005-0046-5. doi:10.1007/s00251-005-0046-5 [DOI] [PubMed] [Google Scholar]
- Borghans J.A.M., De Boer R.J. Diversity in the immune system. In: Segel L.A., Cohen I.R., editors. Design principles for the immune system and other distributed autonomous systems. Oxford University Press; New York, NY: 2001. pp. 161–184. [Google Scholar]
- Borghans J.A.M., Noest A.J., De Boer R.J. Thymic selection does not limit the individual MHC diversity. Eur. J. Immunol. 2003;33:3353–3358. doi: 10.1002/eji.200324365. doi:10.1002/eji.200324365 [DOI] [PubMed] [Google Scholar]
- Borghans J.A.M., Beltman J.B., De Boer R.J. MHC polymorphism under host–pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. doi:10.1007/s00251-003-0630-5 [DOI] [PubMed] [Google Scholar]
- Carrington M., et al. HLA and HIV-1: heterozygote advantage and B*35–Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. doi:10.1126/science.283.5408.1748 [DOI] [PubMed] [Google Scholar]
- Casrouge A., Beaudoing E., Dalle S., Pannetier C., Kanellopoulos J., Kourilsky P. Size estimate of the α β TCR repertoire of naive mouse splenocytes. J. Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- Center for Disease Control 1992 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morbidity and mortality weekly report, no. 41(RR-17). [PubMed]
- Daniels M.A., et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. doi:10.1038/nature05269 [DOI] [PubMed] [Google Scholar]
- Decamposlima P.O., Gavioli R., Zhang Q.J., Wallace L.E., Dolcetti R., Rowe M., Rickinson A.B., Masucci M.G. HLA-A11 epitope loss isolates of Epstein-Barr-virus from a highly A11+ population. Science. 1993;260:98–100. doi: 10.1126/science.7682013. doi:10.1126/science.7682013 [DOI] [PubMed] [Google Scholar]
- Doherty P.C., Zinkernagel R.M. Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. doi:10.1038/256050a0 [DOI] [PubMed] [Google Scholar]
- Edwards S.V., Hedrick P.W. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol. Evol. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. doi:10.1016/S0169-5347(98)01416-5 [DOI] [PubMed] [Google Scholar]
- Falk K., Rotzschke O., Stevanovic S., Jung G., Rammensee H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. doi:10.1038/351290a0 [DOI] [PubMed] [Google Scholar]
- Flajnik M.F. The immune system of ectothermic vertebrates. Vet. Immunol. Immunopathol. 1996;54:145–150. doi: 10.1016/s0165-2427(96)05685-1. doi:10.1016/S0165-2427(96)05685-1 [DOI] [PubMed] [Google Scholar]
- Forsberg L.A., Dannewitz J., Petersson E., Grahn M. Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout—females fishing for optimal MHC dissimilarity. J. Evol. Biol. 2007;20:1859–1869. doi: 10.1111/j.1420-9101.2007.01380.x. doi:10.1111/j.1420-9101.2007.01380.x [DOI] [PubMed] [Google Scholar]
- Fung-Leung W.P., Surh C.D., Liljedahl M., Pang J., Leturcq D., Peterson P.A., Webb S.R., Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. doi:10.1126/science.271.5253.1278 [DOI] [PubMed] [Google Scholar]
- George A.J.T., Stark J., Chan C. Understanding specificity and sensitivity of T-cell recognition. Trends Immunol. 2005;26:653–659. doi: 10.1016/j.it.2005.09.011. doi:10.1016/j.it.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Germain R.N. T-cell development and the CD4–CD8 lineage decision. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. doi:10.1038/nri798 [DOI] [PubMed] [Google Scholar]
- Grimholt U., Larsen S., Nordmo R., Midtlyng P., Kjoeglum S., Storset A., Saebo S., Stet R.J.M. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. doi:10.1007/s00251-003-0567-8 [DOI] [PubMed] [Google Scholar]
- Harf R., Sommer S. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol. Ecol. 2005;14:85–91. doi: 10.1111/j.1365-294X.2004.02402.x. doi:10.1111/j.1365-294X.2004.02402.x [DOI] [PubMed] [Google Scholar]
- Hill A.V.S., et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. doi:10.1038/352595a0 [DOI] [PubMed] [Google Scholar]
- Huseby E.S., Crawford F., White J., Kappler J., Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc. Natl Acad. Sci. USA. 2003;100:11 565–11 570. doi: 10.1073/pnas.1934636100. doi:10.1073/pnas.1934636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby E.S., White J., Crawford F., Vass T., Becker D., Pinilla C., Marrack P., Kappler J.W. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. doi:10.1016/j.cell.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Ignatowicz L., Kappler J., Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. doi:10.1016/S0092-8674(00)81028-4 [DOI] [PubMed] [Google Scholar]
- Ihara Y., Aoki K., Tokunaga K., Takahashi K., Juiji T. HLA and human mate choice: tests on Japanese couples. Anthropol. Sci. 2000:108. [Google Scholar]
- Ilmonen P., Penn D.J., Damjanovich K., Morrison L., Ghotbi L., Potts W.K. Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice. Genetics. 2007;176:2501–2508. doi: 10.1534/genetics.107.074815. doi:10.1534/genetics.107.074815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S., McClintock M.K., Zelano B., Ober C. Paternally inherited HLA alleles are associated with women's choice of male odor. Nat. Genet. 2002;30:175–179. doi: 10.1038/ng830. doi:10.1038/ng830 [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Travers P., Walport M., Schlomchik M. 6th edn. Garland Science Publishing; New York, NY; Abingdon, UK: 2005. The immune system in health and disease. [Google Scholar]
- Jeffery K.J.M., et al. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type T infection. J. Immunol. 2000;165:7278–7284. doi: 10.4049/jimmunol.165.12.7278. [DOI] [PubMed] [Google Scholar]
- Jones E.Y., Fugger L., Strominger J.L., Siebold C. MHC class II proteins and disease: a structural perspective. Nat. Rev. Immunol. 2006;6:271–282. doi: 10.1038/nri1805. doi:10.1038/nri1805 [DOI] [PubMed] [Google Scholar]
- Kast W.M., Brandt R.M.P., Sidney J., Drijfhout J.W., Kubo R.T., Grey H.M., Melief C.J.M., Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type-16 E6 and E7 proteins. J. Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- Kobel H.R., Dupasquier L. Genetics of polyploid Xenopus. Trends Genet. 1986;2:310–315. doi:10.1016/0168-9525(86)90286-6 [Google Scholar]
- Krogsgaard M., Davis M.M. How T cells ‘see’ antigen. Nat. Immunol. 2005;6:239–245. doi: 10.1038/ni1173. doi:10.1038/ni1173 [DOI] [PubMed] [Google Scholar]
- Langefors A., Lohm J., Grahn M., Andersen O., von Schantz T. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. B. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. doi:10.1098/rspb.2000.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor D.A., Zemmour J., Ennis P.D., Parham P. Evolution of class-I MHC genes and proteins—from natural selection to thymic selection. Annu. Rev. Immunol. 1990;8:23–63. doi: 10.1146/annurev.iy.08.040190.000323. doi:10.1146/annurev.iy.08.040190.000323 [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Bergstrom C.T., Antia R. Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med. Genet. 2003;4:2. doi: 10.1186/1471-2350-4-2. doi:10.1186/1471-2350-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohm J., Grahn M., Langefors A., Andersen O., Storset A., von Schantz T. Experimental evidence for major histocompatibility complex-allele-specific resistance to a bacterial infection. Proc. R. Soc. B. 2002;269:2029–2033. doi: 10.1098/rspb.2002.2114. doi:10.1098/rspb.2002.2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen T., Ujvari B. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 2006;19:1973–1978. doi: 10.1111/j.1420-9101.2006.01158.x. doi:10.1111/j.1420-9101.2006.01158.x [DOI] [PubMed] [Google Scholar]
- Martin M.P., Carrington M. Immunogenetics of viral infections. Curr. Opin. Immunol. 2005;17:510–516. doi: 10.1016/j.coi.2005.07.012. doi:10.1016/j.coi.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Martin W.D., Hicks G.G., Mendiratta S.K., Leva H.I., Ruley H.E., VanKaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. doi:10.1016/S0092-8674(00)81030-2 [DOI] [PubMed] [Google Scholar]
- Matsuyama T., Fujiwara A., Nakayasu C., Kamaishi T., Oseko N., Tsutsumi N., Hirono I., Aoki T. Microarray analyses of gene expression in Japanese flounder Paralichthys olivaceus leucocytes during monogenean parasite Neoheterobothrium hirame infection. Dis. Aquat. Organ. 2007;75:79–83. doi: 10.3354/dao075079. doi:10.3354/dao075079 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M., Graf D., Lovatt M., Bommhardt U., Zamoyska R., Fisher A.G. How many thymocytes audition for selection? J. Exp. Med. 1997;186:1149–1158. doi: 10.1084/jem.186.7.1149. doi:10.1084/jem.186.7.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 2006;37:159–186. doi:10.1146/annurev.ecolsys.37.091305.110242 [Google Scholar]
- Milinski M., Griffiths S., Wegner K.M., Reusch T.B.H., Haas-Assenbaum A., Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. doi:10.1073/pnas.0408264102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., Kaukinen K.H., Schulze A.D. Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics. 2002;53:941–963. doi: 10.1007/s00251-001-0398-4. doi:10.1007/s00251-001-0398-4 [DOI] [PubMed] [Google Scholar]
- Monteiro M.C., Couceiro S., Penha-Goncalves C. The multigenic structure of the MHC locus contributes to positive selection efficiency: a role for MHC class II gene-specific restriction. Eur. J. Immunol. 2005;35:3622–3630. doi: 10.1002/eji.200535190. doi:10.1002/eji.200535190 [DOI] [PubMed] [Google Scholar]
- Munks M.W., Gold M.C., Zajac A.L., Doom C.M., Morello C.S., Spector D.H., Hill A.B. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J. Immunol. 2006;176:3760–3766. doi: 10.4049/jimmunol.176.6.3760. [DOI] [PubMed] [Google Scholar]
- Nikolich-Zugich J., Slifka M.K., Messaoudi I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 2004;4:123–132. doi: 10.1038/nri1292. doi:10.1038/nri1292 [DOI] [PubMed] [Google Scholar]
- Nowak M.A., Tarczyhornoch K., Austyn J.M. The optimal number of major histocompatibility complex-molecules in an individual. Proc. Natl Acad. Sci. USA. 1992;89:10 896–10 899. doi: 10.1073/pnas.89.22.10896. doi:10.1073/pnas.89.22.10896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A., May R.M., Sigmund K. Immune responses against multiple epitopes. J. Theor. Biol. 1995;175:325–353. doi: 10.1006/jtbi.1995.0146. doi:10.1006/jtbi.1995.0146 [DOI] [PubMed] [Google Scholar]
- Oksenberg J.R., et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 2004;74:160–167. doi: 10.1086/380997. doi:10.1086/380997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone M.B.A. Molecular mimicry and immune-mediated diseases. FASEB J. 1998;12:1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholczyk R., Kern J., Singh N., Iwashima M., Kraj P., Lgnatowicz L. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. doi:10.1016/j.immuni.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S., Wilson K., Pemberton J.M. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.) Proc. Natl Acad. Sci. USA. 1998;95:3714–3719. doi: 10.1073/pnas.95.7.3714. doi:10.1073/pnas.95.7.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D.J. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology. 2002;108:1–21. doi:10.1046/j.1439-0310.2002.00768.x [Google Scholar]
- Penn D.J., Damjanovich K., Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA. 2002;99:11 260–11 264. doi: 10.1073/pnas.162006499. doi:10.1073/pnas.162006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- Pitcher T.E., Neff B.D. MHC class IIB alleles contribute to both additive and nonadditive genetic effects on survival in Chinook salmon. Mol. Ecol. 2006;15:2357–2365. doi: 10.1111/j.1365-294X.2006.02942.x. doi:10.1111/j.1365-294X.2006.02942.x [DOI] [PubMed] [Google Scholar]
- Potts W.K., Slev P.R. Pathogen-based models favoring MHC genetic diversity. Immunol. Rev. 1995;143:181–197. doi: 10.1111/j.1600-065x.1995.tb00675.x. doi:10.1111/j.1600-065X.1995.tb00675.x [DOI] [PubMed] [Google Scholar]
- Rammensee H.G., Bachmann J., Stevanovic S. Springer; Berlin, Germany: 1997. MHC ligands and peptide motifs. [DOI] [PubMed] [Google Scholar]
- Reusch T.B.H., Haberli M.A., Aeschlimann P.B., Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. doi:10.1038/35104547 [DOI] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., Marsh S.G.E. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. doi:10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal K.L., Zinkernagel R.M. Inability of mice to generate cytotoxic lymphocytes-T to vesicular stomatitis-virus restricted to H-2kk or H-2dk. J. Immunol. 1981;126:446–451. [PubMed] [Google Scholar]
- Santori F.R., et al. Rare, structurally homologous self-peptides promote thymocyte positive selection. Immunity. 2002;17:131–142. doi: 10.1016/s1074-7613(02)00361-8. doi:10.1016/S1074-7613(02)00361-8 [DOI] [PubMed] [Google Scholar]
- Schad J., Ganzhorn J.U., Sommer S. Parasite burden and constitution of major histocompatibility complex in the malagasy mouse lemur, Microcebus murinus. Evolution. 2005;59:439–450. doi:10.1554/04-312 [PubMed] [Google Scholar]
- Sercarz E.E., Lehmann P.V., Ametani A., Benichou G., Miller A., Moudgil K. Dominance and crypticity of T-cell antigenic determinants. Annu. Rev. Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. doi:10.1146/annurev.iy.11.040193.003501 [DOI] [PubMed] [Google Scholar]
- Siebold C., et al. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc. Natl Acad. Sci. USA. 2004;101:1999–2004. doi: 10.1073/pnas.0308458100. doi:10.1073/pnas.0308458100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A. New perspectives on a developmental dilemma: the kinetic signalling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. doi:10.1016/S0952-7915(02)00323-0 [DOI] [PubMed] [Google Scholar]
- Snell G.D. H-2 locus of mouse—observations and speculations concerning its comparative genetics and its polymorphism. Folia Biol. 1968;14:335–358. [PubMed] [Google Scholar]
- Stet R.J.M., de Vries B., Mudde K., Hermsen T., van Heerwaarden J., Shum B.P., Grimholt U. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics. 2002;54:320–331. doi: 10.1007/s00251-002-0477-1. doi:10.1007/s00251-002-0477-1 [DOI] [PubMed] [Google Scholar]
- Stock A.T., Jones C.M., Heath W.R., Carbone F.R. CTL response compensation for the loss of an immunodominant class I-restricted HSV-1 determinant. Immunol. Cell Biol. 2006;84:543–550. doi: 10.1111/j.1440-1711.2006.01469.x. doi:10.1111/j.1440-1711.2006.01469.x [DOI] [PubMed] [Google Scholar]
- Sylwester A.W., et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. doi:10.1084/jem.20050882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz M.R., Thomas H.C., Greenwood B.M., Hill A.V.S. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 1997;17:11–12. doi: 10.1038/ng0997-11. doi:10.1038/ng0997-11 [DOI] [PubMed] [Google Scholar]
- Todd J.A., Bell J.I., McDevitt H.O. HLA-DQ-β gene contributes to susceptibility and resistance to insulin-dependent diabetes-mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. doi:10.1038/329599a0 [DOI] [PubMed] [Google Scholar]
- van den Berg H.A., Rand D.A. Antigen presentation on MHC molecules as a diversity filter that enhances immune efficacy. J. Theor. Biol. 2003;224:249–267. doi: 10.1016/s0022-5193(03)00162-0. doi:10.1016/S0022-5193(03)00162-0 [DOI] [PubMed] [Google Scholar]
- van Meerwijk J.P.M., Marguerat S., Lees R.K., Germain R.N., Fowlkes B.J., MacDonald H.R. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 1997;185:377–383. doi: 10.1084/jem.185.3.377. doi:10.1084/jem.185.3.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic D., Matzinger P. Unresponsiveness to a foreign antigen can be caused by self-tolerance. Nature. 1988;336:222–225. doi: 10.1038/336222a0. doi:10.1038/336222a0 [DOI] [PubMed] [Google Scholar]
- Viney M.E., Riley E.M., Buchanan K.L. Optimal immune responses: immunocompetence revisited. Trends Ecol. Evol. 2005;20:665–669. doi: 10.1016/j.tree.2005.10.003. doi:10.1016/j.tree.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Wallace M.E., Keating R., Heath W.R., Carbone F.R. The cytotoxic T-cell response to herpes simplex virus type 1 infection of C57BL/6 mice is almost entirely directed against a single immunodominant determinant. J. Virol. 1999;73:7619–7626. doi: 10.1128/jvi.73.9.7619-7626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind C., Seebeck T., Bettens F., Paepke A.J. MHC-dependent mate preferences in humans. Proc. R. Soc. B. 1995;260:245–249. doi: 10.1098/rspb.1995.0087. doi:10.1098/rspb.1995.0087 [DOI] [PubMed] [Google Scholar]
- Wedekind C., Walker M., Little T.J. The course of malaria in mice: major histocompatibility complex (MHC) effects, but no general MHC heterozygote advantage in single-strain infections. Genetics. 2005;170:1427–1430. doi: 10.1534/genetics.105.040683. doi:10.1534/genetics.105.040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner, K. M. 2004 Major histocompatibility genes, polymorphism and balancing selection—the case of parasites and sticklebacks. PhD thesis, Christian Albrechts University, Kiel.
- Wegner K.M., Kalbe M., Kurtz J., Reusch T.B.H., Milinski M. Parasite selection for immunogenetic optimality. Science. 2003a;301:1343. doi: 10.1126/science.1088293. doi:10.1126/science.1088293 [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Reusch T.B.H., Kalbe M. Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 2003b;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. doi:10.1046/j.1420-9101.2003.00519.x [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Kalbe M., Rauch G., Kurtz J., Schaschl H., Reusch T.B.H. Genetic variation in MHC class II expression and interactions with MHC sequence polymorphism in three-spined sticklebacks. Mol. Ecol. 2006;15:1153–1164. doi: 10.1111/j.1365-294X.2006.02855.x. [DOI] [PubMed] [Google Scholar]
- Wordsworth B.P., Lanchbury J.S.S., Sakkas L.I., Welsh K.I., Panayi G.S., Bell J.I. HLA-DR4 subtype frequencies in rheumatoid-arthritis indicate that DRB1 is the major susceptibility locus within the HLA class-II region. Proc. Natl Acad. Sci. USA. 1989;86:10 049–10 053. doi: 10.1073/pnas.86.24.10049. doi:10.1073/pnas.86.24.10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J.W. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. doi:10.1016/j.immuni.2006.09.005 [DOI] [PubMed] [Google Scholar]
- Zerrahn J., Held W., Raulet D.H. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. doi:10.1016/S0092-8674(00)81905-4 [DOI] [PubMed] [Google Scholar]
- Zhang X.M., Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhao Z.S., Granucci F., Yeh L., Schaffer P.A., Cantor H. Molecular mimicry by herpes simplex virus type 1: autoimmune disease after viral infection. Science. 1998;279:1344–1347. doi: 10.1126/science.279.5355.1344. doi:10.1126/science.279.5355.1344 [DOI] [PubMed] [Google Scholar]