Abstract
An organism's fitness is critically reliant on its immune system to provide protection against parasites and pathogens. The structure of even simple immune systems is surprisingly complex and clearly will have been moulded by the organism's ecology. The aim of this review and the theme issue is to examine the role of different ecological factors on the evolution of immunity. Here, we will provide a general framework of the field by contextualizing the main ecological factors, including interactions with parasites, other types of biotic as well as abiotic interactions, intraspecific selective constraints (life-history trade-offs, sexual selection) and population genetic processes. We then elaborate the resulting immunological consequences such as the diversity of defence mechanisms (e.g. avoidance behaviour, resistance, tolerance), redundancy and protection against immunopathology, life-history integration of the immune response and shared immunity within a community (e.g. social immunity and microbiota-mediated protection). Our review summarizes the concepts of current importance and directs the reader to promising future research avenues that will deepen our understanding of the defence against parasites and pathogens.
Keywords: ecological immunology, coevolution, adaptive immune system, innate immune system, microbiota, trade-off
1. Timeliness of a merger between ecology and immunology
Ecology is the study of the distribution and abundance of organisms and their interactions with their environment, including parasites and pathogens. Immunology is the study of the physiological functioning of the immune system in states of health and disease. The former discipline implicitly acknowledges the importance of the latter but treats it as a black box. Likewise, the latter discipline implicitly acknowledges the importance of variation between individuals, but relies on a logistic foundation that necessarily removes individual variation from its empirical approach and so avoids the complex interactions which determine an organism's life history in its natural environment.
Over the past decade, new molecular information and techniques have become available, which have facilitated a combination of the two fields and thus placed mechanistic understanding into an ecological context and vice versa. Examples include the inference of natural selection on the immune system by quantitative trait loci mapping of defence genes in natural insect populations such as fruitflies (e.g. Bangham et al. 2007, 2008; Dubuffet et al. 2007), mosquitoes (e.g. Menge et al. 2006; Riehle et al. 2006) or bumblebees (e.g. Wilfert et al. 2007; Wilfert & Schmid-Hempel 2008); the analysis of naturally occurring polymorphism in insect immunity genes (Lazzaro et al. 2004, 2006; Jiggins & Kim 2006, 2007; Obbard et al. 2006) or the comparison of whole genomes of different Drosophila species for reconstruction of immune system evolution (Sackton et al. 2007). Perhaps most important is the realization that although substantial basic information has accumulated independently in both disciplines, full understanding of the complexity encountered demands a perspective that can only be provided by a joint cross-disciplinary approach.
Our review provides an overview of the recently expanding research field of ecological immunology. This research field is concerned with the ecological factors (biotic as well as abiotic) which determine the evolution of the immune system and which may therefore provide the key to understanding its structure and enormous complexity. Here, we will contextualize the main categories of ecological factors and the resulting immunological consequences, and will support our perspective with selected case examples: we make no excuse for cherry-picking key examples rather than attempting a full coverage of the field. Based on this information, we will highlight promising topics for future research that we believe will further our understanding of both ecological and immunological mechanisms. More specific aspects will be addressed in the different reviews of this themed issue.
2. Ecological inputs
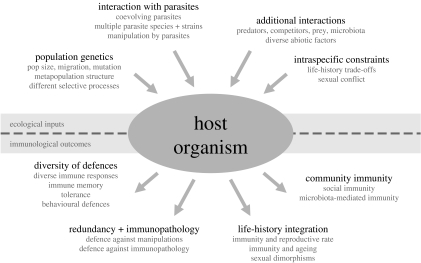
There are four main categories of ecological factors that influence the diversity and complexity of the immune system: interactions with parasites; other types of biotic as well as abiotic interactions; intraspecific selective constraints; and population genetic processes that influence the evolution of immune components (figure 1).
Figure 1.
Ecological inputs and immunological outcomes.
(a) Interactions with parasites
Interactions with parasites (here used in its wide sense, thereby including viruses, bacteria, protists and eukaryotes) will affect the evolution of immune systems, which directly serve to protect the host from parasitic infections and the associated damage. Certain types of interactions are believed to have particular impact on immune systems. The first of these is coevolution with parasites, where parasites in close and long-term association with a particular host species may continuously adapt to the current as well as newly evolving host defences. Parasite adaptation is usually faster than host counter-adaptation due to the parasite's comparatively shorter generation time, its usually larger population size and its often haploid genome (cf. Hamilton et al. 1990). The speed and perseverance of parasite adaptations produce one of the strongest selective pressures known in evolution—with direct relevance for host immunity. Examples for rapidly evolving parasites are found among human viruses such as influenza and HIV (reviewed in Arien et al. 2007; Nelson & Holmes 2007), the well-documented interactions between snail hosts and their trematode parasites in New Zealand lakes (reviewed by Jokela et al. 2003), or rabbit hosts and the myxoma virus (reviewed in Fenner & Fantini 1999).
The second factor impacting on the evolution of immune systems is the necessity to deal with an unpredictable set of many parasites, either different parasite species or different strains of the same species. In both cases, the different parasite ‘types’ are likely to attempt infection with distinct ‘attack’ mechanisms, thus requiring diversity and flexibility in host recognition, processing and effector mechanisms. One of the few systematic studies of parasite diversity and its immunogenetic consequences was performed in lake and river ecotypes of the three-spined sticklebacks Gasterosteus aculeatus in northern Germany, which contain approximately 15 different eukaryotic parasite species (Kalbe et al. 2002; Scharsack et al. 2007). Parasite species richness within a population affects relevant components of both the innate and adaptive immune systems (Scharsack et al. 2007), such as the diversity of major histocompatibility complex (MHC) genes (Wegner et al. 2003a,b). The general importance of interactions with multiple parasite species or strains on host evolution has been emphasized in several theoretical articles. Heterogeneous infections may lead to higher virulence (here defined as reduction in host fitness, resulting from the combination of parasite pathogenicity and host immunity, thus representing a joint character of both host and parasite) if there is competition among parasites (e.g. Antia et al. 1994; Bonhoeffer & Nowak 1994; Nowak & May 1994; Frank 1996; De Roode et al. 2005). Lower virulence may result if parasites are closely related (thus being subject to kin selection), if they cooperate in host exploitation, and/or if they get the same share irrespective of their actions (e.g. Frank 1996; Brown 1999; Brown et al. 2002; Schjorring & Koella 2003; Rauch et al. 2008). In turn, the former scenario should increase, whereas the latter decreases selection on host immunity.
Finally, manipulation by parasites is another potentially relevant factor in many host–parasite associations. In general, the immune system aims at preventing invasion and damage through the recognition and elimination of parasites. In a simple world, parasites adapt to host defences by escaping detection or elimination, e.g. by changing surface molecules. In this case, the most direct evolutionary response of a host consists of changing the efficiency of recognition and immune effector mechanisms. However, if parasites attack by manipulating host defence reactions, the selective pressure on the host becomes more complex. Such interference strategy is widespread among parasites and may affect different levels of the host immune response, as summarized by Schmid-Hempel (2009; see also Schmid-Hempel 2008). In this case, selection no longer acts exclusively on host recognition and elimination mechanisms, but also on protection against manipulations—either by directly targeting such parasite manipulator molecules or by indirectly enhancing robustness of defence responses, e.g. through redundancy in immune signalling.
(b) Additional interactions
Abiotic and biotic ecological factors are major driving forces for the evolution of host–parasite interaction and immune defence. For example, the availability of food organisms biases anti-parasite defence in the nematode host Caenorhabditis elegans (Schulenburg & Müller 2004; Sicard et al. 2007). Similarly, nutrient availability altered parasite resistance of mosquitoes (Ferguson & Read 2002; Bedhomme et al. 2004; Lambrechts et al. 2006). The presence of symbiotic micro-organisms in aphids strongly influences immunity against parasitoid wasps, as well as fungal parasites (Oliver et al. 2003; Scarborough et al. 2005; von Burg et al. 2008), and the composition and diversity of the mammalian gut microbiota affects resistance against pathogens and activity of different components of the immune system (reviewed in O'Hara & Shanahan 2006; Sansonetti & Di Santo 2007). Abiotic factors such as temperature have a severe effect on parasite resistance in the nematode C. elegans (Schulenburg & Müller 2004), the water flea Daphnia magna (Mitchell et al. 2005) and the fruitfly Drosophila melanogaster (Lazzaro et al. 2008).
However, as discussed by Lazzaro & Little (2009), environmental factors will influence immune system evolution only if their impact varies with host genotype, i.e. some host genotypes are more immunocompetent in one environment, whereas others perform better in another environment (genotype×environment interaction; Lazzaro & Little 2009). In this case, the environmental factor may act as a potent selective force that directly determines the distribution of host genotypes and thus the evolution of defence in host populations. Among the above examples, such genotype×environment effects were found for food quality and temperature (Ferguson & Read 2002; Schulenburg & Müller 2004; Mitchell et al. 2005; Lambrechts et al. 2006; Lazzaro et al. 2008).
(c) Intraspecific constraints
Immunity comes at a cost. Since resources are usually limited, these costs should have an important influence on immune system evolution. This section is concerned with such immunity costs that become manifest in the form of trade-offs between immunity and other fitness-related traits. There are three non-exclusive forms in which costs of immunity arise: genetic costs (i.e. genetically fixed higher investment in immunity); usage costs (i.e. energetic costs that only apply upon activation of the immune system); and immunopathology (i.e. tissue damage caused by the immune system). The importance of the resulting life-history trade-offs has been indicated in numerous studies and a large diversity of organisms (reviewed by Sheldon & Verhulst 1996; Schmid-Hempel 2003; Siva-Jothy et al. 2005), and it is also discussed in this issue's review by Lazzaro & Little (2009). The most important trade-off should be between immunity, which ensures survival, and reproductive rate, which directly correlates with fitness. Reduced reproductive rates were indeed observed for immunocompetent individuals/strains (high genetic immunity costs), e.g. in mosquitoes or fruitflies (e.g. Ferdig et al. 1993; Yan et al. 1997; McKean et al. 2008). Similar reductions were found after experimental activation of the immune system (high usage costs; reviewed in Schmid-Hempel 2003), e.g. in mosquitoes, fruitflies or birds (e.g. Bonneaud et al. 2003; Ahmed & Hurd 2006; McKean et al. 2008). One study in insects has provided unequivocal experimental evidence for a cost of immunopathology, namely on tissue integrity in the mealworm beetle (Sadd & Siva-Jothy 2006). Interestingly, immunity costs can often only be demonstrated when organisms are examined in stressful environments, further highlighting the potentially strong influence of the environment (discussed in Lazzaro & Little 2009). For example, fruitflies selected for increased parasitoid resistance showed reduced larval competitiveness only under food limitation (Kraaijeveld & Godfray 1997).
Another constraint ultimately originates from Bateman's principle, i.e. one sex (typically females) invests more in individual offspring than the other (typically males). As a consequence, the two sexes differ in their evolutionary interests, leading to various sex-related selective processes usually summarized under the header ‘sexual selection’. The consequences on the immune system are manifold. As highlighted in this issue's study by Nunn et al. (2009), intra-sexual selection among the limited sex (usually females) favours female individuals with a high investment in immunity if this investment increases not only lifetime per se but also particularly lifetime reproductive success (see also Rolff 2002). By contrast, intra-sexual selection among the less limited sex (usually males) may provide an advantage to male individuals that mainly invest into competition with other males, resulting in the reduced availability of resources for immune functions. Moreover, selection that acts on the interaction between the two sexes (inter-sexual selection) may result in mate choice, usually female choice, where an important criterion for choice is immunocompetence (i.e. the ability to prevent or limit parasitic infections). In this context, choice may be more direct, such that females choose uninfected males or those that indicate high resistance with the expression of other costly traits (e.g. immunocompetence handicap hypothesis; Hamilton & Zuk 1982; Folstad & Karter 1992). A non-exclusive alternative is that choice is more indirect, favouring advantageous immunity allele combinations among the offspring (reviewed in Milinski 2006). The latter has been investigated in much detail in sticklebacks, where an intermediate level of allele diversity was found to be optimal (Reusch et al. 2001; Wegner et al. 2003a; Kurtz et al. 2004), as further elaborated by Woelfing et al. (2009).
(d) Population genetic processes
Evolutionary dynamics in populations are not only affected by particular selective pressures, but also depend on population genetic characteristics, including population size, migration, mutation frequency, number of genes involved in trait expression and metapopulation structure. These population genetic characteristics are of major importance for host–parasite coevolutionary dynamics (e.g. Ebert et al. 2002; Forde et al. 2004; Cooper et al. 2005; Morgan et al. 2005; Brockhurst et al. 2006; Vogwill et al. 2008), which in turn implies that they also affect evolution of the immune system. In fact, Boots et al. (2009) review a number of theoretical articles which use population genetic models in order to evaluate evolution of different aspects of immune defence (e.g. avoidance, resistance or tolerance). These models are either based on a few defence loci (gene-for-gene and matching allele models) or many loci (quantitative genetic models and game theoretical approaches). They emphasize the importance of life-history trade-offs for the evolution of resistance versus either tolerance or immune memory (reviewed in Boots et al. 2009).
The particular combination of selective constraints and population genetic characteristics determines the dynamics of evolutionary change (Woolhouse et al. 2002). Five different non-exclusive forms of such selective dynamics are expected in the context of the evolution of host–parasite interactions and thus host immunity.
Directional selection if the host is confronted with a predictable set of parasites or parasite attack mechanisms (i.e. no large changes in the encountered pathogenicity mechanisms over time); the relevant resistant genes should then spread through the population to fixation.
Negative frequency-dependent dynamics may be observed when host and parasite coevolve; in this case, parasites are likely to adapt to the most common host resistance allele, thus favouring rare alleles, which then spread through the population to high frequency, which in turn leads to a selective advantage of a then rare allele and so on; ultimately this leads to repeated cycles of increase and decrease in particular allele frequencies (e.g. Dybdahl & Lively 1998; Decaestecker et al. 2007; Gandon et al. 2008).
Host–parasite coevolution may also be associated with repeated selective sweeps (e.g. Buckling & Rainey 2002; Bangham et al. 2007; Gandon et al. 2008); this selective process is related to the last point, in that selection favours rare resistance alleles because parasites are likely to evolve to the most common alleles; in this case, however, the rare allele is produced by mutation, recombination or migration, followed by its spread through the population to fixation (similar to directional selection, point (i)); a new sweep is then initiated by a new mutation, recombination event or migrant and so on; this selective process could additionally be initiated if a new variant allele at a particular locus is duplicated at a new locus, where it is present in homozygous condition, facilitating its fast spread through the population, as is possibly true for the numerous plant R resistance genes (Bergelson et al. 2001; Bakker et al. 2006; reviewed in Mitchell-Olds et al. 2007; Salvaudon et al. 2008).
The presence of coevolving or large numbers of different parasites may favour hosts with high diversity at particular immunity genes (e.g. recognition receptors), because these are better able to fend off the diverse set of parasites; such heterozygote advantage or overdominant selection may directly select for high population-wide diversity in immunity genes, as inferred for human innate immunity genes (Ferrer-Admetlla et al. 2008), a mouse virus resistance gene (Ferguson et al. 2008) or possibly mammalian MHC genes (e.g. Apanius et al. 1997; Penn & Potts 1999; but see De Boer et al. 2004).
Neutral processes such as genetic drift may also affect immune system evolution, particularly in small populations and in the absence of selection and migration.
It should be noted that these evolutionary dynamics either operate on standing genetic variation or require new mutations or a combination thereof (e.g. through the recombination of short sequence motifs of variable alleles; see Reusch & Langefors 2005). Therefore, they may entail different time scales of evolutionary change and thus become manifested in gene sequence evolution at either population or species level, as discussed for RNA interference genes by Obbard et al. (2009).
3. Immunological outcomes
High diversity in ecological processes should favour high diversity in immunological mechanisms, since these are more likely to mediate an effective response against different parasites under different environmental conditions. In the following, we discuss the impact on four aspects of immune defence (figure 1): diversity of alternative mechanisms; protection against complications such as parasite manipulations or immunopathology; life-history integration of the immune response; and shared immunity within a community.
(a) Diversity of defences
The availability of a diversity of immune defence options should increase an organism's survival in a variable world. It is likely to be enhanced in the presence of immunity costs: the alternatives are then not only more effective under certain environmental conditions but—as a direct consequence—also allow a more economic usage of resources. Alternative defences may occur at several different levels as follows.
behavioural avoidance of parasites, which limits exposure to parasites and thus minimizes infection risk,
physical barriers that minimize invasion rates of parasites,
other boundary defence mechanisms that reduce the likelihood of infection, e.g. alteration of certain cell surface molecules, which are exploited by parasites for invasion, or constitutively expressed antimicrobial genes on the skin as part of the immediate innate immune system,
control of parasite invasion and replication with the help of the immediate inducible immune response (inducible innate immune system) and/or the delayed adaptive immune response based on, e.g. antibodies, T and B cells,
a learned immune response based on immunological memory mediated by the adaptive immune system, and
tolerance, i.e. the compensation/attenuation of parasite damage without restriction of parasite invasion and growth.
We would like to draw particular attention to some of these defence strategies, which we think deserve further research effort. If a first encounter with a parasite comes with a high likelihood of a second encounter with the same parasite, then selection should favour both high immune specificity and immunological memory in order to increase protection of the host upon secondary exposures, irrespective of the host's phylogenetic affiliation (Moret & Siva-Jothy 2003; Kurtz 2004; Schulenburg et al. 2007). This verbal argument is supported by theoretical models, which additionally emphasize the importance of disease characteristics for the evolution of immunological memory (discussed in this issue's review of Boots et al. 2009). Specific memory is very well characterized for the adaptive immune system, which not only provides higher protection during an individual's lifespan but also enhances offspring immunity through the maternal transfer of antibodies, as detailed in this issue's review by Hasselquist & Nilsson (2009). Importantly, a similar priming of the immune system has now been repeatedly observed in invertebrates that lack the ‘adaptive’ system, e.g. the crustacean copepod Macrocyclops albicans (Kurtz & Franz 2003), the crustacean water flea D. magna (Little et al. 2003), the bumblebee Bombus terrestris (Sadd et al. 2005; Sadd & Schmid-Hempel 2006), the beetles Tenebrio molitor (Moret 2006) and Tribolium castaneum (Roth et al. 2008) and the fruitfly D. melanogaster (Pham et al. 2007). The underlying molecular mechanisms are currently unknown. They appear to require phagocytes and the Toll pathway (Pham et al. 2007). They may also involve highly diverse recognition receptors, e.g. those generated through alternative splicing of the Dscam immunoglobulin in arthropods (Watson et al. 2005; Dong et al. 2006; Brites et al. 2008), or the synergistic interaction of different immune components (reviewed in Schmid-Hempel 2005; Du Pasquier 2006; Kurtz & Armitage 2006; Schulenburg et al. 2007).
Tolerance is defined as the ability to limit the damage caused by a given parasite burden. As elaborated in detail in this issue by Råberg et al. (2009), the concept of tolerance towards parasites is widely applied in the plant literature, but currently ignored in animal studies. To date, there is only one conclusive demonstration of animal tolerance, namely in mice infected with rodent malaria (Råberg et al. 2007). Tolerance may represent a highly economic strategy, since it may be energetically cheaper to limit the damage rather than invest in parasite elimination mechanisms. Moreover, tolerance should be evolutionarily advantageous since it may also favour less pathogenic parasites. In fact, theoretical models suggest that immunity costs as well as high parasite virulence favour the evolution of tolerance (reviewed in this issue by Boots et al. 2009).
Parasite avoidance behaviours are widespread among animals (reviewed in Moore 2002) and are particularly common among social organisms (this issue's review by Cremer & Sixt 2009; see also Cremer et al. 2007). They most likely provide a highly economic defence, since they minimize both the exposure to parasites (thus potential damage) and possibly the energetic costs required for activation of the physiological immune system (discussed in Schulenburg & Ewbank 2007). To date, the underlying genetics are not well understood. The most comprehensive data have been generated for the nematode C. elegans. Avoidance behaviours appear to be central in the nematode's defence against parasites. Genetic analysis implicated the involvement of three main mechanisms: physical pathogen avoidance based on G-protein signalling in chemosensory neurons (Pradel et al. 2007); learning of pathogen avoidance behaviour through serotonin signalling in the nervous system (Zhang et al. 2005); and physical avoidance as well as reduced oral uptake of pathogens as part of a general stress response mediated by insulin-like signalling (Hasshoff et al. 2007). These mechanisms may provide several links to other life-history functions including the physiological immune system, longevity and reproduction (reviewed in Schulenburg & Ewbank 2007). Physical avoidance behaviours may also show a high level of specificity, since nematodes responded differently to distinct Serrawettin surfactant molecules from the pathogen Serratia marcescens (Pradel et al. 2007). This specificity may be mediated by the highly numerous (more than 1000 genes) G-protein-coupled chemoreceptors (review by Schulenburg & Ewbank 2007).
Diversity in defence options may be fixed genetically or it may be generated phenotypically during an individual's lifespan. The latter may be favoured by selection if it enables a more specific (and thus potentially more economic) response to the parasite repertoire encountered by the individual. This strategy is manifested in the adaptive immune system of the higher vertebrates: somatic recombination results in highly diversified antibodies and B/T cell receptors which can then be activated specifically upon exposure to non-self-peptides, leading to specific immune memory (e.g. Janeway et al. 2001). Importantly, this particular system highlights that diversity per se is not necessarily advantageous. Even though it enhances efficiency of parasite detection, high receptor diversity also increases the risk of a misinterpretation of self-molecules as being foreign, potentially causing autoimmune responses. Therefore, specific selective processes act within the immune system to ensure that self-reacting receptors are eliminated. The exact dynamics of these selective processes as well as their consequences on other components of the immune system are as yet not fully understood. They are believed to be responsible for various phenomena, such as an intermediate optimal level of MHC receptor diversity, as discussed in this issue by Woelfing et al. (2009).
(b) Redundancy and immunopathology
We would like to point to two complications that represent a particular challenge for host organisms. One of them results from the widespread strategy of parasites to interfere with host defence signalling (see this issue's reviews by Obbard et al. (2009) and Schmid-Hempel (2009)). Such parasite manipulation could be particularly problematic for the host: even if it shows 100 per cent efficiency in parasite recognition, this will not help if the recognition signal cannot be transferred to the immune effectors. One possible solution is the availability of several redundant immune signalling pathways. These pathways could act independently, thus making it difficult for the parasite, which would have to target all of them. However, in this case, the host response may not be entirely efficient, since it may overreact if there is no parasite manipulation or it could even be very weak if parasites successfully interfere with one of the pathways. Therefore, an interacting network of separate immunity pathways could be advantageous. Such a network would have two important effects. It would minimize the consequences of parasite manipulations, and it would also allow for compensatory actions if it enables the host to detect parasite-mediated disruption of host signalling. To date, the importance of redundancy as well as interconnections among immunity signalling pathways as a defence against parasite manipulations is unexplored.
Another important complication is the risk of immunopathology (this issue's review by Sorci & Faivre 2009; see also Graham et al. 2005). Parasite elimination often relies on cytotoxic compounds, e.g. reactive oxygen and nitrogen species (ROS and RNS), which may be activated in the immediate inflammatory response in vertebrates. These compounds are very efficient at destroying parasites. However, they are generic cytotoxins. Therefore, their activation automatically leads to the damage of host tissue. In turn, selection should favour a very strict regulation of these substances, including their deactivation upon usage. In fact, activation of cytotoxic compounds is usually followed by secretion of anti-inflammatory molecules such as interleukin-10 or transforming growth factor-β (review by Sorci & Faivre 2009). Furthermore, we also expect evolution of specific protective mechanisms within the host, such as expression of antioxidant enzymes such as superoxide dismutase or catalase (e.g. Sorci & Faivre 2009). The amount of oxidative damage, mainly resulting from respiratory burst during innate immune defence, may also depend on the effectiveness of other immune functions, such as MHC-mediated adaptive immunity (Kurtz et al. 2006), even though the exact underlying mechanism is as yet unknown. Immunopathology also represents an important complication in invertebrate innate immune systems: activation of cytotoxic compounds leads to measurable damage of self-tissue in the mealworm beetle T. molitor (Sadd & Siva-Jothy 2006). Similarly, in many organisms, such as the nematode C. elegans, ROS are produced as a defence against pathogens, but may also cause oxidative stress leading to protein damage in the host (Chavez et al. 2007; Mohri-Shiomi & Garsin 2008). In this case, pathogen exposure not only leads to activation of ROS but also antioxidant enzymes such as superoxide dismutase, which significantly minimizes oxidative damage thereby increasing nematode survival (Chavez et al. 2007). Consequently, such a protective mechanism may represent a universal component of animal immune systems.
(c) Life-history integration
The immune system has evolved, and still is evolving, in the context of other life-history requirements and environmental conditions (i.e. not in isolation). Two processes are likely to be of major importance.
The most relevant environmental information (e.g. parasite abundance and diversity, food availability) should be filtered and integrated in order to determine the most efficient response.
Activation of the immune system must take into account developmental stage and nutritional state of the organism in order to minimize negative effects on other life-history functions, which may result from autoimmune damage or resource competition. In this context, we particularly expect a trade-off between immunity and reproduction.
It is as yet unclear how life-history integration of immune defence is mediated at the molecular level. Within the innate immune system, an interesting candidate is the insulin-like signalling cascade. This pathway shows a high degree of conservation across animals. To date, its functions are best understood in the nematode C. elegans. Pathway activation depends on external cues such as food availability or environmental stress (heat, oxidative stress, crowding). Its normal activity in the wild-type associates with high reproductive rates (e.g. Gems et al. 1998; Houthoofd et al. 2005). By contrast, its downregulation in mutant strains leads to activation of many stress resistance genes, including components of the immune system such as antimicrobial peptides and ROS-protective antioxidant enzymes (Murphy et al. 2003; McElwee et al. 2004), ultimately leading to high pathogen resistance (Garsin et al. 2003; Chavez et al. 2007; Hasshoff et al. 2007).
Aspects of mating are expected to be of special importance, since they have a direct effect on fitness (i.e. reproductive success). We would like to point to the following three aspects:
Based on Bateman's principle, the two sexes have different evolutionary interests, and therefore they appear to vary in their investment in immune defence (reviewed in Rolff 2002; Nunn et al. 2009). The underlying mechanisms are currently unknown.
Mate choice may serve to optimize offspring immunity, either by choice of immunocompetent males displaying costly ornaments or by choice of males with complementary immune genes, e.g. for MHC receptor loci. In vertebrates, the relationship between immunocompetence and male ornaments could possibly be mediated by testosterone (see discussion in Nunn et al. 2009). In insects, this relationship could be influenced by the prophenoloxidase cascade, which contributes to both immunity (e.g. the melanization response, production of cytotoxic compounds; Cerenius et al. 2008) and epigamic traits (Siva-Jothy 2000). Choice of mate partners with complementary immunity alleles was shown in sticklebacks to be mediated by chemosensory perception of the diversity of peptides bound to MHC receptors (Milinski et al. 2005).
Since finding a mate as well as all the following steps finally leading to the fertilization of eggs is energetically costly, we also expect a direct trade-off for resources between mating and immunity. Such a trade-off may be mediated by testosterone in the vertebrates (Folstad & Karter 1992) and juvenile hormone in insects (Rolff & Siva-Jothy 2002).
(d) Community immunity
Organisms do not live in isolation, but are usually part of a community. If composition of the community is comparatively stable over time, selection should favour comprehensive co-adaptations among its members, which may also extend to immune defence. Such stability is most convincing for social organisms. Indeed, animal societies have evolved several mechanisms to protect the community from parasite attack, ranging from specific behaviours against parasite-rich material or infected members of the society, structure and organization of the nest, as well as a systemic society-wide activation of physiological immunity, as discussed in this issue's review by Cremer & Sixt (2009; see also Cremer et al. 2007). The underlying molecular mechanisms for such social immunity are as yet unexplored. Considering that humans also evolved to live within social groups, detailed understanding of these processes will be of considerable medical interest.
We further expect the relationship between multicellular organisms and their associated microbes to be of particular importance for immune defence as well as a particular challenge for future research (box 1). In humans, the adult body comprises 10 times more microbial cells than human cells, mainly due to the large number of microbes in the gastrointestinal tract (more than 1010 cells ml−1 intestinal lumen; see O'Hara & Shanahan 2006). If the associations are long lasting, then evolution of mutualistic interactions should be advantageous. In fact, the human microbial ecosystem contributes to numerous body functions, e.g. nutrient processing, regulation of fat storage and also defence against pathogens (reviewed in Backhed et al. 2005; O'Hara & Shanahan 2006; Sansonetti & Di Santo 2007). Similar relationships appear to be widespread among other animal hosts (Oliver et al. 2003; Scarborough et al. 2005; Fraune & Bosch 2007; Grozdanov & Hentschel 2007; Ikeda-Ohtsubo et al. 2007; Ley et al. 2008). Since the community of a host and its symbionts can essentially be viewed as an ecosystem, ecological immunologists should be in the ‘pole position’ to study such interactions (box 1).
Box 1. The role of associated microbes for evolution of immunity.
How does the immune system distinguish between ‘friend’ and ‘foe’? Although this is one of the oldest questions for immunologists, there is now fresh impetus from ecological studies, which have directed attention to hosts that are commonly disregarded by immunological research. A prime example is the highly specialized system of the squid light organ and its symbiont Vibrio fischeri (McFall-Ngai & Ruby 1991; Nyholm & McFall-Ngai 2004). Based on recent technological innovations, it is now possible to characterize the genetic underpinnings of such host–microbe interactions, including full genome sequencing (Ruby et al. 2005). In this mutualistic symbiosis, Vibrio releases molecular patterns that have long been known as immune response-inducing pathogen-associated molecular patterns (PAMPs), e.g. peptidoglycan and lipopolysaccharide. In this case, however, these molecules trigger the establishment of a mutually beneficial symbiosis (Koropatnick et al. 2004). These findings have modified our view of the patterns and receptors regulating host–symbiont relationships, and led to the distinction of PAMPs and context-dependent microbe-associated molecular patterns.
More generally, most animal bodies seem inhabited by a hitherto underestimated diversity of microbes, especially in the gut (e.g. O'Hara & Shanahan 2006; Dethlefsen et al. 2007; Ley et al. 2008). How the immune system manages to learn which of those are harmful and which are not is still a puzzle. It may well be that the necessity to evaluate, and then appropriately respond to, this microbial diversity contributed to the evolution of the adaptive immune system or the repertoire of antimicrobial peptides (e.g. McFall-Ngai 2007; Anselme et al. 2008; see also Reynolds & Rolff 2008). At the same time, these associated microbes appear to play an important direct role in pathogen defence (e.g. Oliver et al. 2003; Scarborough et al. 2005; O'Hara & Shanahan 2006; Sansonetti & Di Santo 2007), including, among others, the development of the B-cell repertoire (reviewed in Lanning et al. 2005). The underlying molecular genetics of microbe-mediated immunity are just being addressed in research. One recent example demonstrates that a single molecule, polysaccharide A, from the intestinal symbiont Bacteroides fragilis, acts as an anti-inflammatory factor, thereby preventing the development of Helicobacter pylori-induced disease (Mazmanian et al. 2008).
Dealing with communities of organisms and explaining their relationships is the core competence of ecologists. Moreover, evolutionary biologists may give further insights by explaining the evolution of such associations. Here, the community of a host and all its symbionts' genomes may be viewed as a ‘hologenome’, such that selection shapes the genomes of the symbionts faster than the genome of the longer lived host, an ecological view originally brought up for corals and their symbionts, facing the risk of coral bleaching (Rosenberg et al. 2007). Since it is more widely applicable, this view might revolutionize our current understanding of evolution and immunity. Molecular immunologists may then help us understand how this community of organisms interacts and coordinates important life-history functions such as defence.
4. Conclusions
An organism's immune defence is an extraordinarily complex, continuously evolving system. It is characterized by high levels of diversity, redundancy and mechanisms for life-history integration. Consequently, a full functional and mechanistic understanding will require consideration of the processes that determine its evolution, i.e. its ecological context. Similarly, the dynamics of ecological processes in natural populations, including those related to host–parasite interactions, depend on molecular genetic mechanisms such as those related to host immunity. Consequently, the merger of these disciplines is likely to be very instructive for practitioners in both parental fields.
Ecological immunology is a young field producing exciting empirical and conceptual insights into the way physiology and ecology interact in the context of disease evolution and host population responses. It clearly has a raft of applied implications, but is also exciting because it is one of the few fields that allows evolutionary ecologists to open up a well-studied black box and begin to understand the evolutionary context of a hugely important functional system. At a more proximate level, ecological immunology will help us put a meaning to the ever-expanding banks of comprehensive—omics data from both laboratory and field environments. Our intention with this review and the themed issue is to provide a framework of the ecological processes and immunological consequences thereof which are of importance in this context.
Acknowledgments
We are very grateful to all contributors to this issue for stimulating discussions and advice, two referees for valuable comments on the manuscript and Claire Rawlinson and James Joseph for editorial support. H.S. acknowledges support from the German Science Foundation (grant SCHU 1415/5-1) and the Wissenschaftskolleg zu Berlin, J.K. from the Swiss National Foundation (3100A0-112992) and the Wissenschaftskolleg zu Berlin and Y.M. from the Centre National de la Recherche Scientifique (CNRS).
Footnotes
One contribution of 11 to a Theme Issue ‘Ecological immunology’.
References
- Ahmed A.M., Hurd H. Immune stimulation and malaria infection impose reproductive costs in Anopheles gambiae via follicular apoptosis. Microbes Infect. 2006;8:308–315. doi: 10.1016/j.micinf.2005.06.026. doi:10.1016/j.micinf.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Anselme, C., Perez-Brocal, V., Vallier, A., Vincent-Monegat, C., Charif, D., Latorre, A., Moya, A. & Heddi, A. 2008 Identification of the Weevil immune genes and their expression in the bacteriome tissue. BMC Biol.6 (doi:10.1186/1741-7007-6-43) [DOI] [PMC free article] [PubMed]
- Antia R., Levin B.R., May R.M. Within-host population dynamics and the evolution and maintenance of microparasite virulence. Am. Nat. 1994;144:457–472. doi:10.1086/285686 [Google Scholar]
- Apanius V., Penn D., Slev P.R., Ruff L.R., Potts W.K. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- Arien K.K., Vanham G., Arts E.J. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev. Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. doi:10.1038/nrmicro1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. doi:10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- Bakker E.G., Toomajian C., Kreitman M., Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. doi:10.1105/tpc.106.042614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangham J., Obbard D.J., Kim K.W., Haddrill P.R., Jiggins F.M. The age and evolution of an antiviral resistance mutation in Drosophila melanogaster. Proc. R. Soc. B. 2007;274:2027–2034. doi: 10.1098/rspb.2007.0611. doi:10.1098/rspb.2007.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangham J., Kim K.W., Webster C.L., Jiggins F.M. Genetic variation affecting host–parasite interactions: different genes affect different aspects of sigma virus replication and transmission in Drosophila melanogaster. Genetics. 2008;178:2191–2199. doi: 10.1534/genetics.107.085449. doi:10.1534/genetics.107.085449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme S., Agnew P., Sidobre C., Michalakis Y. Virulence reaction norms across a food gradient. Proc. R. Soc. B. 2004;271:739–744. doi: 10.1098/rspb.2003.2657. doi:10.1098/rspb.2003.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J., Kreitman M., Stahl E.A., Tian D.C. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. doi:10.1126/science.1061337 [DOI] [PubMed] [Google Scholar]
- Bonhoeffer S., Nowak M.A. Mutation and the evolution of virulence. Proc. R. Soc. B. 1994;258:133–140. doi:10.1098/rspb.1994.0153 [Google Scholar]
- Bonneaud C., Mazuc J., Gonzalez G., Haussy C., Chastel O., Faivre B., Sorci G. Assessing the cost of mounting an immune response. Am. Nat. 2003;161:367–379. doi: 10.1086/346134. doi:10.1086/346134 [DOI] [PubMed] [Google Scholar]
- Boots M., Best A., Miller M.R., White A. The role of ecological feedbacks in the evolution of host defence: what does theory tell us? Phil. Trans. R. Soc. B. 2009;364:27–36. doi: 10.1098/rstb.2008.0160. doi:10.1098/rstb.2008.0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites D., et al. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol. Biol. Evol. 2008;25:1429–1439. doi: 10.1093/molbev/msn087. doi:10.1093/molbev/msn087 [DOI] [PubMed] [Google Scholar]
- Brockhurst M.A., Buckling A., Rainey P.B. Spatial heterogeneity and the stability of host–parasite coexistence. J. Evol. Biol. 2006;19:374–379. doi: 10.1111/j.1420-9101.2005.01026.x. doi:10.1111/j.1420-9101.2005.01026.x [DOI] [PubMed] [Google Scholar]
- Brown S.P. Cooperation and conflict in host-manipulating parasites. Proc. R. Soc. B. 1999;266:1899–1904. doi:10.1098/rspb.1999.0864 [Google Scholar]
- Brown S.P., Hochberg M.E., Grenfell B.T. Does multiple infection select for raised virulence? Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. doi:10.1016/S0966-842X(02)02413-7 [DOI] [PubMed] [Google Scholar]
- Buckling A., Rainey P.B. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. B. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. doi:10.1098/rspb.2001.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerenius L., Lee B.L., Söderhäll K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. doi:10.1016/j.it.2008.02.009 [DOI] [PubMed] [Google Scholar]
- Chavez V., Mohri-Shiomi A., Maadani A., Vega L.A., Garsin D.A. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176:1567–1577. doi: 10.1534/genetics.107.072587. doi:10.1534/genetics.107.072587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T.F., Lenski R.E., Elena S.F. Parasites and mutational load: an experimental test of a pluralistic theory for the evolution of sex. Proc. R. Soc. B. 2005;272:311–317. doi: 10.1098/rspb.2004.2975. doi:10.1098/rspb.2004.2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer S., Sixt M. Analogies in the evolution of individual and social immunity. Phil. Trans. R. Soc. B. 2009;364:129–142. doi: 10.1098/rstb.2008.0166. doi:10.1098/rstb.2008.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer S., Armitage S.A., Schmid-Hempel P. Social immunity. Curr. Biol. 2007;17:R693–R702. doi: 10.1016/j.cub.2007.06.008. doi:10.1016/j.cub.2007.06.008 [DOI] [PubMed] [Google Scholar]
- De Boer R.J., Borghans J.A., van Boven M., Kesmir C., Weissing F.J. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics. 2004;55:725–731. doi: 10.1007/s00251-003-0629-y. doi:10.1007/s00251-003-0629-y [DOI] [PubMed] [Google Scholar]
- Decaestecker E., Gaba S., Raeymaekers J.A., Stoks R., Van Kerckhoven L., Ebert D., De Meester L. Host–parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. doi:10.1038/nature06291 [DOI] [PubMed] [Google Scholar]
- De Roode J.C., et al. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. doi:10.1073/pnas.0500078102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L., McFall-Ngai M., Relman D.A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. doi:10.1038/nature06245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Taylor H.E., Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. doi:10.1371/journal.pbio.0040229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuffet A., Dupas S., Frey F., Drezen J.M., Poirie M., Carton Y. Genetic interactions between the parasitoid wasp Leptopilina boulardi and its Drosophila hosts. Heredity. 2007;98:21–27. doi: 10.1038/sj.hdy.6800893. doi:10.1038/sj.hdy.6800893 [DOI] [PubMed] [Google Scholar]
- Du Pasquier L. Germline and somatic diversification of immune recognition elements in Metazoa. Immunol. Lett. 2006;104:2–17. doi: 10.1016/j.imlet.2005.11.022. doi:10.1016/j.imlet.2005.11.022 [DOI] [PubMed] [Google Scholar]
- Dybdahl M.F., Lively C.M. Host–parasite coevolution: evidence for rare advantage and time-lagged selection in a natural population. Evolution. 1998;52:1057–1066. doi: 10.1111/j.1558-5646.1998.tb01833.x. doi:10.2307/2411236 [DOI] [PubMed] [Google Scholar]
- Ebert D., Haag C., Kirkpatrick M., Riek M., Hottinger J.W., Pajunen V.I. A selective advantage to immigrant genes in a Daphnia metapopulation. Science. 2002;295:485–488. doi: 10.1126/science.1067485. doi:10.1126/science.1067485 [DOI] [PubMed] [Google Scholar]
- Fenner F., Fantini B. CABI Publishing; Wallingford, UK; New York, NY: 1999. Biological control of vertebrate pests. The history of myxomatosis, an experiment in evolution. [Google Scholar]
- Ferdig M.T., Beerntsen B.T., Spray F.J., Li J., Christensen B.M. Reproductive costs associated with resistance in a mosquito-filarial worm system. Am. J. Trop. Med. Hyg. 1993;49:756–762. doi: 10.4269/ajtmh.1993.49.756. [DOI] [PubMed] [Google Scholar]
- Ferguson H.M., Read A.F. Genetic and environmental determinants of malaria parasite virulence in mosquitoes. Proc. R. Soc. B. 2002;269:1217–1224. doi: 10.1098/rspb.2002.2023. doi:10.1098/rspb.2002.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson W., Dvora S., Gallo J., Orth A., Boissinot S. Long-term balancing selection at the West Nile Virus resistance gene, Oas1b, maintains transspecific polymorphisms in the house mouse. Mol. Biol. Evol. 2008;25:1609–1618. doi: 10.1093/molbev/msn106. doi:10.1093/molbev/msn106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., et al. Balancing selection is the main force shaping the evolution of innate immunity genes. J. Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- Folstad I., Karter A.J. Parasites, bright males, and the immunocompetence handicap. Am. Nat. 1992;139:603–622. doi:10.1086/285346 [Google Scholar]
- Forde S.E., Thompson J.N., Bohannan B.J. Adaptation varies through space and time in a coevolving host–parasitoid interaction. Nature. 2004;431:841–844. doi: 10.1038/nature02906. doi:10.1038/nature02906 [DOI] [PubMed] [Google Scholar]
- Frank S.A. Models of parasite virulence. Q. Rev. Biol. 1996;71:37–78. doi: 10.1086/419267. doi:10.1086/419267 [DOI] [PubMed] [Google Scholar]
- Fraune S., Bosch T.C. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl Acad. Sci. USA. 2007;104:13 146–13 151. doi: 10.1073/pnas.0703375104. doi:10.1073/pnas.0703375104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon S., Buckling A., Decaestecker E., Day T. Host–parasite coevolution and patterns of adaptation across time and space. J. Evol. Biol. 2008;21:1861–1866. doi: 10.1111/j.1420-9101.2008.01598.x. doi:10.1111/j.1420-9101.2008.01598.x [DOI] [PubMed] [Google Scholar]
- Garsin D.A., Villanueva J.M., Begun J., Kim D.H., Sifri C.D., Calderwood S.B., Ruvkun G., Ausubel F.M. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. doi:10.1126/science.1080147 [DOI] [PubMed] [Google Scholar]
- Gems D., Sutton A.J., Sundermeyer M.L., Albert P.S., King K.V., Edgley M.L., Larsen P.L., Riddle D.L. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A.L., Allen J.E., Read A.F. Evolutionary causes and consequences of immunopathology. Annu. Rev. Ecol. Evol. Syst. 2005;36:373–397. doi:10.1146/annurev.ecolsys.36.102003.152622 [Google Scholar]
- Grozdanov L., Hentschel U. An environmental genomics perspective on the diversity and function of marine sponge-associated microbiota. Curr. Opin. Microbiol. 2007;10:215–220. doi: 10.1016/j.mib.2007.05.012. doi:10.1016/j.mib.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D., Zuk M. Heritable true fitness and bright birds: a role for parasites. Science. 1982;218:384–387. doi: 10.1126/science.7123238. doi:10.1126/science.7123238 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D., Axelrod R., Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc. Natl Acad. Sci. USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. doi:10.1073/pnas.87.9.3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselquist D., Nilsson J.-A. Maternal transfer of antibodies in vertebrates: trans-generational effects on offspring immunity. Phil. Trans. R. Soc. B. 2009;364:51–60. doi: 10.1098/rstb.2008.0137. doi:10.1098/rstb.2008.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasshoff M., Boehnisch C., Tonn D., Hasert B., Schulenburg H. The role of Caenorhabditis elegans insulin-like signalling in the behavioural avoidance of pathogenic Bacillus thuringiensis. FASEB J. 2007;21:1801–1812. doi: 10.1096/fj.06-6551com. doi:10.1096/fj.06-6551com [DOI] [PubMed] [Google Scholar]
- Houthoofd K., et al. Metabolism, physiology and stress defense in three aging Ins/IGF-1 mutants of the nematode Caenorhabditis elegans. Aging Cell. 2005;4:87–95. doi: 10.1111/j.1474-9726.2005.00150.x. doi:10.1111/j.1474-9726.2005.00150.x [DOI] [PubMed] [Google Scholar]
- Ikeda-Ohtsubo W., Desai M., Stingl U., Brune A. Phylogenetic diversity of ‘Endomicrobia’ and their specific affiliation with termite gut flagellates. Microbiology. 2007;153:3458–3465. doi: 10.1099/mic.0.2007/009217-0. doi:10.1099/mic.0.2007/009217-0 [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Travers P., Walport M., Shlomchik M. 5th edn. Garland Publishing; New York, NY: 2001. Immunobiology. [Google Scholar]
- Jiggins F.M., Kim K.W. Contrasting evolutionary patterns in Drosophila immune receptors. J. Mol. Evol. 2006;63:769–780. doi: 10.1007/s00239-006-0005-2. doi:10.1007/s00239-006-0005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins F.M., Kim K.W. A screen for immunity genes evolving under positive selection in Drosophila. J. Evol. Biol. 2007;20:965–970. doi: 10.1111/j.1420-9101.2007.01305.x. doi:10.1111/j.1420-9101.2007.01305.x [DOI] [PubMed] [Google Scholar]
- Jokela J., Lively C.M., Dybdahl M.F., Fox J.A. Genetic variation in sexual and clonal lineages of a freshwater snail. Biol. J. Linn. Soc. 2003;79:165–181. doi:10.1046/j.1095-8312.2003.00181.x [Google Scholar]
- Kalbe M., Wegner K.M., Reusch T.B.H. Dispersion patterns of parasites in 0+ year three-spined sticklebacks: a cross population comparison. J. Fish Biol. 2002;60:1529–1542. doi:10.1111/j.1095-8649.2002.tb02445.x [Google Scholar]
- Koropatnick T.A., Engle J.T., Apicella M.A., Stabb E.V., Goldman W.E., McFall-Ngai M.J. Microbial factor-mediated development in a host–bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. doi:10.1126/science.1102218 [DOI] [PubMed] [Google Scholar]
- Kraaijeveld A.R., Godfray H.C. Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature. 1997;389:278–280. doi: 10.1038/38483. doi:10.1038/38483 [DOI] [PubMed] [Google Scholar]
- Kurtz J. Memory in the innate and adaptive immune systems. Microbes Infect. 2004;6:1410–1417. doi: 10.1016/j.micinf.2004.10.002. doi:10.1016/j.micinf.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Kurtz J., Armitage S.A. Alternative adaptive immunity in invertebrates. Trends Immunol. 2006;27:493–496. doi: 10.1016/j.it.2006.09.001. doi:10.1016/j.it.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Kurtz J., Franz K. Evidence for memory in invertebrate immunity. Nature. 2003;425:37–38. doi: 10.1038/425037a. doi:10.1038/425037a [DOI] [PubMed] [Google Scholar]
- Kurtz J., Kalbe M., Aeschlimann P.B., Haberli M.A., Wegner K.M., Reusch T.B., Milinski M. Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc. R. Soc. B. 2004;271:197–204. doi: 10.1098/rspb.2003.2567. doi:10.1098/rspb.2003.2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz J., Wegner K.M., Kalbe M., Reusch T.B.H., Schaschl H., Hasselquist D., Milinski M. MHC genes and oxidative stress in sticklebacks: an immuno-ecological approach. Proc. R. Soc. B. 2006;273:1407–1414. doi: 10.1098/rspb.2005.3450. doi:10.1098/rspb.2005.3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L., Chavatte J.M., Snounou G., Koella J.C. Environmental influence on the genetic basis of mosquito resistance to malaria parasites. Proc. R. Soc. B. 2006;273:1501–1506. doi: 10.1098/rspb.2006.3483. doi:10.1098/rspb.2006.3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning D.K., Rhee K.J., Knight K.L. Intestinal bacteria and development of the B-lymphocyte repertoire. Trends Immunol. 2005;26:419–425. doi: 10.1016/j.it.2005.06.001. doi:10.1016/j.it.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Lazzaro B.P., Little T.J. Immunity in a variable world. Phil. Trans. R. Soc. B. 2009;364:15–26. doi: 10.1098/rstb.2008.0141. doi:10.1098/rstb.2008.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B.P., Sceurman B.K., Clark A.G. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. doi:10.1126/science.1092447 [DOI] [PubMed] [Google Scholar]
- Lazzaro B.P., Sackton T.B., Clark A.G. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics. 2006;174:1539–1554. doi: 10.1534/genetics.105.054593. doi:10.1534/genetics.105.054593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B.P., Flores H.A., Lorigan J.G., Yourth C.P. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 2008;4:e1000025. doi: 10.1371/journal.ppat.1000025. doi:10.1371/journal.ppat.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. doi:10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little T.J., O'Connor B., Colegrave N., Watt K., Read A.F. Maternal transfer of strain-specific immunity in an invertebrate. Curr. Biol. 2003;13:489–492. doi: 10.1016/s0960-9822(03)00163-5. doi:10.1016/S0960-9822(03)00163-5 [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. doi:10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- McElwee J.J., Schuster E., Blanc E., Thomas J.H., Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004;279:44 533–44 543. doi: 10.1074/jbc.M406207200. doi:10.1074/jbc.M406207200 [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. doi:10.1038/445153a [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M.J., Ruby E.G. Symbiont recognition and subsequent morphogenesis as early events in an animal–bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. doi:10.1126/science.1962208 [DOI] [PubMed] [Google Scholar]
- McKean K.A., Yourth C.P., Lazzaro B.P., Clark A.G. The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 2008;8:76. doi: 10.1186/1471-2148-8-76. doi:10.1186/1471-2148-8-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menge D.M., Zhong D., Guda T., Gouagna L., Githure J., Beier J., Yan G. Quantitative trait loci controlling refractoriness to Plasmodium falciparum in natural Anopheles gambiae mosquitoes from a malaria-endemic region in western Kenya. Genetics. 2006;173:235–241. doi: 10.1534/genetics.105.055129. doi:10.1534/genetics.105.055129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 2006;37:159–186. doi:10.1146/annurev.ecolsys.37.091305.110242 [Google Scholar]
- Milinski M., Griffiths S., Wegner K.M., Reusch T.B.H., Haas-Assenbaum A., Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA. 2005;102:4414–4418. doi: 10.1073/pnas.0408264102. doi:10.1073/pnas.0408264102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.E., Rogers E.S., Little T.J., Read A.F. Host–parasite and genotype-by-environment interactions: temperature modifies potential for selection by a sterilizing pathogen. Evolution. 2005;59:70–80. doi:10.1554/04-526 [PubMed] [Google Scholar]
- Mitchell-Olds T., Willis J.H., Goldstein D.B. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 2007;8:845–856. doi: 10.1038/nrg2207. doi:10.1038/nrg2207 [DOI] [PubMed] [Google Scholar]
- Mohri-Shiomi A., Garsin D.A. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J. Biol. Chem. 2008;283:194–201. doi: 10.1074/jbc.M707956200. doi:10.1074/jbc.M707956200 [DOI] [PubMed] [Google Scholar]
- Moore J. Oxford University Press; Oxford, UK: 2002. Parasites and the behaviour of animals. [Google Scholar]
- Moret Y. ‘Trans-generational immune priming’: specific enhancement of the antimicrobial immune response in the mealworm beetle, Tenebrio molitor. Proc. R. Soc. B. 2006;273:1399–1405. doi: 10.1098/rspb.2006.3465. doi:10.1098/rspb.2006.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret Y., Siva-Jothy M.T. Adaptive innate immunity? Responsive-mode prophylaxis in the mealworm beetle, Tenebrio molitor. Proc. R. Soc. B. 2003;270:2475–2480. doi: 10.1098/rspb.2003.2511. doi:10.1098/rspb.2003.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A.D., Gandon S., Buckling A. The effect of migration on local adaptation in a coevolving host–parasite system. Nature. 2005;437:253–256. doi: 10.1038/nature03913. doi:10.1038/nature03913 [DOI] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. doi:10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Nelson M.I., Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. doi:10.1038/nrg2053 [DOI] [PubMed] [Google Scholar]
- Nowak M.A., May R.M. Superinfection and the evolution of parasite virulence. Proc. R. Soc. B. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. doi:10.1098/rspb.1994.0012 [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Lindenfors P., Pursall E.R., Rolff J. On sexual dimorphism in immune function. Phil. Trans. R. Soc. B. 2009;364:61–69. doi: 10.1098/rstb.2008.0148. doi:10.1098/rstb.2008.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm S.V., McFall-Ngai M.J. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. doi:10.1038/nrmicro957 [DOI] [PubMed] [Google Scholar]
- Obbard D.J., Jiggins F.M., Halligan D.L., Little T.J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. doi:10.1016/j.cub.2006.01.065 [DOI] [PubMed] [Google Scholar]
- Obbard D.J., Gordon K.H.J., Buck A.H., Jiggins F.M. The evolution of RNAi as a defence against viruses and transposable elements. Phil. Trans. R. Soc. B. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. doi:10.1098/rstb.2008.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. doi:10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K.M., Russell J.A., Moran N.A., Hunter M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl Acad. Sci. USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. doi:10.1073/pnas.0335320100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D.J., Potts W.K. The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 1999;153:145–164. doi: 10.1086/303166. doi:10.1086/303166 [DOI] [PubMed] [Google Scholar]
- Pham L.N., Dionne M.S., Shirasu-Hiza M., Schneider D.S. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. doi:10.1371/journal.ppat.0030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E., Zhang Y., Pujol N., Matsuyama T., Bargmann C.I., Ewbank J.J. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. doi:10.1073/pnas.0610281104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Råberg L., Sim D., Read A.F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. doi:10.1126/science.1148526 [DOI] [PubMed] [Google Scholar]
- Råberg L., Graham A.L., Read A.F. Decomposing health: tolerance and resistance to parasites in animals. Phil. Trans. R. Soc. B. 2009;364:37–49. doi: 10.1098/rstb.2008.0184. doi:10.1098/rstb.2008.0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch G., Kalbe M., Reusch T.B.H. Partitioning average competition and extreme-genotype effects in genetically diverse infections. Oikos. 2008;117:399–405. doi:10.1111/j.2007.0030-1299.16301.x [Google Scholar]
- Reusch T.B., Langefors A. Inter- and intralocus recombination drive MHC class IIB gene diversification in a teleost, the three-spined stickleback Gasterosteus aculeatus. J. Mol. Evol. 2005;61:531–541. doi: 10.1007/s00239-004-0340-0. doi:10.1007/s00239-004-0340-0 [DOI] [PubMed] [Google Scholar]
- Reusch T.B.H., Häberli M.A., Aeschlimann P.B., Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. doi:10.1038/35104547 [DOI] [PubMed] [Google Scholar]
- Reynolds, S. & Rolff, J. 2008 Immune function keeps endosymbionts under control. J. Biol 7, (doi:10.1186/jbiol88) [DOI] [PMC free article] [PubMed]
- Riehle M.M., et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. doi:10.1126/science.1124153 [DOI] [PubMed] [Google Scholar]
- Rolff J. Bateman's principle and immunity. Proc. R. Soc. B. 2002;269:867–872. doi: 10.1098/rspb.2002.1959. doi:10.1098/rspb.2002.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolff J., Siva-Jothy M.T. Copulation corrupts immunity: a mechanism for a cost of mating in insects. Proc. Natl Acad. Sci. USA. 2002;99:9916–9918. doi: 10.1073/pnas.152271999. doi:10.1073/pnas.152271999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. doi:10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Roth O., Sadd B.M., Schmid-Hempel P., Kurtz J. Strain-specific priming of resistance in the red flour beetle, Tribolium castaneum. Proc. R. Soc. B. 2008;276:145–151. doi: 10.1098/rspb.2008.1157. doi:10.1098/rspb.2008.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby E.G., et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. doi:10.1073/pnas.0409900102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T.B., Lazzaro B.P., Schlenke T.A., Evans J.D., Hultmark D., Clark A.G. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. doi:10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- Sadd B.M., Schmid-Hempel P. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr. Biol. 2006;16:1206–1210. doi: 10.1016/j.cub.2006.04.047. doi:10.1016/j.cub.2006.04.047 [DOI] [PubMed] [Google Scholar]
- Sadd B.M., Siva-Jothy M.T. Self-harm caused by an insect's innate immunity. Proc. R. Soc. B. 2006;273:2571–2574. doi: 10.1098/rspb.2006.3574. doi:10.1098/rspb.2006.3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd B.M., Kleinlogel Y., Schmid-Hempel R., Schmid-Hempel P. Trans-generational immune priming in a social insect. Biol. Lett. 2005;1:386–388. doi: 10.1098/rsbl.2005.0369. doi:10.1098/rsbl.2005.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvaudon L., Giraud T., Shykoff J.A. Genetic diversity in natural populations: a fundamental component of plant–microbe interactions. Curr. Opin. Plant Biol. 2008;11:135–143. doi: 10.1016/j.pbi.2008.02.002. doi:10.1016/j.pbi.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Sansonetti P.J., Di Santo J.P. Debugging how bacteria manipulate the immune response. Immunity. 2007;26:149–161. doi: 10.1016/j.immuni.2007.02.004. doi:10.1016/j.immuni.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Scarborough C.L., Ferrari J., Godfray H.C. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. doi:10.1126/science.1120180 [DOI] [PubMed] [Google Scholar]
- Scharsack J.P., Kalbe M., Harrod C., Rauch G. Habitat-specific adaptation of immune responses of stickleback (Gasterosteus aculeatus) lake and river ecotypes. Proc. R. Soc. B. 2007;274:1523–1532. doi: 10.1098/rspb.2007.0210. doi:10.1098/rspb.2007.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjorring S., Koella J.C. Sub-lethal effects of pathogens can lead to the evolution of lower virulence in multiple infections. Proc. R. Soc. B. 2003;270:189–193. doi: 10.1098/rspb.2002.2233. doi:10.1098/rspb.2002.2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. Variation in immune defence as a question of evolutionary ecology. Proc. R. Soc. B. 2003;270:357–366. doi: 10.1098/rspb.2002.2265. doi:10.1098/rspb.2002.2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. Natural insect host–parasite systems show immune priming and specificity: puzzles to be solved. Bioessays. 2005;27:1026–1034. doi: 10.1002/bies.20282. doi:10.1002/bies.20282 [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P. Parasite immune evasion: a momentous molecular war. Trends Ecol. Evol. 2008;23:318–326. doi: 10.1016/j.tree.2008.02.011. doi:10.1016/j.tree.2008.02.011 [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P. Immune defence, parasite evasion strategies and their relevance for ‘macroscopic phenomena’ such as virulence. Phil. Trans. R. Soc. B. 2009;364:85–98. doi: 10.1098/rstb.2008.0157. doi:10.1098/rstb.2008.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., Ewbank J.J. The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol. 2007;66:563–570. doi: 10.1111/j.1365-2958.2007.05946.x. doi:10.1111/j.1365-2958.2007.05946.x [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Müller S. Natural variation in the response of Caenorhabditis elegans towards Bacillus thuringiensis. Parasitology. 2004;128:433–443. doi: 10.1017/s003118200300461x. doi:10.1017/S003118200300461X [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Boehnisch C., Michiels N.K. How do invertebrates generate a highly specific innate immune response? Mol. Immunol. 2007;44:3338–3344. doi: 10.1016/j.molimm.2007.02.019. doi:10.1016/j.molimm.2007.02.019 [DOI] [PubMed] [Google Scholar]
- Sheldon B.C., Verhulst S. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 1996;11:317–321. doi: 10.1016/0169-5347(96)10039-2. doi:10.1016/0169-5347(96)10039-2 [DOI] [PubMed] [Google Scholar]
- Sicard M., Hering S., Schulte R., Gaudriault S., Schulenburg H. The effect of Photorhabdus luminescens (Enterobacteriaceae) on the survival, development, reproduction and behaviour of Caenorhabditis elegans (Nematoda: Rhabditidae) Environ. Microbiol. 2007;9:12–25. doi: 10.1111/j.1462-2920.2006.01099.x. doi:10.1111/j.1462-2920.2006.01099.x [DOI] [PubMed] [Google Scholar]
- Siva-Jothy M.T. A mechanistic link between parasite resistance and expression of a sexually selected trait in a damselfly. Proc. R. Soc. B. 2000;267:2523–2527. doi: 10.1098/rspb.2000.1315. doi:10.1098/rspb.2000.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva-Jothy M.T., Moret Y., Rolff J. Insect immunity: an evolutionary ecology perspective. Adv. Insect Physiol. 2005;32:1–48. [Google Scholar]
- Sorci G., Faivre B. Inflammation and oxidative stress in vertebrate host–parasite systems. Phil. Trans. R. Soc. B. 2009;364:71–83. doi: 10.1098/rstb.2008.0151. doi:10.1098/rstb.2008.0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T., Fenton A., Brockhurst M.A. The impact of parasite dispersal on antagonistic host–parasite coevolution. J. Evol. Biol. 2008;21:1252–1258. doi: 10.1111/j.1420-9101.2008.01574.x. doi:10.1111/j.1420-9101.2008.01574.x [DOI] [PubMed] [Google Scholar]
- von Burg S., Ferrari J., Muller C.B., Vorburger C. Genetic variation and covariation of susceptibility to parasitoids in the aphid Myzus persicae: no evidence for trade-offs. Proc. R. Soc. B. 2008;275:1089–1094. doi: 10.1098/rspb.2008.0018. doi:10.1098/rspb.2008.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson F.L., Püttmann-Holgado R., Thomas F., Lamar D.L., Hughes M., Kondo M., Rebel V.I., Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. doi:10.1126/science.1116887 [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Kalbe M., Kurtz J., Reusch T.B., Milinski M. Parasite selection for immunogenetic optimality. Science. 2003a;301:1343. doi: 10.1126/science.1088293. doi:10.1126/science.1088293 [DOI] [PubMed] [Google Scholar]
- Wegner K.M., Reusch T.B., Kalbe M. Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 2003b;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. doi:10.1046/j.1420-9101.2003.00519.x [DOI] [PubMed] [Google Scholar]
- Wilfert L., Schmid-Hempel P. The genetic architecture of susceptibility to parasites. BMC Evol. Biol. 2008;8:187. doi: 10.1186/1471-2148-8-187. doi:10.1186/1471-2148-8-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert L., Gadau J., Baer B., Schmid-Hempel P. Natural variation in the genetic architecture of a host–parasite interaction in the bumblebee Bombus terrestris. Mol. Ecol. 2007;16:1327–1339. doi: 10.1111/j.1365-294X.2007.03234.x. doi:10.1111/j.1365-294X.2007.03234.x [DOI] [PubMed] [Google Scholar]
- Woelfing B., Traulsen A., Milinski M., Boehm T. Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil. Trans. R. Soc. B. 2009;364:117–128. doi: 10.1098/rstb.2008.0174. doi:10.1098/rstb.2008.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E., Webster J.P., Domingo E., Charlesworth B., Levin B.R. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. doi:10.1038/ng1202-569 [DOI] [PubMed] [Google Scholar]
- Yan G., Christensen B.M., Severson D.W. Comparisons of genetic variability and genome structure among mosquito strains selected for refractoriness to a malaria parasite. J. Hered. 1997;88:187–194. doi: 10.1093/oxfordjournals.jhered.a023087. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu H., Bargmann C.I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. doi:10.1038/nature04216 [DOI] [PubMed] [Google Scholar]