Abstract
Purpose
Myopia (shortsightedness) is one of the most common ocular conditions worldwide and results in blurred distance vision. It is a complex trait influenced by both genetic and environmental factors. We have previously reported linkage of myopia to a 13.01 cM region of chromosome 2q37 in three large multigenerational Australian families that initially overlapped with the known myopia locus, MYP12. The purpose of this study was to perform fine mapping of this region and identify single nucleotide polymorphisms (SNPs) associated with myopia.
Methods
Fine mapping linkage analysis was performed on three multigenerational families with common myopia to refine the previously mapped critical interval. SNPs in the region were also genotyped to assess for association with myopia using an independent case-control cohort.
Results
The disease interval was refined to a 1.83 cM region that is adjacent to rather than overlapping with the MYP12 locus. Subsequent sequencing of all known and hypothetical genes as well as an association study using an independent myopia case-control cohort showed suggestive but not statistically significant association to two intronic SNPs.
Conclusions
We have identified a novel locus for common myopia on chromosome 2q37.
Introduction
Shortsightedness is one of the most common eye conditions clinically manifesting as blurred distance vision. It results when light rays entering the eye are focused in front of rather than on the retina. Prevalence rates for myopia range from 20%–25% in Western countries to over 80% in the urbanized populations of Singapore, China, Japan, and Taiwan [1-6]. Myopia can be clinically defined using spherical equivalent (SphE) measurements that are quantitated using diopter (D) measurements. Individuals having readings of −0.5 D or less in both eyes are considered to be myopic [7]. Clinically, myopia can be classified as common (SphE ≤−0.5 to <−6 D) or high grade (SphE ≤−6 D). The more severe grades of myopia are associated with an increased risk of sight threatening complications such as glaucoma, retinal changes, and cataract [7,8]. Due to the high prevalence of myopia worldwide together with the increased risk of visual morbidity from associated complications, myopia is a significant public health problem. As a consequence, the search for risk factors involved in myopia is paramount if we are to gain insights into its pathogenesis and reduce its burden on health.
Myopia is a complex trait influenced by both genetic and environmental factors [9]. Environmental factors such as reading (near work) are known to influence the development of myopia but appear to account for only 12% of the observed phenotypic variance [10]. The remaining influences on myopia have been suggested to relate to genes as heritability studies indicate that between 50%–94% of population variance is accounted for by genetic factors [11-15]. In support of these heritability studies, familial correlation studies have shown that children with myopic parents have a four times greater risk of developing myopia than children with non-myopic parents [16]. In addition, genetic mapping studies have identified at least 18 chromosomal regions suspected of harboring a myopia gene (MYP1–MYP18). Of these regions, 11 have been implicated in high myopia (MYP1– MYP5, MYP11, MYP12, MYP13, MYP15, MYP16, MYP18) [17-28] and seven in common myopia (MYP6–MYP10, MYP14, MYP17) [29-31]. Five of these loci (MYP2, MYP3, MYP6, MYP10, MYP13) have been confirmed through replication studies in independent family studies [32-38]. Recently, we reported replication of the MYP12 locus using three multigenerational Australian families [39]. The MYP12 locus was initially reported as harboring a gene for high grade myopia whereas our replication study indicated that this locus may also harbor a gene for milder forms of myopia.
We now present fine mapping data for these three Australian myopia families that has refined the mapped myopia interval to a region on chromosome 2q37 that is adjacent to but not overlapping the MYP12 locus. Our findings suggest this is a novel locus for myopia that is distinct from MYP12. Furthermore, we have undertaken an independent association study to identify potential genetic variants that may be associated with myopia.
Methods
Subjects
The three families used in this study were recruited as part of the Genes In Myopia (GEM) Study (Melbourne, Australia), and details regarding their ocular examination have been previously reported [40]. These families had previously undergone a 10 cM genome-wide scan that identified a myopia susceptibility locus to chromosome 2q37 [39]. DNA from all consenting family members was collected from venous blood samples as previously described [41]. For the current study, we defined myopic individuals as family members with spherical equivalent refraction equal to or less than −0.5 diopter sphere (DS) in both eyes and controls as family members with measurements greater than −0.5 DS in both eyes [42-47].
For the case-control association study, we used unrelated individuals also recruited through the GEM Study [40], the Blue Mountains Eye Study (BMES) [48], and the Melbourne Visual Impairment Project (VIP) [49]. Individuals with a history of eye diseases such as keratoconus, age related macular degeneration (AMD), or a history of genetic disorders known to predispose to myopia such as Stickler syndrome or Marfan syndrome were excluded. Using the same definition of what constituted myopia as used in the linkage study, we analyzed 300 myopic and 291 control individuals in a case-control cohort.
This study adhered to the tenets of the Declaration of Helsinki and was approved by the Royal Victoria Eye and Ear Hospital (Melbourne, Australia) and Westmead Hospital at the University of Sydney (Sydney, Australia). All individuals were of Caucasian descent and provided informed written consent before commencement of the study.
Fine mapping linkage analysis
For fine mapping linkage analysis, the starting interval was defined by critical recombination events observed in the three families during the genome-wide linkage analysis [39]. This interval spanned markers D2S396 and D2S338 on chromosome 2q37. For fine mapping, polymorphic microsatellite markers spanning this mapped interval were chosen from the deCODE genetic maps [50]. Genotyping was undertaken by the Australian Genome Research Facility (AGRF; Melbourne, Australia) using a model 377 automated DNA sequencer (Applied Biosystems, Melbourne, Australia). All available family members were included in the genotyping. Genotype error checking was performed using PedManager version 0.9. Multipoint parametric and non-parametric linkage analyses were performed using MERLIN (version 1.1.2) [51]. In the case of parametric linkage analysis, two autosomal dominant models were used with phenocopy rates of 0.1 (model 1) or 0.2 (model 2), a penetrance of 0.9, and disease allele frequency of 0.0133. The choice of these models was based on those described by Chen et al. [15] in the initial linkage paper for these families. Haplotypes were generated using MERLIN (version 1.1.2) [51] and visualized using HaploPainter (version 027beta) [52].
Candidate gene sequencing in the fine mapped region
Identification of all known and hypothetical genes in the refined interval was achieved by mining Ensembl [53], National Centre for Biotechnology Information (NCBI), and University of California Santa Cruz (UCSC) Genome Browser [54]. Bidirectional DNA sequencing was undertaken for all predicted and known exonic regions of these genes and at least 20 bp into each adjacent intron. DNA sequencing was performed in six myopic individuals (two randomly chosen from each of the three families) as well as six unrelated controls (married-in individuals). Primer design, template amplification, and DNA sequencing was performed by the Australian Genome Research Facility (Brisbane, Australia). Sequencing data was visualized using Chromas Lite (version 2.01) and ClustalX (version 2.0) [55] to identify any variations from the reference sequence deposited in the Ensembl database (reference sequence) [53]. The reference sequence for the known single nucleotide polymorphisms (SNPs) was defined by the common allele for the CEU (Utah residents with ancestry from northern and western Europe) population.
Following candidate gene screening, a subset of SNPs of interest were identified and chosen for further analysis in an independent case-control association study. SNPs that were present in only one of the six samples were not present in each of the three families or were present in an equal number of myopic individuals and controls were excluded from subsequent genotype analysis.
Case-control association study
The subset of SNPs that fulfilled the above criteria was genotyped in an independent cohort of unrelated individuals of Caucasian descent. A total of 300 individuals with myopia and 291 controls were genotyped at the Australian Genome Research Facility (Brisbane, Australia) using the MassArray platform (Sequenom Inc., San Diego, CA) and matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) analysis (Sequenom, San Diego, CA). To provide more extensive coverage of intronic and intergenic regions, additional SNPs that were evenly spaced across the entire refined linkage interval were identified using HapMap (Build #36) [56] and also genotyped in the case-control cohort.
Genotyping data was assessed for deviations from the Hardy–Weinberg equilibrium using PLINK [57]. Any SNPs not passing this test (p<0.05 in the controls) were noted and excluded from further analysis. Association tests were also performed using PLINK, and adjustments for multiple testing were made using the Bonferroni correction. Prospective power calculations were performed using Episheet.
Results
Fine mapping linkage analysis
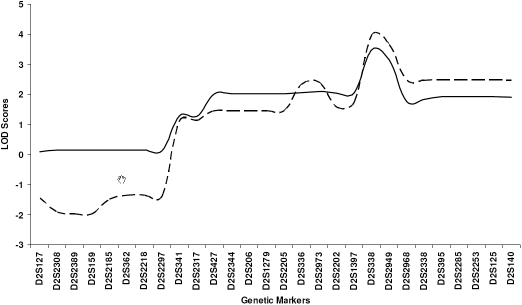
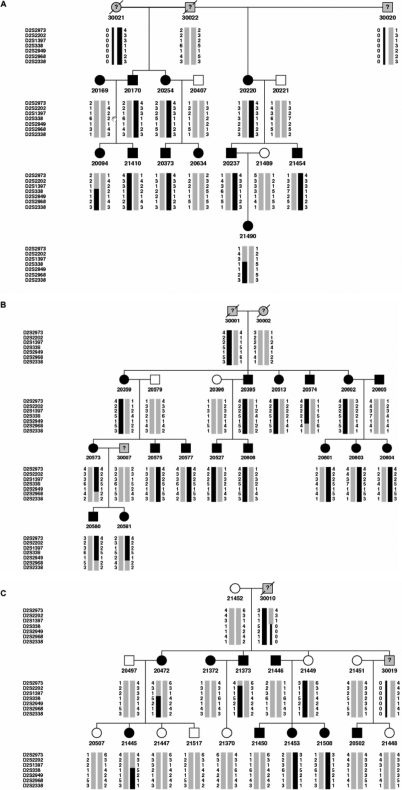
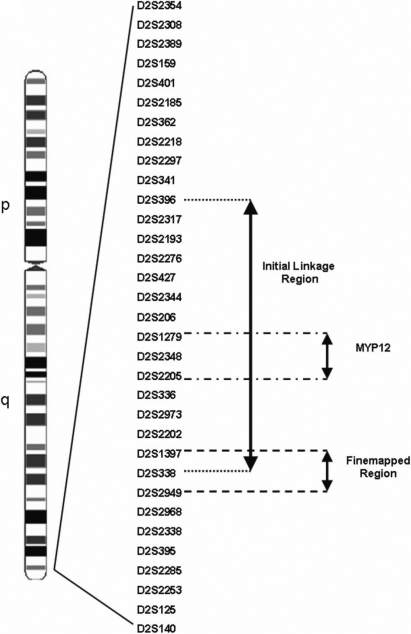
Previous linkage mapping in three myopia families (GEMF0046, GEMF0206, and GEMF0251) identified a 13.01 cM region at 2q37.1 that overlapped with the high myopia locus, MYP12 [39]. A total of 37 myopic and 14 control individuals (including three additional myopic individuals that were not previously available) from these three families were used for fine mapping. Fine mapping of this region resulted in a peak parametric and non-parametric LOD score of 3.97 and 3.48, respectively, at marker D2S338 (Figure 1). This was in agreement with the location of the peak LOD scores reported from our initial genome-wide scan. Haplotype analysis of this region and adjacent regions (6.02 cM proximal and 11.06 cM distal) using 30 polymorphic microsatellite markers at an average spacing of 1.2 cM in the families allowed significant narrowing of the myopia disease gene region (Figure 2). A critical recombination event in individuals 20573, 20580, and 20581 in GEMF0206 (Figure 2A) defined the distal end of the interval to marker D2S2968. A critical recombination event in individuals 20094 and 21490 from GEMF0046 (Figure 2B) and in individuals 21445 and 20472 from GEMF0251 (Figure 2C) defined the proximal end of the refined critical interval to marker D2S1397. The refined interval was identified as a 1.83 cM region on chromosome 2q37.2-2q37.3 that was distal to but did not overlap with the adjacent MYP12 locus (Figure 3).
Figure 1.
LOD Scores for the fine mapping linkage analysis on chromosome 2q37. Parametric (solid line) and nonparametric (dashed line) LOD scores are shown for the three GEM families (GEM0046, GEM0251, and GEM0206).
Figure 2.
Haplotype analysis in the chromosome 2q37 linkage region for the three GEM families. Haplotype analysis in the chromosome 2q37 linkage region is shown for GEM0206 (A), GEM0046 (B), and GEM0251 (C).
Figure 3.
Ideogram of human chromosome 2 showing the location of the newly fine mapped region relative to the original linkage region and MYP12. Fine mapping linkage analysis now clearly indicates that the region of interest is adjacent to but does not overlap MYP12.
Candidate gene sequencing
To identify a potential causative myopia gene or variant within the refined linkage interval, we identified all known and hypothetical genes in this 1.83 cM interval. A total of eight hypothetical (BX647589, AK00798, AK056246, AK023507, LOC728074, LOC728087, LOC728067, Q9UFF6) and six known (ankyrin repeat and SOCS box-containing [ASB18], centaurin gamma 2 [CENTG2], COP9 constitutive photomorphogenic homolog subunit 8 [Arabidopsis; COPS8], chemokine (C-X-C) motif receptor 7 [CXCR7], IQ motif containing with AAA domain [IQCA], and gastrulation brain homeobox 2 [GBX2]) genes were identified.
DNA sequencing was undertaken on all 67 exons and exon-intron boundaries in the known and hypothetical genes that were localized to the refined linkage interval. The intron-exon structure for the hypothetical genes was defined by aligning predicted mRNA sequences with genomic DNA using the BLAST algorithm. The exons were divided into 109 amplicons and sequenced bidirectionally in six myopic individuals from the three GEM families and six controls (no myopia). Overall sequencing success was 94% with seven amplicons located in genes CXCR7 (exon 1 and 2), IQCA (exon 17), and GBX2 (exon 1) proving difficult to sequence. From the successfully sequenced amplicons, a total of 77 SNPs were identified where at least one individual had a genotype that varied from the Ensembl reference sequence. Through the use of our exclusion criteria listed under Methods, this list of SNPs was narrowed to a total of 38. To allow for complete SNP coverage of the refined interval, we also identified all the known HapMap (Build 23a) SNPs located in the difficult to sequence amplicons and added these to our list of candidate SNPs. This brought the total number of SNPs from the hypothetical and known genes to 39.
In addition to the 39 SNPs in the known and hypothetical genes, information from HapMap was used to identify additional intronic and intergenic SNPs in the refined interval. Using this approach, 120 additional SNPs across the entire refined interval as well as the region 561 kb proximal and 835 kb distal to the critical recombination events were chosen. These were selected to ensure they were evenly spaced and covered the entire region. There was an approximate spacing of 4–55 kb (average 51 kb) between each SNP.
Case-control association study
A total of 159 (39+120) SNPs were genotyped in an independent case-control cohort consisting of 300 myopic and 291 control individuals from Australia. Power calculations were undertaken and suggested that a cohort of this size will have 80% power to detect an odds ratio of 1.78. The average genotyping success rate for these SNPs was 98.1%. A total of eight SNPs (5.0%) were found not to be in Hardy–Weinberg equilibrium and so were excluded from further analysis. An additional 14 SNPs (8.8%) were not polymorphic in this cohort, and they were also excluded from further analysis leaving 137 SNPs for the case-control association analysis. The 22 excluded SNPs were scattered throughout the linked region with no bias for one gene or genomic segment. All p values from the case-control association study for the remaining 137 SNPs underwent a Bonferroni correction. Using a conservative significance level of 5×10−4, we observed no statistically significant association to the genotyped SNPs [58].
Discussion
Using a fine mapping linkage based approach, we have been able to identify a novel locus for myopia on chromosome 2q37. Initially, this region was described as overlapping with the known high grade myopia locus, MYP12. However, further fine mapping and haplotype analyses have enabled us to better refine this region to a smaller 1.83 cM segment on chromosome 2q37. Hence, the locus harboring the causative myopia gene in these GEM families is novel and distinct from the MYP12 locus, indicating a degree of genetic heterogeneity in this region of chromosome 2. Detailed SNP analysis and DNA sequencing of all known and hypothetical genes in the refined interval provided no evidence of a causative variant in the coding region of these genes. The best evidence for a causative variant in the region was provided by the two intronic SNPs of rs1986830 and rs4663724, but these only showed suggestive rather than statistically significant association.
Despite strong evidence for a hereditary component influencing myopia, identification of causative genes or DNA variants has so far proven difficult to achieve. To date, five genes, transforming growth factor beta induced factor homeobox 1 (TGIF) [59], paired box 6 (PAX6) [60], collagen, type 1, alpha 1 (COL1A1) [61], hepatocyte growth factor (HGF) [62], and uromodulin-like 1 (UMODL1) [27], have been positively associated with high grade myopia and collagen, type 2, alpha 1 (COL2A1) [63] with common myopia. Of these six candidates, TGIF and COL1A1 can be excluded as candidates as subsequent replication studies have been negative [64-67]. COL2A1, HGF, and UMODL1 have each been reported in single studies and await replication. The final gene, PAX6, has been positively associated with high grade myopia in two independent studies, but results have been negative for common myopia [29,63,68,69]. Hence, to date, PAX6, HGF, and UMODL1 remain the strongest candidates for high grade myopia and collagen, type 2, alpha 1 (COL2A1) for common myopia.
The methodological approach that we have taken to gene identification, namely linkage analysis and DNA sequencing, mirrors the approach that has been used in many other studies mapping myopia loci [70-73]. Unfortunately, these studies have also failed to find a causative variant in the coding region of candidate genes. Given that myopia is a complex disease, it is possible that causative variants are located in intron or flanking regulatory regions as has been demonstrated for other complex diseases such as breast cancer, type 2 diabetes, and chronic kidney disease [74-76]. More detailed assessment of non-coding regions in these linked regions may provide more clues as to the genetic drivers of myopia. SNPs located in the non-coding sequence may affect gene/protein expression indirectly by affecting gene regulation and hence may be important drivers of the disease process. Given this, we extended our assessment of the candidate region to include intronic and intergenic SNPs spanning the linkage interval.
Our study has identified a relatively small linkage region on the long arm of chromosome 2 that represents a novel locus for common myopia. Further analysis of this region failed to convincingly identify genetic variants associated with myopia. However, there are a total of 1,048 known SNPs in the refined linkage interval, and we cannot rule out the possibility that additional SNPs in this region are associated with myopia. There are also several other issues that should be considered when interpreting our findings. One could argue that the cohort we used for the case-control study was underpowered to detect all variants that might be positively associated with myopia. While this is a possibility, the variants that are most likely to be missed are those that have small effect sizes. Although such variants may still contribute to myopia, they are likely to be only minor players. Furthermore, it is also important to be able to assess segregation of SNPs in the original GEM families used in the linkage analysis. However, the relatively small number of families used for this study would make segregation analysis statistically underpowered, and consequently, reliable data cannot be generated. We were aware of this study limitation from the onset and as a consequence, opted instead to validate SNPs in an independent case-control cohort to replicate the initial linkage result.
The findings presented here do not represent the conclusion of this study but do provide ongoing data for further investigation into the exact genetic causes of myopia. Further work needs to be undertaken to extend these findings to ensure complete coverage of this region. SNP genotyping also needs to be confirmed in a larger case-control cohort and also replicated in additional cohorts of both Caucasian and different ethnicities. Clearly, much more work is needed to further elucidate the underlying genetic influences on the development of myopia.
Acknowledgments
The authors wish to thank participants from the GEM, BMES, and VIP studies who made this work possible. This work was supported by the Australian Federal Government through the Cooperative Research Centres Program, the Joan and Peter Clemenger Trust, the Angior Family Foundation, the Myra Stoicescu Trust, the William Buckland Foundation, the Eye Research Australia Foundation, the Sunshine Foundation, the Helen Macpherson Smith Trust, the National Health and Medical Research Council (grant number 529912), and the L.E.W. Carty Trust. The authors wish to state that there are no conflicts of interest.
References
- 1.Wang Q, Klein BEK, Klein R, Moss SE. Refractive Status in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 1994;35:4344–7. [PubMed] [Google Scholar]
- 2.He M, Huang W, Zheng Y, Huang L, Ellwein LB. Refractive error and visual impairment in school children in rural southern China. Ophthalmology. 2007;114:374–82. doi: 10.1016/j.ophtha.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Katz J, Tielsch JM, Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997;38:334–40. [PubMed] [Google Scholar]
- 4.Raju P, Ramesh SV, Arvind H, George R, Baskaran M, Paul PG, Kumaramanickavel G, McCarty C, Vijaya L. Prevalence of refractive errors in a rural South Indian population. Invest Ophthalmol Vis Sci. 2004;45:4268–72. doi: 10.1167/iovs.04-0221. [DOI] [PubMed] [Google Scholar]
- 5.Saw SM, Gazzard G, Koh D, Farook M, Widjaja D, Lee J, Tan DTH. Prevalence rates of refractive errors in Sumatra, Indonesia. Invest Ophthalmol Vis Sci. 2002;43:3174–80. [PubMed] [Google Scholar]
- 6.Wu SY, Nemesure B, Leske MC. Refractive errors in a black adult population: The Barbados Eye Study. Invest Ophthalmol Vis Sci. 1999;40:2179–84. [PubMed] [Google Scholar]
- 7.Curtin BJ. The Myopias: basic science and clinical management. Philadelphia: Harper and Row; 1985. [Google Scholar]
- 8.Mitchell P, Wang JJ. Diabetes, fasting blood glucose and age-related maculopathy: The Blue Mountains Eye Study. Aust N Z J Ophthalmol. 1999;27:197–9. doi: 10.1046/j.1440-1606.1999.00211.x. [DOI] [PubMed] [Google Scholar]
- 9.Mutti DO, Zadnik K, Adams AJ. Myopia. The nature versus nurture debate goes on. Invest Ophthalmol Vis Sci. 1996;37:952–7. [PubMed] [Google Scholar]
- 10.Saw SM, Tan SB, Fung D, Chia KS, Koh D, Tan DTH, Stone RA. IQ and the association with myopia in children. Invest Ophthalmol Vis Sci. 2004;45:2943–8. doi: 10.1167/iovs.03-1296. [DOI] [PubMed] [Google Scholar]
- 11.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: The twin eye study. Invest Ophthalmol Vis Sci. 2001;42:1232–6. [PubMed] [Google Scholar]
- 12.Dirani M, Chamberlain M, Shekar SN, Islam AF, Garoufalis P, Chen CY, Guymer RH, Baird PN. Heritability of refractive error and ocular biometrics: the Genes in Myopia (GEM) twin study. Invest Ophthalmol Vis Sci. 2006;47:4756–61. doi: 10.1167/iovs.06-0270. [DOI] [PubMed] [Google Scholar]
- 13.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–6. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojciechowski R, Congdon N, Bowie H, Munoz B, Gilbert D, West SK. Heritability of refractive error and familial aggregation of myopia in an elderly American population. Invest Ophthalmol Vis Sci. 2005;46:1588–92. doi: 10.1167/iovs.04-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CY, Scurrah KJ, Stankovich J, Garoufalis P, Dirani M, Pertile KK, Richardson AJ, Mitchell P, Baird PN. Heritability and shared environment estimates for myopia and associated ocular biometric traits: the Genes in Myopia (GEM) family study. Hum Genet. 2007;121:511–20. doi: 10.1007/s00439-006-0312-0. [DOI] [PubMed] [Google Scholar]
- 16.Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K. Parental myopia, near work, school achievement, and children's refractive error. Invest Ophthalmol Vis Sci. 2002;43:3633–40. [PubMed] [Google Scholar]
- 17.Nallasamy S, Paluru P, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–36. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 19.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–24. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–6. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 23.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 24.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–19. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young TL, Atwood LD, Ronan SM, Dewan AT, Alvear AB, Peterson J, Holleschau A, King RA. Further refinement of the MYP2 locus for autosomal dominant high myopia by linkage disequilibrium analysis. Ophthalmic Genet. 2001;22:69–75. doi: 10.1076/opge.22.2.69.2233. [DOI] [PubMed] [Google Scholar]
- 27.Nishizaki R, Ota M, Inoko H, Meguro A, Shiota T, Okada E, Mok J, Oka A, Ohno S, Mizuki N. New susceptibility locus for high myopia is linked to the uromodulin-like 1 (UMODL1) gene region on chromosome 21q22.3. Eye. 2009;23:222–9. doi: 10.1038/eye.2008.152. [DOI] [PubMed] [Google Scholar]
- 28.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. Genome-wide Scan Maps a Novel High Myopia Locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 29.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: A genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey-Wilson JE. Genomewide Linkage Scan for Myopia Susceptibility Loci among Ashkenazi Jewish Families Shows Evidence of Linkage on Chromosome 22q12. Am J Hum Genet. 2004;75:448–59. doi: 10.1086/423789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008;32:454–63. doi: 10.1002/gepi.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Li S, Xiao X, Guo X. Confirmation of a genetic locus for X-linked recessive high myopia outside MYP1. J Hum Genet. 2007;52:469–72. doi: 10.1007/s10038-007-0130-9. [DOI] [PubMed] [Google Scholar]
- 33.Lam DSC, Tam POS, Fan DS, Baum L, Leung YF, Pang CP. Familial high myopia linkage to chromosome 18p. Ophthalmologica. 2003;217:115–8. doi: 10.1159/000068554. [DOI] [PubMed] [Google Scholar]
- 34.Stambolian D, Ciner EB, Reider LC, Moy C, Dana D, Owens R, Schlifka M, Holmes T, Ibay G, Bailey-Wilson JE. Genome-wide scan for myopia in the Old Order Amish. Am J Ophthalmol. 2005;140:469–76. doi: 10.1016/j.ajo.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 UK families. Invest Ophthalmol Vis Sci. 2004;45:2879–85. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- 36.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes TM, Ciner E, Bailey-Wilson JE. Genome-wide scan of additional Jewish families confirms linkage of a myopia susceptibility locus to chromosome 22q12. Mol Vis. 2006;12:1499–505. [PubMed] [Google Scholar]
- 37.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Confirmation of linkage to ocular refraction on chromosome 22q and identification of a novel linkage region on 1q. Arch Ophthalmol. 2007;125:80–5. doi: 10.1001/archopht.125.1.80. [DOI] [PubMed] [Google Scholar]
- 38.Nurnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–38. [PubMed] [Google Scholar]
- 39.Chen CY, Stankovich J, Scurrah KJ, Garoufalis P, Dirani M, Pertile KK, Richardson AJ, Baird PN. Linkage replication of the MYP12 locus in common myopia. Invest Ophthalmol Vis Sci. 2007;48:4433–9. doi: 10.1167/iovs.06-1188. [DOI] [PubMed] [Google Scholar]
- 40.Garoufalis P, Chen CYC, Dirani M, Couper TA, Taylor HR, Baird PN. Methodology and recruitment of probands and their families for the Genes in Myopia (GEM) Study. Ophthalmic Epidemiol. 2005;12:383–92. doi: 10.1080/09286580500281222. [DOI] [PubMed] [Google Scholar]
- 41.Richardson AJ, Narendran N, Guymer RH, Vu H, Baird PN. Blood storage at 4 degrees C-factors involved in DNA yield and quality. J Lab Clin Med. 2006;147:290–4. doi: 10.1016/j.lab.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Wong TY, Foster PJ, Hee J, Ng TP, Tielsch JM, Chew SJ, Johnson GJ, Seah SKL. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000;41:2486–94. [PubMed] [Google Scholar]
- 43.Guzowski M, Wang JJ, Rochtchina E, Rose KA, Mitchell P. Five-year refractive changes in an older population - The Blue Mountains Eye Study. Ophthalmology. 2003;110:1364–70. doi: 10.1016/S0161-6420(03)00465-2. [DOI] [PubMed] [Google Scholar]
- 44.Mutti DO, Zadnik K. Age-related decreases in the prevalence of myopia: Longitudinal change or cohort effect? Invest Ophthalmol Vis Sci. 2000;41:2103–7. [PubMed] [Google Scholar]
- 45.Wensor M, McCarty CA, Taylor HR. Prevalence and risk factors of myopia in Victoria, Australia. Arch Ophthalmol. 1999;117:658–63. doi: 10.1001/archopht.117.5.658. [DOI] [PubMed] [Google Scholar]
- 46.Rose K, Smith W, Morgan I, Mitchell P. The increasing prevalence of myopia: implications for Australia. Clin Experiment Ophthalmol. 2001;29:116–20. doi: 10.1046/j.1442-9071.2001.00389.x. [DOI] [PubMed] [Google Scholar]
- 47.Junghans B, Kiely PM, Crewther DP, Crewther SG. Referral rates for a functional vision screening among a large cosmopolitan sample of Australian children. Ophthalmic Physiol Opt. 2002;22:10–25. doi: 10.1046/j.1475-1313.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- 48.Attebo K, Ivers RQ, Mitchell P. Refractive errors in an older population - The blue mountains eye study. Ophthalmology. 1999;106:1066–72. doi: 10.1016/S0161-6420(99)90251-8. [DOI] [PubMed] [Google Scholar]
- 49.Taylor HR, Livingston PM, Stanislavsky YL, McCarty CA. Visual impairment in Australia: Distance visual acuity, near vision, and visual field findings of the Melbourne Visual Impairment Project. Am J Ophthalmol. 1997;123:328–37. doi: 10.1016/s0002-9394(14)70128-x. [DOI] [PubMed] [Google Scholar]
- 50.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 51.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 52.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–2. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 53.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Eyre T, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Holland R, Howe KL, Howe K, Johnson N, Jenkinson A, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Slater G, Smedley D, Spudich G, Trevanion S, Vilella AJ, Vogel J, White S, Wood M, Birney E, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Hubbard TJ, Kasprzyk A, Proctor G, Smith J, Ureta-Vidal A, Searle S. Ensembl 2008. Nucleic Acids Res. 2008;36(Database issue):D707–14. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–72. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 59.Lam DSC, Lee WS, Leung YF, Tam POS, Fan DSP, Fan BJ, Pang CP. TGF beta-induced factor: A candidate gene for high myopia. Invest Ophthalmol Vis Sci. 2003;44:1012–5. doi: 10.1167/iovs.02-0058. [DOI] [PubMed] [Google Scholar]
- 60.Tsai YY, Chiang CC, Lin HJ, Lin JM, Wan L, Tsai FJA. PAX6 gene polymorphism is associated with genetic predisposition to extreme myopia. Eye. 2008;22:576–81. doi: 10.1038/sj.eye.6702982. [DOI] [PubMed] [Google Scholar]
- 61.Inamori Y, Ota M, Inoko H, Okada E, Nishizaki R, Shiota T, Mok J, Oka A, Ohno S, Mizuki N. The COL1A1 gene and high myopia susceptibility in Japanese. Hum Genet. 2007;122:151–7. doi: 10.1007/s00439-007-0388-1. [DOI] [PubMed] [Google Scholar]
- 62.Han W, Yap MKH, Wang J, Yip SP. Family-based association analysis of hepatocyte growth factor (HGF) gene polymorphisms in high myopia. Invest Ophthalmol Vis Sci. 2006;47:2291–9. doi: 10.1167/iovs.05-1344. [DOI] [PubMed] [Google Scholar]
- 63.Mutti DO, Cooper ME, O'Brien S, Jones LA, Mariazita ML, Murray JC, Zadnik K. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–9. [PMC free article] [PubMed] [Google Scholar]
- 64.Pertile KK, Schache M, Islam FM, Chen CY, Dirani M, Mitchell P, Baird PN. Assessment of TGIF as a candidate gene for myopia. Invest Ophthalmol Vis Sci. 2008;49:49–54. doi: 10.1167/iovs.07-0896. [DOI] [PubMed] [Google Scholar]
- 65.Hasumi Y, Inoko H, Mano S, Ota M, Okada E, Kulski JK, Nishizaki R, Mok J, Oka A, Kumagai N, Nishida T, Ohno S, Mizuki N. Analysis of single nucleotide polymorphisms at 13 loci within the transforming growth factor-induced factor gene shows no association with high myopia in Japanese subjects. Immunogenetics. 2006;58:947–53. doi: 10.1007/s00251-006-0155-9. [DOI] [PubMed] [Google Scholar]
- 66.Li J, Zhang QJ, Xiao XS, Li JZ, Zhang FS, Li SQ, Li W, Li T, Jia XY, Guo L, Guo XM. The SNPs analysis of encoding sequence of interacting factor gene in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:454–6. [PubMed] [Google Scholar]
- 67.Liang CL, Hung KS, Tsai YY, Chang W, Wang HS, Juo SH. Systematic assessment if the tagged polymorphisms of the COL1A1 gene for high myopia. J Hum Genet. 2007;52:374–7. doi: 10.1007/s10038-007-0117-6. [DOI] [PubMed] [Google Scholar]
- 68.Simpson CL, Hysi P, Bhattacharya SS, Hammond CJ, Webster A, Peckham CS, Sham PC, Rahi JS. The Roles of PAX6 and SOX2 in Myopia: lessons from the 1958 British Birth Cohort. Invest Ophthalmol Vis Sci. 2007;48:4421–5. doi: 10.1167/iovs.07-0231. [DOI] [PubMed] [Google Scholar]
- 69.Hewitt AW, Kearns LS, Jamieson RV, Williamson KA, van Heyningen V, Mackey DA. PAX6 mutations may be associated with high myopia. Ophthalmic Genet. 2007;28:179–82. doi: 10.1080/13816810701356676. [DOI] [PubMed] [Google Scholar]
- 70.Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence variants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci. 2004;45:2091–7. doi: 10.1167/iovs.03-0933. [DOI] [PubMed] [Google Scholar]
- 71.Scavello GS, Jr, Paluru PC, Zhou J, White PS, Rappaport EF, Young TL. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- 72.Zhou J, Young TL. Evaluation of Lipin 2 as a candidate gene for autosomal dominant 1 high-grade myopia. Gene. 2005;352:10–9. doi: 10.1016/j.gene.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 73.Paluru PC, Scavello GS, Ganter WR, Young TL. Exclusion of lumican and fibromodulin as candidate genes in MYP3 linked high grade myopia. Mol Vis. 2004;10:917–22. [PubMed] [Google Scholar]
- 74.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 75.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kottgen A, Kao WH, Hwang SJ, Boerwinkle E, Yang Q, Levy D, Benjamin EJ, Larson MG, Astor BC, Coresh J, Fox CS. Genome-wide association study for renal traits in the Framingham Heart and Atherosclerosis Risk in Communities Studies. BMC Med Genet. 2008;9:49. doi: 10.1186/1471-2350-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]