Abstract
Background
Chitinolytic enzymes play important roles in the pathophysiology of allergic airway responses in mouse models of asthma. Acidic mammalian chitinase (AMCase) and chitotriosidase (CHIT1) have chitinolytic activity, but relatively little is known about their expression in human asthma.
Objective
To determine the expression and activity of AMCase and CHIT1 in health, asthma, and chronic obstructive pulmonary disease (COPD) (disease control), taking account of the null 24bp duplication in the CHIT1 gene.
Methods
We measured chitinase activity in bronchoalveolar lavage (BAL) at multiple pH’s using a synthetic chitin substrate. We also determined AMCase and CHIT1 gene expression in epithelial brushings and BAL macrophages by real time RT-PCR. Paired DNA samples were genotyped for the CHIT1 duplication.
Results
In all subgroups, the pH profile of chitinase activity in BAL matched that of chitotriosidase, not AMCase, and chitinase activity was absent in subjects genetically deficient in active chitotriosidase. Although AMCase protein was detectable in lavage, AMCase transcripts in macrophages were consistent with an isoform lacking enzymatic activity. Median chitinase activity in BAL tended to be lower than normal in asthma, but was increased 7-fold in COPD, where CHIT1 gene expression in macrophages was increased.
Conclusions
Chitinase activity in the lung is the result of CHIT1 activity. Although AMCase protein is detectable in the lung, our data indicates that it is inactive. Chitinase activity is not increased in asthma and in fact tends to be decreased. The high levels of chitinase activity in COPD result from upregulation of CHIT1 gene expression, especially in macrophages.
Keywords: AMCase, CHIT1, chitinase, asthma, smokers, bronchoalveolar lavage
Introduction
Chitin, a linear polymer of β(1–4)-linked N-acetyl-D-glucosamine, is a component of the exoskeletons of mites and other arthropods, the lining of the insect gut, and the microfilarial sheath of parasitic nematodes 1–4. Chitin is also an important structural polymer in fungal cell walls, where it is functionally analogous to peptidoglycan in bacteria.
Chitinases are the enzymes that digest the chitin polymer, and humans have two chitinases encoded in their genome: chitotriosidase (CHIT1) and acidic mammalian chitinase (AMCase) 5. The substrate for these chitinases is presumably environmental chitin, because all mammals lack a chitin synthase. CHIT1 is prominently expressed in macrophages, but the biological consequences of its overexpression in diseases associated with macrophage activation (lysosomal lipid storage disorders, thalassemia, sarcoidosis, and visceral Leishmaniasis) are unknown 6, 7. A relatively common 24 base pair duplication in exon-10 of CHIT1 activates a cryptic 3′ splice site and results in an enzymatically inactive protein deficient in 29 amino acids 8. The relatively high prevalence of chitotriosidase deficiency from this duplication prompted a search for other chitinases that may compensate for the lack of functional CHIT1. This led to the discovery of AMCase, named for its pronounced pH optimum at pH 2.3 9. Like CHIT1, AMCase is capable of cleaving artificial chitin-like substrates as well as natural substrates. The full-length cDNA for human AMCase is almost identical to TSA1902-L and TSA1902-S - two isoforms previously described as splice variants of a gene encoding a chitinase-like protein in man 10. These TSA transcripts are actually splice variants of the AMCase gene, but the proteins they encode do not have chitinolytic activity 10. CHIT1 and AMCase belong to the Family 18 of glycosyl hydrolases, which also include other proteins structurally related to chitinases but lacking in chitinolytic activity 5. These “chi-lectins” include chitinase-3-like-1 (CHI3L1), a protein of uncertain function, which is elevated in serum in a variety of inflammatory diseases, including severe asthma 11. Genetic variation in the CHI3LI gene has also been associated with asthma 12.
Insights into the role of chitinases and chitin in allergy and asthma have been greatly advanced by two recent studies. Zhu et al 13 showed that AMCase expression and activity is upregulated in an ovalbumin mouse model of asthma and that it’s expression is dependent on IL-13. Furthermore, they found that inhibition of AMCase enzymatic activity prevents much of the airway hyperresponsiveness and inflammation present in these mice after challenge. In stark contrast, Reese et al 14 showed that polymeric chitin administrated to the lungs of mice could induce the recruitment of immune cells associated with allergy and asthma, such as eosinophils and basophils. Moreover, AMCase mediated degradation of chitin acted as a negative regulator of this process. Despite these important but contrasting roles for chitinase activity in the development of allergic inflammation in the lungs of mice, only sparse clinical data is available to guide opinion on which of these conflicting roles operates in human asthma. Although Zhu et al 13 used in situ hybridization to show that AMCase gene expression is increased in airway mucosal biopsies and in small airway tissues from asthmatics, these authors did not report on CHIT1 expression or on levels of chitinase activity in lung secretions. Therefore, in this study, we set out to determine the relative contributions of AMCase and CHIT1 to lung chitinase activity and to determine if chitinase activity differs from normal in asthma or in a disease control (habitual smokers with mild COPD).
METHODS
Subjects and clinical samples
We studied biological samples stored in the Airway Tissue Bank at the University of California, San Francisco (UCSF) that had been collected during research bronchoscopy from 40 non-smoking subjects with asthma, 25 current smokers without asthma, and 26 healthy non-smoking controls (Table 1). Asthmatic subjects had a prior physician diagnosis of asthma, PC20 methacholine <8 mg/mL, and were using only inhaled beta-agonist medications for therapy. Current smokers had been smoking at least 10 cigarettes per day and had at least a 10 pack-year total consumption. Healthy controls were non-smokers with no history of lung disease and PC20 methacholine >16 mg/mL. The samples withdrawn from the tissue bank for this study were aliquots of bronchoalveolar lavage (BAL) fluid supernatant, RNA from BAL macrophages, RNA from epithelial brushings, and DNA (extracted from either venous blood cells [n=40] or from frozen unfixed bronchial mucosal biopsies [n=18]). Epithelial brushings and macrophages from some of the subjects in all subject subgroups have been used in previously reported studies 15–19. Two of the smoker subjects contributed bronchoscopy samples to the bank from two bronchoscopies done in a one year period. In one of these subjects, we used RNA from epithelial brushings from the first bronchoscopy and BAL supernatant from the second bronchoscopy. In the second subject, we used epithelial cell RNA from the first bronchoscopy and BAL supernanant and BAL macrophage RNA from the second bronchoscopy. For both of these subjects we provide clinical data in Table 1 from the first bronchoscopy.
Table 1.
Clinical characteristics of subject groups.
| Characteristic | Controls | Asthmatics | Smokers |
|---|---|---|---|
| N | 26 | 40 | 25 |
| Gender | 13F/13M | 22F/28M | 6F/19M* |
| Age (years) | 40 ± 9 | 36 ± 12 | 51 ± 10* |
| FEV1 (% predicted) | 103 ± 12 | 85 ± 11* | 83±16* |
| FEV1/FVC | 0.80 ± 0.05 | 0.85 ± 0.11* | 0.66 ± 0.12* |
| PC20 (mg/dl methacholine) | 64.0 (20.4, 64.0) | 0.3 (0.1, 1.0)* | 26.8 (6.9, 64.0)* |
| Pack Years Smoking | 0 (0, 0) | 0 (0, 0) | 38 (27, 44) |
| GOLD Classification | |||
| Class 0 | - | - | 11 |
| Class 1 | - | - | 5 |
| Class 2 | - | - | 9 |
| DLCO (% predicted)† | - | - | 86 ± 19% |
| No. with DLCO < 80% predtcted | - | - | 8 |
FOOTNOTE: Data are presented as mean +/− SD or median (interquartile range).
Abbreviations: FEV1, volume of air exhaled in the first second of a forced exhalation;
FVC, forced vital capacity (volume of air exhaled in an entire forced exhalation);
PC20, the concentration of methacholine that caused a 20% decline in FEV1;
DLCO, diffusing capacity to carbon monoxide (†data for n = 24).
GOLD classification denotes presence and severity of COPD 36.
No subjects with asthma were using corticosteroids or long-acting β-agonists prior to enrollment.
, P < 0.05 compared with nonsmoking healthy control subjects based on Mann-Whitney two-sample ranksum test for quantitative traits and χ2 analysis for gender.
Clinical procedures
Spirometry, methacholine challenge, measurement of diffusing capacity and bronchoscopy had been performed using methods described previously 15. At bronchoscopy, bronchoalveolar lavage was performed in either the right middle lobe or lingula before brushings were obtained from lower lobe bronchi in the ipsilateral lung; the bronchoscope was then moved to the contralateral lung where bronchial biopsies were taken from 2nd through 4th order carinae of lower lobe, middle lobe, and upper segments.
Chitinase Activity Assays
Chitinase activity was determined using the synthetic chitin substrate 4-MU-(4-deoxy)chitobiose 20. All chitinase assays were performed in a total volume of 100 microliters containing 15 microliters of unconcentrated BAL and 75 micromolar 4-MU-(4-deoxy)chitobiose in McIlvaine’s buffer at three different pH values (pH 2.2, 4.6, or 7.0). Reactions were incubated in the dark for 2 hours at 37 degrees Celcius. All reactions were conducted in duplicate in 96 well fluorescent spectroscopy plates (Sigma) with 4-Methylumbelliferone also loaded as a standard. Reactions were stopped by the addition of 120 μl of 1M glycine/NaOH pH 10.5 to reaction wells. Plates were then immediately read on the SpectraMax Gemini XS fluorescence plate reader at an excitation wavelenght of 365nm and an emission wavelength of 460nm. For determination of BAL samples velocities, the mean fluorescence units detected from the duplicate assays were converted to nanomoles of product produced by extrapolation using the 4-MU standard curves generated on each plate, and divided by reaction time and volume of BAL loaded in the assay (15 μl).
Gene Profiling
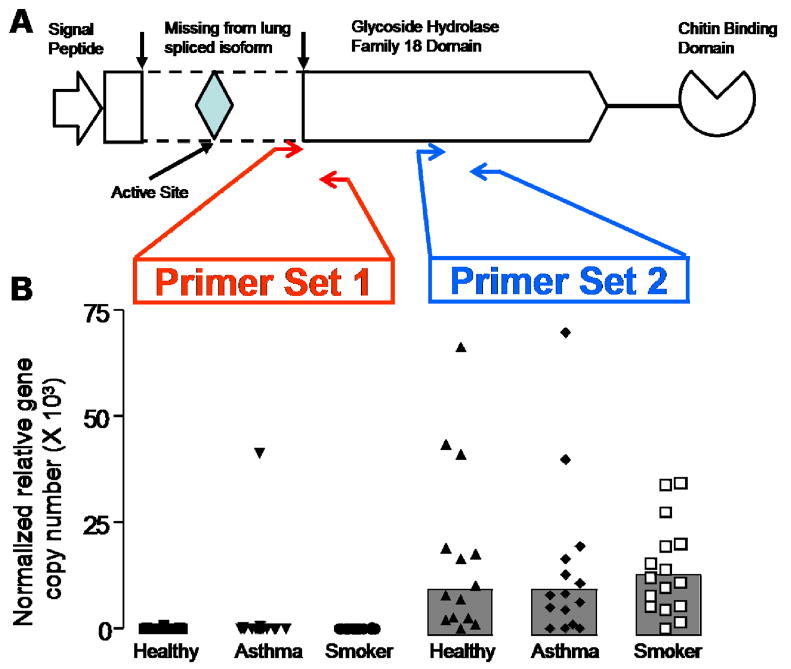
Gene expression for CHIT1 and AMCase in epithelial cells and macrophages was measured using methods of real time RT-PCR described previously 15, 17, 18, 21. For CHIT1, we designed one primer set against a region common to all known variants (Table E1). For AMCase, we designed two primer sets (Table E1 and Figure 3A). The first primer set (primer set 1) was designed to include a region across exon 6 that is known to result in an inactive variant when it is spliced out 10. The second primer set (primer set 2) was designed against a portion of the AMCase mRNA contained in all known transcripts.
Figure 3. AMCase gene expression in lavage macrophages from healthy subjects, asthmatics, and habitual smokers.
Panel A - AMCase protein domain structure, with region spliced out in TSA1902S transcript denoted by arrows and dotted lines. Red arrows denote region amplified by RT-PCR primer set 1, and blue arrows denote region amplified by RT-PCR primer set 2.
Panel B - AMCase gene expression stratified by red primer set 1 (active site-containing transcripts) and blue primer set 2 (all AMCase transcripts)
Genotyping
The CHIT1 24bp duplication (rs3831317) was genotyped using the AcycloPrime-FP-TDI (PerkinElmer) method 22. The PCR cocktail included: 3.0–5.0 ng genomic DNA, 0.1–0.2 μM primers, 2.5 mM MgCl2, 50 μM dNTPs, 6 μl volume with Platinum Taq PCR buffer and 0.1–0.2 units Platinum Taq (Invitrogen) plus 1 μl extra water to counteract evaporation. PCR cycling conditions were as follows: 95°C for 2 minutes, 40 cycles of 92°C for 10 seconds, 57°C for 20 seconds, 68°C for 30 seconds, and final extension at 68°C for 10 minutes. We used AcycloPrime-FP kits for enzymatic cleanup and single base extension genotyping reactions. Plates were read on an EnVision fluorescence polarization plate reader (PerkinElmer). PCR primers and allele-specific FP primers and Fluorescent tagged terminator nucleotides are listed in Table E2.
SDS polyacrylamide gel electrophoresis for AMCase in BAL
To detect AMCase in BAL, we used a polyclonal anti-mouse YM1 antibody (Stem Cell Technologies), because this antbody was generated against a petide sequence that is also present in human AMCase and there is no other orthologue to YM1 in humans 23. Using purified human recombinant AMCase as a control, we confirmed that the YM1 antibody detects human AMCase (Figure 4).
Figure 4. AMCase protein in BAL from healthy subjects (n = 13), asthmatics (n = 16), and habitual smokers (n = 19).
Y-axis values generated from densitometry analysis of protein bands relative to intensity of control AMCase protein. *Indicates significantly less than healthy, p<0.05.
Statistical Analysis
All two-way subject group comparisons of gene expression data and chitinase activity were performed by using the Mann-Whitney rank-sum test to account for the non-normally distributed nature of the tested variables. Correlations between chitinase activity at pH 4.6 and 7.0, and between chitinase activity and gene expression data was performed by Spearman’s rank correlation test. Multiple linear regression analysis was used to test the association of CHIT1 duplication genotype on chitinase activity at pH 4.6. Chitinase activity was log transformed for use in the regression model. All statistical analysis was performed using STATA 8.0 S/E statistical software (College Station, TX).
Results
Lung chitinase activity is modulated by pH and disease
We measured total chitinase activity in the bronchoalveolar lavage (BAL) of 77 subjects including 31 asthmatics, 24 healthy subjects, and 22 smokers using a synthetic chitin substrate, 4-MU-(4-deoxy)chitobiose 20. This substrate is digested by both AMCase and CHIT1, but the pH profile for the activity of these two chitinolytic enzymes is distinct and can be used to infer whether CHIT1 or AMCase is the responsible enzyme for chitinase activity in a biological sample. For example, previous studies have shown that optimal enzymatic activity of AMCase and CHIT1 occurs at a pH of 4.6, but that AMCase enzyme retains 30% of it’s activity at pH of 2.2, whereas CHIT is inactive at this pH 9, 24. In contrast, CHIT1 retains 75% of its enzymatic activity at pH of 7.0, whereas AMCase is only 30% active 9, 24. We found that chitinase activity in BAL was undetectable at pH 2.2 in the large majority of samples; lavage from only one subject (a smoker) had activity above the lower assay limits (Figure 1A). In contrast, chitinase activity was easily detectable in BAL from all subject groups at pH’s of 4.6 and 7.0, although values at pH 7.0 were ~20% lower than at pH 4.6 (Figure 1A). In addition, chitinase activity at pH 4.6 and pH 7.0 were strongly correlated (rho=0.99, p <.0.0001). This pH profile for chitinase activity in lavage is consistent with the activity of CHIT1 in lung secretions and inconsistent with significant AMCase activity 9, 24.
Figure 1. Effect of pH and disease status on chitinase activity in 77 bronchoalveolar lavage (BAL) samples.
Panel A – Points represent chitinase activity measured for each BAL sample using the synthetic chitin substrate 4-MU-(4-deoxy)chitobiose in McIlvaine’s buffer at pH 2.2, pH 4.6, and pH 7.0. Line represents the median value.
Panel B – Points represent chitinase activity measured for each BAL sample stratified by disease status. *Indicates significantly greater than healthy, p<0.05.
Comparison of chitinase activity at pH 4.6 between subject groups revealed that activity was lower than normal in the asthma subgroup and markedly higher than normal in COPD (Figure 1B). Specifically, median chitinase activity was 60% lower than normal in the asthma subgroup (0.38 nmoles/ml/hr [interquartile range 0.2–1.77] vs 0.95 nmoles/ml/hr [0.57 – 1.47], p=0.077)(Figure 1B). In contrast, median chitinase activity was 7-fold higher than normal in the COPD subgroup (6.64 [1.90 – 17.48], p<0.0001)(Figure 1B).
Chitotriosidase is the primary active lung chitinase
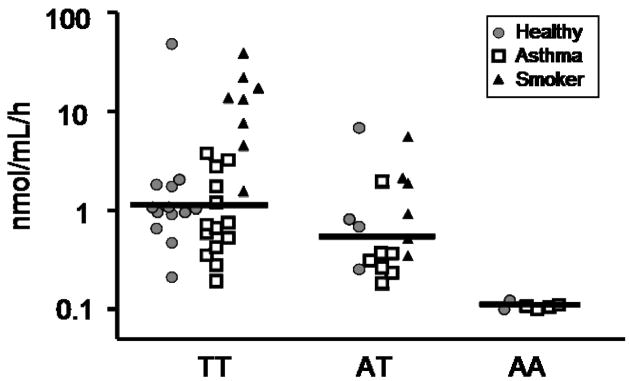
The absence of BAL chitinase activity at pH 2.2 in nearly all subjects coupled with high activity at pH 7.0 suggested that CHIT1 was the primary source of chitinase activity in the lungs. To examine this hypothesis further, we exploited knowledge of a common null genetic variant in the CHIT1 gene. Namely, there is a well-characterized 24bp duplication polymorphism in exon 10 of the CHIT1 gene, resulting in abnormally spliced mRNA and completely inactive CHIT1 protein 8. Subjects homozygous for this duplication should not display significant chitinase activity at either pH 4.6 or 7.0 if CHIT1 is the only source of chitinase activity in the BAL fluid. DNA was available on 58 of the original 77 subjects used to measure chitinase activity, including 25 asthmatics, 19 healthy subjects, and 14 smokers. Genotyping the 24bp duplication revealed 2 of the healthy subjects (10.5%), 4 of the asthmatic subjects (16%), and none of the smokers were homozygous for the duplication. Strikingly, all six subjects homozygous for the CHIT1 duplication displayed no chitinase activity at either pH 4.6 or pH 7.0 (Figure 2), a result which strongly implicates CHIT1 as the primary source of chitinase activity in human lung secretions. Indeed, no subjects genotyped lacked chitinase activity at either pH 4.6 or pH 7.0, other than those homozygous for the CHIT1 duplication polymorphism. Furthermore, we found median chitinase activity levels to be higher in subjects not carrying the duplication versus subjects heterozygous for the duplication, implying a gene dosage effect (Figure 2). This additive genotypic effect of the CHIT1 duplication on chitinase activity was tested by linear regression and persisted after correcting for age, gender, and race (P < 0.0001).
Figure 2. Chitinase activity in BAL stratified by CHIT1 24bp duplication genotype status.
Absence of AMCase enzymatic activity is due to inactive splice variant
The pH profile of chitinase activity in BAL samples and the effect of the CHIT1 duplication on BAL chitinase activity argue against AMCase activity in human lung secretions. However, previous reports have found AMCase mRNA expression in both lung epithelial and macrophage cells 13. Moreover, another study found expression of multiple splice variants of the AMCase mRNA in human lungs 10. Interestingly, one of these splice variants (TSA1902S) results in the removal of the sixth exon which contains the conserved active site residues required for enzymatic activity of the AMCase protein (Figure 3A) 10. Based on this report, we hypothesized that the TSA1902S transcript was the primary splice variant of AMCase mRNA expressed in the lung. To test this hypothesis, we designed two sets of quantitative RT-PCR primers. The first primer set was designed against a portion of the AMCase mRNA contained in all known transcripts (Figure 3A). The second primer set was designed against a region within the spliced out Exon 6 of the inactive TSA1902S variant (Figure 3A). We used these two primer sets to measure AMCase transcripts in the two main cell types with the potential to secrete chitinase proteins into the airway; epithelial cells from 76 bronchial brushing samples and airway macrophages purifed from a subset (n=45) of the subjects. As a positive control, we also measured AMCase transcripts in human stomach poly A RNA (pooled from 7 subjects, Clontech, Mountain View, CA). Epithelial gene expression of AMCase examined with either primer was low or absent (data not shown); this was not a result of assay failure, because abundant AMCase transcripts were detected in stomach tissue using both primer types. Using the first primer set, we detected “total” AMCase expression in macrophages, but we found no difference in expression between subject groups (Figure 3B). Using the second primer set, we found absent or very low expression of AMCase in macrophages from all subject groups (Figure 3B). These data indicate that the AMCase mRNA expressed in lung macrophages is primarily a splice variant lacking exon 6 and thus enzymatically inactive.
To examine AMCase at the protein level, we probed BAL from a subset of subjects for AMCase using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Figure 4). Semi-quantitative densitometry analysis of the protein blots showed that AMCase protein levels were lower than normal in asthmatic subjects (P = 0.023) (Figure 4). BAL AMCase protein data was available for three of the six subjects homozygous for the null CHIT1 duplication. AMCase protein was easily detectable in BAL from these three subjects, even though, as described above, all three had no chitinase activity in their BAL. Taken together, these results suggest that the AMCase protein detectable in lung secretions is an isoform lacking enzymatic activity.
CHIT1 gene expression is strongly correlated with lung chitinase activity
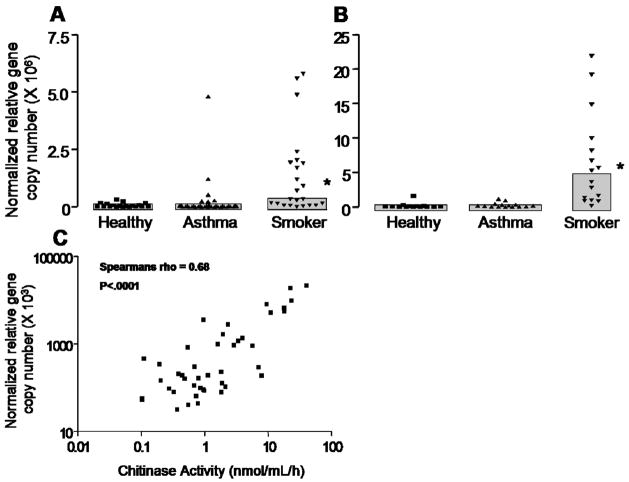
CHIT1 gene expression was most abundant in macrophages but was also observed in epithelial cells (Figure 5A & 5B). Expression was markedly increased in both epithelial cells and in macrophages from the smoker subgroup (Figure 5A & 5B). Moreover, CHIT1 gene expression in macrophages was positively correlated with lung chitinase activity ( rho = 0.68, P < 0.0001) (Figure 5C). We also found a positive correlation between CHIT1 gene expression in epithelial cells and lavage chitinase activity, although the association was not as strong as in macrophages (rho = 0.52, P < 0.0001) (data not shown).
Figure 5. CHIT1 gene expression and correlation with BAL chitinase activity.
Panel A – CHIT1 gene expression in bronchial brushings stratified by disease status. *Indicates significantly greater than healthy, p<0.05.
Panel B – CHIT1 gene expression in lavage macrophages stratified by disease status. *Indicates significantly greater than healthy, p<0.05.
Panel C– Scatterplot of CHIT1 gene expression by BAL chitinase activity at pH 4.6.
DISCUSSION
We characterized the relative contribution of both active human chitinases, CHIT1 and AMCase, to chitinase activity in the lung from healthy subjects, asthmatics, and habitual smokers with mild COPD. We determined chitotriosidase to be the primary active chitinase in the lung, and we found that it’s expression is strongly dependent on genetics and on smoking habit.
We used a multi-faceted approach to establish that CHIT1, not AMCase, is the principal active chitinase in the human lung, including determination of enzymatic activity, gene expression, and protein expression. We found that the pH profile for lung chitinase activity was consistent with the activity of CHIT1, not AMCase, showing high activity at near neutral pHs values and a complete absence of activity at low pH values 9, 24. In addition, we found a lack of chitinase activity in BAL from all six subjects genetically deficient in CHIT1 activity. Indeed, there appeared to be an additive effect of the CHIT1 duplication on lung chitinase activity, with carriers of the duplication variant having reduced chitinase activity compared to non-carriers. In addition, CHIT1 gene expression in both macrophages and epithelial cells was strongly correlated with chitinase activity in lung secretions, further solidifying the primacy of CHIT1 in contribution to lung chitinase activity.
AMCase transcript numbers have been reported to be increased in asthmatic airways 13, but our data do not confirm this. Rather, we find that inactive AMCase variants are expressed in the lung and that these variants are not differentially expressed in asthmatics or in smokers. Specifically, our data show that the numbers of “total” AMCase trancripts are higher than the numbers of active AMCase transcripts in epithelial cells and macrophages in all subject groups. This finding agrees with a previous report showing expression of splice variants of AMCase in the lung, variants which lack regions of exon-six containing the conserved active site residues required for enzymatic activity 10. Consistent with this is our finding that BAL samples which had no chitinase activity still had detectable AMCase protein, indicating that the protein lacks chitinase activity.
Our results have important implications for the role of chitinases in asthma. Paradoxically, mouse studies have implicated lung chitinase activity in both the promotion of allergen-induced airway inflammation and in reduction of chitin-induced airway eosinophilia 13, 14. Our results indicate that the upregulation in lung chitinase activity in mouse models of asthma does not extend to human asthmatics. In fact, our results do not support a pro-inflammatory role for lung chitinase activity in asthma pathology; rather, we find that chitinase activity tends to be lower than normal in asthmatics. Furthermore, we find that AMCase protein levels in BAL are lower than normal in asthma. These results are more supportive of the proposed protective role of chitinase activity and chitin-binding proteins in asthma and allergy, as evidenced by the inhibition of polymeric chitin’s Th2 inflammatory effects after digestion with AMCase 14. It is important to note several limitations with regard to interperation of this data. First, our observations were restricted to stable mild-moderate adult asthmatics. It will be important to determine whether our observations extend to subjects with more severe disease or in response to acute allergen challenge. Second, although the lung expressed AMCase is inactive, it does retain an intact chitin-binding domain, and as such it could bind chitin-containing environmental allergens and affect airway inflammation. The function of chi-lectins is unknown but these molecules have been implicated in mechanisms of tissue remodeling and inflammation 25–28..
Chitinase activity was markedly higher than normal in bronchoalveolar lavage from the subgroup of habitual smokers with mild COPD. To our knowledge, this is the first report of increased chitinase activity in airway secretions from smokers. CHIT1 gene expression was much higher in macrophages than in epithelial brushings, and we found a positive correlation between lavage chitinase activity and CHIT1 gene expression in macrophages, findings which point to macrophages as the main cellular source of the chitinase activity in these subjects. The mechanism by which cigarette smoke induces CHIT1 expression remains to be determined. Plant chitinases have well defined roles in pathogen response 29 and the role of human chitinases has generally been assumed to be in defense against chitin containing pathogens 30, 31. One possibility is that there are chitin particles in inhaled tobacco smoke, which occur as a consequence of fungal infection of tobacco leaf. It is estimated that as many as 270 fungal spores may be present in a single cigarette, and fungal spores from A. alternata have been detected in cigarette ash 32–34. Polymeric chitin is known to be pro-inflammatory 14, and if present in cigarette smoke, may activate macrophages and airway epithelial cells to increase chitinase expression and/or secretion. Other fungal-derived compounds such as beta glucans can also induce chitinase production from plants and are potent macrophage stimulators 35.
In conclusion, we have used multiple lines of evidence including biochemical, genetic, and gene and protein expression data in determining that chitotriosidase, not acidic mammalian chitinase, is the primary chitinase responsible for chitinase activity in the lung. We found that chitinase activity tended to be lower than normal in asthma, a finding which supports a protective role for chitinolytic activity in allergic inflammation. In contrast, chitinase activity and CHIT1 gene expression are increased in habitual smokers, probably because cigarette smoke induces activation of pulmonary macrophages. Taken together, our findings show that chitotriosidase activity in lung disease is modulated in ways which reflect underlying disease susceptibilities and specific environmental exposures.
Acknowledgments
FUNDING: Research support was from NIH AI077439 (JVF and EGB), HL080414 (JVF), HL078885 (EGB), RR17002 (PGW), from the Sandler Asthma Basic Research Center (JVF and EGB) and the Sandler Program for Asthma Research (EGB).
ABBREVIATIONS
- AMCase
acidic mammalian chitinase
- CHIT1
chitotriosidase
- BAL
bronchoalveolar lavage
Footnotes
KEY MESSAGE: Chitotriosidase not AMCase is the primary active chitinase in the human lung and its activity is upregulated in habitual smokers but not asthmatics.
CAPSULE SUMMARY: Macrophage and epithelial expressed chitotriosidase is responsible for chitinase activity in the lung, whereas enzymatically inactive AMCase is expressed in the lung. Chitinase activity is upregulated in habitual smokers and possibly downregulated among asthmatics.
References
- 1.Araujo AC, Souto-Padron T, de Souza W. Cytochemical localization of carbohydrate residues in microfilariae of Wuchereria bancrofti and Brugia malayi. J Histochem Cytochem. 1993;41:571–8. doi: 10.1177/41.4.8450196. [DOI] [PubMed] [Google Scholar]
- 2.Debono M, Gordee RS. Antibiotics that inhibit fungal cell wall development. Annu Rev Microbiol. 1994;48:471–97. doi: 10.1146/annurev.mi.48.100194.002351. [DOI] [PubMed] [Google Scholar]
- 3.Naclerio RM, Durham SR, Mygind N. 123. Rhinitis : Mechanisms and Management. New York: 1999. 1999. [Google Scholar]
- 4.Neville AC, Parry DA, Woodhead-Galloway J. The chitin crystallite in arthropod cuticle. J Cell Sci. 1976;21:73–82. doi: 10.1242/jcs.21.1.73. [DOI] [PubMed] [Google Scholar]
- 5.Bussink AP, Speijer D, Aerts JM, Boot RG. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics. 2007;177:959–70. doi: 10.1534/genetics.107.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aerts JM, Hollak CE. Plasma and metabolic abnormalities in Gaucher’s disease. Baillieres Clin Haematol. 1997;10:691–709. doi: 10.1016/s0950-3536(97)80034-0. [DOI] [PubMed] [Google Scholar]
- 7.Barone R, Malaguarnera L, Angius A, Musumeci S. Plasma chitotriosidase activity in patients with beta-thalassemia. Am J Hematol. 2003;72:285–6. doi: 10.1002/ajh.10294. [DOI] [PubMed] [Google Scholar]
- 8.Boot RG, Renkema GH, Verhoek M, Strijland A, Bliek J, de Meulemeester TM, et al. The human chitotriosidase gene. Nature of inherited enzyme deficiency. J Biol Chem. 1998;273:25680–5. doi: 10.1074/jbc.273.40.25680. [DOI] [PubMed] [Google Scholar]
- 9.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, et al. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 2001;276:6770–8. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 10.Saito A, Ozaki K, Fujiwara T, Nakamura Y, Tanigami A. Isolation and mapping of a human lung-specific gene, TSA1902, encoding a novel chitinase family member. Gene. 1999;239:325–31. doi: 10.1016/s0378-1119(99)00394-7. [DOI] [PubMed] [Google Scholar]
- 11.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–27. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 12.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–91. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–82. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 14.Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447:92–6. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Innes AL, Woodruff PG, Ferrando RE, Donnelly S, Dolganov GM, Lazarus SC, et al. Epithelial mucin stores are increased in the large airways of smokers with airflow obstruction. Chest. 2006;130:1102–8. doi: 10.1378/chest.130.4.1102. [DOI] [PubMed] [Google Scholar]
- 16.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, et al. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol. 2005;116:305–11. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 17.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodruff PG, Dolganov GM, Ferrando RE, Donnelly S, Hays SR, Solberg OD, et al. Hyperplasia of smooth muscle in mild to moderate asthma without changes in cell size or gene expression. Am J Respir Crit Care Med. 2004;169:1001–6. doi: 10.1164/rccm.200311-1529OC. [DOI] [PubMed] [Google Scholar]
- 19.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, et al. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–92. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguilera B, Ghauharali-van der Vlugt K, Helmond MT, Out JM, Donker-Koopman WE, Groener JE, et al. Transglycosidase activity of chitotriosidase: improved enzymatic assay for the human macrophage chitinase. J Biol Chem. 2003;278:40911–6. doi: 10.1074/jbc.M301804200. [DOI] [PubMed] [Google Scholar]
- 21.Dolganov GM, Woodruff PG, Novikov AA, Zhang Y, Ferrando RE, Szubin R, et al. A novel method of gene transcript profiling in airway biopsy homogenates reveals increased expression of a Na+-K+-Cl− cotransporter (NKCC1) in asthmatic subjects. Genome Res. 2001;11:1473–83. doi: 10.1101/gr.191301. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–8. [PMC free article] [PubMed] [Google Scholar]
- 23.Boot RG, Bussink AP, Aerts JM. Human acidic mammalian chitinase erroneously known as eosinophil chemotactic cytokine is not the ortholog of mouse YM1. J Immunol. 2005;175:2041–2. doi: 10.4049/jimmunol.175.4.2041-a. [DOI] [PubMed] [Google Scholar]
- 24.Chou YT, Yao S, Czerwinski R, Fleming M, Krykbaev R, Xuan D, et al. Kinetic characterization of recombinant human acidic mammalian chitinase. Biochemistry. 2006;45:4444–54. doi: 10.1021/bi0525977. [DOI] [PubMed] [Google Scholar]
- 25.De Ceuninck F, Gaufillier S, Bonnaud A, Sabatini M, Lesur C, Pastoureau P. YKL-40 (cartilage gp-39) induces proliferative events in cultured chondrocytes and synoviocytes and increases glycosaminoglycan synthesis in chondrocytes. Biochem Biophys Res Commun. 2001;285:926–31. doi: 10.1006/bbrc.2001.5253. [DOI] [PubMed] [Google Scholar]
- 26.Ling H, Recklies AD. The chitinase 3-like protein human cartilage glycoprotein 39 inhibits cellular responses to the inflammatory cytokines interleukin-1 and tumour necrosis factor-alpha. Biochem J. 2004;380:651–9. doi: 10.1042/BJ20040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malinda KM, Ponce L, Kleinman HK, Shackelton LM, Millis AJ. Gp38k, a protein synthesized by vascular smooth muscle cells, stimulates directional migration of human umbilical vein endothelial cells. Exp Cell Res. 1999;250:168–73. doi: 10.1006/excr.1999.4511. [DOI] [PubMed] [Google Scholar]
- 28.Recklies AD, White C, Ling H. The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem J. 2002;365:119–26. doi: 10.1042/BJ20020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasprzewska A. Plant chitinases--regulation and function. Cell Mol Biol Lett. 2003;8:809–24. [PubMed] [Google Scholar]
- 30.van Eijk M, van Roomen CP, Renkema GH, Bussink AP, Andrews L, Blommaart EF, et al. Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity. Int Immunol. 2005;17:1505–12. doi: 10.1093/intimm/dxh328. [DOI] [PubMed] [Google Scholar]
- 31.Labadaridis I, Dimitriou E, Theodorakis M, Kafalidis G, Velegraki A, Michelakakis H. Chitotriosidase in neonates with fungal and bacterial infections. Arch Dis Child Fetal Neonatal Ed. 2005;90:F531–2. doi: 10.1136/adc.2004.051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verweij PE, Kerremans JJ, Voss A, Meis JF. Fungal contamination of tobacco and marijuana. Jama. 2000;284:2875. doi: 10.1001/jama.284.22.2875. [DOI] [PubMed] [Google Scholar]
- 33.Salo PM, Arbes SJ, Yin M, Cohn R, Sever M, Muilenberg M. Dustborne Alternaria alternata allergens in US beds; results from the first National Survey of Lead and Allergens in Housing. American Journal of Respiratory and Critical Care Medicine. 2004;169(suppl 7):A 368. [Google Scholar]
- 34.Bogden JD, Kemp FW, Buse M, Thind IS, Louria DB, Forgacs J, et al. Composition of tobaccos from countries with high and low incidences of lung cancer. I. Selenium, polonium-210, Alternaria, tar, and nicotine. J Natl Cancer Inst. 1981;66:27–31. [PubMed] [Google Scholar]
- 35.Jamois F, Ferrieres V, Guegan JP, Yvin JC, Plusquellec D, Vetvicka V. Glucan-like synthetic oligosaccharides: iterative synthesis of linear oligo-beta-(1,3)-glucans and immunostimulatory effects. Glycobiology. 2005;15:393–407. doi: 10.1093/glycob/cwi020. [DOI] [PubMed] [Google Scholar]
- 36.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–76. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]