Abstract
ATP: Co(I)rrinoid adenosyltransferases (ACAs) catalyze the transfer of the adenosyl moiety from ATP to cob(I)alamin via a four-coordinate cob(II)alamin intermediate. At present, it is unknown how ACAs promote the formation of the four-coordinate corrinoid species needed for activity. The published high-resolution crystal structure of the ACA from Lactobacillus reuteri (LrPduO) in complex with ATP and cob(II)alamin shows that the environment around the α face of the corrin ring consists of bulky hydrophobic residues. To understand how these residues promote the generation of the four-coordinate cob(II)alamin, variants of the human-type ACA enzyme from Lactobacillus reuteri (LrPduO) were kinetically and structurally characterized. These studies revealed that residue Phe112 is critical in the displacement of 5,6-dimethylbenzimidazole (DMB) from its coordination bond with the Co ion of the ring, resulting in the formation of the four-coordinate species. An F112A substitution resulted in a 80% drop in the catalytic efficiency of the enzyme. The explanation for this loss of activity was obtained from the crystal structure of the mutant protein, which showed cob(II)alamin bound in the active site with DMB coordinated to the cobalt ion. The crystal structure of an LrPduOF112H variant showed a DMB-off/His-on interaction between the corrinoid and the enzyme, whose catalytic efficiency was four orders of magnitude lower than that of the wild-type protein. The analysis of the kinetic parameters of LrPduOF112H suggests that the F112H substitution negatively impacts product release. Substitutions of other hydrophobic residues in the Cbl binding pocket did not result in significant defects in catalytic efficiency in vitro, however, none of the variant enzymes analyzed in this work supported AdoCbl biosynthesis in vivo.
Keywords: corrinoid adenosylation, 4-coordinate corrinoids, coenzyme B12 synthesis, human-type corrinoid adenosyltransferases, coenzyme metabolism, B12 biosynthesis enzymology
The chemistry of B12-dependent reactions and the biosynthesis of this complex coenzyme have been an area of intense investigation for decades. Coenzyme B12 (adenosylcobalamin, AdoCbl) and methylcobalamin (MeCbl) are the two biologically active forms of B12. AdoCbl participates in radical-based intramolecular rearrangements (1–3), deaminations (4), dehydrations (5), reductions (6, 7), and reductive dehalogenations (8), while Cbl serves as a transient methyl carrier in methylation reactions (9–11). Despite the range of chemical reactions carried out by these forms of B12, MeCbl and, in some cases, AdoCbl bind to enzymes in very similar conformations. For example, the crystal structures of methylmalonyl-CoA mutase (MMCM, AdoCbl-dependent) and methionine synthase (MeCbl-dependent) revealed Cbl bound in the active site in a base-off/His-on conformation (12, 13). In contrast, the active sites of the bacterial and human ATP:Co(I)rrinoid adenosyltransferases do not have a histidinyl residue in close proximity to the cobalt ion, suggesting that catalysis does not occur via a DMB-off/His-on intermediate.
One fascinating aspect of the corrinoid adenosylation reaction is the reduction of the Co(II) corrinoid substrate. While the environment inside the cell is sufficiently reductive to drive the Co3+/2+ reduction, (14), it is not low enough to drive the Co2+/1+ reduction (15, 16). ACA enzymes bind cob(II)alamin and facilitate its reduction by generating a four-coordinate cob(II)alamin species in the active site (17–20). The four-coordinate species lacks axial ligands, the absence of which stabilizes the 3dz2 orbital and raises the Co2+/1+ reduction midpoint potential to within the range of reducing agents inside the cell (17). By binding cob(II)alamin in the active site, ACA enzymes generate a Co1+ “super-nucleophile” and prevent its quenching by deleterious side reaction. At present, however, there is no direct evidence that ACA enzymes deficient in their ability to generate the four-coordinate cob(II)alamin are impaired in catalyzing the adenosylation reaction.
The crystal structure of the Lactobacillus reuteri PduO enzyme in complex with ATP and cob(II)alamin confirmed the existence of the four-coordinate cob(II)alamin intermediate (21). The protein environment around the vacant α-axial ligand region consists of several bulky and hydrophobic residues positioned in the proximity of the cob(II)alamin substrate (21). The role of these residues in the generation of the four-coordinate intermediate remains unclear. Kinetic and structural analyses reported here provide insights into the role of hydrophobic residues in the generation of the four-coordinate cob(II)alamin intermediate. We also report the structure and kinetic behavior of LrPduO with Cbl bound in the base-off/His-on conformation, and discuss possible reasons why ACA enzymes do not bind Cbl in the His-on form.
EXPERIMENTAL PROCEDURES
Protein production and purification
Variants of LrPduO were generated using the QuickChange® XL site-directed mutagenesis kit (Stratagene). The pTEV3 plasmid (22) carrying the wild-type Lr pduO+ allele (23) was used as a template for polymerase chain reaction (PCR)-based site-directed mutagenesis as per the manufacturer’s instructions. The presence and nature of the mutations were verified by sequencing the plasmids using non-radioactive BigDye® (ABI-PRISM) protocols. Sequencing reactions were resolved at the DNA sequence facility of the Biotechnology Center of the University of Wisconsin-Madison. Recombinant LrPduO variants were overexpressed in Escherichia coli strain BL21 (DE3). Derivatives of plasmid pTEV3 carrying L.r. pduO alleles encoding LrPduO variants with a rTEV protease-cleavable N-terminal His6 tag were constructed, genes were overexpressed, proteins were isolated using Ni2+ affinity chromatography as described elsewhere (23); N-terminal tags were cleaved using rTEV protease (24). Flavodoxin (FldA) and ferredoxin (flavodoxin):NADP+ reductase (Fpr) were produced and purified as described (25, 26).
Crystallization and data collection
LrPduO variant proteins were produced and purified using described protocols (23). All crystals of tag-less LrPduO proteins were grown using the vapor diffusion method in an anoxic chamber at 25°C. The protein concentration in the crystallization solutions was 12 mg/ml, 15 mg/ml and 11 mg/ml for the LrPduOF112A, LrPduOF112H, and LrPduOΔ183 variants, respectively. Crystals of the pre-catalytic Cbl complex for the LrPduOF112A variant (LrPduOF112A:ATP:cob(II)alamin) were grown by mixing 3.5 μl of protein solution containing E. coli FMN reductase (Fre; 30 μg/ml), NADH (50 mM), FMN (10 mM), HOCbl (10 mM), ATP (10 mM), MgCl2 (10 mM), and NaCl (300 mM) with 3.5 μl of reservoir solution [polyethylene glycol 8000 (14%(w/v)); KCl (200 mM); 2-(N-morpholino)ethanesulfonic acid (MES, 100 mM, pH 6). Crystals of the pre-catalytic Cbl complex for the LrPduOΔ183–188 protein were grown by mixing 3.5 μl of protein solution containing Fre (30 μg/ml), NADH (10 mM), FMN (2 mM), HOCbl (2 mM), ATP (2.5 mM), MgCl2 (2.5 mM), and NaCl (300 mM) with 3.5 μl of reservoir solution [polyethylene glycol 8000 (14%(w/v)); KCl (200 mM); MES (100 mM, pH 6)]. Crystals of the pre-catalytic Cbl complex for the LrPduOF112H protein were grown by mixing 3.5 μl of protein solution containing Fre (30 μg/ml), NADH (10 mM), FMN (2 mM), HOCbl (2 mM), ATP (2.5 mM), MgCl2 (2.5 mM), and NaCl (300 mM) with 3.5 μl of reservoir solution [polyethylene glycol (PEG) 8000 (14%(w/v)]; KCl (200 mM); 3-[4-(2-hydroxyethylpiperazin-1-yl]propane-1-sulfonic acid (HEPPS, 100 mM, pH 8.6). Several large, brown-colored, cubic crystals (~200 μm3) appeared spontaneously after 24 h and were grown for an additional 48 h. The crystals for each of the LrPduO proteins were transferred to an anoxic synthetic mother liquor solution [glycerol (2% v/v); PEG 8000 (10%(w/v)]; MES (100 mM, pH 6) or, for LrPduOF112H, HEPPS (100 mM, pH 8.6); KCl (200 mM), Fre (35 μg/ml); NADH (10 mM); FMN (2 mM); HOCbl (2 mM); ATP (2 mM); MgCl2 (2.5 mM)] and, in an anoxic chamber, incrementally transferred in five steps to an anoxic cryoprotectant solution [glycerol (20%(v/v)), PEG 8000 (12%(w/v)); MES (100 mM, pH 6) or, for LrPduOF112H, HEPPS (100 mM, pH 8.6); KCl (300 mM); Fre (35 μg/ml ); NADH (10 mM); FMN (2 mM); HOCbl (2 mM); ATP (2 mM); MgCl2 (2.5 mM)]. The crystals were briefly exposed to oxygen (≤5 s) while they were flash frozen in liquid nitrogen.
All crystals belong to the space group R3 with one subunit in the asymmetric unit. Data sets were collected at the Advanced Photon Source in Argonne, IL on the 19BM beamline. Diffraction data were integrated and scaled with the program HKL2000 (27). Data collection statistics are summarized in table 1.
Table 1.
Data collection and refinement statistics
| LrPduOF112A | LrPduOF112H | LrPduOΔ183 | |
|---|---|---|---|
| Space Group | R 3 | R 3 | R 3 |
| Cell Dimensions | |||
| a, b, c (Å) | 80.7, 80.7, 89.7 | 80.9, 80.9, 90.0 | 67.7, 67.7, 111.2 |
| α, β, γ(°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution range, Å | 30.0–1.48 (1.52–1.48)a | 30.0–1.17 (1.20–1.17)a | 30.0–1.38 (1.43–1.38)a |
| Redundancy | 5.6 (5.0) | 4.4 (2.0) | 6.3 (3.1) |
| Completeness (%) | 99.7 (98.6) | 99.6 (97.3) | 97.3 (82.0) |
| Unique Reflections | 36 186 | 73 398 | 38 038 |
| Rmerge (%) | 5.7 (12.8) | 7.0 (17.9) | 6.3 (9.7) |
| Average I/σ | 46.2 (13.4) | 41.4 (6.2) | 47.7 (14.3) |
| Rcryst | 0.156 (0.150) | 0.161 (0.202) | 0.162 (0.154) |
| Rfree | 0.174 (0.172) | 0.182 (0.203) | 0.181 (0.169) |
| No. protein atoms | 1543 | 1619 | 1517 |
| No. water molecules | 158 | 211 | 167 |
| Wilson B-value (Å2) | 23.4 | 15.1 | 14.3 |
|
| |||
| Average B-factors (Å2) | |||
| Protein | 17.8 | 13.4 | 12.9 |
| Ligands | 20.0 | 14.7 | 14.9 |
| Solvent | 32.3 | 29.0 | 26.9 |
|
| |||
| Ramachandran (%) | |||
| Most favored | 96.4 | 96.5 | 97.6 |
| Additionally allowed | 3.6 | 3.5 | 2.4 |
| Generously allowed | 0 | 0 | 0 |
| Disallowed | 0 | 0 | 0 |
|
| |||
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.014 | 0.022 | 0.012 |
| Bond angles (°) | 2.20 | 2.85 | 2.09 |
Values in parentheses are for the highest resolution bin
Structure determination and refinement
The structures were determined by molecular replacement with the program MOLREP (28) starting from the model of the wild-type LrPduO protein in complex with ATP (PDB 2NT8). Final refinement was carried out with the program REFMAC (29). Heteroatoms, water molecules and multiple conformations were built using the program COOT (30). The final models for (LrPduOF112A:ATP:cob(II)alamin), (LrPduOF112H:ATP:cob(II)alamin), and (L r P d u OΔ183–188: ATP:cob(II)alamin) were refined to 1.5Å, 1.2Å, and 1.4Å, respectively. The models include residues 2–182, 1–188 and 2–181 for LrPduOF112A, LrPduOF112H and LrPduOΔ183, respectively. Ramachandran plots for all models show that greater than 96% of residues are in the most favored region with no residues falling in the disallowed region. Refinement statistics are listed in table 1.
In vitro adenosylation activity assays
The Co+ assays were performed using the continuous spectrophotometric method described previously, without modifications (23). In this assay, the cobalt ion of Cbl is chemically reduced in solution to cob(I)alamin by Ti(III)citrate. The Co+ assay was performed under anoxic condition and the adenosylation reaction was initiated by the addition of LrPduO. The reaction mixture included 2-amino-2-hydroxymethylpropane-1,3-diol hydrochloride (Tris-HCl, 0.2 M; pH 8 at 37 °C), MgCl2 (1.5 mM), HOCbl (0.1 – 20 μM), and ATP (1 μM-1 mM). The Co2+ assay was modified from a previously used end-point assay (26) to allow the appearance of product (AdoCbl) to be continuously monitored in the presence of a protein reducing system. The Co2+ assays were performed under anoxic conditions at 37 °C. Empty quartz cuvettes were flushed with oxygen-free N2 for 5 min. Under a stream of O2-free N2, 2-amino-2-hydroxymethylpropane-1,3-diol hydrochloride (Tris-HCl, 0.2 M; pH 8 at 37 °C), KCl (0.1 M), MgCl2 (1.5 mM), HOCbl (4 μM-0.2 mM), Fpr (73 μg/ml), FldA (0.6 mg/ml), NADPH (1 mM), and ATP (1 μM-1 mM) were added to the cuvette in the order stated. To ensure that all cob(III)alamin was reduced to cob(II)alamin, reaction mixtures were incubated at 37°C for 10 min. The adenosylation reaction was initiated by the addition of LrPduO protein. The appearance of AdoCbl was monitored at 525 nm. To minimize the contribution from photolysis, initial velocities were measured within the first five min after reaction was initiated. The photolysis of AdoCbl at 525 nm over the first 5 min was determined to be insignificant (data not shown). The reduction of cob(II)alamin was monitored at 473 nm using the Co2+ assay.
In vivo assessment of function
The function of LrPduO proteins was also assessed in vivo. For this purpose we used a strain of Salmonella enterica serovar Typhimurium LT2 that carried a chromosomal deletion of the cobA that encodes the housekeeping ATP:Co(I)rrinoid adenosyltransferase. CobA is needed for salvaging cobinamide (Cbi), a precursor of AdoCbl (31, 32). As shown elsewhere, the L.r. pduO+ gene substitutes for cobA+ during growth of a cobA strain on minimal medium supplemented with glycerol (30 mM) and dicyano-Cbi [(CN)2Cbi, 150 nM] (23, 33).
RESULTS AND DISCUSSION
Spectroscopic and structural analyses have shown that PduO-type ACA enzymes facilitate the thermodynamically unfavorable Co2+/1+ reduction of the cobalt ion in corrinoids by generating a four-coordinate Co(II) corrinoid species with no axial (α and β) ligands (17, 18). To gain insights into the molecular basis of the formation of the four-coordinate Co(II) corrinoid species in PduO-type ACA enzymes, we performed structural and kinetic analysis of LrPduO enzyme variants.
We used two assays to distinguish between the two functions of LrPduO, namely, assistance in the Co2+/1+ reduction, and Co(I) corrinoid adenosylation. In one assay, we used the NADPH-dependent flavodoxin protein reductase (Fpr)/flavodoxin (FldA) system to reduce Co2+ to Co1+; hereafter this assay is referred to as Co2+ assay. In the Co2+ assay, the PduO enzyme must bind cob(II)alamin and facilitate the generation of cob(I)alamin in its active site. Conversely, in the second assay, referred to as Co+ assay, we used Ti(III)citrate to reduce Co2+ to Co+ in solution, allowing the cob(I)alamin adenosylation reaction to be measured directly. Cobalamin was used as the corrinoid substrate in both assays. Co2+ assay allowed us to indirectly assess the formation of the four-coordinate cob(II)alamin species, whereas Co+ assay, allowed us to assess the catalytic competency of the variants. We used both assays in combination with high-resolution crystal structures to identify residues involved in the formation of the four-coordinate cob(II)alamin species.
Residue Phe112 displaces the lower ligand
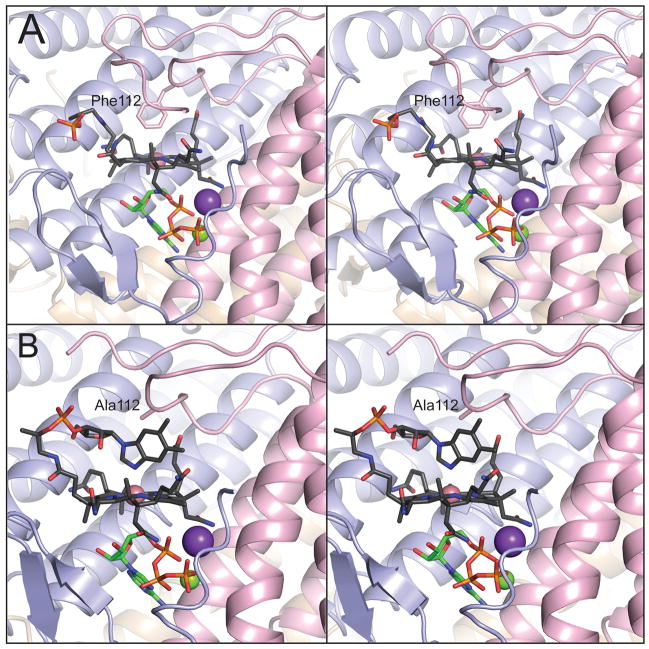
The crystal structure of LrPduO in complex with cob(II)alamin and ATP revealed cob(II)alamin bound in a conformation with the lower axial ligand (DMB) excluded from the active site (21). Residue Phe112 is positioned 3.8 Å away from the cobalt ion, suggesting that this residue plays an important role in the formation of the four-coordinate cob(II)alamin intermediate (21, 34, 35). To further investigate the role of Phe112, we changed the coding sequence of the L.r. pduO+ gene to encode variant protein LrPduOF112A. The latter was crystallized under anoxic conditions and its X-ray structure was determined at 1.5 Å resolution. The crystal structure of LrPduOF112A revealed cob(II)alamin bound to the active site as a five-coordinate species with DMB serving as a fifth axial ligand (Fig. 1). The LrPduOF112A variant displayed extremely low activity in Co2+ assay. However, in the Co+ assay (adenosylation of cob(I)alamin) LrPduOF112A was catalytically competent and displayed only a slight decrease in (~4-fold) (Table 3). This result is consistent with the idea that, in the absence of a four-kcat coordinate cob(II)alamin species, the enzyme is inactive. The kinetic parameters of the LrPduOF112A enzyme support the idea that the steric bulk presented by the phenyl moiety of the side chain of Phe112 is critical for the formation of the four-coordinate species and hence to the reactivity of the enzyme.
FIGURE 1.
Phe112 displaces the lower ligand of cob(II)alamin in LrPduO to generate a four-coordinate intermediate. Stereoviews of the active sites of (A) wild-type LrPduO (PDB ID: 3CI1, St. Maurice et al., 2008) and (B) LrPduOF112A with the individual subunits of the trimer represented as ribbons colored in blue, red and brown. For clarity, only the active site at the interface of the red and blue subunits is shown. The carbon atoms of cob(II)alamin are colored in black while the carbon atoms of ATP are colored in green. The potassium (purple sphere) and magnesium (green sphere) atoms are also displayed. The electron density corresponding to the DMB portion of cob(II)alamin is disordered in the wild-type structure.
Table 3.
Kinetic parameters of LrPduO variants using the Co+ assay. The substrate cob(I)alamin was generated chemically using Ti(III)citrate
| Enzyme | ATP | Cob(I)alamin | ||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| wild type | 2.2 ± 0.1 | (2.6 ± 0.1) × 10−2 | (1.2 ± 0.1) × 104 | 0.13 ± 0.01 | (2.4 ± 0.1) × 10−2 | (1.8 ± 0.2) × 105 |
| F112A | 34.6 ± 8.8 | (5.6 ± 0.0) × 10−3 | (1.7 ± 0.4) × 102 | 1.9 ± 0.5 | (6.6 ± 0.5) × 10−3 | (3.6 ± 1.0) × 103 |
| F112H | 2.6 ± 0.0 | (3.8 ± 0.1) × 10−4 | (1.4 ± 0.1) × 102 | 2.8 ± 0.4 | (4.2 ± 0.1) × 10−4 | (1.5 ± 0.2) × 102 |
| F112Y | 1.2 ± 0.1 | (1.6 ± 0.0) × 10−3 | (1.4 ± 0.1) × 103 | 0.55 ± 0.05 | (1.7 ± 0.0) × 10−3 | (3.1 ± 0.3) × 103 |
| F112W | 2.0 ± 0.6 | (1.8 ± 0.2) × 10−3 | (9.8 ± 3.4) × 102 | 0.77 ± 0.12 | (2.0 ± 0.2) × 10−3 | (2.6 ± 0.5) × 103 |
| F163A | 15.9 ± 6.8 | (1.5 ± 0.1) × 10−2 | (1.8 ± 0.4) × 103 | 0.19 ± 0.03 | (1.5 ± 0.3) × 10−2 | (7.9 ± 1.9) × 104 |
| F187A | 1.2 ± 0.4 | (2.1 ± 0.4) × 10−2 | (1.7 ± 0.6) × 104 | 0.11 ± 0.01 | (1.8 ± 0.1) × 10−2 | (1.6 ± 0.2) × 105 |
| ΔS183 | 3.1 ± 0.2 | (2.3 ± 0.6) × 10−2 | (7.5 ± 2.2) × 103 | 0.07 ± 0.06a | (2.0 ± 0.2) × 10−2 | 3.95 ± × 105 |
significant error due to absorbance near detection limit
Effects of a F112H substitution
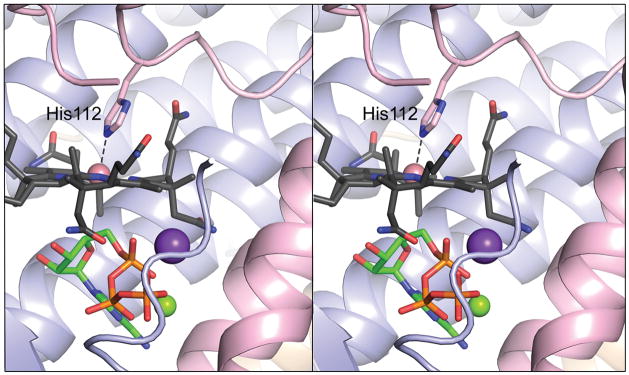
Several Cbl-dependent enzymes share the Cbl-binding motif “DXHXXG”, in which the histidinyl residue coordinates to the cobalt ion (36) generating a DMB-off/His-on conformation of the cofactor (12, 37); the DXHXXG motif is not present in any ACA enzyme. The proximity of residue Phe112 to the cobalt ion of Cbl offered an opportunity to generate a DMB-off/His-on ACA enzyme by site-directed mutagenesis, allowing us to assess the effect of a DMB-off/His-on complex on the activity of the enzyme. The LrPduOF112H variant was constructed, and its X-ray crystal structure was determined to 1.5 Å resolution. The structure of LrPduOF112H confirmed that cob(II)alamin was bound in a DMB-off/His-on conformation (Fig. 2). The F112H substitution resulted in a substantial drop in kcat (~60-fold) in assays Co2+ and Co+ (Tables 2, 3), suggesting that LrPduOF112H was impaired in its ability to catalyze the adenosylation of cob(I)alamin and/or release the AdoCbl product. Notably, the specific activity of LrPduOF112H for the reduction of cob(II)alamin to cob(I)alamin was only 19-fold lower than that of the wild-type LrPduO protein (1.5 vs. 28 mM Co1+ min−1 mg−1, respectively).
FIGURE 2.
His112 acts as a lower ligand to five-coordinate cob(II)alamin in LrPduOF112H. Stereoview of LrPduOF112H with the individual subunits of the trimer represented as ribbons colored in blue, red and brown. For clarity, only the active site at the interface of the red and blue subunits is shown. The carbon atoms of cob(II)alamin are colored in black while the carbon atoms of ATP are colored in green. The potassium (purple sphere) and magnesium (green sphere) atoms are also displayed. The electron density corresponding to the DMB portion of cob(II)alamin is disordered and, therefore, DMB could not be modeled.
Table 2.
Kinetic parameters of LrPduO wild type and variants using the Co2+ assay. The substrate cob(II)alamin was generated using a protein reducing system
| Enzyme | ATP | Cob(II)alamin | ||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| wild type | 5.5 ± 1.1 | (2.9 ± 0.1) × 10−2 | (5.5 ± 1.1) × 103 | 7.8 ± 1.1 | (3.8 ± 0.5) × 10−2 | (4.8 ± 0.9) × 103 |
| F112A | UDa | UD | UD | UD | ||
| F112H | 10.4 ± 0.8 | (5.9 ± 0.2) × 10−4 | (5.7 ± 0.5) × 101 | 53.0 ± 8.6 | (6.7 ± 0.2) × 10−4 | (1.3 ± 0.2) × 101 |
| F112Y | 9.4 ± 0.4 | (2.3 ± 0.1) × 10−3 | (2.5 ± 0.2) × 102 | 34.2 ± 10.7 | (2.7 ± 0.4) × 10−3 | (8.3 ± 2.8) × 101 |
| F112W | 7.3 ± 0.1 | (2.8 ± 0.1) × 10−3 | (3.9 ± 0.2) × 102 | 28.8 ± 5.7 | (3.2 ± 0.1) × 10−3 | (1.1 ± 0.2) × 102 |
| F163A | 96.1 ± 7.1b | (1.5 ± 0.1) × 10−2 b | (1.6 ± 0.2) × 102 b | 134 ± 13 | (2.7 ± 0.4) ×10−2 | (2.0 ± 0.3) × 102 |
| F187A | 7.9 ± 1.0 | (1.4 ± 0.1) × 10−2 | (1.8 ± 0.3) × 103 | 15.8 ± 3 | (1.5 ± 0.1) × 10−2 | (9.3 ± 1.9) × 102 |
| ΔS183 | 9.9 ± 0.9 | (1.7 ± 0.1) × 10−2 | (1.7 ± 0.2) × 103 | 16.3 ± 1.7 | (1.8 ± 0.1) × 10−2 | (1.1 ± 0.1) × 103 |
Unable to determine constants due to extremely low activity
Kinetic parameters obtained under sub-saturating concentrations of cob(II)alamin
Saturating ATP was at least 100-fold the KM, however saturating cob(II)alamin was limited to 3- to 10- fold the KM as a result of assay sensitivity.
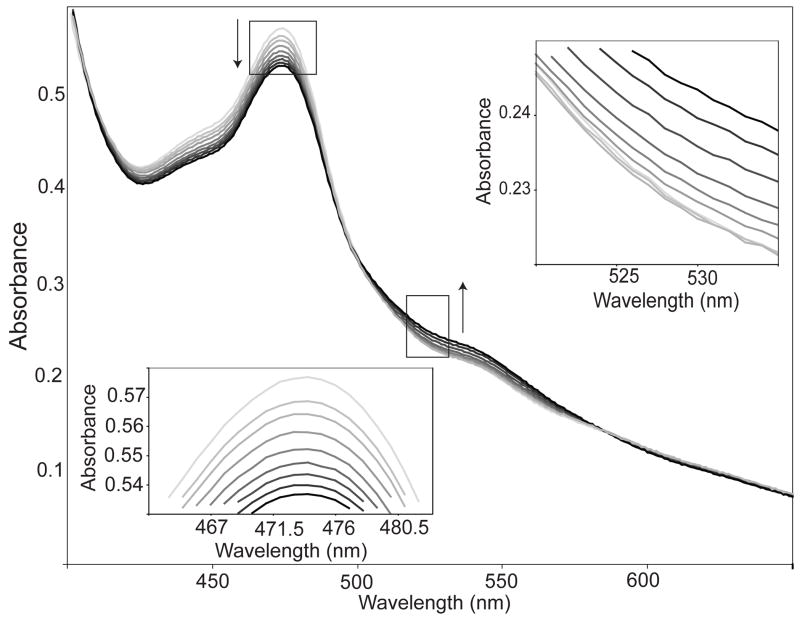
We also measured a ~5-min lag in the reduction of cob(II)alamin (Co2+ assay), but not in the adenosylation reaction (Co+ assay); the observed lag did not depend on the concentration of the reagents or substrates (data not shown). Overlays of spectral scans (from 300 nm to 700 nm) obtained at different times showed a lag in the generation of product (i.e., AdoCbl; 525 nm), but not in the reduction of the substrate (Fig. 3). Since Co2+ assay is essentially a coupled assay (i.e., reduction followed by adenosylation), the observed lag is consistent with cob(I)alamin adenosylation being the rate-limiting step in the reaction catalyzed by LrPduOF112H. Reaction lags are typically observed in coupled assays where the second step in the reaction is rate limiting (38).
FIGURE 3.
Spectral changes associated with the enzymic conversion of cob(II)alamin to AdoCbl by LrPduOF112H. Arrows and increasing darkness in the spectra represent successive time points after reaction was initiated by the addition of ATP (light gray, 1min; black, 11 min). LrPduOF112H facilitates the reduction of cob(II)alamin (as evidenced by the decrease in absorbance at 473 nm) however there is a lag in the generation of AdoCbl (as evidenced in initial lag in absorbance at 525 nm).
The severity of the effect of different α ligands on LrPduO activity varies substantially
Even though LrPduOF112A and LrPduOF112H proteins bound cob(II)alamin as a five-coordinate species (Figs. 1B, 2), their enzymatic activity was vastly different. The critical distinction between these two variants lies in the nature of the α ligand. In LrPduOF112H, the α ligand is a histidinyl side chain of the enzyme, whereas in LrPduOF112A the α ligand is DMB. The explanation for the kinetic differences is not immediately obvious since the Co-N coordination bond in both variants is very similar in the crystal structures. The apparent similarity, however, may be imposed by the packing in the crystals. Indeed, recent spectroscopic analysis of these variants revealed the coordination-bond between Co-N(His112) to be longer than the Co-N(DMB) (Kiyoung et al. manuscript in preparation). A longer coordination bond of Co-N(His112) in LrPduOF112H would result in a faster displacement of the lower ligand, therefore a higher kcat in Co2+ assay.
To explain the data shown in figure 3, we propose that the reduced kcat of LrPduOF112H may be the result of a slower nucleophilic attack by the Co ion of cob(I)alamin on the 5′-carbon of ATP, and/or a slow product release. Considering that the Co-C bond in AdoCbl is formed with cob(I)alamin (a 4-coordinate Cbl species), wild-type LrPduO may first generate an AdoCbl/DMB-off product, a cob(III)alamin species that is energetically unstable in the absence of an α axial ligand. Consequently, a DMB-on conformation would be favored prior to, or concomitant with, product release. In LrPduOF112H, the coordination bond with N(His) may stabilize Co3+ of AdoCbl, resulting in an AdoCbl/His-on species. For product to be released, AdoCbl/His-on must first undergo the thermodynamically unfavorable step that generates AdoCbl/DMB-off. This scenario would be consistent with a slow product-release step and would explain the equivalent drop in kcat of LrPduOF112H with cob(I)alamin and cob(II)alamin relative to the wild-type enzyme. Similarly, other Phe112 variants (i.e., LrPduOF112Y, LrPduOF112W) retained their ability to adenosylate cob(II)alamin albeit not as efficiently as the wild-type protein (Table 2). The kcat of variants LrPduOF112W and LrPduOF112Y decreased ~10-fold in both assays, suggesting a problem with product realease as the one observed with LrPduOF112H. This is possible since Trp112 and Tyr112 have functional groups that can interact with the cobalt ion of Cbl.
In contrast with ACA enzymes, Cbl-dependent enzymes that bind Cbl in its base-off/His-on form would not have the problem of product release because Cbl stays bound to the enzyme acting as a radical initiator or methyl carrier. In PduO-type enzymes, phenylalanine is the ideal residue for PduO-type enzymes because it is bulky enough to displace the lower ligand of Cbl, and does not coordinate with the cobalt ion of Cbl, hence product release can be rapid. Further studies of product release in wild-type LrPduO and LrPduOF112H proteins are needed to help understand the effect of the Co-N(His) coordination in LrPduOF112H.
Function of other hydrophobic residues in the Cbl-binding site of LrPduO
The structure of LrPduO in complex with ATP and cob(II)alamin shows a hydrophobic pocket with aromatic residues around the corrin ring. In hemoproteins, interactions between heme and aromatic side chains have been proposed to stabilize the holoprotein fold, and to contribute to the high affinity of the protein for heme (39). To determine whether additional aromatic residues play a role in PduO-mediated catalysis, two phenylalanine residues, Phe163 and Phe187, located inside the hydrophobic pocket were changed by site-directed mutagenesis.
Phe163
Residue Phe163 is conserved in all PduO-type enzymes, except in S. enterica PduO, where tyrosine occupies this position. The function of the structurally equivalent residue in the human ACA enzyme was suggested to be involved in Cbl binding (34, 35). Notably, the Km of LrPduOF163A for cob(II)alamin increased significantly (17-fold), but the kcat remained unchanged (Table 2). In contrast, the Km and the kcat of LrPduOF163A for cob(I)alamin were very similar to those of the wild-type enzyme (Table 3). Together, these results suggest that Phe163 and DMB interact when Cbl binds to the active site of LrPduO. The negative effect of the substitution is not observed with cob(I)alamin because this form of Cbl does not have the axial ligands.
Phe187
Although residue Phe187 is not conserved, it is part of the C-terminal loop (Ser183-Arg188) that becomes ordered upon binding of the corrinoid substrate (21). The kinetic parameters of LrPduOF187A for cob(I)alamin and cob(II)alamin were not significantly different to the parameters of the wild type enzyme (Tables 2, 3).
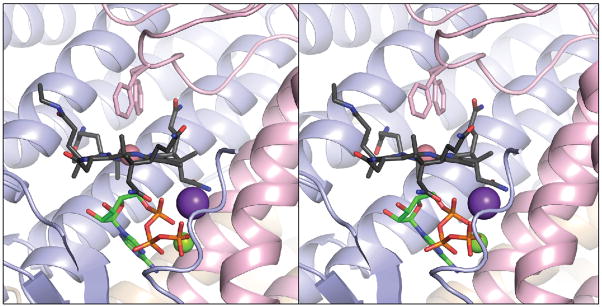
To analyze the contribution of the C-terminal loop to the function of LrPduO, we deleted the last six amino acid residues of the protein (i.e., Ser183 to Arg188). The crystal structure of the LrPduOΔ183–188 protein was determined to 1.4 Å resolution (Fig. 4). The absence of the C-terminal residues resulted in greater conformational freedom for residue Phe112 in the active site, with the electron density suggesting two alternative conformations for the side chain of Phe112. The crystal structure of the LrPduOΔ183–188 protein also showed that a four-coordinate cob(II)alamin intermediate could still form in the active site of the enzyme (Fig. 4). Consistent with this finding, the catalytic efficiency of the LrPduOΔ183–188 protein was only modestly reduced (3–4-fold) (Tables 2, 3), suggesting that the C terminus of the proteins can be modified or truncated without profoundly impacting the catalytic efficiency of the enzyme.
FIGURE 4.
The LrPduOΔ183 variant retains the ability to generate a four-coordinate intermediate. Stereoview of LrPduOΔ183 with the individual subunits of the trimer represented as ribbons colored in blue, red and brown. For clarity, only the active site at the interface of the red and blue subunits is shown. The carbon atoms of cob(II)alamin are colored in black while the carbon atoms of ATP are colored in green. The potassium (purple sphere) and magnesium (green sphere) atoms are also displayed. Residue Phe112 adopts two alternate conformations in this structure. Both conformations are displayed. The electron density corresponding to the DMB portion of cob(II)alamin is disordered and, therefore, DMB could not be modeled.
In vivo functionality of LrPduO variants
None of the variants, even those with measurable activity, supported AdoCbl biosynthesis in vivo (data not shown). The fact that LrPduOΔ183–188 protein did not support in vivo biosynthesis of AdoCbl, in spite of its modest decrease in activity, suggest that this part of the protein may be important for protein-protein interactions. ATP corrinoid adenosyltransferases are thought to transport this valuble, but limited coenzyme, to the corresponding AdoCbl-dependent enzyme (40). In the human adenosyltransferase, a truncation of the final 16 residues has been shown to result in an early onset of methylmalonic aciduria (41).
CONCLUSIONS
Here we report structural and kinetic data to support the idea that the inability to generate the four-coordinate cob(II)alamin intermediate renders a human-type ACA enzyme inactive. The side chain of residue Phe112 in the active site of the enzyme is critical to the function of the enzyme for three reasons: i) it is in close proximity to the Co ion of the ring; ii) it is bulky; and iii) it cannot form a coordination bond with the Co ion. Hence, when cob(II)alamin binds to the enzyme, the lower ligand is displaced and the resulting four-coordinate cob(II)alamin species remains as such because the phenyl side chain cannot form a coordination bond with the Co ion.
Acknowledgments
We thank Thomas Brunold and Kiyoung Park (UW-Madison, Chemistry Department) for helpful discussions and critical review of the manuscript. The Structural Biology BM19 beamline Argonne National Laboratory Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Footnotes
Work in the Escalante-Semerena lab was supported by NIH grant R01-GM40313 to J.C.E.-S. Work in the Rayment lab was supported by NIH grant AR35186. P.E.M. was supported in part by NIH grant NRSA F31-GM081979.
Abbreviations: ACA, ATP:Co(I)rrinoid adenosyltransferase; LrPduO, Lactobacillus reuteri PduO; SePduO, Salmonella enterica PduO; Cbl, cobalamin; cbi, cobinamide; AdoCbl, adenosylcobalamin; HOCbl, hydroxycobalamin; DMB, 5,6-dimethylbenzimidazole; MMCM, methylmalonyl-CoA mutase; Tris-HCl, tris(hydroxymethyl)aminomethane hydrochloride; FMN, flavin mononucleotide; FldA, flavodoxin; Fpr, ferredoxin (flavodoxin):NADPH reductase; Fre, FMN reductase; MES, morpholinoethanefulfonic acid; NADH, reduced nicotinamide adenine dinucleotide.
The atomic coordinates and structure factors for the complexes of LrPduOF112H, LrPduOF112A, and LrPduOΔS183 in complex with ATP and cob(II)alamin have been deposited to the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rscb.org) under the PDB accession numbers 3GAH, 3GAI, and 3GAJ, respectively.
References
- 1.Halpern J. Mechanisms of coenzyme B1 2-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R, Chowdhury S. Methylmalonyl-CoA mutase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 707–729. [Google Scholar]
- 3.Buckel W, Bröker G, Bothe H, Pierik A. Glutamate mutase and 2-Methylglutarate mutase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 757–782. [Google Scholar]
- 4.Bandarian V, Reed GH. Ethanolamine ammonia-lyase. In: Banerjee R, editor. Chemistry ad Biochemistry of B12. John Wiley & Sons, Ic; New York: 1999. pp. 811–833. [Google Scholar]
- 5.Toraya T. Diol dehydratase and glycerol dehydratase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 783–809. [Google Scholar]
- 6.Fontecave M. Ribonucleotide reductases and radical reactions. Cell Mol Life Sci. 1998;54:684–695. doi: 10.1007/s000180050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontecave M, Mulliez E. Ribonucleotide Reductases. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 731–756. [Google Scholar]
- 8.Wohlfarth G, Diekert G. Reductive dehalogenases. In: Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons, Inc; New York: 1999. pp. 871–893. [Google Scholar]
- 9.Gärtner P, Ecker A, Fischer R, Linder D, Fuchs G, Thauer RK. Purification and properties of N5- methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1993;213:537–545. doi: 10.1111/j.1432-1033.1993.tb17792.x. [DOI] [PubMed] [Google Scholar]
- 10.Taylor RT, Weissbach H. N5-methylenetetrahydrofolate-homocysteine methyltransferases. In: Boyer PD, editor. The Enzymes. Academic Press, Inc; New York: 1973. pp. 121–165. [Google Scholar]
- 11.van der Meijden P, Brommelstroet BWt, Poiroit CM, van der Drift C, Vogels GD. Purification and properties of methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J Bacteriol. 1984;160:629–635. doi: 10.1128/jb.160.2.629-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Bosecke P, Diat O, Evans PR. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2Å resolution. Structure. 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 13.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B1 2: A 3.0Å X-ray structure of B1 2-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 14.Fonseca MV, Escalante-Semerena JC. Reduction of cob(III)alamin to cob(II)alamin in Salmonella enterica Serovar Typhimurium LT2. J Bacteriol. 2000;182:4304–4309. doi: 10.1128/jb.182.15.4304-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver L, Leadbeater C, Campopiano DJ, Baxter RL, Daff SN, Chapman SK, Munro AW. Characterisation of flavodoxin NADP+ oxidoreductase and flavodoxin; key components of electron transfer in Escherichia coli. Eur J Biochem. 1998;257:577–585. doi: 10.1046/j.1432-1327.1998.2570577.x. [DOI] [PubMed] [Google Scholar]
- 16.Olteanu H, Wolthers KR, Munro AW, Scrutton NS, Banerjee R. Kinetic and thermodynamic characterization of the common polymorphic variants of human methionine synthase reductase. Biochemistry. 2004;43:1988–1997. doi: 10.1021/bi035910i. [DOI] [PubMed] [Google Scholar]
- 17.Stich TA, Yamanishi M, Banerjee R, Brunold TC. Spectroscopic evidence for the formation of a four-coordinate Co(2+)cobalamin species upon binding to the human ATP:Cobalamin adenosyltransferase. J Am Chem Soc. 2005;127:7660–7661. doi: 10.1021/ja050546r. [DOI] [PubMed] [Google Scholar]
- 18.Stich TA, Buan NR, Escalante-Semerena JC, Brunold TC. Spectroscopic and computational studies of the ATP:Corrinoid adenosyltransferase (CobA) from Salmonella enterica: Insights into the mechanism of adenosylcobalamin biosynthesis. J Am Chem Soc. 2005;127:8710–8719. doi: 10.1021/ja042142p. [DOI] [PubMed] [Google Scholar]
- 19.Park K, Mera PE, Escalante-Semerena JC, Brunold TC. Kinetic and spectroscopic studies of the ATP:corrinoid adenosyltransferase PduO from Lactobacillus reuteri: substrate specificity and insights into the mechanism of Co(II)corrinoid reduction. Biochemistry. 2008;47:9007–9015. doi: 10.1021/bi800419e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanishi M, Labunska T, Banerjee R. Mirror “base-off” conformation of coenzyme B12 in human adenosyltransferase and its downstream target, methylmalonyl-CoA mutase. J Am Chem Soc. 2005;127:526–527. doi: 10.1021/ja044365l. [DOI] [PubMed] [Google Scholar]
- 21.St Maurice M, Mera P, Park K, Brunold TC, Escalante-Semerena JC, Rayment I. Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate. Biochemistry. 2008;47:5755–5766. doi: 10.1021/bi800132d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St Maurice M, Mera PE, Taranto MP, Sesma F, Escalante-Semerena JC, Rayment I. Structural characterization of the active site of the PduO-type ATP:Co(I)rrinoid adenosyltransferase from Lactobacillus reuteri. J Biol Chem. 2007;282:2596–2605. doi: 10.1074/jbc.M609557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall DA, Jordan-Starck TC, Loo RO, Ludwig ML, Matthews RG. Interaction of flavodoxin with cobalamin-dependent methionine synthase. Biochemistry. 2000;39:10711–10719. doi: 10.1021/bi001096c. [DOI] [PubMed] [Google Scholar]
- 26.Fonseca MV, Escalante-Semerena JC. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J Biol Chem. 2001;276:32101–32108. doi: 10.1074/jbc.M102510200. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the Maximum-Likelihood Method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Escalante-Semerena JC, Suh SJ, Roth JR. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh S, Escalante-Semerena JC. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J Bacteriol. 1995;177:921–925. doi: 10.1128/jb.177.4.921-925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schubert HL, Hill CP. Structure of ATP-bound human ATP:cobalamin adenosyltransferase. Biochemistry. 2006;45:15188–15196. doi: 10.1021/bi061396f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan C, Bobik TA. Functional characterization and mutation analysis of human ATP:Cob(I)alamin adenosyltransferase. Biochemistry. 2008;47:2806–2813. doi: 10.1021/bi800084a. [DOI] [PubMed] [Google Scholar]
- 36.Marsh EN, Holloway DE. Cloning and sequencing of glutamate mutase component S from Clostridium tetanomorphum. Homologies with other cobalamin-dependent enzymes. FEBS Lett. 1992;310:167–170. doi: 10.1016/0014-5793(92)81321-c. [DOI] [PubMed] [Google Scholar]
- 37.Reitzer R, Gruber K, Jogl G, Wagner UG, Bothe H, Buckel W, Kratky C. Glutamate mutase from Clostridium cochlearium: the structure of a coenzyme B12-dependent enzyme provides new mechanistic insights. Structure Fold Des. 1999;7:891–902. doi: 10.1016/s0969-2126(99)80116-6. [DOI] [PubMed] [Google Scholar]
- 38.Cook FC, Cleland WW. Enzyme Kinetics and Mechanism. Taylor & Francis Group, LLC; New York: 2007. [Google Scholar]
- 39.Liu D, Williamson DA, Kennedy ML, Williams TD, Morton MM, Benson DR. Aromatic side chain-porphyrin interactions in designed hemoproteins. J Amer Chem Soc. 1999;121:11798–11812. [Google Scholar]
- 40.Padovani D, Labunska T, Palfey BA, Ballou DP, Banerjee R. Adenosyltransferase tailors and delivers coenzyme B12. Nat Chem Biol. 2008;4:194–196. doi: 10.1038/nchembio.67. [DOI] [PubMed] [Google Scholar]
- 41.Lerner-Ellis JP, Gradinger AB, Watkins D, Tirone JC, Villeneuve A, Dobson CM, Montpetit A, Lepage P, Gravel RA, Rosenblatt DS. Mutation and biochemical analysis of patients belonging to the cblB complementation class of vitamin B12-dependent methylmalonic aciduria. Mol Genet Metab. 2006;87:219–225. doi: 10.1016/j.ymgme.2005.11.011. [DOI] [PubMed] [Google Scholar]