Abstract
Summary: We have developed ClueGO, an easy to use Cytoscape plug-in that strongly improves biological interpretation of large lists of genes. ClueGO integrates Gene Ontology (GO) terms as well as KEGG/BioCarta pathways and creates a functionally organized GO/pathway term network. It can analyze one or compare two lists of genes and comprehensively visualizes functionally grouped terms. A one-click update option allows ClueGO to automatically download the most recent GO/KEGG release at any time. ClueGO provides an intuitive representation of the analysis results and can be optionally used in conjunction with the GOlorize plug-in.
Availability: http://www.ici.upmc.fr/cluego/cluegoDownload.shtml
Contact: jerome.galon@crc.jussieu.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Since the number of genes that can be analyzed by high-throughput experiments by far exceeded what can be interpreted by a single person, different attempts have been initiated in order to capture biological information and systematically organize the wealth of data. For example Gene Ontology (GO) (Ashburner et al., 2000) annotates genes to biological/cellular/molecular terms in a hierarchically structured way, whereas Kyoto encyclopedia of genes and genomes (KEGG) (Kanehisa et al., 2002) and BioCarta assigns genes to functional pathways. Several functional enrichment analysis tools (e.g. Boyle et al., 2004; Huang et al., 2007; Maere et al., 2005; Ramos et al., 2008; Zeeberg et al., 2003) and algorithms (e.g. Li et al., 2008) were developed to enhance data interpretation.
As most of these tools mainly present their results as long lists or complex hierarchical trees, we aimed to develop ClueGO a Cytoscape (Shannon et al., 2003) plug-in to facilitate the biological interpretation and to visualize functionally grouped terms in the form of networks and charts. Other tools like BiNGO (Maere et al., 2005) or PIPE (Ramos et al., 2008) assess overrepresented GO terms and reconstruct the hierarchical ontology tree, whereas ClueGO uses kappa statistics to link the terms in the network. Compared with the approach of Ramos et al. (2008) which creates an in silico annotation network based on pathways and protein interaction data and maps the gene list of interest afterwards, ClueGO generates a dynamical network structure by already initially considering the gene lists of interest. ClueGO integrates GO terms as well as KEGG/BioCarta pathways and creates a functionally organized GO/pathway term network. A variety of flexible restriction criteria allow for visualizations in different levels of specificity. In addition, ClueGO can compare clusters of genes and visualizes their functional differences. ClueGO takes advantage of Cytoscape's versatile visualization framework and can be used in conjunction with the GOlorize plug-in (Garcia et al., 2007).
2 METHODS AND IMPLEMENTATION
ClueGO has two major features: it can be either used for the visualization of terms corresponding to a list of genes, or the comparison of functional annotations of two clusters.
2.1 Data import
Gene identifier sets can be directly uploaded in simple text format or interactively derived from gene network graphs visualized in Cytoscape. ClueGO supports several gene identifiers and organisms by default and is easy extendable for additional ones in a plug-in like manner (Supplementary Material).
2.2 Annotation sources
To allow a fast analysis, ClueGO uses precompiled annotation files including GO, KEGG and BioCarta for a wide range of organisms. A one-click update feature automatically downloads the latest ontology and annotation sources and creates new precompiled files that are added to the existing ones. This ensures an up-to-date functional analysis. Additionally ClueGO can easily integrate new annotation sources in a plug-in like way (Supplementary Material).
2.3 Enrichment tests
ClueGO offers the possibility to calculate enrichment/depletion tests for terms and groups as left-sided (Enrichment), right-sided (Depletion) or two-sided (Enrichment/Depletion) tests based on the hypergeometric distribution. Furthermore it provides options to calculate mid-P-values and doubling for two-sided tests to deal with discreetness and conservatism effects as suggested by (Rivals et al., 2007). To correct the P-values for multiple testing several standard correction methods are proposed (Bonferroni, Bonferroni step-down and Benjamini-Hochberg).
2.4 Network generation and visualization
To create the annotations network ClueGO provides predefined functional analysis settings ranging from general to very specific ones. Furthermore, the user can adjust the analysis parameters to focus on terms, e.g. in certain GO level intervals, with particular evidence codes or with a certain number and percentage of associated genes. An optional redundancy reduction feature (Fusion) assesses GO terms in a parent–child relation sharing similar associated genes and preserves the more representative parent or child term. The relationship between the selected terms is defined based on their shared genes in a similar way as described by Huang et al. (2007). ClueGO creates first a binary gene-term matrix with the selected terms and their associated genes. Based on this matrix, a term–term similarity matrix is calculated using chance corrected kappa statistics to determine the association strength between the terms. Since the term–term matrix is of categorical origin, kappa statistic was found to be the most suitable method. Finally, the created network represents the terms as nodes which are linked based on a predefined kappa score level. The kappa score level threshold can initially be adjusted on a positive scale from 0 to 1 to restrict the network connectivity in a customized way. The size of the nodes reflects the enrichment significance of the terms. The network is automatically laid out using the Organic layout algorithm supported by Cytoscape. The functional groups are created by iterative merging of initially defined groups based on the predefined kappa score threshold. The final groups are fixed or randomly colored and overlaid with the network. Functional groups represented by their most significant (leading) term are visualized in the network providing an insightful view of their interrelations. Also other ways of selecting the group leading term, e.g. based on the number or percentage of genes per term are provided. As an alternative to the kappa score grouping the GO hierarchy using parent–child relationships can be used to create functional groups.
When comparing two gene clusters, another original feature of ClueGO allows to switch the visualization of the groups on the network to the cluster distribution over the terms. Besides the network, ClueGO provides overview charts showing the groups and their leading term as well as detailed term histograms for both, cluster specific and common terms.
Like BiNGO, ClueGO can be used in conjuntion with GOlorize for functional analysis of a Cytoscape gene network. The created networks, charts and analysis results can be saved as project in a specified folder and used for further analysis.
3 CASE STUDY
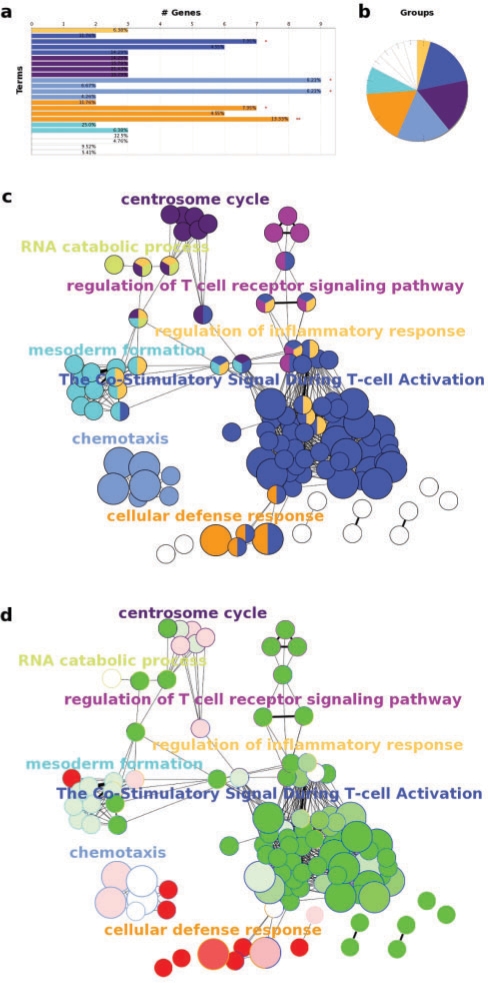
To demonstrate how ClueGO assesses and compares biological functions for clusters of genes we selected up- and down-regulated natural killer (NK) cell genes in healthy donors from an expression profile of human peripheral blood lymphocytes (GSE6887, Gene Expression Omnibus). For upregulated NK genes ClueGO revealed specific terms like ‘Natural killer cell mediated cytotoxicity’ in the group ‘Cellular defense response’. Downregulated in NK cells compared with the reference (a pool of all immune cell types) were genes involved in the innate immune response (Macrophages), but also in the adaptive immune response (T and B cell). The common functionality refers to characteristics of leukocytes (chemotaxis), besides other terms involved in cell division and metabolism (Fig. 1).
Fig. 1.
ClueGO example analysis of up- and down-regulated NK cell genes in peripheral blood from healthy human donors. (a) GO/pathway terms specific for upregulated genes. The bars represent the number of genes associated with the terms. The percentage of genes per term is shown as bar label. (b) Overview chart with functional groups including specific terms for upregulated genes. (c) Functionally grouped network with terms as nodes linked based on their kappa score level (≥0.3), where only the label of the most significant term per group is shown. The node size represents the term enrichment significance. Functionally related groups partially overlap. Not grouped terms are shown in white. (d) The distribution of two clusters visualized on network (c). Terms with up/downregulated genes are shown in red/green, respectively. The color gradient shows the gene proportion of each cluster associated with the term. Equal proportions of the two clusters are represented in white.
4 SUMMARY
ClueGO is a user friendly Cytoscape plug-in to analyze interrelations of terms and functional groups in biological networks. A variety of flexible adjustments allow for a profound exploration of gene clusters in annotation networks. Our tool is easily extendable to new organisms and identifier types as well as new annotation sources which can be included in a transparent, plug-in like manner. Furthermore, the one-click update feature of ClueGO ensures an up-to-date analysis at any time.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A Van Cortenbosch for the name of the tool.
Funding: INSERM; Ville de Paris; INCa; the Austrian Ministry for Science and Research, Project GEN-AU; BINII; the European 7FP Grant Agreement 202230 (GENINCA).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, et al. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia O, et al. GOlorize: a cytoscape plug-in for network visualization with Gene Ontology-based layout and coloring. Bioinformatics. 2007;23:394–396. doi: 10.1093/bioinformatics/btl605. [DOI] [PubMed] [Google Scholar]
- Huang DW, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183–R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. A global pathway crosstalk network. Bioinformatics. 2008;24:1442–1447. doi: 10.1093/bioinformatics/btn200. [DOI] [PubMed] [Google Scholar]
- Maere S, et al. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Ramos H, et al. The protein information and property explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data. Bioinformatics. 2008;24:2110–2111. doi: 10.1093/bioinformatics/btn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals I, et al. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23:401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28–R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.