Abstract
Despite the large amount of research and reporting on potential biomarkers in cancer, very few markers have been brought to use in the clinic. Disorganization plays a large part in this low yield. The Early Detection Research Network (EDRN) of the National Cancer Institute has been initiated to foster collaboration among independent institutions/ laboratories to facilitate, standardize, and centralize discovery and validation of candidate biomarkers. EDRN comprises four components: biomarker reference laboratories; biomarker developmental laboratories; clinical epidemiology and validation centers; and a data management and coordinating center. Biomarker validation proceeds through five phases—the preclinical exploratory, clinical assay and validation, retrospective longitudinal, prospective screening, and cancer control phases. A number of candidate markers in colon cancer, esophageal adenocarcinoma, and hepatocellular carcinoma (HCC) currently are moving through the developmental process. Ongoing EDRN collaborations assessing the potential utility of des-gamma carboxyprothrombin (DCP) in discriminating early HCC in patients with cirrhosis and the ability of DNA methylation analysis to predict progression from Barrett’s esophagus to esophageal cancer are summarized. EDRN welcomes collaboration in biomarker validation and assembly of sample reference libraries.
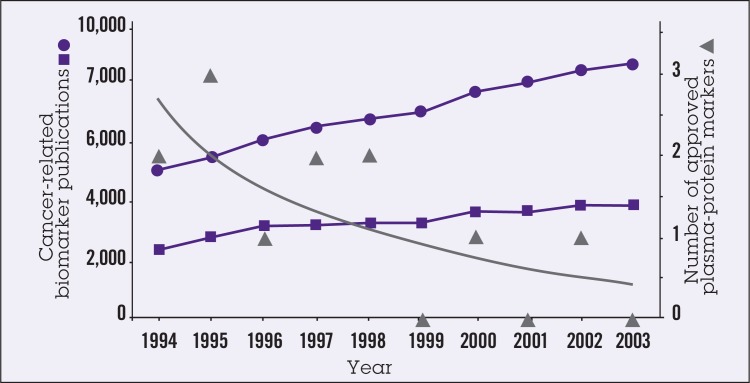
Despite the large amount of research that has been devoted to identifying cancer biomarkers since the mid-1990s and the large numbers of publications in the area, only a handful of cancer biomarkers have been approved for use (Figure 1).1 There are many reasons for the slow progress of biologic/molecular markers from laboratory to clinic, although most probably can be grouped under the heading of “disorganization.” Early findings on marker identification from individual laboratories often are fragmented and disconnected; reported detection assays often are not reproducible, and generalizability of the findings often is questionable. Research on validation is not considered glamorous and suffers from lack of funding. Overall, there is an absence of a systems approach and established network pathways for discovery, validation, and clinical development of markers. In short, collaboration is needed.
Figure 1.
Number of publications under Medline heading “biomarker” (squares) and text word ‘biomarker’ (circles) and number of plasma-protein markers approved by the US Food and Drug Administration (FDA) (triangles) each year from 1994 to 2003. Reprinted from Ludwig and Weinstein,1 with permission from Macmillan Publishers Ltd: Nature Reviews Cancer, copyright 2005.
EARLY DETECTION RESEARCH NETWORK MISSION
The Early Detection Research Network (EDRN), administered by the Cancer Biomarkers Research Group in the Division of Cancer Prevention of the National Cancer Institute, has been instituted to provide “infrastructure” for supporting collaborative research on molecular, genetic, and other biomarkers in human cancer detection and risk assessment. EDRN comprises four components: biomarker reference laboratories; biomarker developmental laboratories; clinical epidemiology and validation centers; and a data management and coordinating center. In addition to providing a national infrastructure supporting a vertical collaborative approach to move promising biomarkers/ technologies to clinical validation, the charges of the EDRN are the following: establish guidelines and criteria for marker validation; develop and institute quality assurance regimens, standard operating procedures, etc; conduct early clinical and epidemiologic studies to evaluate predictive value of markers; and foster public-private partnerships. EDRN welcomes collaboration of independent laboratories in identifying and developing potentially useful markers/technologies.
TECHNOLOGIES
Technologies being used by EDRN in biomarker development include genomics, proteomics (including protein pattern analysis, autoantigens, and autoantibodies), epigenomics, metabolomics, glycomics, and imaging (eg, spectral imaging). Glycomics and spectral imaging are relatively newer technologies. The former exploits the fact that many current cancer biomarkers are glycoproteins and that the glycan structures can be used to improve diagnostic performance. For example, in a recent study reported by Block et al,2 the glycoprotein GP73 was found to be a serum biomarker for hepatocellular carcinoma (HCC). Use of levels of fucosylated oligosaccharides of GP73 vs. total GP73 protein levels improved marker sensitivity from 67% to 89% and specificity from 90% to 100% (M. Mehta and T.M. Block, personal cummunication, June 2007).3
Biophotonics can be used to detect the genetic/epigenetic correlates of field carcinogenesis in the histologically normal mucosa, and thus allows risk stratification for the presence of precancerous or cancerous lesions in the organ of interest. This approach to assessing readily accessible mucosa to sense neoplastic transformation elsewhere is applicable to virtually all types of epithelia.
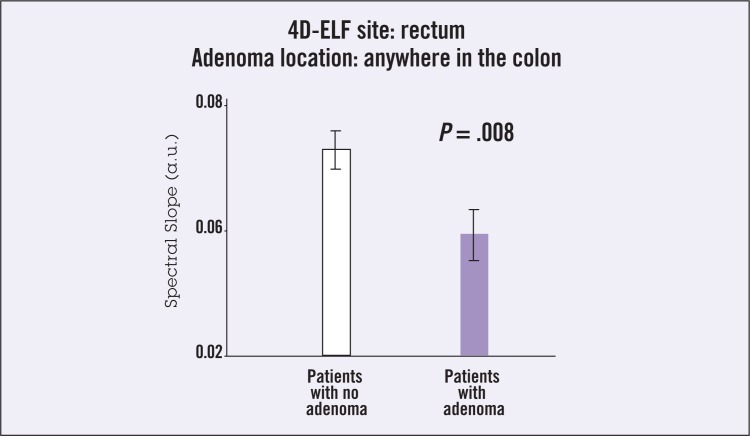
Using a biophotonics technique of low-coherence enhanced backscattering (LEBS), investigators were able to significantly discriminate patients without dysplasia from those harboring adenomas by LEBS analysis of biopsies from histologically and endoscopically normal colonic mucosa obtained from any portion of the colon. A study was performed in 63 subjects undergoing screening colonoscopy.4 This study demonstrated the feasibility of detecting the presence of neoplasia based on the optical analysis of tissue outside the extent of a histologically defined neoplasia. In a subsequent study, performed in collaboration with the EDRN, these investigators obtained two rectal biopsies from endoscopically normal mucosa in 100 patients without coagulopathy or colitis undergoing screening colonoscopy in a managed-care setting.5 Four-dimensional elastic light scattering “fingerprinting” (4D-ELF) was performed on fresh samples within 1 hour of biopsy using the validated spectral markers. The technique was shown to discriminate absence of neoplasia from past/current adenoma to a highly significant degree (P < .008) (Figure 2).
Figure 2.
Ability of light-scattering spectroscopy (LSS) imaging to discriminate absence of neoplasia from adenoma (current or past) in endoscopically normal mucosa from mid-transverse colon biopsies in 100 patients undergoing screening colonoscopy. Abbreviations: a.u. = absorbance unit; 4D-ELF = four-dimensional elastic light scattering fingerprinting. Data from Roy et al.5
BIOMARKER VALIDATION
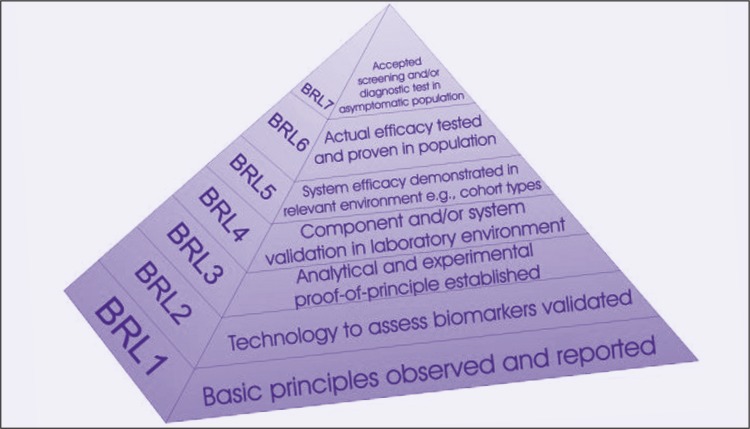
Within EDRN, there are five phases of biomarker development.6 Phase 1 is the Preclinical Exploratory phase, comprising exploratory studies to identify useful biomarkers. Phase 2 is the Clinical Assay and Validation phase, comprising studies to determine the capacity of biomarkers to distinguish among people with and without cancer or cancer risk. Phase 3 is the Retrospective Longitudinal phase, in which it is determined how well biomarkers detect preclinical disease by testing the markers in tissues prospectively collected from research cohorts. Phase 4 is the Prospective Screening phase, in which the extent and characteristics of disease detected by the test are identified and the false referral rate is determined. Phase 5 is the Cancer Control phase, comprising large-scale population studies to evaluate both the role of the biomarkers in detecting disease and the overall impact of screening in the population. We have teamed with the National Aeronautics and Space Administration (NASA) Jet Propulsion Laboratory for informatics support, and have adapted their engineering approach in ascertaining biomarker readiness level, as shown in Figure 3.
Figure 3.
With the assistance of the National Aeronautics and Space Administration (NASA) Jet Propulsion Laboratory, the Early Detection Research Network (EDRN) has adapted an engineering approach to ascertaining biomarker readiness level (BRL) in biomarker discovery and development.
The progress of a number of candidate biomarkers in colon cancer, esophageal cancer, and hepatocellular carcinoma (HCC) is shown in Tables 1 to 3. Although none of the candidate-markers has yet entered development Phase 3, a relatively large number of markers are progressing through the evaluation system. A major impediment in moving from Phase 1 to Phase 2 is the lack of adequate sample reference libraries for evaluating potential markers. EDRN is in the process of developing sets of case and control samples that are statistically powered to permit rapid assessment of biomarkers/ technologies across the range of cancer-related settings. Collaboration with independent institutions and laboratories is welcome in this regard, as well.
Table 1.
Evaluation stages of various candidate biomarkers for colon cancer.
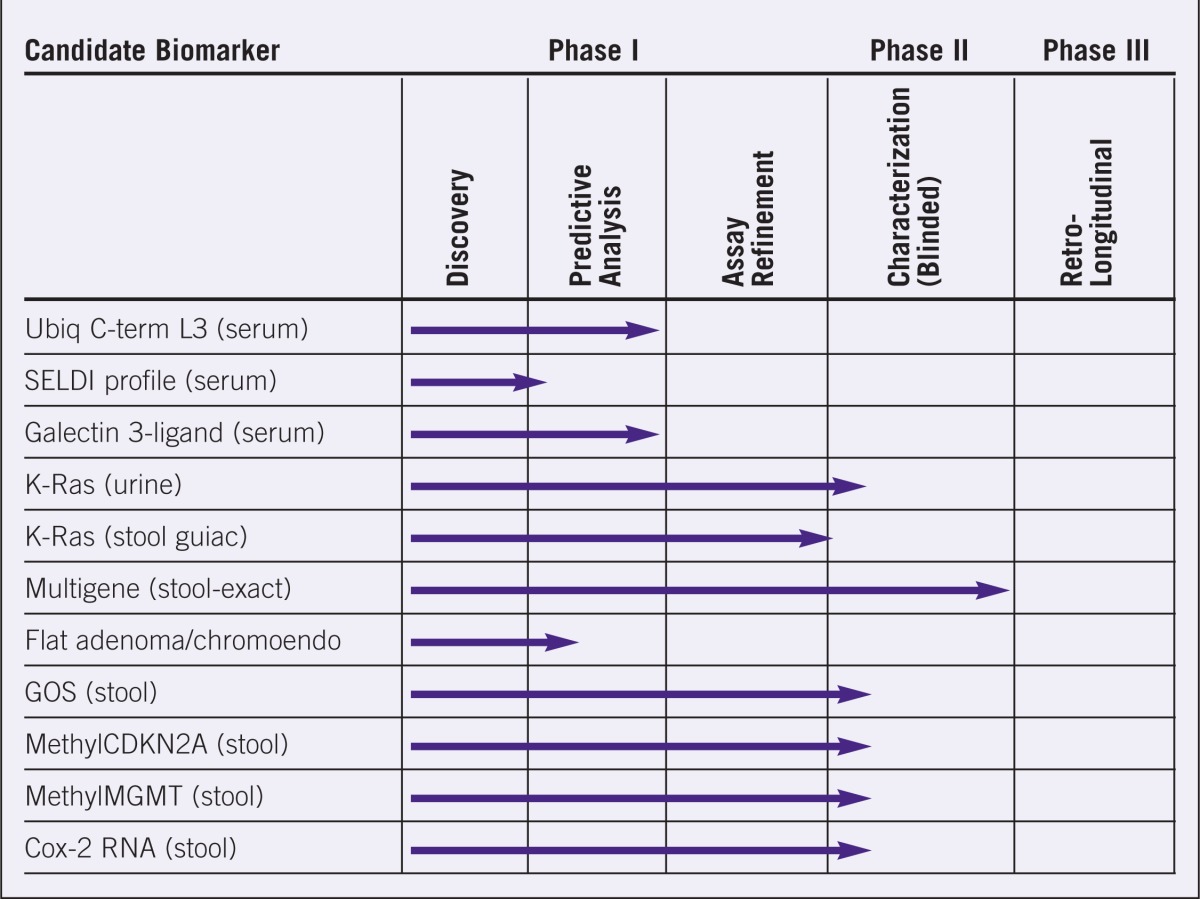
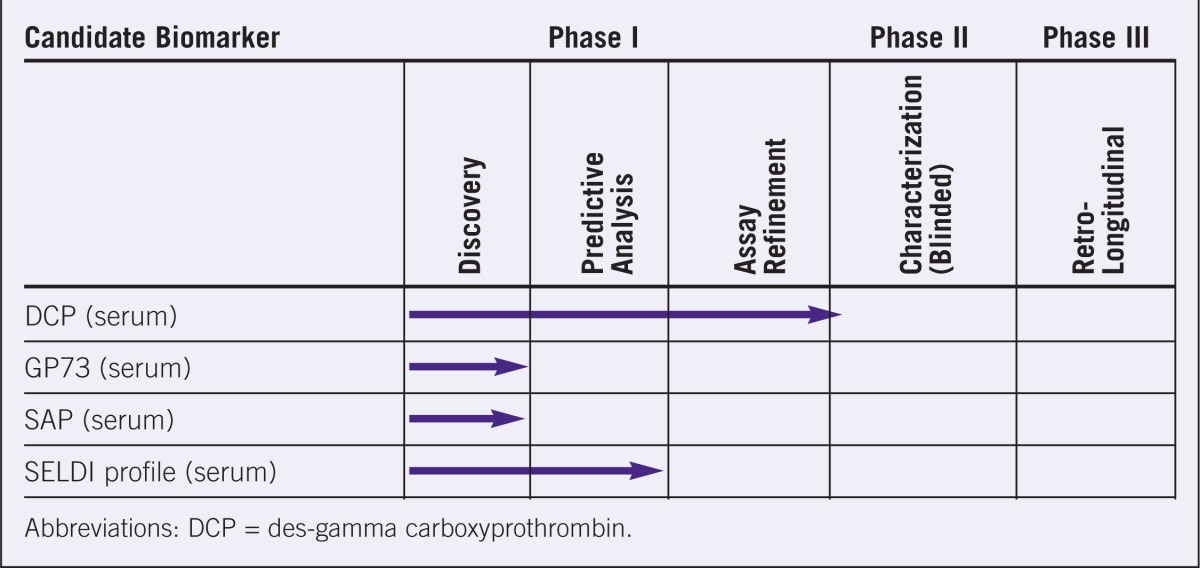
Table 3.
Evaluation stages of various candidate biomarkers for liver carcinomas.
EXAMPLES OF CURRENT WORK
Examples of ongoing work in EDRN include studies assessing the utility of desgamma carboxyprothrombin (DCP) in replacing or complementing alpha-fetoprotein (AFP) in early identification of HCC in patients with cirrhosis (the primary risk factor for HCC), and studies assessing DNA methylation in prediction of progression from Barrett’s esophagus (BE) to esophageal adenocarcinoma.
DCP in Hepatocellular Carcinoma
DCP is a prothrombin precursor that is increased in HCC due to decreased activity of gamma-glutamyl-carboxylase. The validation studies for DCP in HCC are designed to determine the sensitivity and specificity of DCP for diagnosis of early HCC, compare the performance characteristics of DCP and AFP in diagnosis, and to determine whether demographics or etiology of underlying liver disease alter the expression of DCP or AFP. Participating centers include the University of Michigan Medical Center (Ann Arbor), University of Pennsylvania (Philadelphia), Mount Sinai Hospital (New York, NY), Mayo Clinic (Rochester, MN), Stanford University (Palo Alto, CA), and St. Louis University (St. Louis, MO). A case-control study compared DCP and AFP in patients with modified TNM stage I and II HCC (eligible for liver transplant) prior to any cancer therapy and control patients with cirrhosis without HCC.7 Results showed that DCP levels but not AFP levels were significantly higher in HCC vs. cirrhosis. Comparison of the area under the receiver operating characteristic (AUROC) curve showed that DCP was significantly more accurate than AFP in distinguishing HCC from cirrhosis (P = .0022). For identifying HCC, a DCP cutoff of 150 mAU/mL had sensitivity of 89%, specificity of 96%, positive predictive value of 91%, and negative predictive value of 88%; at a cutoff of 13 ng/mL, AFP had sensitivity of 62%, specificity of 76%, positive predictive value of 78%, and negative predictive value of 71%.8
Currently, we are collecting plasma, serum, and peripheral blood cell samples, both for additional testing of DCP and for future use in testing other candidate markers. Investigators at University of California, Los Angeles, currently are assessing DCP and AFP in a sandwich enzyme-linked immunosorbent assay in a blinded manner, with 10% of serum samples also being assessed in a blinded manner at the University of Michigan for quality control purposes.
DNA Methylation in Esophageal Cancer
Barrett’s esophagus is a premalignant lesion for esophageal adenocarcinoma; patients with BE may undergo surveillance endoscopy every 2 or 3 years, but the diagnostic yield of surveillance is low due to the low rate of progression to adenocarcinoma (<1 per 100 patients per year). Three genes (HPP1, p16, and RUNX3) have been found to be differentially methylated in progressors vs. nonprogressors, and BE tissue from progressors has significantly more methylated genes than tissue from nonprogressors.9,10 A multicenter retrospective study is being performed to determine whether DNA methylation analysis involving these three genes and/or fluorescence in situ hybridization in biopsies from patients with BE can discriminate nonprogressors from patients likely to progress to high-grade dysplasia or adenocarcinoma. The study involves samples from 100 BE patients (progressors) who had no or lowgrade dysplasia on surveillance endoscopy with a biopsy protocol over 6 months to 6 years prior to development of high-grade dysplasia or adenocarcinoma, and samples from 200 BE patients (nonprogressors) with no or low-grade dysplasia who have not progressed beyond low-grade dysplasia during a similar time period including at least three endoscopies with a biopsy protocol. This Inter-SPORE (Specialized Programs of Research Excellence)/ EDRN initiative involves investigators from the University of Arizona (Tucson, AZ), Johns Hopkins University (Baltimore, MD), Mayo Clinic (Rochester, MN), Mayo Clinic (Jacksonville, FL), University of North Carolina (Chapel Hill, NC), Vanderbilt University (Nashville, TN), University of Texas (Houston, TX), and the Fred Hutchinson Cancer Research Center (Seattle, WA).
CONCLUSION
The Early Detection Research Network has a straightforward mission: to translate newly emerging molecular knowledge into practical clinical tests to detect cancer and cancer risk. For most cancers, successful treatment depends on early detection, and successful prevention depends on the accurate evaluation of risk. EDRN seeks to give treatments the opportunity to work and to make prevention possible. To be successful, this enterprise requires team science—effective interdisciplinary communication and collaboration in the fields of clinical/medical oncology, molecular biology, observational epidemiology, and biostatistics. The EDRN presents an opportunity and a challenge for the scientific community—an opportunity to make science work for people and a challenge to make this new model of collaboration a productive scientific construct. We welcome and value your participation and collaboration in identifying and validating biomarkers, developing sample reference libraries, and moving candidate markers into the clinical arena.
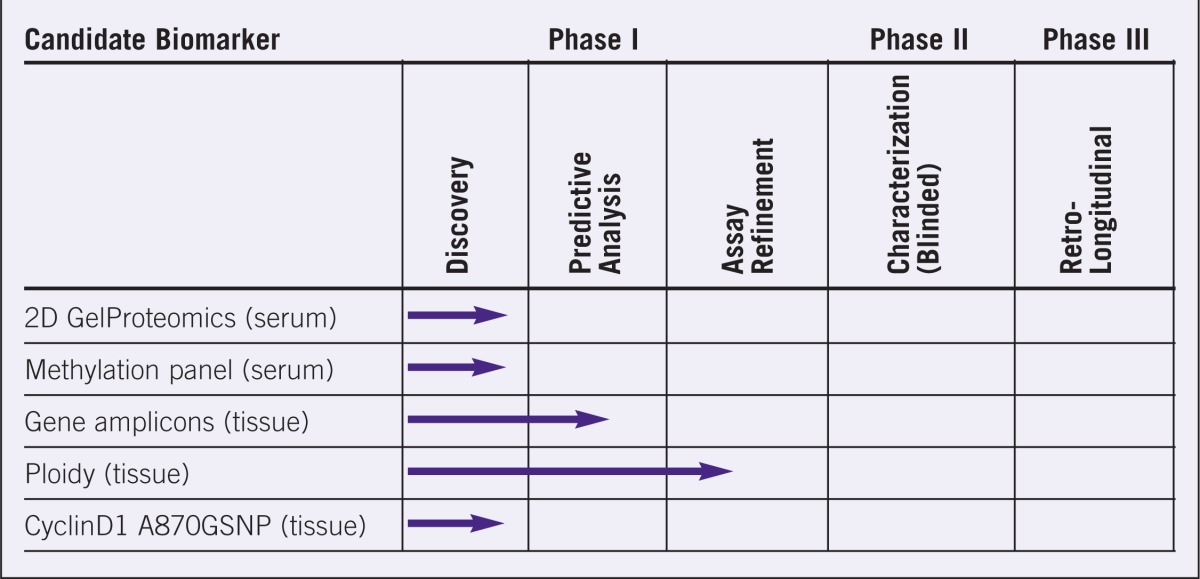
Table 2.
Evaluation stages for various candidate biomarkers for esophageal carcinomas.
Acknowledgments
The author would like to thank Drs. Hemant Roy, Jorge Marrero, and John Weinstein for providing figures and for use of their research in writing this manuscript.
Footnotes
Disclosures of Potential Conflicts of Interest
Dr. Srivastava has no potential conflicts of interest to disclose.
REFERENCES
- 1.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 2.Block TM, Communale MA, Lowman M, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrero JA, Romano PR, Steel LF, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Roy HK, Liu Y, Wali RK, et al. Four-dimensional elastic light-scattering fingerprints as preneoplastic markers in the rat model of colon carcinogenesis. Gastroenterology. 2004;126:1071–1081. doi: 10.1053/j.gastro.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Roy HK, Kim YL, Liu Y, et al. Risk stratification of colon carcinogenesis through enhanced backscattering spectroscopy analysis of the uninvolved colonic mucosa. Clin Cancer Res. 2006;12:961–968. doi: 10.1158/1078-0432.CCR-05-1605. [DOI] [PubMed] [Google Scholar]
- 6.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Acad Sci. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 7.Volk ML, Hernandez JC, Su GL, et al. Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L31. Cancer Biomark. 2007;3:79–87. doi: 10.3233/cbm-2007-3202. [DOI] [PubMed] [Google Scholar]
- 8.Marrero JA, Su GL, Wei W, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in American patients. Hepatology. 2003;37:1114–1121. doi: 10.1053/jhep.2003.50195. [DOI] [PubMed] [Google Scholar]
- 9.Schulmann K, Sterian A, Berki A, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–4148. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton JP, Sato F, Greenwald BD, et al. Promoter methylation and response to chemotherapy and radiation in esophageal cancer. Clin Gastroenterol Hepatol. 2006;4:701–708. doi: 10.1016/j.cgh.2006.03.007. [DOI] [PubMed] [Google Scholar]