Abstract
Hepatocellular carcinoma (HCC) is one of the most lethal cancers. Surgical intervention is the only curative option, with only a small fraction of patients being eligible. Conventional chemotherapy and radiotherapy have not been effective in treating this disease, thus leaving patients with an extremely poor prognosis. In viral, alcoholic, and other chronic hepatitis, it has been shown that there is an activation of the progenitor/stem cell population, which has been found to reside in the canals of Hering. In fact, the degree of inflammation and the disease stage have been correlated with the degree of activation. Dysregulation of key regulatory signaling pathways such as transforming growth factor-beta/transforming growth factor-beta receptor (TGF-β/TBR), insulin-like growth factor/IGF-1 receptor (IGF/IGF-1R), hepatocyte growth factor (HGF/MET), Wnt/β-catenin/FZD, and transforming growth factor-α/epidermal growth factor receptor (TGF-α/EGFR) in this progenitor/stem cell population could give rise to HCC. Further understanding of these key signaling pathways and the molecular and genetic alterations associated with HCC could provide major advances in new therapeutic and diagnostic modalities.
Hepatocellular carcinoma (HCC) is the fifth most common solid malignancy worldwide and causes more than 600,000 deaths annually.1 Current data indicate that the incidence of HCC is steadily increasing in the United States.2,3 Prognosis remains extremely poor with a 5-year survival rate of less than 5% without treatment.3 Currently, the only curative therapeutic option for early-stage HCC is surgical intervention, including percutaneous ablation, hepatic resection, and liver transplantation. However, only 12% of diagnosed HCC patients are deemed eligible for curative therapy.4,5
Accumulating evidence suggests that development of HCC is a multistep process associated with changes in host gene expression, altered DNA methylation, and point mutations or loss of heterozygosity (LOH) in selected cellular genes.6 The dynamics of these cellular changes remain unclear, and it is still a challenge to identify the rate-limiting steps in initiation and progression of HCC. However, a number of molecular changes occur in high frequency in preneoplastic tissues, such as cirrhotic tissue, hepatic adenomas and dysplastic nodules. For example, chronic hepatitis B virus (HBV) infection, which is one of the most prominent risk factors for hepatocarcinogenesis, appears to disrupt senescence-related pathways by different mechanisms. These include inactivation of p53, p55sen and hyperphosphorylation of the retinoblastoma protein (pRb), as well as down-regulation of sui1 (a translational factor) and the cyclin-dependent kinase inhibitor, p21WAF1/CIP1/SDI1.7–9 Perturbation of several signaling pathways such as wingless (Wnt/β-catenin/FZD), JAK/STAT, MAPK, insulin-like growth factor 2 (IGF-2), and transforming growth factor-beta (TGF-β) have also been identified.10 Approximately 40% of HCCs display chromosomal abnormalities.11–14 In addition, microsatellite instability (MSI) and dysfunction of the mismatch repair genes, hMSH2 and hMLH1, are present in up to 11% of HCCs.15,16 These are in turn associated with mutations in TGFβRII, M6P/IGFIIR, and BAX genes.17 Among proto-oncogenes, c-myc is upregulated in approximately 50% of HCCs,18,19 and cyclin D1 is overexpressed in approximately 40% of HCCs.20,21 Dysregulation of these positive mediators of cellular proliferation promotes autonomous and unregulated cellular growth in HCC.
STEM CELLS AND HEPATOCARCINOGENESIS
Both epigenetic and genomic alterations that accumulate in prolonged chronic inflammatory states, such as in cirrhosis and chronic hepatitis, compromise an intricate balance of various regulatory pathways, resulting in accelerated proliferation of hepatocytes and development of monoclonal hepatocyte populations. These populations harbor dysplastic hepatocytes that will eventually evolve to dysplastic nodules, which are true preneoplastic lesions, 30% of which will develop into HCCs within 5 years.22 Though these dysplastic lesions derive from monoclonal cell populations, different lesions from the same liver possess different combinations of genomic aberrations. This heterogeneity in genomic changes supports the view that diverse combinations of cellular alterations can sufficiently transformnormal hepatocytes intomalignant ones.
In continuously renewing systems such as gastrointestinal, hematopoietic, and epidermal tissues, it is widely accepted that stem cells are the only cells with the potential to acquire sufficient genetic alterations for malignant transformation because of their long lifespan as compared to shortlived differentiated cells. However, the liver has several cell types that have the longevity to acquire the requisite number of genetic changes for neoplastic development. Hepatocytes, cholangiocytes, and progenitor/stem cells are normally relatively dormant, but they possess enormous proliferative capacity under certain stimuli. In rodents, the liver can be restored to its original volume within 10 days even after a two-thirds partial hepatectomy.23,24 Transplantation of normal mature wild-type hepatocytes into transgenic mice with urokinase plasminogen activator, in which the transgenic hepatocytes undergo massive necrosis, results in 12 or more cell divisions and most of the transgenic liver is replaced.25 Serial transplantation experiments also have shown a near infinite proliferative capacity of hepatocytes.26 It has also been shown that when there is massive bile duct injury, hepatocytes can differentiate into cholangiocytes.27 Thus, mature hepatocytes have stem cell properties. However, in prolonged chronic inflammatory diseases, such as viral hepatitis and cirrhosis, hepatocytes actually undergo replicative senescence because of telomerase shortening.26 Another cell population, a progenitor/stem cell, is activated under these circumstances and it is thus possible that progenitor/stem cells could be at least partly responsible for HCCs.
In general, stem cells are undifferentiated cells with the ability to undergo asymmetric cell division producing two different daughter cells: a stem cell and a committed progenitor cell. In the adult gastrointestinal system, stem cells generally considered to be tissue-specific are able to give rise only to progeny cells corresponding to their tissue of origin. During liver development, the liver bud is seen around embryonic day (ED) 8.5 as endodermal stem cells begin to proliferate and differentiate under the influence of fibroblast growth factor signaling from the cardiogenic mesoderm28 and bone morphogenetic protein signaling from the septum transversum mesenchyme.29 This region then produces cells destined to become hepatoblasts, bipotential cells committed to fetal hepatocytes, and biliary epithelial cells. Hepatoblasts begin expressing alpha-fetoprotein (AFP) and albumin (Alb) and later express cytokeratins (CKs) 7 and 19. Just before ED16, hepatoblasts diverge along two cell lineages: hepatocytes (AFP+/Alb+) and cholangiocytes (CK19+).30,31 In adult human tissues, these hepatic progenitor cells (or mouse oval cells) are immature epithelial cells found residing in the smallest terminal branches of the biliary tree called the canals of Hering.32 This compartment is also known as the progenitor cell compartment in humans and oval cell compartment in rodents.33 In this compartment, the immature epithelial cells, which express both bile ductular and hepatocyte markers (CK19 and AFP), are in direct physical continuity with hepatocytes at one membrane boundary and bile duct cells at another boundary and are considered to represent hepatic stem cells.34,35
The progenitor cell compartment (oval cell compartment in rodents) can be activated when the mature hepatocytic or cholangiocytic compartments are damaged or their replication is inhibited.32,36 Such circumstances can be observed in cirrhosis and chronic inflammatory liver diseases when hepatocytes undergo senescence owing to telomere shortening after 20 to 30 years of continuous replication.26 Activation of the progenitor cell compartment, also known as “ductular reaction” (“oval cell reaction” in rodents), is merely an expansion of transit amplifying progenitor cells, which can differentiate into hepatocytes and biliary cells. Intermediate hepatocytes, which have an intermediate phenotype between progenitor cells/biliary ductular cells and mature hepatocytes, are seen in moderateto-severe inflammatory hepatitis. In fact, the degree of progenitor cell activation and the number of these intermediate hepatocytes correlate with the degree of inflammation and fibrosis in diseases like chronic hepatitis, hemochromatosis, and nonalcoholic steatohepatitis.37,38 It has been demonstrated that sequestered hepatocytes in cirrhosis are in continuity with reactive ductules.39
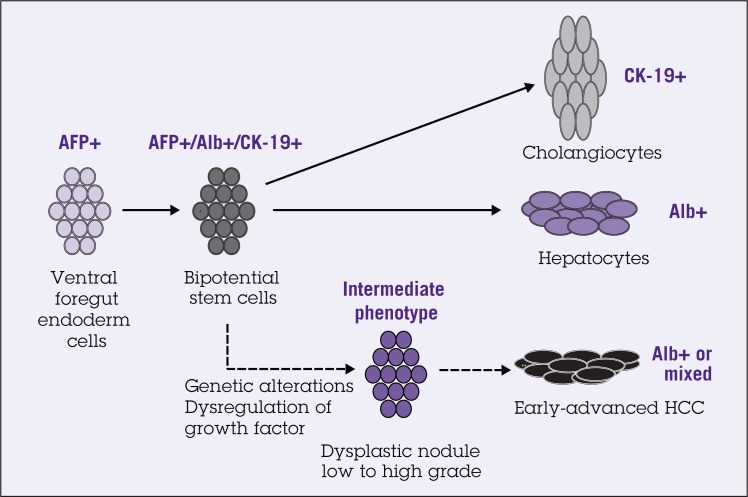
Several studies have shown a progenitor cell phenotype in a substantial number of HCCs. Detailed immunophenotyping of HCCs revealed that 28% to 50% of HCCs express markers of progenitor/biliary cells such as CK7 and CK19.40–43 These tumors also consist of cells that have an intermediate phenotype between progenitors and mature hepatocytes. In fact, HCCs with CK19 expression have a significantly worse prognosis and higher recurrence after surgical resection and liver transplantation than CK19-negative HCCs.40 Nevertheless, the question remains whether this immature intermediate phenotype represents progenitor cell differentiation arrest or dedifferentiation of mature hepatocytes. The histologic and immunophenotyping studies favor the progenitor cell differentiation arrest model. Small dysplastic foci (less than 1 mm in size) represent the earliest premalignant lesions, and 55% of them are comprised of progenitor cells and intermediate hepatocytes, instead of senescent hepatocytes.26 A recent study also identified a side population of cells (SP), which have characteristics of both hepatocytic and cholangiocytic lineages, in human HCC cell lines, Huh7 and PLC/PRF/5. Importantly, injection of SP cells into nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice leads to tumor formation. Tumor-initiating potential is maintained in these SP cells after serial transplantations.44 These results suggest that hepatic progenitor/stem cells could account for human hepatocarcinogenesis (Figure 1).
Figure 1.
Schematic representation of fetal liver development and hepatocarcinogenesis process based on stem cell model. About half of the small cell dysplasic lesions consist of progenitor cells and intermediate cells. Abbreviations: AFP = alpha-fetoprotein; Alb = albumin; HCC = hepatocellular carcinoma.
MOLECULAR SIGNALING PATHWAYS AND HEPATOCARCINOGENESIS
Normal embryogenesis and organ development as well as tissue regeneration and repair require an intricate balance of various molecular growth factor signals. Dysregulation of these signaling pathways and their components is the central principle in human tumorigenesis. Recent studies have identified regulatory pathways including TGF-β/TBR, IGF/IGF-1R, HGF/MET, Wnt/β-catenin/FZD, and TGF-α/EGFR as key contributory factors to the transformation, proliferation, antiapoptosis, and invasive behaviors of human HCCs.
Transforming Growth Factor-Beta Signaling
Initially described for its transforming capability, TGF-β plays an important role in a wide range of cellular responses, including cellular homeostatsis, cell differentiation, proliferation, migration, and apoptosis. The TGF-β superfamily comprises more than 30 members, including the TGF-βs, bone morphogenetic proteins, activins, and nodal. The basic signaling cascade of TGF-β involves type I and type II transmembrane serine/threonine kinase receptors (TBRI and TBRII). Cellular responses are mediated by the intracellular signaling proteins, Smads.45 Vertebrates possess at least nine Smad proteins categorized into three functional classes: (1) receptor activated Smads (R-Smads): Smad1, Smad2, Smad3, Smad5, and Smad8; (2) co-mediator Smads: Smad4 and Smad10; and (3) inhibitory Smads: Smad6 and Smad7.46 Activation of Smads by TGF-β results in association of R-Smads with Smad4. This Smad complex then translocates to the nucleus where it participates in regulation of TGF-β target gene expression such as p15, p21, and E-cadherin by interacting with various transcriptional factors such as CBP/p300 and SKI/SNO47,48 (Figure 2).
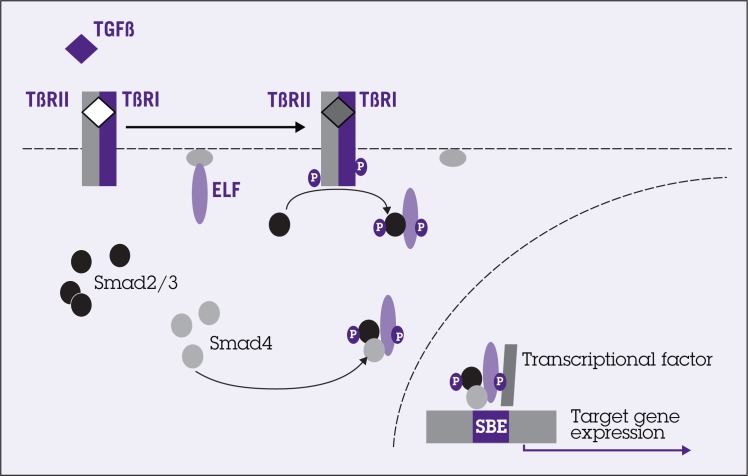
Figure 2.
Transforming growth factor-beta (TGF-β) signals through distinct receptors and Smads, which are modulated by β-spectrin embryonic liver fodrin protein (ELF). TGF-β binds to serine/threonine kinase receptor complexes I and II (TBRI and TBRII), which subsequently phosphorylates receptor-associated Smad proteins, such as Smad2 and Smad3. Smad2/3 then forms heterometric complex with Smad4 and ELF proteins and translocates to the nucleus, interacting with transcriptional factor and activating target genes. Abbreviation: SBE = Smad binding element.
Recent studies show that adaptor proteins such as embryonic liver fodrin protein (ELF), the Smad anchor for receptor activation (SARA), filamin, and microtubules play critical roles in modulating TGF-β signaling. Genetic studies have highlighted that ELF, a major dynamic scaffolding protein, is required for Smad3 and Smad4 co-localization.49–51 Disruption of ELF results in mislocalization of Smad3 and Smad4 leading to loss of the TGF-β–dependent transcriptional response.49,52,53 Interestingly, we have also demonstrated in previous work that ELF is crucial for multiple developmental processes. ELF-/- mutant mice share strikingly similar phenotypes with double heterozygous Smad2/Smad3 mutants, including profound defects in gut, liver, and cardiovascular and nervous systems.54–58 Livers from these mutants display distorted liver architecture and scant early intrahepatic bile duct development.49 Interestingly, by 15 months of age, 40% of heterozygous ELF mice spontaneously developed HCC and also showed additional phenotypic changes such as increased centrilobular steatosis and highgrade dysplasia. A marked attenuation of the TGF-β –mediated antiproliferative response has also been shown in several human HCC cell lines.53 Functional inactivation of TGF-β signaling via expression of a dominant-negative mutant TBRII in transgenic mice treated with the carcinogen diethylnitrosamine resulted in higher rates of preneoplastic lesions and HCCs as compared with similarly treated wildtype mice.6 In addition, MSI or inactivation of the mismatch repair genes, hMSH2 and hMLH1, is present in up to 11% of HCCs.15,16 and is in turn associated with mutations in TGFβRII, M6P/IGFIIR, and BAX genes.17 Most studies document a reduction of TGF-β receptors in up to 70% of HCCs.59 However, Smad proteins shown to be impaired in other cancers appear to play a minor role in HCCs. Smad4, which is mutated in 50% of pancreatic cancers, is also mutated in 10% of HCCs. Similarly, Smad2 mutations are identified in fewer than 5% of HCCs.60,61 Finally, inhibitory Smad7 is upregulated in 60% of advanced HCCs.62
Yet, TGF-β levels in serum and urine are increased in HCC patients.63,64 In addition, up to 40% of HCCs have increased TGF-β based on immunohistochemical studies.65,66 High TGF-β expression levels have been correlated with advanced clinical stages of HCC.67 The dual role of TGF-β signaling in HCCs may be explained by its effect on the tumor tissue microenvironment and on selective loss of the TGF-β–induced antiproliferative pathway. Tumorderived TGF-β could contribute to tumor growth indirectly by suppressing immune surveillance or stimulating production of angiogenic factors. Tumor cells that have selectively lost their growth-inhibitory responsiveness to TGF-β but retain an otherwise functional TGF-β signaling pathway may exhibit enhanced migration and invasive behavior in response to TGF-β stimulation.68,69 TGF-β signaling also has been shown to induce an epithelial to mesenchymal transition (EMT) in these cells.69 This EMT process is characterized by decreased cell-cell adhesion, through the decrease in E-cadherin, leading to enhanced migration and invasiveness.
Wingless/β-Catenin Signaling
The Wnt signaling pathway is highly conserved evolutionarily. It plays an important role in cell proliferation, cell/cell interactions, motility, tissue development and modeling, as well as axis formation.70,71 Initiation of Wnt signaling involves the binding of Wnt proteins to a receptor complex consisting of the frizzled receptor family (Fz) and a member of the low-density lipid receptor family, Lrp5 or Lrp6. The key intracellular component is cytoplasmic β-catenin protein.72,73 Under normal circumstances, when the Wnt signaling pathway is inactive, the tumor suppressor APC forms a trimeric complex, known as the “destruction complex,” with glycogen synthase kinase-3β (GSK3β) and axin/conductin. This complex interacts with and serine phosphorylates β-catenin, thus targeting it for degradation by the ubiquitin-proteasome pathway. In addition to this canonical degradation, β-catenin can be degraded without phosphorylation through a pathway involving a p53-inducible E3-ubiquitin ligase, seven in absentia homolog (SIAH).74 When Wnt ligand is present, it binds to Fz and Lrp5 or Lrp6. Ligand binding results in accumulation of cytoplasmic β-catenin by recruiting the cytoplasmic protein Dishevelled (Dsh), which disrupts the destruction complex. β-catenin then interacts with a member of the TCF/LEF (T-cell factor/lymphocyte enhancer factor) family of DNA-binding proteins, resulting in activation of Wnt target genes to increase cell proliferation.75,76 Wnt/β-catenin target genes include cell-cycle promoting genes such as c-MYC and cyclin D1, the antiapoptotic gene survivin, and pro-invasive genes such as MMP.59 Therefore, a mutation in Wnt signaling pathway components results in the accumulation of β-catenin, and predisposes tissues to tumorigenesis (Figure 3).
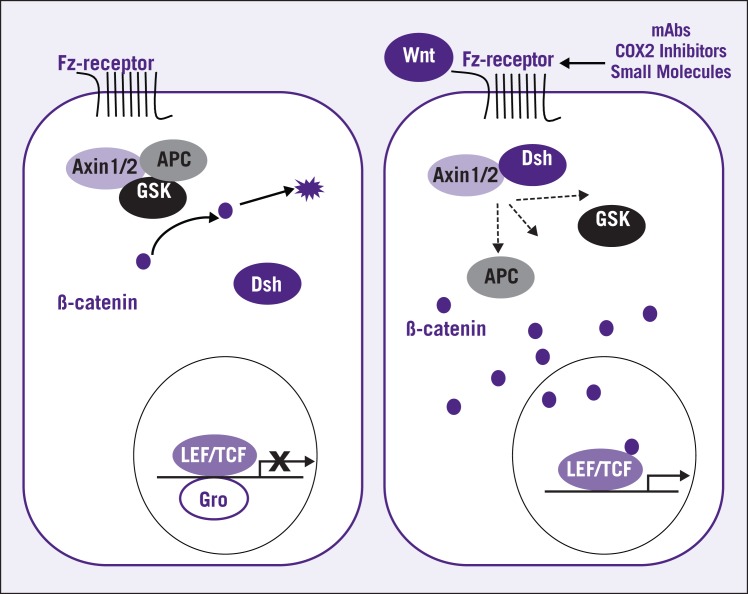
Figure 3.
In the absence of Wnt stimulation, the APC (adenomatous polyposis coli) forms a trimeric complex, known as the “destruction complex,” with glycogen synthase kinase-3β (GSK) and Axin. This complex then interacts with β-catenin and degrades by the ubiquitin-proteasome pathway. When Wnt ligands bind to the seven-transmembrane receptor, the cytoplasmic protein Dishevelled (Dsh) is recruited to the membrane and binds to Axin1 and Axin2. The mechanism of Dsh-mediated inhibition of Axin is not well understood, but it has been suggested that Dsh might disrupt the destruction complex. Inhibition of Axin results in accumulation of β-catenin, which subsequently translocates into the nucleus. β-catenin interacts with LEF/TCF (lymphocyte enhancer factor/T cell factor) proteins and serves as a coactivator of LEF/TCFs to stimulate transcription of Wnt target gene. Grouch protein (Gro) acts as corepressor of LEF/TCFs and normally binds to LEF/TCFs in the absence of β-catenin. New therapeutic treatments aimed at this pathway include monoclonal antibodies, cyclooxygenase (COX)-2 inhibitors, and several small molecules.
Alterations in Wnt signaling components have been described in HCCs. Up to 40% of HCCs exhibit accumulation of nuclear β-catenin. Several mechanisms contribute to β-catenin accumulation in HCC, such as downregulation of the Dshinhibitor, dapper homolog 1 (HDPR1), and upregulation of PIN1 (a prolyl cis/trans isomerase), a β-catenin/APC destabilizer.77–79 Axin-2 has been found to be mutationally inactivated in 3% to 14% of HCCs.80,81 In addition, the Fz-7 receptor is frequently overexpressed.82 Taken together, these results suggest that more than one portion of the Wnt signaling pathway has to be dysregulated in liver tumors to achieve aberrant β-catenin nuclear accumulation.
Insulin-Like Growth Factor Signaling
IGF signaling plays a central role in embryogenesis, lifespan regulation, and cell proliferation. IGF signaling consists of IGF ligands (IGF-I and IGF-II), IGF binding proteins (IGFBP 1–6), and membrane-bound IGF receptors (IGF-1R, IGF-II/M6PR, and IGF-2R). IGF ligands can also bind to the insulin receptor. IGF signaling is initiated by the binding of IGF ligands, which results in phosphorylation of intracellular target proteins. These proteins then convey the signal to specific downstream effectors such as INSR-substrate IR5 leading to activation of, for example, phosphatidylinositol 3-kinase (PI3K) and protein kinase B (AKT/PKB). Binding of growth factor receptor-bound protein 2 (Grb2) to the receptor can lead to activation of mitogenactivated protein kinase family (MAPK) signaling. The result is transcriptional activation of various target genes such as p27, c-myc, c-FOS, cyclin B and vascular endothelial growth factor (VEGF)83 (Figure 4).
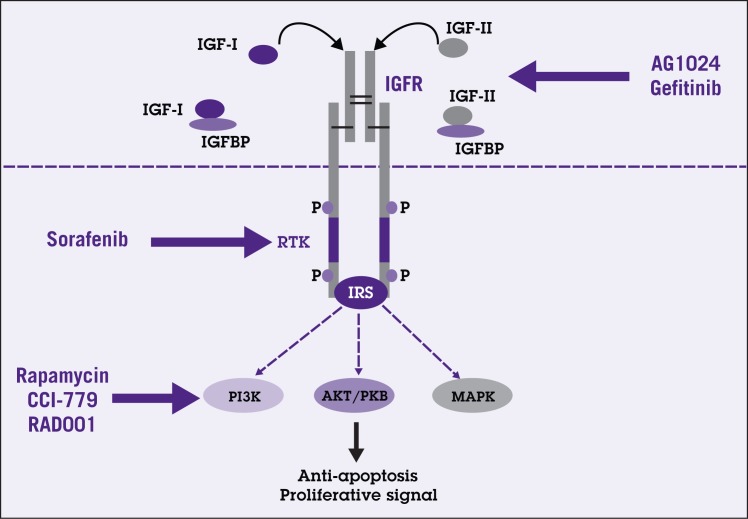
Figure 4.
Simplified schematic diagram of insulin-like growth factor (IGF) signaling. IGF-I and IGF-II bind to IGF receptor (IGFR), a receptor tyrosine kinase (RTK), with high affinity resulting in phosphorylation of intracellular proteins including insulin receptor substrate (IRS). The signal is then conveyed to specific downstream effectors such as phosphatidylinositol 3-kinase (PI3K), protein kinase B (AKT/PKB), and mitogen-activated protein kinase (MAPK) pathways. These pathways play crucial roles in antiapoptosis as well as cell proliferation. Bioavailability of both IGFs is influenced by the presence of IGF binding proteins (IGFBP). New therapeutic agents such as AG1024 and gefitinib aim to block this signaling pathway at level of the receptor. Downstream targeting agents such as rapamycin, CCI-779, and RAD001 are also under investigation.
Dysregulation of IGF signaling in HCC occurs predominantly at the level of IGF-II bioavailbility. Overall, increased levels of IGF-II are found in 16% to 40% of HCCs.59 In chronic viral hepatitis B, HBV-derived HBx protein contributes to overexpression of IGF-II through SP1-mediated reactivation of fetal-type IGF-II.84,85 In chronic hepatitis C, the HCV-derived core gene product also induces overexpression of IGF-II by acting as a transactivator through SP1 and EGR-1 binding sites.86 IGF-II bioavailability is inversely dependent on IGF-2R expression level. A reduced IGF-2R level is associated with less ligand-receptor binding and thus a relative increase in local IGF-II bioavailability. In fact, IGF-2R level is reduced in 63% of HCCs87 and loss of heterozygosity at the IGF-2R locus has been implicated in HCCs and in preneoplastic lesions.88,89 Similarly, reduced expression of IGFBP also results in a relative increase in IGF-II concentration.
Hepatocyte Growth Factor/MET Signaling
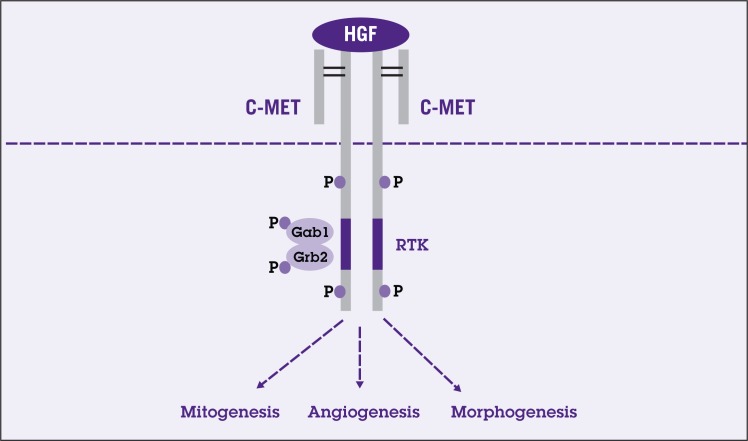
HGF is one of the most potent growth factors for hepatocytes and plays crucial roles in proliferation, migration, cell survival, morphogenesis, angiogenesis, and tissue regeneration.90 HGF binds the tyrosine kinase receptor, c-MET, which is expressed in epithelial and endothelial cells. Binding of HGF to the c-MET receptor results in receptor autophosphorylation as well as phosphorylation of adaptor proteins such as Gab-1 and Grb2. The HGF signaling is then conveyed to activation of various downstream effectors such as phospholipase C (PLC), Stats, PI3K, and extracellular signalingregulated kinase (ERK1/2).91 Specificity of HGF signaling is achieved through different membranous binding partners and the adaptor proteins. Examples of HGF target genes are MMP and urokinase-type plasminogen activator (uPA) (Figure 5).
Figure 5.
Simplified schematic of hepatocyte growth factor (HGF)/MET signaling pathway. Activation of phosphorylation of the kinase domain in cMET, a receptor tyrosine kinase (RTK), by HGF results in activation of adaptor proteins: growth factor receptor-bound protein2 (Grb2) and Grb2-associated binding protein (Grb2 and Gab1). Activation of these adaptor proteins leads to activation of various downstream effectors resulting in transcription of various genes important in mitogenesis, angiogenesis, and morphogenesis.
HCC has been shown to release tumor cell products inducing stellate cells and myofibroblasts to secrete HGF. The increased HGF from these cells in turn promotes tumor cell invasiveness.92–94 Furthermore, c-MET is also overexpressed in HCC as compared with normal liver. c-MET is overexpressed in 20% to 48%of HCCs. The increase in c-MET could be partly due to genomic alterations such as 7q gains or to growth factor-dependent transcriptional activation of c-MET.95–97 However, increased c-MET levels do not appear to correlate with tumor size or invasiveness.96
Transforming Growth Factor-α/Epidermal Growth Factor Signaling
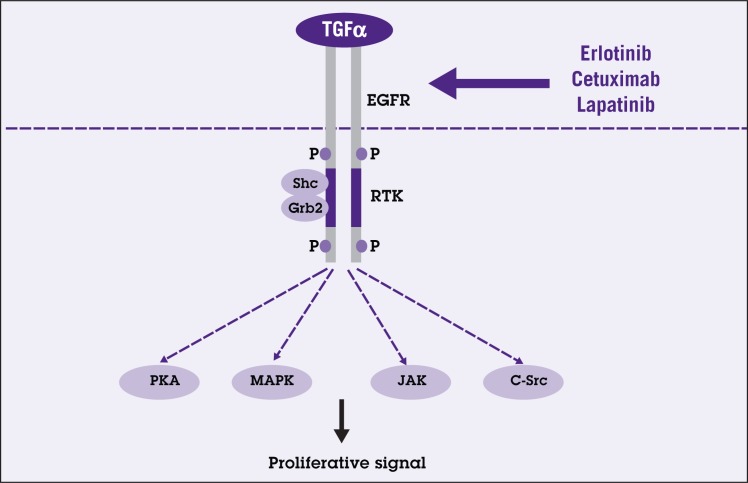
TGF-α/EGF signaling comprises at least eight ligands including TGF-α, EGF, heparin-binding EGF, amphiregulin, betacellulin, epiregulin, epigen, and crypto. TGF-α/EGF signaling conveys its signal through the receptor tyrosine kinase family: EGFR (Her1/ErbB-1), neu/Her2/ErbB-2, Her3/ErbB-3, and Her4/ErbB-4. Different ligand specificities and concentrations lead to differential phosphorylation of multiple tyrosine residues at the cytoplasmic portion of the molecule.98 Phosphorylation of the receptor’s cytoplasmic portion serves as a docking site for recruitment of proteins with Src homology 2 domains, such as Grb2 and Shc.99 The signal will then activate multiple downstream pathways, which can directly or indirectly interact with each other (Figure 6).
Figure 6.
Simplified schematic diagram of transforming growth factor-alpha/epidermal growth factor receptor signaling pathway (TGF-α/EGFR). TGF-α binding to EGFR results in stimulation of the endogenous receptor tyrosine kinase (RTK). The activated membrane-bound EGFR serves as a docking site for recruitment of proteins such as Src homology 2 domain containing (Shc) and growth factor receptor-bound protein2 (Grb2). The signal then activates one of several intracellular signal transduction pathways including protein kinase A (PKA), mitogen-activated protein kinase (MAPK), Jak/Stat, and C-src pathways, which play important roles in cell proliferation. New therapeutic agents such as erlotinib, cetuximab, and lapatinib aim to block this signaling cascade.
TGF-α and EGF act as potent mitogens for hepatocytes and stimulate DNA synthesis.100 TGF-α has been shown to be overexpressed in human HCCs as well as in HCC cell lines.101 TGF-α appears to act during the early stages of hepatocarcinogenesis and its level is correlated with tumor differentiation and proliferation.102,103 The pro-TGF-α/EGF signaling level is increased from normal liver to preneoplastic lesions and to HCCs. Interestingly, TGF-α levels in HCCs correlate with the presence of viral polypeptides (HBS and HBC) in the adjacent non-cancerous liver tissue; and in HCC cells, HBV-DNA induces TGF-α expression.104 Heparin-binding EGF, which can be used as a prognostic marker for diseasefree survival, is also markedly increased in 59% to 100% of HCCs as compared with surrounding normal tissue by immunohistochemical staining.105
NEW THERAPEUTIC TARGETS
The only curative options for earlier stages of HCC are surgical resection, transplantation, or percutaneous ablation. However, only 12% of patients are eligible for surgical intervention because most HCCs are diagnosed at a late stage and the majority develop in diseased livers with poor hepatic functional reserve. Numerous experimental strategies are aimed at the aforementioned growth factor molecular pathways. A major signaling pathway in HCC is TGF-β, and serine/threonine kinase inhibitors (RSTK) have been developed. SB-431542, a small RSTK, has been shown to block TGF-β– and activin-mediated signaling. Reduced phosphorylation and decreased nuclear translocation of the Smads are observed with this inhibitor. More important, TGF-β– evoked protumorigenic cellular effects are diminished.106 SD-208 RSTK has also been shown to inhibit TGF-β– mediated migration and invasion in tumor cell models. However, viability and proliferation are not decreased with this compound.107
Therapeutic options targeting Wnt signaling are limited; however, receptor tyrosine kinase inhibitors (RTK) have also been shown to reduce receptor signaling. The crucial consideration is to create a highly selective inhibitor that will not interfere with other receptor tyrosine kinases such as INSR, which can lead to diabetogenic effects. Highly selective inhibitors such as tyrphostins, NVP-AEW541, and cyclolignans are able to reduce activation of IGF-1R and downstream AKT/PKB. In vitro and in vivo studies have shown a reduction in tumor cell growth especially in combination with chemotherapeutic agents.108,109 Novel neutralizing antibodies against IGF-1R such as IR3 which has been shown to reduce receptor autophosphorylation and signaling are also under active investigation.110–112 Antisense RNA and antisense oligodeoxynucleotide techniques to block IGF-1R synthesis are another approach to increase apoptosis and reduce proliferation.113
Inhibitors of the c-MET receptor such as PHA-665752, SU11271, SU11274, and SU11606 have been shown to reduce c-MET phosphorylation, resulting in subsequent inactivation of downstream effectors such as AKT/PKB, Gab-1, PLC, and Stat3. By blocking the c-MET receptor, decreases in proliferation, cell motility, and invasion of different types of tumor cells have been achieved. In a tumor xenograft mouse model, PHA-665752 caused a reduction in tumor volume and intensive cell death.114 Treatment of HCC with SU5416, an RTK inhibitor, was also shown to reduce activation of ERK1/2 and AKT/PKB effectors. The Ras/VEGF-R inhibitor BAY 43-9006 (sorafenib) reduced proliferation and angiogenesis in a promising phase II clinical trial. Treatment of HCC patients with BAY 43-9006 resulted in stable disease or tumor shrinkage in 43% of cases.59
CONCLUSIONS
Emerging studies regarding the role of hepatic progenitor/stem cells are only now being described. By far, the major underlying etiology of HCCs is chronic inflammatory diseases from viral hepatitis or cirrhosis. Under these circumstances, the progenitor cell compartment is activated as the mature hepatocytes become senescent. Substantial numbers of HCCs display an intermediate phenotype of progenitor and hepatocyte cells. Current studies favor the differentiation arrest model, in which the maturation process of the progenitor/stem cell is disrupted. Many growth factor signaling pathways play crucial roles in the maturation of liver cells such as TGF-β/TBR, IGF/IGF-1R, HGF/MET, Wnt/β-catenin/FZD, and TGF-α/EGFR signaling. Dysregulation of these pathways represents a crucial role in hepatocarcinogenesis. Further studies are necessary to elucidate cross-talk between the different regulatory pathways. The understanding of these interdependent regulatory pathways holds promise for the development of new therapeutic approaches to this devastating disease.
Acknowledgments
2005–2010: PHS (NIH) RO1CA106614-01A2, “Role of elf/Smad4 in GI cell proliferation and cell cycle regulation”;
1996–2006: PHS (NIH) R01DK56111, “PRAJA-1 a novel anti-apoptosis RIN finger protein”;
2006–2011: PHS (NIH) R01CA042857-18A1, “Metastatic Potential of Colorectal Carcinoma”;
2002–2007: Co-PI: PHS (NIH) RO1DK58637, “Regulation of gastrointestinal and neural epithelial cell proliferation”;
2004–2008 VA Merit Review “TGF-beta Signaling in Gastrointestinal and Liver Diseases.”
Footnotes
Disclosures of Potential Conflicts of Interest
Dr. Mishra has no potential conflicts of interest to disclose.
References
- 1.Bosch FX, Erdinger L, Ingel F, et al. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, et al. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 4.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127(suppl 1):S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Kanzler S, Meyer E, Lohse AW, et al. Hepatocellular expression of a dominant-negative mutant TGF-beta type II receptor accelerates chemically induced hepatocarcinogenesis. Oncogene. 2001;20:5015–5024. doi: 10.1038/sj.onc.1204544. [DOI] [PubMed] [Google Scholar]
- 7.Huo TI, Wang XW, Forgues M, et al. Hepatitis B virus X mutants derived from human hepatocellular carcinoma retain the ability to abrogate p53-induced apoptosis. Oncogene. 2001;20:3620–3628. doi: 10.1038/sj.onc.1204495. [DOI] [PubMed] [Google Scholar]
- 8.Ueda H, Ullrich SJ, Gangemi JD, et al. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 9.Feitelson MA. Hepatitis B virus in hepatocarcinogenesis. J Cell Physiol. 1999;181:188–202. doi: 10.1002/(SICI)1097-4652(199911)181:2<188::AID-JCP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci U S A. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mise K, Tashiro S, Yogita S, et al. Assessment of the biological malignancy of hepatocellular carcinoma: relationship to clinicopathological factors and prognosis. Clin Cancer Res. 1998;4:1475–1482. [PubMed] [Google Scholar]
- 12.Kato A, Kubo K, Kurokawa F, et al. Numerical aberrations of chromosomes 16, 17, and 18 in hepatocellular carcinoma: a FISH and FCM analysis of 20 cases. Dig Dis Sci. 1998;43:1–7. doi: 10.1023/a:1018838731634. [DOI] [PubMed] [Google Scholar]
- 13.Coleman WB. Mechanisms of human hepatocarcinogenesis. Curr Mol Med. 2003;3:573–588. doi: 10.2174/1566524033479546. [DOI] [PubMed] [Google Scholar]
- 14.Kusano N, Shiraishi K, Kubo K, et al. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858–1862. doi: 10.1002/hep.510290636. [DOI] [PubMed] [Google Scholar]
- 15.Macdonald GA, Greenson JK, Kato K, et al. Microsatellite instability and loss of heterozygosity at DNA mismatch repair gene loci occurs during hepatic carcinogenesis. Hepatology. 1998;28:90–97. doi: 10.1002/hep.510280114. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Itoh F, Fukushima H, et al. Infrequent widespread microsatellite instability in hepatocellular carcinomas. Int J Oncol. 2000;16:543–547. doi: 10.3892/ijo.16.3.543. [DOI] [PubMed] [Google Scholar]
- 17.Yakushiji H, Mukai S, Matsukura S, et al. DNA mismatch repair deficiency in curatively resected sextuple primary cancers in different organs: a molecular case report. Cancer Lett. 1999;142:17–22. doi: 10.1016/s0304-3835(99)00110-x. [DOI] [PubMed] [Google Scholar]
- 18.Tiniakos D, Spandidos DA, Kakkanas A, et al. Expression of ras and myc oncogenes in human hepatocellular carcinoma and nonneoplastic liver tissues. Anticancer Res. 1989;9:715–721. [PubMed] [Google Scholar]
- 19.Tiniakos D, Spandidos DA, Yiagnisis M, et al. Expression of ras and c-myc oncoproteins and hepatitis B surface antigen in human liver disease. Hepatogastroenterology. 1993;40:37–40. [PubMed] [Google Scholar]
- 20.Ito Y, Matsuura N, Sakon M, et al. Expression and prognostic roles of the G1-S modulators in hepatocellular carcinoma: p27 independently predicts the recurrence. Hepatology. 1999;30:90–99. doi: 10.1002/hep.510300114. [DOI] [PubMed] [Google Scholar]
- 21.Ito Y, Sasaki Y, Horimoto M, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951–958. doi: 10.1002/hep.510270409. [DOI] [PubMed] [Google Scholar]
- 22.Borzio M, Fargion S, Borzio F, et al. Impact of large regenerative, low grade and high grade dysplastic nodules in hepatocellular carcinoma development. J Hepatol. 2003;39:208–214. doi: 10.1016/s0168-8278(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 23.Fausto N. Liver regeneration and repair: hepatocytes progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 24.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 25.Rhim JA, Sandgren EP, Degen JP, et al. Replacement of diseased mouse liver by hepatic cell transplantation. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 26.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 27.Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung J, Zheng M, Goldfarb M, et al. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 29.Rossi JM, Dunn NR, Hogan BL, et al. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiojiri N, Lemire JM, Fausto N. Cell lineages and oval cell progenitors in rat liver development. Cancer Res. 1991;51:2611–2620. [PubMed] [Google Scholar]
- 31.Hixson DC, Faris RA, Thompson NL. An antigenic portrait of the liver during carcinogenesis. Pathobiology. 1990;58:65–77. doi: 10.1159/000163565. [DOI] [PubMed] [Google Scholar]
- 32.Roskams TA, Libbrecht L, Desmet VJ. Progenitor cells in diseased human liver. Semin Liver Dis. 2003;23:385–396. doi: 10.1055/s-2004-815564. [DOI] [PubMed] [Google Scholar]
- 33.Roskams T, Cassiman D, De Vos R, et al. Neuroregulation of the neuroendocrine compartment of the liver. Anat Rec A Discov Mol Cell Evol Biol. 2004;280:910–923. doi: 10.1002/ar.a.20096. [DOI] [PubMed] [Google Scholar]
- 34.Newsome PN, Hussain MA, Theise ND. Hepatic oval cells: helping redefine a paradigm in stem cell biology. Curr Top Dev Biol. 2004;61:1–28. doi: 10.1016/S0070-2153(04)61001-5. [DOI] [PubMed] [Google Scholar]
- 35.Theise ND, Saxena R, Portmann BC, et al. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425–1433. doi: 10.1002/hep.510300614. [DOI] [PubMed] [Google Scholar]
- 36.Roskams T, Yang SQ, Koteish A, et al. Oxidative stress and oval cell accumulation in mice and humans with alcoholic and nonalcoholic fatty liver disease. Am J Pathol. 2003;163:1301–1311. doi: 10.1016/S0002-9440(10)63489-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowes KN, Brennan BA, Yeoh GC, et al. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol. 1999;154:537–541. doi: 10.1016/S0002-9440(10)65299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libbrecht L, Desmet V, Van Damme B, et al. Deep intralobular extension of human hepatic ‘progenitor cells’ correlates with parenchymal inflammation in chronic viral hepatitis: can ‘progenitor cells’ migrate? J Pathol. 2000;192:373–378. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH700>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Falkowski O, An HJ, Ianus IA, et al. Regeneration of hepatocyte ‘buds’ in cirrhosis from intrabiliary stem cells. J Hepatol. 2003;39:357–364. doi: 10.1016/s0168-8278(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 40.Wu PC, Lai VC, Fang JW, et al. Hepatocellular carcinoma expressing both hepatocellular and biliary markers also expresses cytokeratin 14, a marker of bipotential progenitor cells. J Hepatol. 1999;31:965–966. doi: 10.1016/s0168-8278(99)80303-1. [DOI] [PubMed] [Google Scholar]
- 41.Van Eyken P, Sciot R, Paterson A, et al. Cytokeratin expression in hepatocellular carcinoma: an immunohistochemical study. Hum Pathol. 1988;19:562–568. doi: 10.1016/s0046-8177(88)80205-3. [DOI] [PubMed] [Google Scholar]
- 42.Uenishi T, Kubo S, Yamamoto T, et al. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851–857. doi: 10.1111/j.1349-7006.2003.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsia CC, Evarts RP, Nakatsukasa H, et al. Occurrence of oval-type cells in hepatitis B virus-associated human hepatocarcinogenesis. Hepatology. 1992;16:1327–1333. doi: 10.1002/hep.1840160604. [DOI] [PubMed] [Google Scholar]
- 44.Chiba T, Kita K, Zheng YW, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 45.Piek E, Heldin CH, Ten Dijke P. Specificity, diversity, and regulation in TGF-beta superfamily signaling. Faseb J. 1999;13:2105–2124. [PubMed] [Google Scholar]
- 46.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 47.Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 48.Liu X, Sun Y, Weinberg RA, et al. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 49.Tang Y, Katuri V, Dillner A, et al. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 50.Tsukazaki T, Chiang TA, Davison AF, et al. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 51.Dong C, Li Z, Alvarez R, Jr, et al. Microtubule binding to Smads may regulate TGF beta activity. Mol Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 52.Mishra B, Tang Y, Katuri V, et al. Loss of cooperative function of transforming growth factorbeta signaling proteins, smad3 with embryonic liver fodrin, a beta-spectrin, in primary biliary cirrhosis. Liver Int. 2004;24:637–645. doi: 10.1111/j.1478-3231.2004.0958.x. [DOI] [PubMed] [Google Scholar]
- 53.Tang Y, Katuri V, Srinivasan R, et al. Transforming growth factor-beta suppresses nonmetastatic colon cancer through Smad4 and adaptor protein ELF at an early stage of tumorigenesis. Cancer Res. 2005;65:4228–4237. doi: 10.1158/0008-5472.CAN-04-4585. [DOI] [PubMed] [Google Scholar]
- 54.Sporn MB, Roberts AB. Transforming growth factor-beta: new chemical forms and new biological roles. Biofactors. 1988;1:89–93. [PubMed] [Google Scholar]
- 55.Mishra L, Cai T, Yu P, et al. Elf3 encodes a novel 200-kD beta-spectrin: role in liver development. Oncogene. 1999;18:353–364. doi: 10.1038/sj.onc.1202313. [DOI] [PubMed] [Google Scholar]
- 56.Sanford LP, Ormsby I, Gittenberger-de Groot AC, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schier AF, Talbot WS. Nodal signaling and the zebrafish organizer. Int J Dev Biol. 2001;45:289–297. [PubMed] [Google Scholar]
- 58.Munoz-Sanjuan I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 59.Breuhahn K, Longerich T, Schirmacher P. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 2006;25: 3787–3800. doi: 10.1038/sj.onc.1209556. [DOI] [PubMed] [Google Scholar]
- 60.Yakicier MC, Irmak MB, Romano A, et al. Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene. 1999;18:4879–4883. doi: 10.1038/sj.onc.1202866. [DOI] [PubMed] [Google Scholar]
- 61.Longerich T, Breuhahn K, Odenthal M, et al. Factors of transforming growth factor beta signalling are co-regulated in human hepatocellular carcinoma. Virchows Arch. 2004;445:589–596. doi: 10.1007/s00428-004-1118-x. [DOI] [PubMed] [Google Scholar]
- 62.Park YN, Chae KJ, Oh BK, et al. Expression of Smad7 in hepatocellular carcinoma and dysplastic nodules: resistance mechanism to transforming growth factor-beta. Hepatogastroenterology. 2004;51:396–400. [PubMed] [Google Scholar]
- 63.Tsai JF, Chuang LY, Jeng JE, et al. Clinical relevance of transforming growth factor-beta 1 in the urine of patients with hepatocellular carcinoma. Medicine (Baltimore) 1997;76:213–226. doi: 10.1097/00005792-199705000-00007. [DOI] [PubMed] [Google Scholar]
- 64.Kim HG, Chung YH, Song BC, et al. Expression of transforming growth factor beta-1 in chronic hepatitis and hepatocellular carcinoma associated with hepatitis C virus infection. Korean J Intern Med. 2000;15:165–170. doi: 10.3904/kjim.2000.15.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bedossa P, Peltier E, Terris B, et al. Transforming growth factor-beta 1 (TGF-beta 1) and TGF-beta 1 receptors in normal, cirrhotic, and neoplastic human livers. Hepatology. 1995;21:760–766. [PubMed] [Google Scholar]
- 66.Abou-Shady M, Baer HU, Friess H, et al. Transforming growth factor betas and their signaling receptors in human hepatocellular carcinoma. Am J Surg. 1999;177:209–215. doi: 10.1016/s0002-9610(99)00012-4. [DOI] [PubMed] [Google Scholar]
- 67.Reiss M. Transforming growth factor-beta and cancer: a love-hate relationship? Oncol Res. 1997;9:447–457. [PubMed] [Google Scholar]
- 68.Cui W, Fowlis DJ, Bryson S, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 69.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 70.Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 71.Kikuchi A. Tumor formation by genetic mutations in the components of the Wnt signaling pathway. Cancer Sci. 2003;94:225–229. doi: 10.1111/j.1349-7006.2003.tb01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller LD, Park KS, Guo QM, et al. Silencing of Wnt signaling and activation of multiple metabolic pathways in response to thyroid hormone-stimulated cell proliferation. Mol Cell Biol. 2001;21:6626–6639. doi: 10.1128/MCB.21.19.6626-6639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moon RT, Shah K. Developmental biology: signalling polarity. Nature. 2002;417:239–240. doi: 10.1038/417239a. [DOI] [PubMed] [Google Scholar]
- 74.Polakis P. More than one way to skin a catenin. Cell. 2001;105:563–566. doi: 10.1016/s0092-8674(01)00379-8. [DOI] [PubMed] [Google Scholar]
- 75.Kishida M, Koyama S, Kishida S, et al. Axin prevents Wnt-3a-induced accumulation of beta-catenin. Oncogene. 1999;18:979–985. doi: 10.1038/sj.onc.1202388. [DOI] [PubMed] [Google Scholar]
- 76.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 77.Yau TO, Chan CY, Chan KL, et al. HDPR1, a novel inhibitor of the WNT/beta-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene. 2005;24:1607–1614. doi: 10.1038/sj.onc.1208340. [DOI] [PubMed] [Google Scholar]
- 78.Ryo A, Nakamura M, Wulf G, et al. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 79.Nhieu JT, Renard CA, Wei Y, et al. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol. 1999;155: 703–710. doi: 10.1016/s0002-9440(10)65168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Satoh S, Daigo Y, Furukawa Y, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virusmediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 81.Taniguchi K, Roberts LR, Aderca IN, et al. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21:4863–4871. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- 82.Merle P, de la Monte S, Kim M, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 83.Foulstone E, Prince S, Zaccheo O, et al. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- 84.Lee YI, Lee S, Lee Y, et al. The human hepatitis B virus transactivator X gene product regulates Sp1 mediated transcription of an insulin-like growth factor II promoter 4. Oncogene. 1998;16:2367–2380. doi: 10.1038/sj.onc.1201760. [DOI] [PubMed] [Google Scholar]
- 85.Kang-Park S, Lee JH, Shin JH, et al. Activation of the IGF-II gene by HBV-X protein requires PKC and p44/p42 map kinase signalings. Biochem Biophys Res Commun. 2001;283:303–307. doi: 10.1006/bbrc.2001.4767. [DOI] [PubMed] [Google Scholar]
- 86.Lee S, Park U, Lee YI. Hepatitis C virus core protein transactivates insulin-like growth factor II gene transcription through acting concurrently on Egr1 and Sp1 sites. Virology. 2001;283:167–177. doi: 10.1006/viro.2001.0892. [DOI] [PubMed] [Google Scholar]
- 87.Sue SR, Chari RS, Kong FM, et al. Transforming growth factor-beta receptors and mannose 6-phosphate/insulin-like growth factor-II receptor expression in human hepatocellular carcinoma. Ann Surg. 1995;222:171–178. doi: 10.1097/00000658-199508000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Souza AT, Hankins GR, Washington MK, et al. Frequent loss of heterozygosity on 6q at the mannose 6-phosphate/insulin-like growth factor II receptor locus in human hepatocellular tumors. Oncogene. 1995;10:1725–1729. [PubMed] [Google Scholar]
- 89.Oka Y, Waterland RA, Killian JK, et al. M6P/IGF2R tumor suppressor gene mutated in hepatocellular carcinomas in Japan. Hepatology. 2002;35:1153–1163. doi: 10.1053/jhep.2002.32669. [DOI] [PubMed] [Google Scholar]
- 90.Efimova EA, Glanemann M, Liu L, et al. Effects of human hepatocyte growth factor on the proliferation of human hepatocytes and hepatocellular carcinoma cell lines. Eur Surg Res. 2004;36:300–307. doi: 10.1159/000079915. [DOI] [PubMed] [Google Scholar]
- 91.Stuart KA, Riordan SM, Lidder S, et al. Hepatocyte growth factor/scatter factor-induced intracellular signalling. Int J Exp Pathol. 2000;81: 17–30. doi: 10.1046/j.1365-2613.2000.00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.D'Errico A, Fiorentino M, Ponzetto A, et al. Liver hepatocyte growth factor does not always correlate with hepatocellular proliferation in human liver lesions: its specific receptor c-met does. Hepatology. 1996;24:60–64. doi: 10.1002/hep.510240112. [DOI] [PubMed] [Google Scholar]
- 93.Neaud V, Faouzi S, Guirouilh J, et al. Human hepatic myofibroblasts increase invasiveness of hepatocellular carcinoma cells: evidence for a role of hepatocyte growth factor. Hepatology. 1997;26:1458–1466. doi: 10.1053/jhep.1997.v26.pm0009397985. [DOI] [PubMed] [Google Scholar]
- 94.Guirouilh J, Le Bail B, Boussarie L, et al. Expression of hepatocyte growth factor in human hepatocellular carcinoma. J Hepatol. 2001;34:78–83. doi: 10.1016/s0168-8278(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 95.Moinzadeh P, Breuhahn K, Stutzer H, et al. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological-grade–results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–941. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boix L, Rosa JL, Ventura F, et al. c-met mRNA overexpression in human hepatocellular carcinoma. Hepatology. 1994;19:88–91. [PubMed] [Google Scholar]
- 97.Tavian D, De Petro G, Benetti A, et al. u-PA and c-MET mRNA expression is co-ordinately enhanced while hepatocyte growth factor mRNA is down-regulated in human hepatocellular carcinoma. Int J Cancer. 2000;87:644–649. [PubMed] [Google Scholar]
- 98.Guo L, Kozlosky CJ, Ericsson LH, et al. Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J Am Soc Mass Spectrom. 2003;14:1022–1031. doi: 10.1016/S1044-0305(03)00206-X. [DOI] [PubMed] [Google Scholar]
- 99.Zhang T, Ma J, Cao X.Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling Biochem J 376(Pt 2)457–464.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rescan C, Coutant A, Talarmin H, et al. Mechanism in the sequential control of cell morphology and S phase entry by epidermal growth factor involves distinct MEK/ERK activations. Mol Biol Cell. 2001;12:725–738. doi: 10.1091/mbc.12.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hisaka T, Yano H, Haramaki M, et al. Expressions of epidermal growth factor family and its receptor in hepatocellular carcinoma cell lines: relationship to cell proliferation. Int J Oncol. 1999;14:453–460. doi: 10.3892/ijo.14.3.453. [DOI] [PubMed] [Google Scholar]
- 102.Kira S, Nakanishi T, Suemori S, et al. Expression of transforming growth factor alpha and epidermal growth factor receptor in human hepatocellular carcinoma. Liver. 1997;17:177–182. doi: 10.1111/j.1600-0676.1997.tb00803.x. [DOI] [PubMed] [Google Scholar]
- 103.Zhang J, Wang WL, Li O, et al. Expression of transforming growth factor-alpha and hepatitis B surface antigen in human hepatocellular carcinoma tissues and its significance. World J Gastroenterol. 2004;10:830–833. doi: 10.3748/wjg.v10.i6.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tabor E, Farshid M, Di Bisceglie A, et al. Increased expression of transforming growth factor alpha after transfection of a human hepatoblastoma cell line with the hepatitis B virus. J Med Virol. 1992;37:271–273. doi: 10.1002/jmv.1890370406. [DOI] [PubMed] [Google Scholar]
- 105.Inui Y, Higashiyama S, Kawata S, et al. Expression of heparin-binding epidermal growth factor in human hepatocellular carcinoma. Gastroenterology. 1994;107:1799–1804. doi: 10.1016/0016-5085(94)90823-0. [DOI] [PubMed] [Google Scholar]
- 106.Hjelmeland MD, Hjelmeland AB, Sathornsumetee S, et al. SB-431542, a small molecule transforming growth factor-beta-receptor antagonist, inhibits human glioma cell line proliferation and motility. Mol Cancer Ther. 2004;3:737–745. [PubMed] [Google Scholar]
- 107.Hayashi T, Hideshima T, Nguyen AN, et al. Transforming growth factor beta receptor I kinase inhibitor down-regulates cytokine secretion and multiple myeloma cell growth in the bone marrow microenvironment. Clin Cancer Res. 2004;10:7540–7546. doi: 10.1158/1078-0432.CCR-04-0632. [DOI] [PubMed] [Google Scholar]
- 108.Mitsiades CS, Mitsiades NS, McMullan CJ, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–230. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 109.Scotlandi K, Manara MC, Nicoletti G, et al. Antitumor activity of the insulin-like growth factor-I receptor kinase inhibitor NVPAEW541 in musculoskeletal tumors. Cancer Res. 2005;65:3868–3876. doi: 10.1158/0008-5472.CAN-04-3192. [DOI] [PubMed] [Google Scholar]
- 110.Scotlandi K, Benini S, Nanni P, et al. Blockage of insulin-like growth factor-I receptor inhibits the growth of Ewing's sarcoma in athymic mice. Cancer Res. 1998;58:4127–4131. [PubMed] [Google Scholar]
- 111.Cohen BD, Baker DA, Soderstrom C, et al. Combination therapy enhances the inhibition of tumor growth with the fully human anti-type 1 insulin-like growth factor receptor monoclonal antibody CP-751,871. Clin Cancer Res. 2005;11:2063–2073. doi: 10.1158/1078-0432.CCR-04-1070. [DOI] [PubMed] [Google Scholar]
- 112.Sachdev D, Li SL, Hartell JS, et al. A chimeric humanized single-chain antibody against the type I insulin-like growth factor (IGF) receptor renders breast cancer cells refractory to the mitogenic effects of IGF-I. Cancer Res. 2003;63:627–635. [PubMed] [Google Scholar]
- 113.Lafarge-Frayssinet C, Duc HT, Frayssinet C, et al. Antisense insulin-like growth factor I transferred into a rat hepatoma cell line inhibits tumorigenesis by modulating major histocompatibility complex I cell surface expression. Cancer Gene Ther. 1997;4:276–285. [PubMed] [Google Scholar]
- 114.Christensen JG, Schreck R, Burrows J, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]