Abstract
Canavan disease is a fatal neurological disorder caused by the malfunctioning of a single metabolic enzyme, aspartoacylase, that catalyzes the deacetylation of N-acetyl-l-aspartate to produce l-aspartate and acetate. The structure of human brain aspartoacylase has been determined in complex with a stable tetrahedral intermediate analogue, N-phosphonomethyl-l-aspartate. This potent inhibitor forms multiple interactions between each of its heteroatoms and the substrate binding groups arrayed within the active site. The binding of the catalytic intermediate analogue induces the conformational ordering of several substrate binding groups, thereby setting up the active site for catalysis. The highly ordered binding of this inhibitor has allowed assignments to be made for substrate binding groups and provides strong support for a carboxypeptidase-type mechanism for the hydrolysis of the amide bond of the substrate, N-acetyl-l-aspartate.
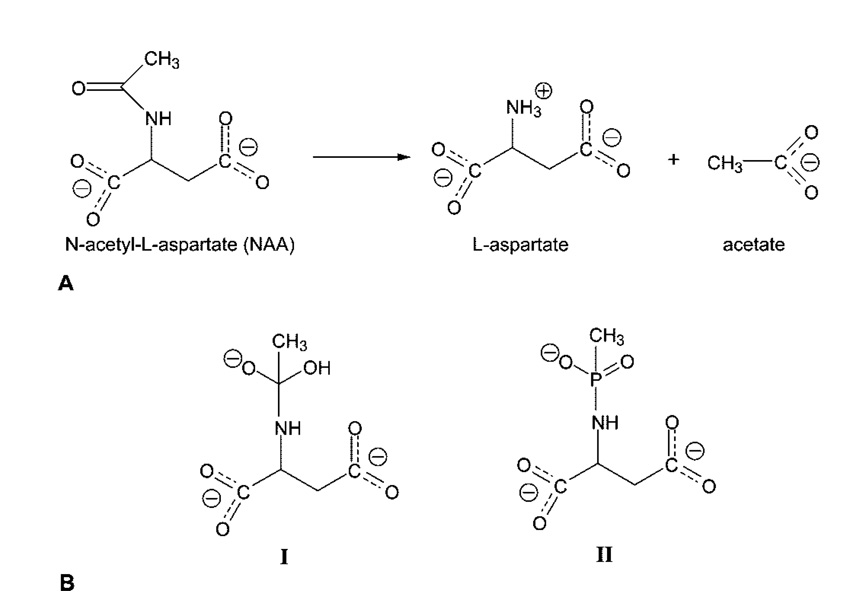
Canavan disease (CD)1 is a fatal autosomal recessive disorder that affects the central nervous system (1) and for which there is currently no effective treatment. The symptoms for CD can be noticed as early as 3–6 months of age and include rapid increase of the head circumference (megalocephaly), lack of head control, reduced visual responsiveness, abnormal muscle tone (i.e., floppiness or stiffness), and mental retardation (2). CD patients usually do not live past the first decade of their lives. Canavan disease is caused by a defect in the capability of the brain to metabolize N-acetyl-l-aspartate (NAA) (3), one of the most abundant amino acids in the brain (4). Aspartoacylase, whose function is to catalyze the deacetylation of NAA (Figure 1), is located primarily in the white matter of the brain, more specifically in the cytosol of the oligodendrocytes (5). Analysis of the acy2 gene that encodes aspartoacylase taken from DNA isolated from CD patients has revealed more than 50 different mutations, including numerous deletions, missense mutations, and premature terminations (6). In most cases, the missense mutations result in nonconservative amino acid substitutions, leading to an altered enzyme that either is not expressed at all or is expressed but has little or no catalytic activity (6).
Figure 1.
(A) Deacetylation reaction catalyzed by aspartoacylase. (B) Structure of the putative reaction intermediate (I) that would be formed in a carboxypeptidase-like mechanism for the deacetylation of N-acetyl-l-aspartate and structure of the phosphonamidate analogue of this intermediate (II) that was cocrystallized with human aspartoacylase.
Aspartoacylase was originally proposed to be member of the esterase family since it was shown to be inactivated by diisopropylfluorophosphate, a classic serine protease inactivator (7). On the basis of these studies, conserved serine, histidine, and glutamate residues were identified and proposed to function as a catalytic triad. However, subsequent alignment studies showed few similarities between aspartoacylase and the esterase family, instead suggesting that aspartoacylase is a member of the zinc peptidase superfamily (8). This classification is supported by recent work in which aspartoacylase was shown to bind one zinc ion per monomer (9), consistent with its assignment to the zinc carboxypeptidase family. Recently, the crystal structures of the rat form and the human form of aspartoacylase have been determined at 1.8 and 2.8 Å resolution, respectively, by the Center for Eukaryotic Structural Genomics (10). Both of these structures are the apo form of the enzyme that was crystallized in the absence of either substrates or products. These structures confirm the presence of zinc in the enzyme, as well as the identity of the zinc binding ligands which had been proposed previously. In addition, the ligands that are potentially involved in substrate binding have been suggested through a comparison of the structure of aspartoacylase with those of the carboxypeptidases (10). However, the sequence of human aspartoacylase is only ~10% identical with that of bovine carboxypeptidase A (CPA) (8). A structural comparison of rat aspartoacylase with CPA found that the C-terminal domain of aspartoacylase is not present in carboxypeptidase, while the N-terminal domain of CPA is approximately 60 residues longer than the corresponding domain of aspartoacylase (10).
No structures had been determined for any complexes of aspartoacylase with bound substrates, products, or analogues to directly examine these active site assignments and their proposed role in the catalytic mechanism. We have now determined the structure of human aspartoacylase (hACY2) complexed with a tetrahedral intermediate analogue bound in the active site. From an examination of this structure, we have made definitive assignments of the key active site functional groups. These structural assignments, combined with kinetic characterization of several active site mutants, provide new insights into the catalytic mechanism of aspartoacylase.
EXPERIMENTAL PROCEDURES
Materials
The N-methyl phosphonamidate derivative of l-aspartic acid was synthesized by a modification of a previously published procedure (11) starting from the dimethyl ester of l-aspartic acid. Methyl methylchlorophosphonate (0.8 g, 6.2 mmol) in 10 mL of chloroform was added dropwise to 1.0 g (5.1 mmol) of dimethyl l-aspartate-HCl and 3 mL of triethylamine in 20 mL of chloroform. The mixture was cooled and stirred for 3 h at room temperature, diluted with 50 mL of chloroform, and then washed twice with 2.0 N hydrochloric acid and then twice with a saturated brine solution. After removal of the solvent, the compound was purified by silica gel column chromatography via elution with an ethyl acetate/methanol mixture (12/1). The dimethyl N-methylmethoxyphosphinyl-l-aspartate product was obtained as an oily product in a yield of 78% (1.0 g). The trisodium salt of N-phosphonamidate-l-aspartate was obtained (0.7 g isolated yield) by hydrolysis of the diester with sodium hydroxide and was characterized by 1H and 31P NMR and by ESI-MS. Chromatography resins were purchased from Amersham Biosciences, and the crystallization screening kits were obtained from Hampton Research and Jena Biosciences.
Cloning, Expression, and Enzyme Purification
The experimental details regarding the cloning, expression, and purification of aspartoacylase have been described in previously published work (9). Briefly, the human acy2 gene was cloned in pPICZ A and then transformed into the KM71H Pichia pastoris cell line. The enzyme was expressed by adding 1% MeOH to the expression medium. Aspartoacylase was then extracted from the cells using a BeadBeater cell disruptor and loaded on a HiTrap Chelating HP column for the initial purification step. The enzyme was further purified by anion exchange using a Source 15Q column on an Äkta chromatography system, followed by a size exclusion step (Superdex 200) to produce the final, highly purified enzyme. The zinc content of the enzyme was determined by inductively coupled plasma (ICP) emission spectroscopy as previously described (9).
Active Site Mutants and Kinetic Studies
The QuikChange II site-directed mutagenesis kit (Stratagene) was used to construct the mutants, and each mutation was confirmed by sequencing. The vector containing each mutated gene was linearized and inserted into the yeast genome of the KM71H cell line and was then expressed and purified under the same conditions as the native enzyme. Kinetic parameters for the mutants were determined by a coupled assay wherein the hydrolyzed l-aspartate product was converted to fumarate using l-aspartase (12). The concentration of the substrate, N-acetyl-l-aspartate was varied from 0.2 to 1.25 mM, and the production of fumarate was assessed at 240 nm (ϵ240 = 2.53 mM−1 cm−1) by using a Cary 50 UV–visible spectrophotometer. The inhibition constant (Ki) was determined at constant NAA levels by varying the intermediate analogue concentration (30–300 μM) and fitting the data using a Dixon plot.
Crystallization and Data Collection
Initial crystallization conditions were identified by sparse matrix screening of hACY2 at a protein concentration of 8–10 mg/mL. Both index screens (Hampton Research) and JB Screen Basic (Jena Bioscience) yielded initial crystals for the apoenzyme. The optimized conditions for the apoenzyme crystals consisted of 50 mM sodium citrate (pH 6.0), 300 mM K2HPO4, and 15–19% polyethylene glycol 3350. Small crystalline rods grew within 2–3 days at 20 °C. The crystals were cryoprotected by mixing the well solution with 20–30% ethylene glycol. The crystals diffracted to ~4 Å on an in-house Rigaku FR-E rotating anode X-ray generator, with the data quality limited primarily by the small crystal size. Somewhat improved crystals were obtained from aspartoacylase that had been treated with neuraminidase, which functions to remove terminal sialic acid groups from glycoproteins. All of the subsequent structural studies were conducted with this neuraminidase-treated enzyme, which has kinetic parameters identical to those of native aspartoacylase. The hACY2–intermediate analog complex crystals were obtained by cocrystallization, incubating the enzyme with 2 mM inhibitor at 4 °C for 1 h prior to setting up crystallization trials. Crystals of the enzyme–intermediate analogue complex were grown under the same conditions as the apoenzyme and had the same diffraction quality on our home source. Both crystal forms, apo and complex, belong to space group P42212 with similar unit cell dimensions. Full crystallographic data were measured on beamline 23ID-B of the GM/CA Collaborative Access Team at the Advanced Photon Source, using a MARmosaic Charge Coupled Device detector with 16 tiled chips in a 4 × 4 array and 300 mm2 sensitive area (MARUSA). HKL2000 (13) was used for indexing and integration and Scalepack for scaling of all diffraction data. Data processing statistics for the apoenzyme and the enzyme–intermediate analogue complex are summarized in Table 1.
Table 1.
Data Collection and Refinement Statistics
| apoenzyme | inhibitor complex | |
|---|---|---|
| Collection Statistics | ||
| wavelength (Å) | 1.033 | 1.283 |
| space group | P42212 | P42212 |
| cell dimensions | ||
| a = b, c (Å) | 143.79, 104.08 | 143.08, 104.6 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 |
| resolution (Å)a | 50.0–2.7 (2.8–2.7) | 50.0–2.7 (2.8–2.7) |
| no. of reflections | 23564 | 27490 |
| redundancya | 3.9 (2.5) | 10.7 (5.3) |
| completeness (%)a | 87.3 (47.2) | 95.3 (62.7) |
| Rsym (%)ab | 6.2 (30.6) | 9.4 (35.4) |
| I/σIa | 17.8 (2.8) | 20.7 (2.9) |
| χ2a | 0.96 (0.91) | 0.99 (0.97) |
| Refinement Statistics | ||
| Rwork, Rfree (%)c | 21.6, 26.9 | 20.2, 27.1 |
| Wilson B factor (Å2) | 65.3 | 72.7 |
| rmsd for bonds (Å), angles (deg) | 0.013, 1.47 | 0.012, 1.64 |
| ESU (Å)d | 0.361 | 0.321 |
| no. of atoms (protein, metal, ligand, water) | 4860, 2, 10, 30 | 4842, 2, 26, 69 |
| B factor (Å2) (protein, metal, ligand, water) | 44, 58, 49, 55 | 60, 83, 70, 62 |
Values in parentheses are for the highest-resolution shell.
Rsym = ΣhΣi|Ii(h) – 〈I(h)〉|/Σ iIi(h), where Ii(h) is the intensity of an individual measurement and 〈I(h)〉 is the mean intensity of the reflection.
Rwork = Σh║Fobs| – |Fcalc║/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated for 5% of the randomly selected unique reflections omitted from structure refinement.
Dispersion precision indicator based on Rfree.
Structure Determination, Refinement, and Analysis
The structures of apo-hACY2 and its complex with the intermediate analogue were refined with REFMAC5 (14) using the atomic coordinates of hACY2 from the Protein Data Bank (entry 2I3C) as a starting model. A single cycle of rigid body refinement at 3.5 Å was followed by restrained refinement at the highest resolution, with TLS refinement used in the final cycles. Revisions of the protein model, water molecules, and inhibitor placement were done with COOT (15). All peaks in the 2Fo – Fc map higher than 1σ and within hydrogen bonding distance of polar protein atoms were examined using the water placement procedure in COOT. Occupancies for the ligand in the complex structure were assigned to 1.0. In the apoenzyme structure, phosphate molecules A and B were assigned occupancies of 0.7 and 0.3, respectively, to accommodate the close contacts between them. The aligned structures were analyzed and the figures rendered with the PyMOL Molecular Graphics System (16).
RESULTS AND DISCUSSION
Verification of a Carboxypeptidase-Type Mechanism
Both sequence alignment (8) and structural analysis (10) have placed aspartoacylase among the members of the carboxypeptidase family. To test this hypothesis, aspartoacylase was examined using a stable analogue that was synthesized to resemble the intermediate that would be produced during a carboxypeptidase-like catalytic cycle. In this proposed reaction scheme, a zinc-bound hydroxide, generated by proton abstraction, would serve as the nucleophile to attack the acetyl group of NAA and generate a tetrahedral intermediate. Subsequent collapse of this intermediate (Figure 1B, compound I) would lead to the products. A methylphos-phonamidate analogue of this putative catalytic intermediate, N-methylphosphono-l-aspartate, has been synthesized (Figure 1B, compound II) and tested as a possible inhibition of aspartoacylase. This analogue was found to be a potent inhibitor, with a Ki value of ~300 nM, providing strong evidence in support of a carboxypeptidase-type mechanism for the hydrolysis of NAA catalyzed by aspartoacylase. To gain further insight into the catalytic mechanism, aspartoacylase was cocrystallized in the presence of saturating levels of this inhibitor with the aim of forming and subsequently determining the structure of a tetrahedral intermediate analogue complex.
Structural Analysis of the Aspartoacylase—Intermediate Analogue Complex
Diffraction quality crystals of human aspartoacylase belonging to the P42212 space group with two molecules in the asymmetric unit were obtained from a potassium phosphate/PEG 3350 grid screen. After optimization, the enzyme in both forms, apo and in complex with its intermediate analogue, yielded crystals that diffracted to 2.7 Å resolution. Fluorescence scans around the zinc absorption edge confirmed the presence of zinc in the hACY2 crystals. However, the anomalous signal in the MAD data was not sufficient for de novo phasing. As a structure of hACY2 (PDB entry 2I3C) (10) became available, these coordinates were used in the initial refinement stages. The resulting electron density maps allowed a complete model to be built for residues 10–310, while no interpretable density was observed for either the first nine amino acids at the amino terminus (residues 1–9) or the final three amino acids at the carboxyl terminus (residues 311–313). The structures of the apoenzyme and the intermediate analogue complex were refined to crystallographic R factors of 21.6 and 20.2%, respectively. The essential data collection parameters and refinement statistics for both structures are presented in Table 1.
Overall Structure of Human Aspartoacylase
Each of the structures of aspartoacylase that has now been determined shows that this enzyme is a dimer with extensive contact surface area between the two subunits, supporting the previous biochemical studies (12). The structures of the hACY2 apoenzyme and the intermediate analogue complex have a nearly identical overall fold, with a rmsd for 301 Cα atoms of only 0.3 Å. Since the protein was incubated with the inhibitor prior to crystallization, the absence of significant conformational rearrangements suggests that side chain movements must be the predominant mode used to accommodate intermediate binding, and presumably substrate binding as well, without requiring substantial structural rearrangements.
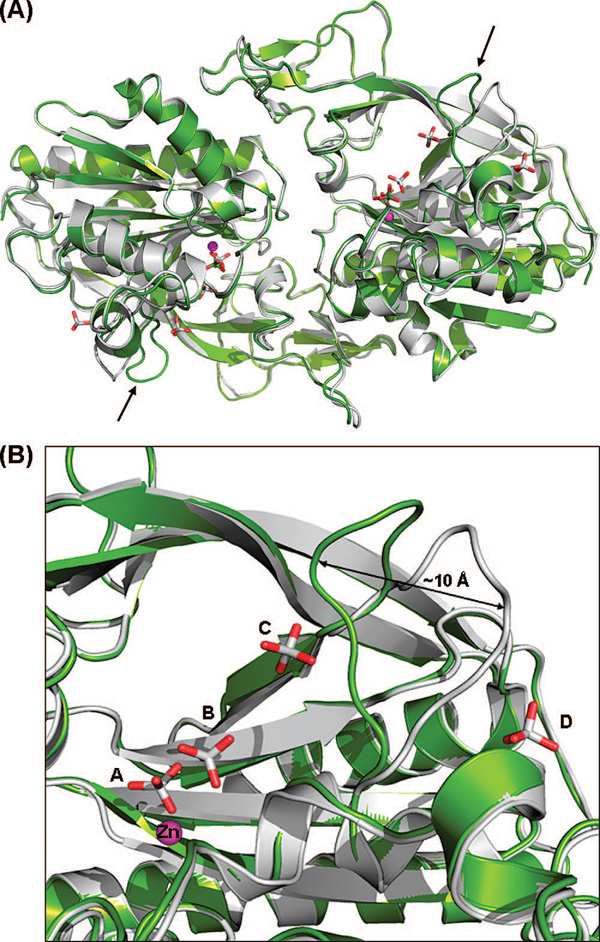
Our apoenzyme structure is similar to the previously determined apoenzyme structure (PDB entry 2I3C) of hACY2 (10), with a rmsd of 0.4 Å between subunits A and 0.36 Å between subunits B. The primary difference between these two structures is the conformation of the loop from amino acid 158 to 164 (Figure 2A). This loop in the 2I3C structure is more solvent-exposed than in our structure, with the position of L162 at the end of the loop undergoing the largest shift of ~10 Å in the Cα atom position between these two structures (Figure 2B). In our structure, this 158–164 loop adopts a more compact and less solvent accessible conformation. The new loop position is stabilized by a water-mediated hydrogen bond between the backbone carbonyl of L162 and the ϵ-amino group of K228. In the 2I3C structure, this position is occupied instead by a phosphate ion.
Figure 2.
(A) Comparison of the human apoenzyme structure of hACY2 (green) with the recently determined structure (PDB entry 2I3C) (10) of the same molecule (gray). The overlay was created by a least-squares superimposition of the Cα atoms of the complete polypeptide chain. The only major difference between the two structures is the position of the loop comprising residues 158–164, shown with black arrows. (B) Close-up view of the 158–164 loop in both structures. The difference in loop conformations between the two molecules is likely caused by the presence of additional phosphate ions (phosphates C and D) bound in the previously determined structure.
A phosphate group is bound in the active site in both hACY2 apoenzyme structures (phosphate A in Figure 2B), with additional electron density indicating the presence of a second phosphate with partial occupancy (phosphate B) adjacent to the active site phosphate. Given their proximity, it is unlikely that both phosphates A and B can be bound simultaneously. These same phosphate binding sites are also present in the 2I3C hACY2 structure along with two additional phosphate groups in this structure located near the flexible loop (Figure 2, phosphates C and D). These additional bound phosphates are a consequence of the higher phosphate concentration (1.4 M) used under these crystallization conditions, compared with our apoenzyme structure which was crystallized from 0.3 M potassium phosphate. The difference in loop conformations observed between the two human apoenzyme structures is likely a consequence of the presence of these additional phosphate ions in the 2I3C structure. One of the extra phosphates in the 2I3C structure (phosphate C) forms a hydrogen bond with the side chain of H159 in this loop, along with an additional interaction with the ϵ-amino group of K228. The other phosphate is on the opposite side of the loop (phosphate D), interacting through a water molecule with the main chain amide of Y164 and also with the side chain of D104. Together, the binding of these additional phosphates prevents this loop, which may serve an important function in controlling access to the active site as described below, from adopting the closed conformation observed in our apoenzyme structure. Except for this difference in loop conformation, these apo-aspartoacylase structures are essentially identical.
Putative Glycosylation Site
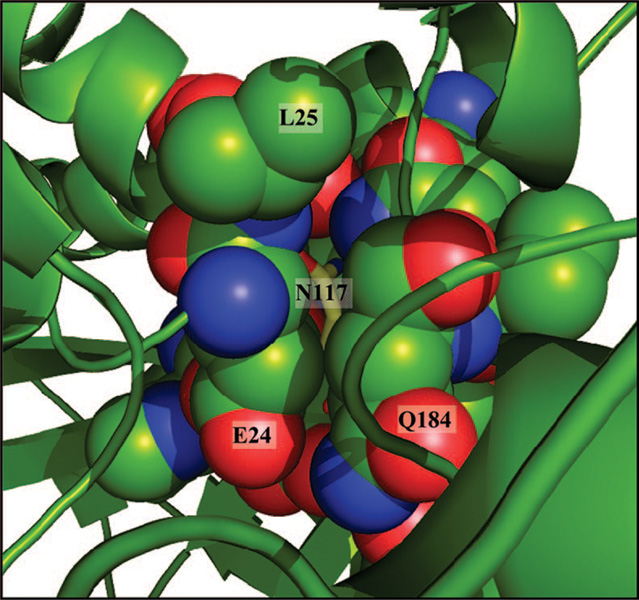
A number of recent experimental results have supported the earlier suggestion that human aspartoacylase is glycosylated at a consensus N×T glycosylation motif at position N117 (7). Our studies of hACY2 showed that this carbohydrate plays a role in maintaining the stability and catalytic activity of the enzyme (12). However, surprisingly, there is no evidence for the presence of a carbohydrate moiety at this position in either of the hACY2 apoenzyme structures. The failure to observe a glycosylated enzyme in the 2I3C apoenzyme structure was not unexpected, since the enzyme was expressed in a prokaryotic organism (Escherichia coli) that does not have the capacity to support such glycosylations. However, there was also no density observed for this glycan in our structure even though the enzyme was produced from the same eukaryotic organism (P. pastoris) that was used previously to produce human aspartoacylase for the carbohydrate characterization studies. Furthermore, the putative glycosylation site, N117, is not situated near the surface of the protein as would be expected but instead is buried within the interior of the protein (Figure 3). This side chain amide forms several hydrogen bonds with side chain and backbone functional groups and also caps the dipole of a helix. In addition, no unassigned density sufficient to fit a carbohydrate moiety was found elsewhere in the electron density maps. To determine if the hACY2 enzyme form that was crystallized is fully active, several crystals of the apoenzyme were washed with buffer and then tested using the previously published assay (12). The catalytic activity of the crystallized enzyme was found to be comparable to that of the freshly purified P. pastoris-expressed hACY2.
Figure 3.
View of the region around the putative glycosylation site. The buried nature of the proposed glycosylated amino acid, N117 (yellow sticks), is shown in the hACY2 structure. The surrounding residues are shown in a space-filling representation with the spheres drawn at 80% of the van der Waals radii to allow a view of N117.
Despite the failure to observe the glycan at this position in the crystal structure, there is ample evidence supporting the presence of a carbohydrate in the fully mature, functional form of human aspartoacylase in solution. The enzyme expressed in P. pastoris is significantly more stable and more active than the E. coli-expressed enzyme (9). Treatment of this active enzyme form with protein N-glycosidase F, an enzyme that cleaves glycans at the point of attachment to the side chain amide group, causes aspartoacylase to revert to a less active and less stable form with properties similar to those of the enzyme which is produced in E. coli (9). In addition, the carbohydrate moiety was removed from P. pastoris-expressed aspartoacylase, chromatographically isolated, and structurally characterized by mass spectrometry (9). Also, a conservative Asn to Gln mutation (N117Q) produced an enzyme form that will not be glycosylated at this position and was found to have significantly lower catalytic activity (9). Glycosylation obviously changes the catalytic properties of aspartoacylase in solution and would necessitate a significant structural rearrangement to expose this putative site for modification. It is not clear why the expected carbohydrate moiety is not observed in these structures. The treatment of the enzyme with neuraminidase that led to improved crystals is expected to remove only the terminal sialic acid moiety but should not affect the remainder of the glycan structure. There are a number of other solvent-exposed asparagine residues on the surface of aspartoacylase, several of which have somewhat disordered side chains. While none of these residues are located within a typical glycosylation sequence, the incorporation of a highly flexible glycan at any of these positions would resolve this apparent discrepancy between the solution properties and the crystal structure of aspartoacylase.
Active Site Structure and Mode of Intermediate Binding
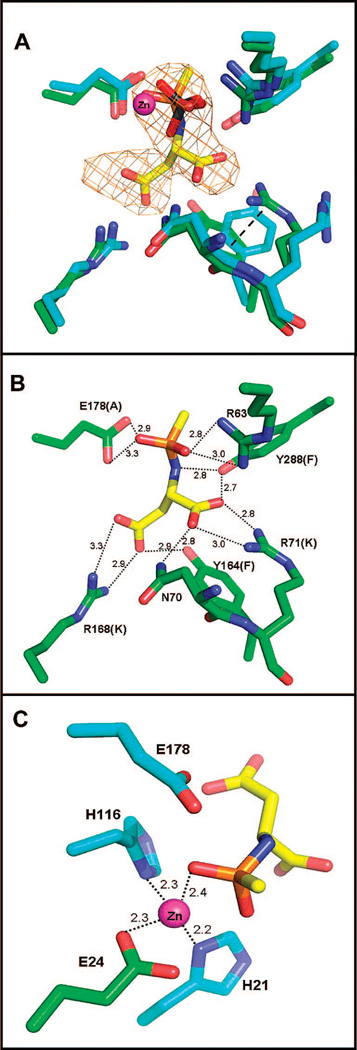
The active site of hACY2 appears to be already formed in the correct conformation for substrate binding, since a comparison of the apoenzyme and the inhibitor complex structures does not indicate any dramatic domain movements associated with intermediate analogue binding. There is, however, a phosphate molecule already bound in active site of the apoenzyme structures, occupying the same position as the phosphonate group of the intermediate analogue in the complex structure. This phosphate molecule interacts directly with the catalytic zinc and is within hydrogen bonding distance of R63, Y288, and E178, the same interactions observed in the complex structure with the phosphonate group of the intermediate analogue. Thus, these apoenzyme structures are not truly in the “apo” form since a portion of the active site is already occupied, and any significant domain movements that might be required to accommodate substrate binding may have already occurred in response to phosphate ion binding.
A number of the residues that can potentially be involved in substrate binding or catalysis have been inferred by sequence alignment studies (8), as well as by analysis of the apoenzyme structure of aspartoacylase (10). Now the identity of these residues and their roles can be assigned with greater certainty, based on our structure of the complex of hACY2 with a catalytic intermediate analogue that was cocrystallized with the enzyme. The Fo – Fc omit map shows the catalytic intermediate analogue bound in both active sites of the functional dimer (shown in Figure 4) with approximately the same occupancies. However, when the active sites of monomers A and B are overlaid using the CR atoms for positioning, the atomic coordinates of the bound intermediates do not exactly superimpose. There is an average difference of ~0.4 Å for the positions of the 13 atoms in the intermediate analogue, with a maximum displacement of 0.7 Å for the position of the α-amino nitrogen. The positional difference within the two active sites can be approximated by a rotation around the center of the phosphonate group resulting in a displacement of the α- and β-carboxylate groups. These differences in positions are relatively small (although larger than the estimate of coordinate errors in Table 1), and they can easily be accommodated by rearrangements in the positions of the active site binding groups as described below. Aspartoacylase shows sigmoidal kinetics at low substrate concentrations (<0.3 mM NAA) that are diagnostic of cooperative substrate binding between the subunits (9). This difference in the binding orientation of the intermediate analogue between the two subunits may be related to the cooperative binding of substrates in the dimer, although a definitive correlation to cooperative substrate binding will require additional structural studies.
Figure 4.
(A) Overlay of the active sites of the apoenzyme (blue gray) and the intermediate analogue complex (green) structures. The catalytic zinc is colored magenta, and the phosphate molecule from the apo structure (light orange representing the phosphorus atom) is located 0.7 Å from the position of the phosphonate group of the catalytic intermediate analogue in the complex structure. Well-defined difference density (omit Fo – Fc map, contoured at 3σ) allows accurate positioning of the intermediate analogue into the active site. There are only minor changes in the positions of the substrate binding groups, except for the conformational rearrangements of the side chains of R71 and Y164 which are now stabilized by a cation-π interaction (dashed line). (B) The heteroatoms of the intermediate are involved in multiple binding interactions with the active site functional groups, with dashed lines showing each interaction and the distances in angstroms listed. The active site mutants produced to examine the role of these functional groups are shown in parentheses. (C) The four-coordinate tetrahedral binding site of zinc with the metal–ligand distances annotatedand the position of the catalytic E178 shown.
The intermediate analogue forms an extensive network of interactions with the enzyme. In addition to direct coordination between the phosphonamidate oxygens and the catalytic zinc, there are several hydrogen bonding interactions to this functional group. One of the oxygens of the phosphonamidate forms an electrostatic interaction with R63, while the other oxygen is within hydrogen bonding distance to the E178 side chain (Figure 4). The nitrogen of the intermediate analogue interacts with the p-hydroxyl group of Y288, while the α-carboxylic group interacts with both N70 and Y288, in addition to forming a bidentate interaction with R71. The β-carboxylic group of the analogue forms a bidentate interaction with R168 and also interacts with the p-hydroxyl group of Y164. Thus, every heteroatom of the intermediate analogue is engaged in one or more favorable contacts, with a total of at least 14 well-defined electrostatic and hydrogen bonding interactions between this intermediate analogue and the enzyme (Table 2). Similar types of interactions are observed in the active site of the second monomer, with only a variation in the orientation of a single side chain leading to an ~150° rotation around the Cδ–Nϵ bond of the guanidino group of R71. This different orientation prevents R71 from forming the same bidentate interaction with the α-carboxylate of the intermediate analogue in this subunit; however, this side chain is still capable of forming a single, well-ordered hydrogen bond with the carboxylate group.
Table 2.
Interactions between Aspartoacylase and the Methylphosphon-amidate Inhibitor
| distance (Å) |
|||
|---|---|---|---|
| inhibitor atom | protein atom | subunit A | subunit B |
| β-carboxyl | |||
| OD2 | NH1 R168 | 3.3 | 3.3 |
| OD1 | NH2 R168 | 2.9 | 2.7 |
| OD1 | OH Y164 | 2.9 | 2.6 |
| α-carboxyl | |||
| OG2 | ND2 N70 | 2.6 | 2.7 |
| OG2 | NH1 R71 | 3.0 | a |
| OG1 | NH2 R71 | 2.8 | 2.7 |
| OG1 | OH Y288 | 2.7 | 2.9 |
| α-amino | |||
| NH | OH Y288 | 2.8 | 2.5 |
| phosphonate | |||
| OAG | NH1 R63 | 2.8 | 3.0 |
| OAG | NH2 R63 | 3.0 | 3.4 |
| OAG | Zn | 2.5 | 2.2 |
| OAD | Zn | 2.4 | 3.0 |
| OAD | OE2 E178 | 2.9 | 3.2 |
| OAD | OE1 E178 | 3.3 | 2.9 |
This interaction is not present in subunit B.
In addition to these polar interactions with each functional group, the methyl group of the analogue binds in a pocket in the active site through hydrophobic interactions with the side chains of F282 and A287 as well as to the methyl group of T118, with distances of 4.4–4.8 Å. The size of this hydrophobic pocket is such that it can easily accommodate the monochloro- and dichloroacetyl analogues that were previously found to be alternative substrates for aspartoacylase (12). However, the depth of the pocket is insufficient to allow the binding of dipeptides or larger substrates, including the trichloromethyl analogue of NAA that was shown not to be a substrate for aspartoacylase (12).
Conservative mutations of any of these substrate binding groups have a significant impact on the catalytic efficiency of aspartoacylase. The Y288F, Y164F, and R168K mutants each have less than 1% of the catalytic activity of the native enzyme (Table 3), while the R71K mutant has ~6% of the native activity. Despite replacement of these functional groups that directly participate in substrate binding, the Km for NAA is essentially unaffected in each of these mutants, except for a relatively modest 6-fold increase in the Y164F mutant (Table 3). This result is not unexpected given the extensive network of interactions that exist between the intermediate analogue and the enzyme. Disruption of any single binding interaction can lead to changes in substrate orientation and rotomer distribution that can have an impact on catalytic efficiency without significantly altering the Michaelis constant for the substrate.
Table 3.
Kinetic Characterization of Aspartoacylase Active Site Mutants
| mutant | kcat (s−1) | % kcat | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| nativea | 12.7 ± 0.05 | 100 | 0.12 ± 0.03 | 1.0 × 105 |
| Y288F | 0.130 ± 0.001 | 0.8 | b | |
| Y164F | 0.020 ± 0.003 | 0.2 | 0.75 ± 0.18 | 2.6 × 102 |
| R168K | 0.13 ± 0.05 | 0.8 | 0.10 ± 0.03 | 1.3 × 103 |
| R71K | 0.73 ± 0.07 | 5.7 | 0.09 ± 0.04 | 7.7 × 103 |
| E178A | 0.46 ± 0.05 | 3.6 | b |
Taken from ref 9.
We have been unable to obtain reproducible Km values with reasonable errors for these mutants.
Structural Changes upon Intermediate Analogue Binding
The apoenzyme and the intermediate analogue complex have nearly identical backbone structures; however, a detailed examination of the active sites does show significant movements in the side chain positions of two amino acids, R71 and Y164. Tyrosine 164 had not been previously suggested as a possible substrate binding group, and there are no clinical mutations identified at this position. There is very low electron density in the apoenzyme structure close to the Cα atom of Y164, suggesting a high degree of rotamer diversity in the side chain in the absence of a bound ligand. In addition, there are no specific interactions with either solvent or protein atoms that contribute to stabilization of a single orientation of this side chain. The R71 side chain is essentially disordered in the apoenzyme structure, and what little density is present in one of the monomers suggests that this side chain is pointing away from the active site.
When the catalytic intermediate analogue binds, the side chains of both of these residues become ordered and are then oriented into the active site. The side chain functional group of R71 shifts by 4–5 Å from its most likely orientation in the apoenzyme into position to form an electrostatic interaction with the α-carboxyl group of the intermediate analogue (Figure 4). A rotation of ~45° around the Cα-Cβ bond of the Y164 side chain is required to move this functional group from the best positioning in the apoenzyme electron density into position to participate in substrate binding. Such a side chain rearrangement results in a 4 Å displacement of the p-hydroxyl group to move into the position that it occupies in the complex structure. In its new orientation in the complex, the side chain of Y164 is well positioned to bond with the β-carboxyl group of the intermediate analogue. Removing the hydroxyl group of Y164 is the only mutation that altered the Km for the substrate. The new positions of R71 and Y164 are further stabilized in the complex through the formation of a stacked cation—π interaction between these two side chains at a distance of 3.7–3.9 Å between the terminal nitrogens and the aromatic ring (Figure 4). This type of electrostatic interaction can be quite strong (17), with either a parallel or perpendicular geometry between binding partners such as arginine or lysine and aromatic side chains at an optimal separation of 3.6–3.8 Å (18). It is now clear, on the basis of this intermediate analogue complex structure, how even the conservative mutations made at either of these positions would affect substrate orientation and catalysis by eliminating hydrogen bond donating groups and by disrupting this stabilizing cation-π interaction (Table 3).
While the conformational differences of the 158–164 loop between the two apoenzyme structures (Figure 2B) appears to be a consequence of the different phosphate levels in the crystallization conditions, by virtue of its location this flexible loop could serve to control access to the active site of aspartoacylase. On the basis of the conformational variation of this loop between the rat and human apoenzymes, the recently published study has speculated that this loop could play such a role (10). We do not yet have a structure of the true apo form of hACY2 in which the active site is completely unoccupied. However, in the absence of bound ligands, this 158–164 loop is likely quite flexible since even in the complex structure the B factors in this region are higher than the average values of the protein backbone. The same closed loop conformation is observed in both subunits of the hACY2 structure with bound phosphate and in the complex with the intermediate analogue, suggesting that occupation of at least a portion of the substrate binding site is required to trigger closure of this loop and is consistent with a possible role in modulating active site access. Thus, the binding of this intermediate analogue (and substrate binding as well) likely triggers an enzyme closure around the substrate with the conformational change in the flexible 158–164 loop coupled with these two mobile residues moving into position to help orient the substrate for catalysis. Tyrosine 288 had been proposed to play a role in substrate binding analogous to that proposed for the highly mobile tyrosine (Y248) in the carboxypeptidases (10). However, it now appears that Y164, through its interaction with the carboxyl rather than the amino group of the intermediate analogue, is actually the corresponding mobile substrate binding residue that plays this role in aspartoacylase.
Proposed Catalytic Mechanism of Aspartoacylase
On the basis of a detailed analysis of the structure of hYCY2 in complex with the tetrahedral intermediate analogue, we can now propose specific assignments for the roles of the active site functional groups. The identity of the zinc ligands, H21, E24, and H116, has been previously suggested (8) and recently confirmed both by structural studies (10) and by mutagenic studies in which several additional mutants of the zinc ligands in aspartoacylase have been reported (19). These mutants, E24A, H21A, and H116A, had no detectable activity, confirming the essential role of zinc in the catalytic mechanism. A clinical mutant has also been identified in which one of these zinc ligands (E24) is replaced with a glycine (20). We constructed this E24G mutant, but no protein expression was observed, presumably a consequence of improper folding and subsequent digestion of this mutated enzyme. The more conservative E24D mutant is found to express in soluble form but has no detectable catalytic activity and slowly precipitates after purification. An ion-coupled plasma emission study of this freshly purified E24D mutant shows the presence of zinc bound to the protein, but at less than stoichiometric levels (0.6 Zn atom/subunit), suggesting that even this conservative mutation compromises the structural integrity of the zinc coordination site.
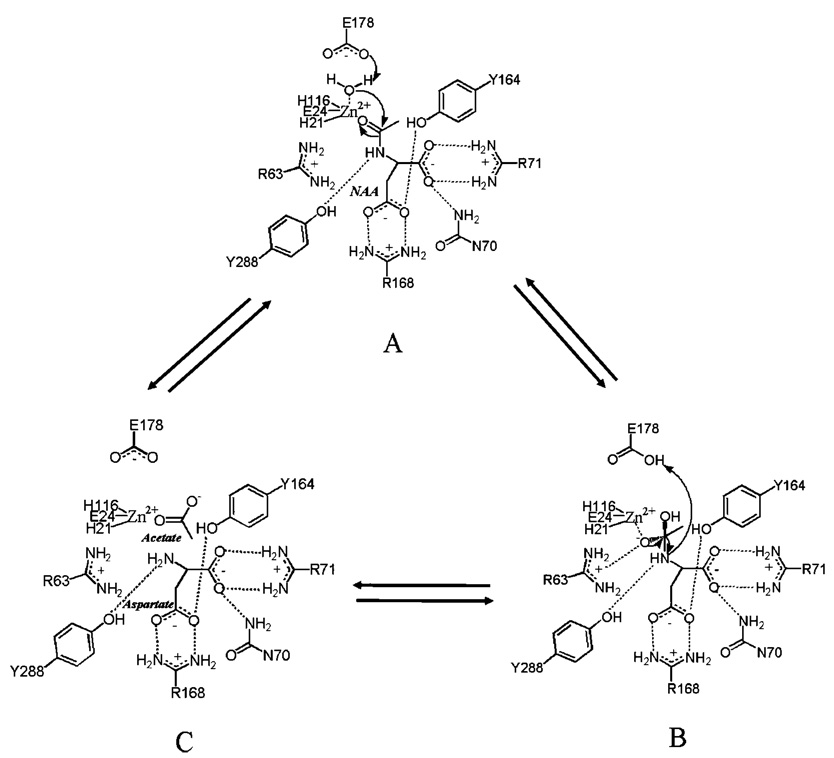
On the basis of the interactions observed in the enzyme–intermediate analogue complex, it is likely that the α-carboxyl group of the substrate is positioned through interactions with the side chain functional groups of N70, R71, and Y288. The β-carboxyl group will be oriented through a bidentate interaction with R168 and also a hydrogen bond to Y164. The α-amino group of the substrate would be positioned to hydrogen bond with Y288, and this will orient the acetyl carbonyl oxygen through a likely interaction with R63 (Figure 5). This would then place the carbonyl carbon of the substrate in position for attack by the zinc-coordinated hydroxyl group formed by proton transfer from the metal-bound water to E178 (Figure 5A). Hydroxide attack generates the tetrahedral intermediate, with the developing negative charge stabilized by the positive charge on the guanidino group of R63 (Figure 5B). It is this proposed intermediate that is represented by our structure of the bound tetrahedral intermediate analogue. Collapse of this intermediate and cleavage of the C–N bond are catalyzed by protonation of the amino group by E178, leading to formation of the acetate and l-aspartate products (Figure 5C). Release of these products and binding of a water molecule in the zinc coordination sphere complete the catalytic cycle. Removal of the functional group at position 178 by mutagenesis leads to an enzyme form (E178A) which has been expressed and purified but has less than 5% of the activity of the native enzyme (Table 3). A more conservative E178D mutant has recently been reported to have ~10% of the catalytic activity of the native enzyme (19), confirming the need to correctly position this carboxyl group to serve an acid–base catalytic function.
Figure 5.
Proposed catalytic mechanism of aspartoacylase. The putative interactions with the substrate binding groups are shown, as well as the nucleophilic attack that must occur (A) to generate the tetrahedral intermediate (B) in the proposed carboxypeptidase-type mechanism of NAA hydrolysis. Collapse of this intermediate leads to the production of l-aspartate and acetate (C).
CONCLUSIONS
This new structure of human aspartoacylase complexed with a catalytic intermediate analogue gives valuable insights into the catalytic mechanism of this important brain enzyme. Roles have been assigned to the residues that are involved in substrate binding and in catalysis. Enzyme conformational changes have been identified that are proposed to be an essential component of the catalytic cycle of aspartoacylase. This improved comprehension of the detailed mechanism of human aspartoacylase can provide the basis for understanding the defects in the clinical mutants that lead to Canavan disease.
ACKNOWLEDGMENT
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38. GM/ CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). We thank Frank Raushel (Texas A&M University) for useful discussions and M. A. Namboodiri for providing the human acy2 gene.
Footnotes
This work was supported by a grant from the National Institutes of Health (NS45664), and C.X. was supported by Grant GM71790.
The atomic coordinates for apo-aspartoacylase (2O53) and the tetrahedral intermediate analogue complex of human aspartoacylase (2O4H) have been deposited at the RCSB Protein Data Bank.
Abbreviations: CD, Canavan disease; hACY2, human aspartoacylase; NAA, N-acetyl-l-aspartate; rmsd, root-mean-square deviation.
REFERENCES
- 1.Canavan MM. Schindler’s Encephalitis Periaxialis Diffusa. Arch. Neurol. Psychiatry. 1931;25:299–308. [Google Scholar]
- 2.Gordon N. Canavan disease: A review of recent developments. Eur. J. Paediatr. Neurol. 2001;5:65–69. doi: 10.1053/ejpn.2001.0467. [DOI] [PubMed] [Google Scholar]
- 3.Matalon R, Michals-Matalon K, Sebesta M, Deanching M, Gashkoff P, Casanova J. Aspartoacylase Deficiency and N-acetylaspartic aciduria in Patients with Canavan Disease. Am. J. Med. Genet. 1988;29:463–471. doi: 10.1002/ajmg.1320290234. [DOI] [PubMed] [Google Scholar]
- 4.Tallan HH, Moore S, Stein WH. N-Acetyl-l-aspartic acid in brain. J. Biol. Chem. 1956;219:257–264. [PubMed] [Google Scholar]
- 5.Madhavarao CN, Moffett JR, Moore RA, Viola RE, Namboodiri MA, Jacobowitz DM. Immunohistochemical Localization of Aspartoacylase in the Rat Central Nervous System. J. Comp. Neurol. 2004;472:318–329. doi: 10.1002/cne.20080. [DOI] [PubMed] [Google Scholar]
- 6.Matalon R, Michals-Matalon K. Recent Advances in Canavan Disease. AdV. Pediatr. 1999;46:493–506. [PubMed] [Google Scholar]
- 7.Kaul R, Gao GP, Balamurugan K, Matalon R. Cloning of the human aspartoacylase cDNA and a common missense mutation in Canavan disease. Nat. Genet. 1993;5:118–123. doi: 10.1038/ng1093-118. [DOI] [PubMed] [Google Scholar]
- 8.Makarova KS, Grishin NV. The Zn-peptidase Superfamily: Functional Convergence After Evolutionary Divergence. J. Mol. Biol. 1999;292:11–17. doi: 10.1006/jmbi.1999.3059. [DOI] [PubMed] [Google Scholar]
- 9.Le Coq J, An HJ, Lebrilla CB, Viola RE. Characterization of Human Aspartoacylase: The Brain Enzyme Responsible for Canavan Disease. Biochemistry. 2006;45:5878–5884. doi: 10.1021/bi052608w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitto E, Bingman CA, Wesenberg GE, McCoy JG, Phillips GN., Jr Structure of Aspartoacylase, the Brain Enzyme Impaired in Canavan Disease. Proc. Natl. Acad. Sci. U.S.A. 2007;104:456–461. doi: 10.1073/pnas.0607817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Hall R, Cummings J, Raushel FM. Tight Binding Inhibitors of N-Acyl Amino Sugar and N-Acyl Amino Acid Deacetylases. J. Am. Chem. Soc. 2006;128:4244–4245. doi: 10.1021/ja0600680. [DOI] [PubMed] [Google Scholar]
- 12.Moore RA, Le Coq J, Faehnle CR, Viola RE. Purification and Preliminary Characterization of Brain Aspartoacylase. Arch. Biochem. Biophys. 2003;413:1–8. doi: 10.1016/s0003-9861(03)00055-9. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Macromolecular Crystallography, Part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 14.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 17.Mecozzi S, West AP, Dougherty DA. Cation-πinteractions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma JC, Dougherty DA. The Cation-π Interaction. Chem. Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 19.Herga S, Berrin JG, Perrier J, Puigserver A, Giardina T. Identification of the zinc binding ligands and the catalytic residue in human aspartoacylase, an enzyme involved in Canavan disease. FEBS Lett. 2006;580:5899–5904. doi: 10.1016/j.febslet.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 20.Zeng BJ, Wang ZH, Ribeiro LA, Leone P, De Gasperi R, Kim SJ, Raghavan S, Ong E, Pastores GM, Kolodny EH. Identification and characterization of novel mutations of the aspartoacylase gene in non-Jewish patients with Canavan disease. J. Inherited Metab. Dis. 2002;25:557–570. doi: 10.1023/a:1022091223498. [DOI] [PubMed] [Google Scholar]