SUMMARY
Prestin (SLC26A5) is the molecular motor responsible for cochlear amplification by mammalian cochlea outer hair cells and has the unique combined properties of energy-independent motility, voltage sensitivity, and speed of cellular shape change. The ion transporter capability, typical of SLC26A members, was exchanged for electromotility function and is a newly derived feature of the therian cochlea. A putative minimal essential motif for the electromotility motor (meEM) was identified through the amalgamation of comparative genomic, evolution, and structural diversification approaches. Comparisons were done among nonmammalian vertebrates, eutherian mammalian species, and the opossum and platypus. The opossum and platypus SLC26A5 proteins were comparable to the eutherian consensus sequence. Suggested from the point-accepted mutation analysis, the meEM motif spans all the transmembrane segments and represented residues 66–503. Within the eutherian clade, the meEM was highly conserved with a substitution frequency of only 39/7497 (0.5%) residues, compared with 5.7% in SLC26A4 and 12.8% in SLC26A6 genes. Clade-specific substitutions were not observed and there was no sequence correlation with low or high hearing frequency specialists. We were able to identify that within the highly conserved meEM motif two regions, which are unique to all therian species, appear to be the most derived features in the SLC26A5 peptide.
INTRODUCTION
The molecular evolution of a protein family involves a combination of gene expansion events that includes gene duplication and duplication of the whole genome (Haldane 1932; Ohno 1970, 1993; Taylor and Raes 2004; Domazet-Loso et al. 2007). The eutherian genome has presumably undergone two rounds of whole genome duplication (2R Hypothesis) occurring in early chordate evolution (Urochordata–Craniata) (Garcia-Fernandez and Holland 1994; Dehal and Boore 2005; Hallbook et al. 2006). Expansion and contraction of the number of gene family members can occur by a number of mechanisms, including but not limited to the acquisition of transposable elements, independent gene duplication through cis- or trans-events, and alternation of chromosomal number with subsequent deletion or translocation of genes, chromosomal segments or whole chromosomes (Furlong and Holland 2002). After duplication these isogenic copies, under selective pressure, can be routed along a number of possible evolutionary pathways that diversify the isogenes through point mutations, insertions or deletions (Indels), and/or conversion events within the coding, noncoding, and/or cis-regulatory regions (King et al. 2007; Prud’homme et al. 2007; Roth et al. 2007; Zheng et al. 2007). These pathways are defined as nonfunctionalization, neofunctionalization, and subfunctionalization of the evolving isogenes (Ohno 1970; Force et al. 1999; Lynch and Conery 2000). There is evidence that formation of paralogs is often due to an initial acceleration of the nonsynonymous substitution rate leading to functional divergence that is then followed by purifying selection, which excludes substitutions that prevent or interfere with refinement of the protein structure–function relationship (Li et al. 1985; Ohta 1993; Lynch and Conery 2000). Metazoan genomic comparisons have recently identified punctuated protein evolution in metazoan lineages that identify evolutionary innovations in protein structure and function (Domazet-Loso, Brajkovic, and Tautz 2007; Putnam et al. 2007). Comparative analysis of proteins and their paralogs provide insights into the diversity of protein structures and, combined with functional studies on engineered proteins, reveal structure–function relationships that contribute to phenotypic and morphological diversification (Marsh and Griffiths 2005). Recently, Ortlund et al. (2007) have combined phylogenic and functional approaches with X-ray crystallography to elucidate the generation of new functional paradigms of proteins.
Here we examined the evolution of an anion transporter molecule to a unique ATP-independent electromotility motor protein, prestin or SLC26A5, that provides sound amplification of the cochlea with rapid conformational changes that occur in kilohertz ranges (Dallos and Fakler 2002; Dallos et al. 2006). Prestin (SLC26A5) is a protein critical for the function of the peripheral auditory system in mammals. Its presence results in outer hair cell (OHC) electromotility, allowing OHCs to elongate and contract in synchronization with sound waves to amplify sounds (Zheng et al. 2000). Electromotility in OHC is characterized by a somatic shape change in length of up to 5%, which occurs with alterations of the membrane potential (Brownell et al. 1985; Ashmore 1987; Santos-Sacchi and Dilger 1988). This motility is voltage dependent and a nonlinear capacitance (NLC) is thought to initiate a bimodal conformational change in the protein itself (Santos-Sacchi 1991; Oliver et al. 2001; Santos-Sacchi et al. 2001). Removal of prestin by targeted mutagenesis in mice (Liberman et al. 2002) results in a 40–60 dB loss of cochlear sensitivity in vivo. In humans a spontaneous 5′-noncoding region (NCR) splice acceptor mutation (IVS2-2A>G) in prestin exon 3 is associated with congenital nonsyndromic deafness in DFNB61 patients (Liu et al. 2003). Thus, prestin is necessary for electromotility and mediate the active amplification, boosting cochlear sensitivity by 100–1000-fold (40–60dB).
The SLC26A family members of anion exchangers are expressed in the luminal or apical membranes of epithelial tissue and are primarily involved in transport of Cl− and (Mount and Romero 2004). However, each member has a distinctive expression distribution in epithelial cells and anion specificity (Mount and Romero 2004; Kere 2006). Cochlear expression analysis of the other SLC26A3–6 transporters has demonstrated that SLC26A4–6 genes are expressed, whereas SLC26A3 is not present. SLC26A4 is present in the spindle cells of stria vascularis, spiral prominence, outer sulcus cells (along with their root processes), and the Claudius and Deiter’s cells of the organ of Corti (Everett et al. 1999; Yoshino et al. 2004). SLC26A6 transcripts are found in the organ of Corti and likely are expressed in the supporting cells. In comparison, SLC26A5 has a limited expression profile that is primarily restricted to OHCs (Zheng et al. 2000), but transcripts can be detected in brain, testis, and vestibular tissue (Adler et al. 2003; Zheng et al. 2003). The biological importance of these four genes is further exemplified by human SLC26A inherited diseases (Kere 2006). Four genetic diseases associated with the SLC26A family are congenital chloridorrhea (SLC26A3), Pendred syndrome (SLC26A4), nonsyndromic deafness (SLC26A5), and calcium oxalate nephrolithiasis (SLC26A6), and are based on their respective unique functional and expression profiles. Genetically engineered null mutant mice exhibit similar diseases (Everett et al. 2001; Liu et al. 2003; Wang et al. 2005b; Jiang et al. 2006; Schweinfest et al. 2006). The physiological importance of these transporters and the integrity of their associated functional protein motifs are demonstrated by the high frequency of pathogenic mutant alleles associated with these functional domains (Kere 2006).
Two major protein motifs represented in prestin were identified as the sulfate transporter and the sulfate transporter and antisigma-factor antagonists (STAS) domains (Zheng et al. 2000). The presence of a sulfate transporter domain placed prestin within the family of sulfate permeases and related transporters (SUL1) and the major facilitator super-family. The SUL1 domain is highly conserved in the SLC26A gene family (Zheng et al. 2000; Weber et al. 2003; Franchini and Elgoyhen 2006; Rajagopalan et al. 2006). Unlike the other members of the SLC26A family, mammalian SLC26A5 has the unique combined properties of the voltage-dependent conformational changes, NLC and the absence of apparent anion transporter capabilities (Zheng et al. 2000; Oliver et al. 2001; Dallos and Fakler 2002; Mount and Romero 2004). NLC represents the “voltage sensor” of prestin, whereas the conformational changes are mediated by the “actuator” (Dallos and Fakler 2002). The voltage sensor was found to interact with chloride and bicarbonate ions at the cytoplasmic side of the membrane (Oliver et al. 2001).
The structural–functional relationship of the SLC26A5 protein with electromotility and NLC has been explored using numerous strategies, including mutational and bioinformatics approaches. Mutational studies have targeted nonconserved charged amino acids in the sulfate transporter motif (Oliver et al. 2001), glycosylation sites (Matsuda et al. 2004), phosphorylation sites (Deak et al. 2005), cysteine residues (McGuire et al. 2007), chimeric (protein segments derived from other Slc26a family members) or domain swapping (Zheng et al. 2005), and terminal deletions of the amino- and carboxy-cytoplasmic tails (Navaratnam et al. 2005; Zheng et al. 2005). Protein sequence comparisons of the Slc26a5 orthologs and paralogs have also permitted the identification of putative amino acid residues that may be involved in electromotile and NLC characteristics (Rajagopalan et al. 2006; Albert et al. 2007). Generally, the results of alteration of the SLC26A5 sequence falls into three categories. These are (1) little to no change in NLC function or electrophysiological properties, (2) the maintenance of NLC with shift in the membrane potential (V1/2) where the two conformation states are equal in proportion, and (3) the loss of NLC (Dallos, Zheng, and Cheatham 2006). Loss of NLC was suggested to be due to failure of SLC26A5 to insert into the plasma membrane and is observed in mutant proteins without the carboxy-cytoplasmic tails (Zheng et al. 2005). Loss of membrane expression is also observed with mutated STAS domains of other SLC26A family members (Taylor et al. 2002; Karniski 2004). Truncation of either the amino- or carboxy-cytoplasmic tails were found to eliminate NLC, in spite of the surface expression of the mutated SLC26A5 (Navaratnam et al. 2005). Attempts to document electromotility in nonmammalian hair cells have been unsuccessful (Adler et al. 2003; He et al. 2003; Weber et al. 2003; Boekhoff-Falk 2005; Albert et al. 2007). Recently the zebrafish (Danio rerio, Dreri) slc26a5 was found to be expressed in hair cells, but displayed a weaker voltage dependence, and slower kinetics (Albert et al. 2007). In addition, no electromotility was observed and the subcellular immunodetection pattern was similar to that described for rodent vestibular hair cells (Adler et al. 2003). Therefore, it appears the evolution of SLC26A5 voltage-dependent capacitance and plasma membrane-based electro-motility parallels that of the mammalian cochlea. This evolution should be reflected in the associated prestin protein sequences.
Comparative genome analysis represents a powerful technique for functional inference of genes (Domazet-Loso, Brajkovic, and Tautz 2007; Putnam et al. 2007). Its foundation is the ability to identify orthologous genes. Several methods have been developed for mapping orthologs through sequence comparison utilizing a combined approach of BLAST searching followed by more accurate sequence-alignment schemes (e.g., CLUSTALW and PAML) (Sonnhammer and Hollich 2005; Cantarel et al. 2006). One of the key issues with all these methods is their underlying assumption that sequence similarity alone contains sufficient information for prediction of orthologs among species. Other additional criteria are needed to distinguish between SLC26A5 orthologs and other SLC26A family members (paralogs), especially in evolutionarily distant nonmammalian species. Herein, we have established more stringent criteria for identification of the SLC26A5 orthologs. These were: (1) maintenance of chromosomal linkage; (2) similar gene organization of exon and exon–intron boundaries; (3) nucleotide and amino acid homology; and (4) maintenance of the SUL and STAS structural motifs. Using this approach we have established the evolution of the SLC26A family of sulfate transporters and the conservation of prestin peptide sequences within the mammalian taxa. Using these criteria our genomic and bioinformatic research objectives were also able to identify and sequence the prestin ortholog of the platypus (Ornithorhynchus anatinus [Oanat]) and the opossum (Monodelphis domestica [Mdome]), belonging to monotremes and marsupials, respectively. Because the modern marsupials and monotremes are the survivors of early branching of the mammalian evolutionary tree (~110 Myr, respectively), we chose the egg-laying prototherian platypus and the meta-therian oppossum as representative of the early divergence from the eutherian mammals. Our initial steps were to identify the platypus and opossum SlC26A5 orthologs and their associated genomic sequence and structure. With the recent expansion of genomic sequencing to include a large number of mammalian and nonmammalian vertebrate (NMV) species, we hypothesized that a significant degree of homology would be preserved among the SLC26A5 orthologs and its electromotility properties would also remain highly conserved among mammalian species, including the monotremes and marsupials.
MATERIALS AND METHODS
Identification and characterization of SLC26A5 containing Oanat BAC clones
BAC Library OA__Bb (Platypus) filters were obtained from the Clemson University Genomics Institute (CUGI) BAC/EST Resource Center (Clemson, SC, USA). The OA_Bb clones in the library had an average insert size of 143 kb and represented 11 genome equivalents. Southern blot analysis was done with the Oanat genomic BAC filters using standard conditions and P-32-labeled probes were derived from cloned, mouse Slc26a5 coding region cDNA that were hybridized overnight at 50°C. Following hybridization, blots were washed under moderately stringent conditions (0.5 × SSC/0.1% SDS at 50°C for 30min) and exposed to Kodak XAR5 film at room temperature for 4–48 h with intensifying screens. Positive clones were obtained from CUGI and BAC DNA was obtained using large-scale BAC purification kit following the manufacturer’s protocol. All BAC genomic fragments were characterized by restriction enzyme digestion, sequence analysis, and Southern blotting analyses. BamHI, EcoRI, and HindIII restriction enzyme digestions were done overnight at 37°C and analyzed by agarose gel electrophoresis.
Gene-specific oligodeoxynucleotide primers (designed with the assistance of the Oligo 4.0 program, Natl. Biosciences, Plymouth, MN, USA) were prepared so that the Oanat genomic sequence could be confirmed. PCR reactions were performed in an MJR thermocycler using 3.0 U Taq Polymerase (PE Biosystems, Foster City, CA, USA), BAC DNA preparation for 35 cycles (94°C for 30 sec, 50–55°C for 30 sec, and 72°C for 3.5 min). The amplified products were purified by gel electrophoresis, and subcloned pCRII-TOPO (Invitrogen Corporation, Carlsbad, CA, USA).
SLC26A paralog identification and sequence annotation
Identification of the genomic sequences of the SLC26A5 orthologs was done using BLAST analysis on Ensembl and NCBI genomic databases. Ortholog and paralog comparisons (Thompson et al., 1994) were done using CLUSTALW and the Clc protein workbench (version 3 by CLC Bio, Cambridge, MA, USA). The parameters used for these alignments were: Gap open cost 1/4 5.0, Gap extension cost 1/4 1.0, and End gap cost 1/4 cheap (all end gaps are treated as gap extensions and any gaps past 10 were free). Additional manual annotation and alignment were performed by the investigators. Bidirectional Blast similarity searching using the tBlastn algorithms was applied to find additional orthologs in the genomic sequences obtained from NCBI (http://www.ncbi.nlm.nih.gov/Genomes/), Ensembl (http://www.ensembl.org/index.html) and UCSC Genome Bioinformatics (http://genome.cse.ucsc.edu/cgi-bin/hgBlat) websites. Ensembl Release 45 was used as the primary database, except platypus and Ciona intestinalis (Cinte) genomic sequences were derived from Release 46 (August, 2007). NCBI and Ensembl annotated SLC26A paralog protein sequences were confirmed by manual inspection of the corresponding BLAST or Blat searches. Unannotated or incorrectly annotated genes were manually determined. Conservation of synteny was determined by comparing adjacent orthologous gene pairs allowing for inversions in gene order between species. If there was a match, the two orthologous gene pairs were considered to be part of a syntenous region. Such regions were extended by directional scanning along the genome. Gene order and inversions were also noted.
Phylogenetic analysis
Orthologous genes were grouped using all-against-all, pairwise BLAST similarity searches at the level of predicted proteins keeping reciprocally best-matching genes. Phylogenetic analyses were carried out in MEGA 4.1 (Tamura et al. 2007) using maximum likelihood, neighbor-joining, minimum evolution, maximum parsimony, and unweighted pair group method with arithmetic mean algorithms with pairwise deletion. Comparisons were made with models using Dayoff PAM and the James, Taylor, Thorton (JTT) matrices. Reliable bootstrap values were obtained for all nodes of the tree except for the terminal nodes linking. Stability of clades was evaluated by 500–1000 bootstrap rearrangements.
RESULTS
Evolution of the SLC26A gene family
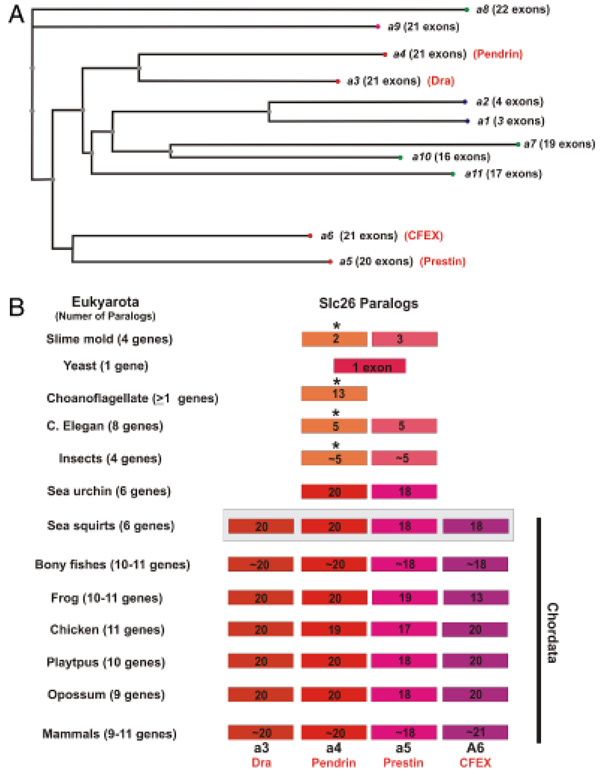
We have used a comparative genomic approach to determine the evolution of SLC26A gene family nucleotide and amino acid homologies and the maintenance of SUL and STAS motifs (Zheng et al. 2000; Weber et al. 2003; Franchini and Elgoyhen 2006; Rajagopalan et al. 2006). Examination of the chromosomal synteny and genomic organization, including exon–intron boundaries, was included to solidify the identification of each ortholog. Based on genomic analysis, the mammalian SLC26A family consists of 9–11 genes (Mount and Romero 2004; Franchini and Elgoyhen 2006). As depicted in Fig. 1A, the mouse (Mus musculus, Mmusc) genome contains 11 genes that can be generally divided into two major groups, which are SLC26A4/11 (nine members) and SLC26A5/6 (two members). The SLC26A4/11 group can be further subdivided into two subfamilies based on sequence homology and genomic organization. The SLC26A4 subfamily is represented by the prototypic Slc26a4 gene and includes a3, a4, a8, and a9, whereas, the SLC26A11 subfamily contains the prototypic Slc26a11 along with a1, a2, and a10 genes.
Fig. 1.
Evolution of the SLC26A paralogs as revealed by mouse protein comparisons. (A) The relationship of the paralogous SLC26A proteins from the nonredundant protein sequence database from Mmusc is depicted. A distance tree was determined using the fast minimum evolution algorithm of BLAST pairwise alignments with a maximum sequence difference of 0.75. Members of the SLC26A4/5 (red dot) and SLC26A1/11 (blue and green dots) subfamilies are indicated. Aliases for SLC26A3–6 members are provided. (B) The evolutionary expansion of the SLC26A4/5 paralogs is illustrated with the number of genes represented in each taxon being provided. The difference shades of red and purple indicate a common ancestral precursor. The number of coding exons associated with each gene is provided. The paralogs designated with the asterisk (*) are associated with the permease-like SLCA1/11 subfamily. The taxa within the chordate are demarcated. The paralogs of the sea squirt (Cinte), as indicated by the gray box, have a high-sequence homology among themselves making precise identification of the orthologous genes difficult. There are no linkage associations to assist in the ortholog assignments.
Similar to the bioinformatic analyses of Franchini and Elgoyhen (2006), we were able to determine that the initial paralogs of the SLC26A family could be separated based on their similarity with the sulfate permeases or transporters, which were represented by the prototypic SLC26A11 and SLC26A4 proteins, respectively. Fig. 1B summarizes the results of the evolution of SLC26 in eukyarotes. Analysis of the yeast (Saccharomyces cerevisiae, Scere) genome identified only one SLC26A-like gene, which had a homology with the SLC26A4 prototype. The Choanoflagellates were selected for analysis, because they are among the closest unicellular relatives of animals (King and Carroll 2001). From the available, but limited choanoflagellate (Monosiga brevicollis, Mbrev) genomic sequence (http://genome.jgi-psf.org/Monbr1/Monbr1.home.html), a single SLC26A gene with 13 putative exons was identified in the Monbr1/scaffold_11:371728-374996 and encoded a 556 amino acid protein (Monbr1:25685). The deduced amino acid sequence had the greatest similarity with the Mmusc SLC26A11 (expected = 2e – 91). Paralogs representing the two major grouping of SLC26A genes, SLC26A11-like and SLC26A4-like, were present in slime molds (Dictyostelium discoideum, Ddisc), worm (Caenorhabditis elegans, Celeg), and a variety of insects (fruit fly, Drosophila melanogaster [Dmela]; honey bee, Apis mellifera [Amell]; mosquito, Anopheles gambiae [Agamb] and Aedes aegypti [Aaegy]; and red flour beetle, Tribolium castaneum [Tcast]). A common feature in the genomic organization of these SLC26A family members was the relatively small number of exons (~ 2–5 exons). BLAST analyses using mouse SLC26A4 and SLC26A5 polypeptide sequences ascertained that the deduced SLC26A4-like proteins in slime mold, worms, and insects have a slightly lower homology with SLC26A5, but were not significantly different in homology with SLC26A4. Annotation of these ancestral genes as orthologs or paralogs of Slc26a4, Slc26a5, or even Slc26a6 genes is at best difficult. This is reflected in the inconsistencies in SLC26A orthologs designations in both the Ensembl and NCBI databases. Variations in both the number of paralogs and the degree of identity and similarity were observed among these species. The degree of completion of the associated genomes also contributed to some of the observed variations. Similar to other gene families, the number and sequence differences in paralogs appear to expand and contract through the evolution of each of these eukaryotic species.
Distinct paralogs of the SLC26A3/4 and SLC26A5/6 ancestral genes became first apparent in deuterostomes, as suggested by genomic analyses of sea urchin (Strongylocentrotus purpuratus [Spurp]) (see Fig. 1B). However, in the invertebrate chordates, represented by two species of sea squirt (Ciona intestinalis [Cinte] and Ciona savignyi [Csavi]), there is an expansion of the SLC26A3/4 and SLC26A5/6 paralogs as well as an increase in exon numbers. Two genes located on chromosome 7q are linked in a head to head orientation, similar to the linkage found with vertebrate SLC26A3 and SLC26A4 loci. The other two paralogs are more similar to the ancestral SLC26A5/6 gene. In the vertebrates the number of exons in each paralog increased to that presently observed in all extant vertebrate species with the exon–intron boundaries, for the most part, remaining consistent among the SLC26A3/4 and SLC26A5/6 orthologs. Distinct SLC26A3 and SLC26A6 genes first appeared in bony fish with their associated linkage relationships. An additional feature in the bony fish was an expansion of the number of SLC26A6-like paralogs. Because SLC26A3 has remained in close linkage with the SLC26A4 locus in all vertebrate species, SLC26A3 gene likely arose from a tandem duplication of the ancestral SLC26A3/4 gene. However, SLC26A6 has neither a linkage with SCL26A5 nor a parallel chromosomal segment synteny. Therefore, genomic duplication event, such as those reflected in other genes, may have led to the eventual divergence of these two genes from the SLC26A5/6 ancestral gene.
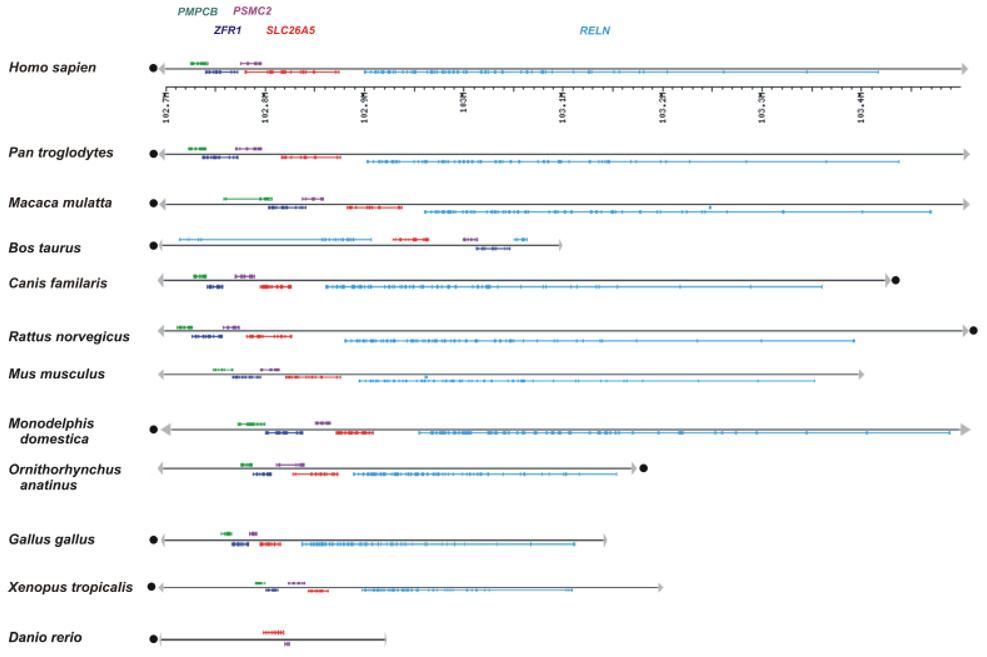
In order to compare only the SLC26A5 orthologs we used chromosomal synteny as our initial step in the classification and exclusion of other SLC26A paralogs. As shown in Fig. 2, we were able to identify that four genes were consistently linked with SLC26A5 and retained synteny in all vertebrates, except for bony fish. Reelin (RELN) was always 5′ to SLC26A5 and both loci were always in the same 5′–3′ orientation. Three genes were present downstream (3′) and retained the same relative order and orientation. The most adjacent was proteasome (prosome, macropain) 26S subunit, ATPase, 2 (PSMC2), next was zuotin related factor 1 (ZFR1), which is also designated as DnaJ (Hsp40) homolog, subfamily C, member 2 (DNAJC2), and peptidase (mitochondrial processing) beta (PMPCB) being the most 3′. Both PSMC2 and PMPCB were in an opposite orientation to SCL26A5, whereas ZFR1 had the same tail to head orientation and in general was physically overlapping with the 5′ PSMC2 and 3′ PMPCB loci. Except for RELN, synteny was maintained in the corresponding chromosomal segments of the Japanese pufferfish Fugu (Takifugu rubripes, Trubr), green spotted pufferfish (Tetraodon nigroviridis, Tnigr), Japanese medaka (Oryzias latipes, Olati) and Stickleback (Gasterosteus aculeatus, Gacul). However, linkage of SLC26A5 with only the PSMC2 locus was observed in zebrafish (Dreri). Chromosomal synteny made misidentification of orthologs of the aminotic SLC26A5 impossible. There were no linkage relationships observed in either the sea urchin or sea squirt genomes.
Fig. 2.
Chromosomal synteny among different vertebrate SLC26A5 loci. The linkage relationship of SLC26A5 (red) with the Reelin (RELN) (blue), proteasome (prosome, macropain) 26S subunit, ATPase, 2 (PSMC2) (green), zuotin related factor 1 (ZFR1) (dark blue) and peptidase (mitochondrial processing) beta (PMPCB) (purple) loci is depicted for 12 vertebrate species with an emphasis on mammals. The chromosomal orientations (dot) and the associated physical length of the corresponding chromosomal segments are indicated.
In general, the vertebrate SLC26A3 and SLC26A4 loci were linked with orthologs representing the BCAP29, CBLL1 and DLD loci, whereas the SLC26A6 locus was linked with CELSR3 and TMEM89 (data not shown). If linkage data was unavailable, then genomic organization of the coding exons for the SLC26A3–6 genes within a BAC clone or contig was used for identification of the respective orthologs. As such, the orthologs for each of the four vertebrate paralogs could be readily identified by the organization and encoded sequences present at the exon 5′ and 3′ boundaries.
Evolution of the vertebrate SLC26A5 peptide
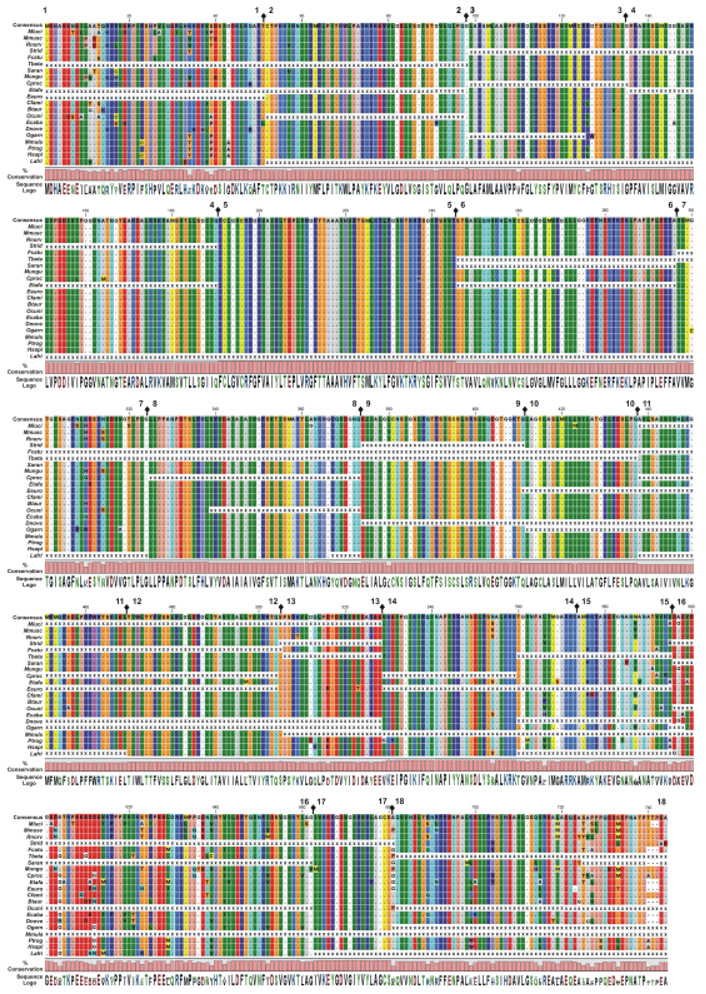
After we had documented the orthologs of the SLC26A5 genes, the amino acid homologies among the vertebrata were then examined. Three regions of heterogeneity were identified, which were in the amino- and carboxy-termini and a highly negatively charged region within or adjacent to the STAS domain. Eight NMV species were evaluated; chicken (Gallus gallus, Ggall), frog (Xenopus tropicalis, Xtrop), zebrafish (Danio rerio, Dreri), fugu (Takifugu rubripes, Trubr), mefugu (Takifugu obscurus, Tobsc), medaka (Oryzias latipes, Olati), stickleback (Gasterosteus aculeatus, Gacul), and tetradon (Tetraodon nigroviridis, Tnigr). In the NMV SLC26A5 peptides, these three regions have a higher degree of sequence variability compared with the SUL and STAS domains (Fig. 3). Because of the increased availability of sequenced mammalian genomes (Hubbard et al. 2007), we were able to compare full and partial protein sequences of up to 22 eutherian species; the armadillo (Dasypus novemcinctus, Dnove), bushbaby (Otolemur garnettii, Ogami), cat (Felis catus, Fcatu), cow (Bos taurus, Btaru), chimpanzee (Pan troglodytes, Ptrog), dog (Canis familiaris, Cfami), elephant (Loxodonta africana, Lafri), gerbil (Meriones unguiculatus, Mungu), guinea-pig (Cavia porcellus, Cproc), European and Madagascar hedgehogs (Echinops telfairi, Etefa and Erinaceus europaeus, Eeuro), horse (Equus caballus, Ecaba), human (Homo sapiens, Hsapi), microbat (Myotis lucifugus, Mluci), mouse (Mus musculus, Mmusc), pig (Sus scrofa, Sscro), rabbit (Oryctolagus cuniculus, Ocuni), rat (Rattus norvegicus, Rnorv), rhesus monkey (Macaca mulatta, Mmula), European shrew (Sorex araneus, Saran), ground squirrel (Spermophilus tridecemlineatus, Strid), and treeshrew (Tupaia belangeri, Tbela). The protein alignment for SLC26A5 is provided in Fig. 4. The eutherian mammal isofunctional SLC26A5 family members were highly conserved (see Fig. 4). Likewise, little if any divergence was observed in the transmembrane encompassing region (SulPtp) of the eutherian species. This high degree of conservation in SulPtp was in stark contrast with the variability in the deduced amino acid sequences from the chicken (Ggall), frog (Xtrop), and the six bony fish SLC26A5 (see Fig. 3). Comparison of the eutherian and NMV SLC26A5 amino acid sequences demonstrated Indels within prestin were primarily restricted to the amino- and carboxy-termini, two regions within SulPtp, and the negatively charged segment. As depicted in Fig. 5, the transition of the NMV to the eutherian sequences within SulPtp differed by deletions within coding exon 4 that reduced the length of a hydrophilic loop and by insertion of a putative transmembrane hydrophobic α-helix within exon 6. Among the SLC26A5 orthologs, the eutherian coding exons 6 and 7 were represented by a single exon in the NMVs (Fig. 5). Beside single residue variations within each paralogs, Indels were observed within the coding exons, whereas the exon boundaries remained conserved. Among the vertebrate SLC26A5 genes, Indels were predominately confined to five coding exons: 1, 4, 6, 16, and 18 (Fig. 3–Fig. 5). Coding exons 1 and 18 represented the amino-terminus and carboxy-terminus, respectively, coding exons 4 and 6 were within the SulPtp region, and exon 16 represented the charged cluster and STAS domains. Indels within coding exon 16 were primarily associated with the charged cluster motif. The prevalence of Indels associated with exons 4 and 6 was also found when the eutherian SLC26A5 consensus protein was compared with the eutherian consensus proteins of SLC26A4 and SLC26A6.
Fig. 3.
SLC26A5 amino acid variations among nonmammalian vertebrates (NMV). Alignment of the NMV prestin polypeptides are shown with the deduced polypeptide sequences being compared with the frog (Xtrop) SLC26A5 sequence. The amino acid alignments are presented with identical residues to the initial polypeptide being represented as dots. Background color of each residue is presented using RasMOL colors designations (Sayle and Milner-White, 1995). Gaps in the aligned sequences are indicated by the dashed line. The consensus sequence, residue conservation, and utilization (Sequence logo) are provided and missing of sequence data is indicated by X. Residue number and the location of each exon boundary ( ) are provided at the top of each row of sequence.
) are provided at the top of each row of sequence.
Fig. 4.
Alignment of the eutherian SLC26A5 proteins. The deduced polypeptide sequences from eutherian species compared with a SLC26A5 eutherian consensus sequence. The eutherian species are arranged from high- to low-frequency specialists. The amino acid alignments are presented with identical residues represented as dots. Background color of each residue is presented using RasMOL colors designations (Sayle and Milner-White, 1995). Gaps in the aligned sequences are indicated by the dashed line. The residue conservation and utilization (Sequence logo) are provided and absence of sequence data is indicated by X. Residue number and the location of each coding exon (➋) are provided at the top of each row of sequence.
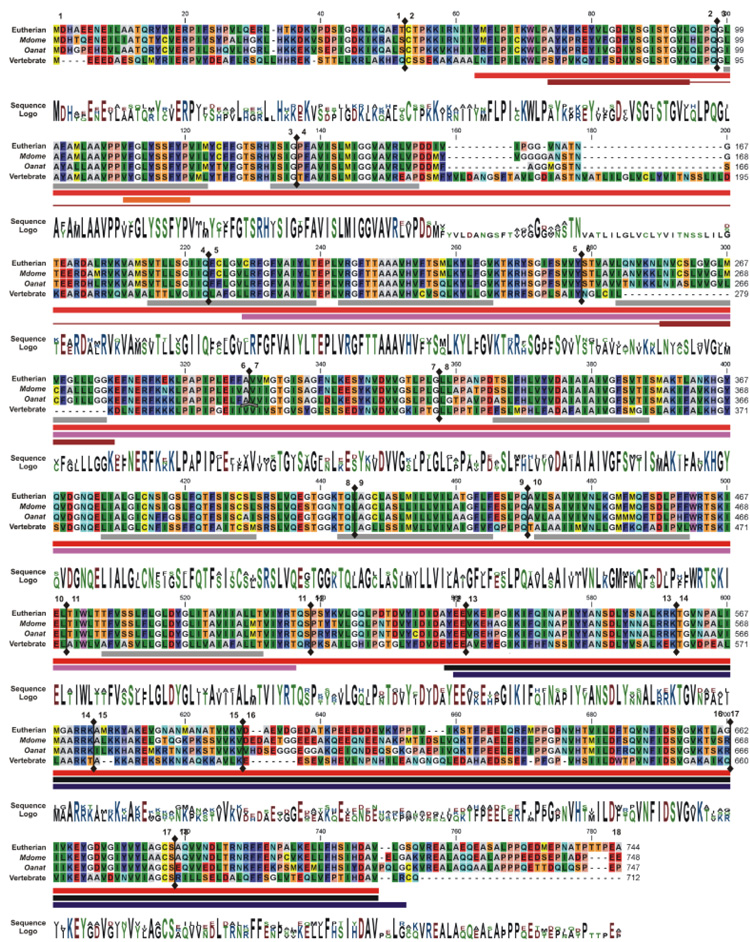
Fig. 5.
Alignment of the eutheria and nonmammalian vertebrate SLC26A5 consensus sequences with opossum and platypus polypeptides. The amino acid alignments are presented with identical residues to the initial polypeptide being shown as dots. Background color of each residue is presented using RasMOL colors designations (Sayle and Milner-White, 1995). Gaps in the aligned sequences are indicated by the dashed line. The residue conservation and utilization (Sequence logo) are provided and absence of sequence data is indicated by X. The coding exon contribution to the deduced amino acid sequences are indicated by ↕ and the number of the exon is indicated. The protein motifs were obtained from Ensembl protein features for mouse (Mmusc) SLC26A5. These motifs, prosites, domains are demarcated by: gray (▪)—hydrophobic transmembrane regions; red (▪)—SUL1 [InterPro: IPR001902], magenta (▪)—Sulph_transpt [Pfam: PF00916; InterPro: IPR011547], orange (▪)—SulP_transpt [Prosite patterns: PS01130; InterPro: IPR001902]; black (▪) [Pfam: PF01740; InterPro: IPR002645] and dark blue (▪) [Prosite profiles: PS50801; InterPro: IPR002645]—STAS motifs; and brown (▪)—Sodium:dicarboxylate symporter [PRINTS: PR00173 and InterPro: IPR001991].
Identification of the platypus and opossum SLC26A5 orthologs
Differences in the consensus peptides of eutherian and NMV SLC26A5 clearly demonstrated that Indels in exons 6 and 8 appear to be a mammalian innovation (see Fig. 5). We predicted that SLC26A5 peptides from primitive prototherian (monotremes) and metatherian (marsupials) species should also exhibit these mammalian adaptations in SLC26A5 peptide sequences. Inner ear morphological data support monotremes as the sister group of extant marsupials and eutherian mammals by the presence of a recognizable organ of Corti (Fernandez and Schmidt 1963; Jorgensen and Locket 1995; Ladhams and Pickles 1996) compared with the basilar papilla of avians, amphibians, reptiles, and bony fish. Like the latter, monotremes have a lagena at the tip of the cochlea that is absent in eutherian cochlea (Ladhams and Pickles 1996; Fritzsch et al. 2002). This cochlear phenotype is shared by both the platypus and the echidna (Ladhams and Pickles 1996).
In order to determine the genomic sequence of the SLC26A5 gene in the platypus (Ornithorhynchus anatinus, Oanat), we had identified and obtained OA_Bb BAC genomic clones that hybridized with a mouse prestin cDNA probe. Four clones, 435A11, 449F06, 506D22, and 556D19, were selected and found to containing the full complement of Oanat SLC26A5 coding exons (see supplementary Fig. S1). Subsequent PCR and sequencing analyses confirmed and corrected the genomic organization and sequence of the platypus prestin provided by Ensembl.
Comparisons of the Oanat and Mdome SLC26A5 proteins showed a homology of 597/724 (82.4%) identity and 652/724 (90.0%) similarity with 3/724 gaps (Fig. 5). Both taxa were more divergent from the NMV consensus sequence (373/743 [50.0%] identity, 483/743 [65.0%] similarity, 70/743 gaps and 376/732 [51.4%] identity, 484/732 [65.0%] similarity, 70/743 gaps, respectively) compared with the eutherian consensus protein (584/747 [78.2%] identity, 652/747 [87.2%] similarity, 10/747 gaps and 614/746 [82.3%] identity, 669/746 [89.7%] similarity, 7/746 gaps, respectively). Within the SulPtp domain, an amino acid homology of 91–93% was found among Oanat, Mdome, and the eutherian consensus proteins. Gaps between the eutherian and the Oanat and Mdome proteins were primarily associated with the amino- and carboxy-cytoplasmic tails. Coding exon 4 has small Indels with the Oanat with one fewer residue (DDMF-AGGMGSTN) and the Mdomi (DDIVIPGGGGNSTN) with an additional residue compared with the shortened eutherian amino acid sequence (DDIVIPGG-VNATN). No sequence gaps among these three proteins were found in the inserted “eutherian” transmembrane hydrophobic α-helix of the exon 6 with Oanat and Mdome having the highest homology and Oanat and the eutherians exhibiting the greatest diversity. A third region of heterogeneity was also observed, but corresponded to the diverse nonhelical segment (residues 308–320) found among the eutherian clades. Based on the homology among the platypus, opossum, and the eutherian SLC26A5 proteins, we conclude that the Indel alternations in exons 4 and 6 were the major changes that likely created the final evolutionary steps in formation of the minimal essential motif for the electromotility motor (meEM) in the mammalian prestin.
Comparison of the mammalian SLC26A4, SLC26A5, and SLC26A6 orthologs
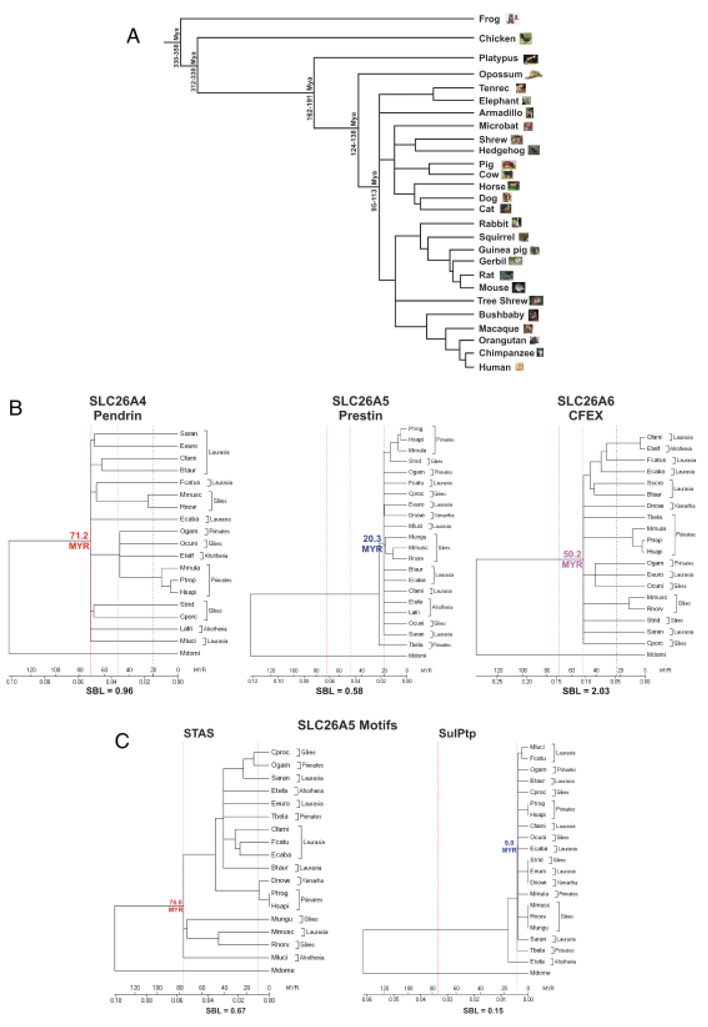
The opossum SLC26A5 genomic sequence was also available for analysis and provided an excellent outgroup of eutherian mammals for phylogenic comparisons (see Fig. 5 and Fig. 6). We selected the extant marsupial clade, since this taxon branched from the eutherian lineage ~ 124–138 Myr ago, as depicted in Fig. 6A. The opossum inner ear has similar anatomical and physiological properties of the eutherian inner ear. The opossum cochlea exhibits a flexible basilar membrane, parallel cellular organization of the organ of Corti (Fernandez and Schmidt 1963), OHC electromotility (R. Hallworth, unpublished data), and an auditory upper frequency sensitivity limit beyond those observed for NMVs (Frost and Masterton 1994; Reimer 1995).
Fig. 6.
Cladograms of the evolution of the SLC26A4, 5, and 6 proteins and the corresponding SulPtp and STAS motifs of the consensus eutherian SLC26A5 polypeptide. (A) The tree of evolutionary relationships of mammalian species used in this analysis with the frog and chicken serving as the outgroups as modified from (Benton and Donoghue, 2007). The branch lengths are proportional to the length in million of years (Myr) at the time of divergence of the taxa. The minimum age constraints are based on the oldest fossil evidence and maximum ages are based on bracketing the ages of sister groups in the fossiliferous formation. (B) Phylogenetic analysis of the SLC26A4 (pendrin), SLC26A5 (prestin), and SLC26A6 (CFEX) are shown using the opossum (Mdome) as the outgroup. Trees were established using MEGA4 program (Tamura et al. 2007) with the following parameters: neighbor-joining, pairwise deletion, constant substitution rate, and the amino acid JTT matrix. The sums of the branch lengths (SBL) are given. The time scale (Myr) is based on the divergence of the marsupials from the eutherian lineage and are represented by red line (71.2 Myr—SLC26A4), a dashed blue line (20.3 Myr—SLC26A5), and a dotted purple line (50.2 Myr—SLC26A6). (C) Phyogenetic trees are shown for the SLC26A5 protein motifs, STAS (red line—76.6 Myr) and SulPtp (blue dotted line—9.0 Myr). Opossum was used as the outgroup and a time scale (Myr) and SBL are indicated.
High protein sequence conservation among orthologs corresponds with the functional motifs or domains, whereas greater variability is observed outside of these domains (Table 1). The proteins representing each gene were divided into six contiguous topological regions, representing the amino-terminus, a segment containing all the putative transmembrane spanning (TM) helices (i.e., SulPtp), the amino portion of the intracellular tail (designated as SUL1′), a charged cluster motif within the intracellular tail, the STAS domain and the carboxy-terminus (Zheng, et al. 2000; Oliver et al. 2001; Deak et al. 2005; Zheng et al. 2005). As summarized in Table 1 and depicted in Fig. 6B, the total variability differed among the mammalian orthologs of these three genes. SLC26A5 orthologs have the lowest frequency of amino acid substitutions with a highly significant increase of 2-fold (P>0.001) in orthologs of SLC26A4 (see supplementary Fig. S2) and 4-fold in SLC26A6 (see supplementary Fig. S3). Of the six comparable regions, only the SUL1′ regions paralleled the increasing change in the amino acid sequence variations among the three paralogs. The greatest variability was observed in the charged cluster and carboxy-terminus regions. In contrast, the substitution rates in the STAS domains were similar for all three paralogs, suggesting this motif is necessary for function of these transporters. Most striking was the low frequency (39/7497 residues, 0.5%) of substitution associated with the SLC26A5 SulPtp region, which was significantly lower than the conserved SUL′ (32/1390 residues, 2.3%) and STAS (52/1411 residues, 5.1%) regions. In addition, the SulPtp region in SLC26A4 and SLC26A6, respectively, had ~11- and 25-fold greater substitution frequencies. Of the 39 residues differing from the consensus SLC26A5 SulPtp sequence, 16 substitutions were confined to a nonhelical stretch, residues 308–320, with the observed K310H and N314S substitutions being primarily associated with the Glires clade (Fig. 3), represented by Ocuni (rabbit), Strid (squirrel), Cporc (guinea-pig), Mungu (gerbil), Rnorv (rat), and Mmusc (mouse). Three additional variants, F1206L, I132V, and L380I, were also observed. Of the remaining substitutions 10/13 had neutral to positive PAML scores with only 3/7497 residues (0.04%) being nonconservative substitutions. This extremely high degree of conservation likely reflects stringent conformational and functional properties associated the SLC26A5 SulPtp region. Of the mammalian SLC26A5 genes 9 of 21 species had incomplete sequence data available and limited the identification of all amino acid variations within prestin. Despite the partial protein sequence information, this sequence data set suggests that the prestin meEM motif comprises the entire SulPtp region.
Table 1.
Regional variations in amino acid substitutions among the SLC26A4–6 orthologs
| Protein domain |
||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Parameters | N-terminus | SulPtp | SUL1′ | Charged cluster | STAS | C-terminus | Total |
| SLC26A4 | Domain Sites | 1–69 | 70–517 | 518–610 | 611–628 | 629–733 | 734–780 | 1–780 |
| (Pendrin) | % substitutions | 12.6 (125/1016) | 5.7 (407/7180) | 5.6 (84/1513) | 7.1 (22/311) | 6.3 (101/1601) | 10.8 (75/694) | 6.6 (814/12315) |
| PAML | 0.217±0.039 | 0.102±0.009 | 0.124±0.019 | 0.132±0.046 | 0.105±0.018 | 0.181±0.039 | 0.115±0.007 | |
| SLC26A5 | Domain Sites | 1–65 | 66–503 | 504–587 | 588–630 | 631–723 | 714–744 | 1–744 |
| (Prestin) | % substitutions | 7.3 (77/1056) | 0.5 (39/7497) | 2.3 (32/1390) | 11.7 (84/718) | 5.1 (52/1411) | 15.9 (76/477) | 3.0 (380/12649) |
| PAML | 0.120±0.025 | 0.012±0.003 | 0.079±0.017 | 0.179±0.036 | 0.086±0.021 | 0.176±0.052 | 0.048±0.003 | |
| SLC26A6 | Domain Sites | 1–112 | 113–508 | 509–630 | 631–651 | 652–738 | 739–759 | 1–759 |
| (CFEX) | % substitutions | 7.8 (139/1774) | 12.8 (811/6330) | 15.8 (252/1594) | 26.5 (89/336) | 7.3 (105/1434) | 24.7 (88/355) | 12.6 (1485/11823) |
| PAML | 0.288±0.036 | 0.192±0.015 | 0.240±0.036 | 0.783±0.212 | 0.163±0.029 | 0.285±0.072 | 0.234±0.072 | |
DISCUSSION
We have used the combined approaches of comparative evolutionary biology and protein structural and functional relationships to help predict the polypeptide motif responsible for the polypeptide motif responsible for electromotility. Evolution of OHC electromotility has required both the molecular evolution of an ATP-independent electrical motor protein and the structural context in which this novel protein can function, the mammalian cochlea. This required anatomical framework includes the evolution of both the unique structure of the OHC lateral wall and the cytoarchitecture of both the organ of Corti and the tectorial membrane. It is within this cellular scaffolding that prestin can mediate the OHC cochlear amplifier function (Dallos and Fakler 2002; Dallos, Zheng, and Cheatham 2006). Use of electromotility as a mechanism for sound amplification is only found in extant eutherian mammals, which include marsupials. The monotremes (echidna and platypus) are unique among vertebrates by sharing many features common to the eutherian mammals and to ancestral mammals, birds and reptiles. Although the monotreme middle ear is typically mammalian and the cochlea has a flexible basilar membrane, it has an atypical organ of Corti. The monotreme organ of Corti has three rows of both inner hair cells (IHCs) and pillar cells and five or more rows of OHCs. Also, it has a comma-shaped, rather than coiled, cochlea that contains at its end a lagenar macula (Fernandez and Schmidt 1963; Ladhams and Pickles 1996). The ancestral mammalian cochlea had already begun the transition from the basilar papilla to the mammalian cochlea around 163–191 Myr with the divergence of the prototherian clade (Benton and Donoghue 2007; Nikolaev et al. 2007). The transition to a eutherian cochlea was completed during the next 39–53 Myr as suggested by the opossum cochlea with the marsupial divergence from the eutherian lineage by 124–138 Myr. The cladogram of the eutherian SulPtp domain demonstrated the stability of the sequence with an extremely low substitution rate with the absence of clade-specific substitution (Fig. 6B) compared with SulPtp domains of SLC26A4 and SLC26A6. In addition there was no correlation in the SulPtp polypeptide sequence variation with high (Mluci [2–120 kHz], Mmusc [1–91 kHz]) or low frequency (Lafri [0.02–12 kHz], Hsapi [0.06–23 kHz]), Ptrog (0.04–28 kHz) specialist (Fay and Popper 1994; Heffner 2004). Our phylogenic analysis supports the idea that two major Indel changes in the SLC26A5 SulPtp motif occurred during the separation of the NMVs from the ancestral mammals. These sites within the SulPtp domain were further modified by substitution mutations to become completely conserved with no additional substitutions occurring in the eutherian lineage, which underwent rapid expansion 95–113 Myr into Afrotheria, Xenarthra, and especially the Boreoeutheria taxon, which subsequently branched into the Laurasiatheria and Euarchontoglires clades (Benton and Donoghue 2007).
Evolution of the meEM motif is related to the improved and subsequently unaltered molecular function of OHC electromotility and this function is in turn directly related to the tertiary and quaternary structure of the protein (Choi and Kim 2006; Fornasari et al. 2007; Ortlund et al. 2007; Sitbon and Pietrokovski 2007). It is our supposition that the meEM motif, because of its structural and functional requirements, must be highly conserved and exhibit little to no sequence variation. In the SulPtp motif of the SLC26A family members, what appears to be a the purifying selection process (Marsh and Griffiths 2005; Choi and Kim 2006), has incorporated both Indel changes and missense substitutions to alter the NMV SLC26A5 protein to provide OHC fast motility. The stringency of the meEM motif is demonstrated by the extremely low substitution rate in the putative meEM motif of eutherian SLC26A5 (see Fig. 6C). Two major functional constraints are: (1) the 10–12 membrane spanning segments in toto that permit the fast transition between two electromotile conformations that affect the intramembrane dimensions; and (2) the interactions among the tightly packed prestin homooligomeric structures in the OHC lateral wall (Zheng et al. 2006; Wu et al. 2007; Zhang et al. 2007). Thus, the major nonsynonymous evolutionary steps, represented by the two SulPtp Indels, and the subsequent purifying selection in the therian SLC26A5 molecular structure is based on the biophysics of its thermodynamic stability and folding to provide the necessary electrogenic conformational changes associated with OHC electromotility (Schaechinger and Oliver 2007; Sitbon and Pietrokovski 2007). Based on our comparisons,the meEM motif spans the amino acid residues 66–503 and encompasses all the TM regions.
At least two topological models of the SLC26A5 have been suggested based on the predicted TM α-helices (Oliver et al. 2001; Santos-Sacchi et al. 2001; Zheng et al. 2001; Deak et al. 2005; Rajagopalan et al. 2006). Secondary structure algorithms predicted that the number of TM regions vary from 10 to 12 regions. Because both the amino- and carboxy-tails are cytoplasmic (Oliver et al. 2001; Zheng et al. 2001; Navaratnam et al. 2005), an even number of TM motifs are required. Presently, a 12 TM topology is the most prevalent model (Oliver et al. 2001; Zheng et al. 2001; Rajagopalan et al. 2006). Deak et al. (2005) further refined this model by proposing the presence of two short, distorted helices that intercalate into the plasma membrane to bring the S238 residue intracellularly to permit phosphorylation by cytoplasmically localized kinases and phosphatases. Short helices that interdigitate into the membrane are present in many ion channels, transporter, and exchanger proteins, and are required elements in the topological and functional properties (Durell et al. 1998; Gouaux and Mackinnon 2005). Presently, the models of prestin topology are controversial and will require X-ray crystallographic analyses to elucidate the compressed and expanded structural forms (Dallos, Zheng, and Cheatham 2006; Muallem and Ashmore 2006).
A predicted hallmark of the ancestral SLC26A4/5 gene would be regulation of Cl− and ion in the gastrointestinal tract or its equivalent. It appears that SLC26A3 has retained this function and is also electrogenic (Shcheynikov et al. 2006). SLC26A4 has evolved both a unique expression in thyrocytes and an anion specificity for iodine, chloride, formate, and nitrate (Scott et al. 1999; Scott and Karniski 2000). Although SLC26A6 has the widest tissue expression pattern, with the highest transcript levels in the intestine, kidney, and pancreas, this transporter has evolved a high affinity for Cl−/oxalate anion exchange along with sulfate and Cl−/formate transport (Jiang et al. 2002). In vitro expression analyses demonstrated that mouse SLC26A6 exhibits electrogenic properties, whereas the human ortholog is an electroneutral Cl−/ exchanger (Chernova et al. 2005). Human and mouse SLC26A6 have an amino acid homology of 76% identity and 86% similarity. SLC26A5 is more highly derived, with the functional characteristics of electrogenicity, NLC, voltage-dependent conformational movement, and a transducer of intracellular Cl− and/or without mediating anion transport (Dallos and Fakler 2002). However, the supposition that prestin does not function as a transporter is controversial and recent experimental data suggest that the mammalian SLC26A5 is a functional anion antiporter (Muallem and Ashmore 2006; Schaechinger and Oliver 2007).
SLC26A5 and SLC26A6 have diverged from the SLC26A5/6 ancestral gene in both their associated functional properties and their expression profiles. Not only does this necessitate changes in the protein sequences, with modification of the associated SulPtp and STAS structure motifs, but would also require changes in the transcription regulation and regulatory elements in the 5′ promoter region. The tail to head orientation and syntenic relationship of Reln with SLC26A5 is maintained in all mammals and provides a stable platform for evolution, likely including primary promoter elements. SLC26A5 and SLC26A6 are differentially expressed in the organ of Corti with restricted expression in OHCs and supporting cells, respectively (Everett et al. 1999; Yoshino et al. 2004). Because both cochlear hair cells and their supporting cells are derived from the same cell lineage, the transcript regulatory mechanism must provide for this dichotomy in cellular expression (Fekete et al. 1998; Fritzsch et al. 2006). Expression of genes in cochlear hair cells is mediated through a gene network involving the bHLH gene, Atoh1, and the other important transcription regulatory factors, BarHL, Pou4f3, and Gfi1, which are involved in cochlear hair cell maturation and maintenance (Bermingham et al. 1999; Keithley et al. 1999; Li et al. 2002; Hertzano et al. 2004; Matei et al. 2005). The developmental upregulation of prestin occurring in prenatal mice (Zheng et al. 2000) parallels maturation of hair cells and suggests it is downstream of Atoh1 regulatory proteins. An additional regulatory element would be required to eliminate SLC26A5 in IHCs. Evidence for a negative regulatory element was the identification of a mouse Slc26a5 promoter fragment that controlled IHC expression (Li et al. 2004). Promoter evolution of SLC26A5 should require positive regulatory elements that are mediated via Atoh1 network process and a negative element to suppress expression in IHCs. Similarly, cochlear expression of SLC26A6 should be mediated by transcription regulatory factors associated with supporting cells cell fate acquisition, maturation, and maintenance. The Delta/Notch/Hes signaling pathway is known to play a prominent role in supporting cell development, maturation, and maintenance (Adam et al. 1998; Haddon et al. 1998; Lewis et al. 1998; Fritzsch, Beisel, and Hansen 2006), but is not involved in cell fate specification (Daudet et al. 2007). Interestingly, extra rows of hair cells are not unique to monotremes, but are found in genetically engineered mutants affecting Delta/Notch/Hes signaling pathway, the PCP pathway for convergent extension, Neurog1, and Foxg1 (Ma et al. 2000; Zine et al. 2001; Matei et al. 2005; Wang et al. 2005a; Brooker et al. 2006; Pauley et al. 2006). Both the Atoh1 and Delta/Notch/Hes gene networks are ancient gene mechanisms involved in neurosensory systems (Fritzsch and Beisel 2001) and it is the transcription factors and elements in which the SLC26A5 promoter evolved for OHC expression. Bioinformatic approaches are now being employed along with promoter dissection to identify these regulatory elements.
With the rapid increase in a plethora of genomic sequences, phylogenic analysis is now providing a viable approach for evolutionary biology, genomic, developmental, proteomic, and biomedical research (Birney et al. 2007; Huttley et al. 2007; Nikolaev et al. 2007). The vertebrate inner ear represents a rich evolutionary change in the morphology and sensory properties to form a single organ comprising of three sensory systems for detection of sound pressure (cochlea), gravity and linear acceleration (saccular and utricular maculae), and angular acceleration (cristae ampullares of the semicircular canals) (Fritzsch and Beisel 2001; Fritzsch et al. 2007). Both novel and co-opted transcription regulatory factors have facilitated these evolving morphological changes along with evolution of novel and co-opted physiological and functional genes to facilitate mechanosensory detection apparati. The present cladogram of SLC26A5 is still incomplete and requires the elucidation of the genomic sequences of several extant aquatic chordates, such as hagfish (Eptatretus burgeri [Eburg]), lamprey (Petromyzon marinus [Pmari]), and latimeria (Latimeria menadoensis [Lmend]). Sequencing efforts are currently in progress (Danke et al. 2004; Miyake and Amemiya 2004; Suzuki et al. 2004). Currently, Cinte is being used as the invertebrate-chordate outgroup. However, Lmena would be the most compatible outgroup for the mammals, including the prototheria and metatheria, because the coel-acanth genome is thought to be stable and evolving neutrally with few major rearrangements and is the most representative of the ancestral tetrapod genomes (Danke et al. 2004; Noonan et al. 2004). In addition, latimeria has a tetrapod-like basilar papilla with hair cells connected to a tectorial membrane like structure (Fritzsch 1988, 2003). We are presently pursuing this avenue of phylogenetic investigation into the sequence and linkage of the Lmena SLC26A3–6 paralogs.
In summary, our investigation suggests that there was a corresponding evolution of the SLC26A5-associated electro-motility with the formation of the mammalian organ of Corti and that the functional requirements of the meEM motif leads to an extraordinarily, highly conserved polypeptide sequence. Evolution of the mammalian SLC26A5 gene was associated with the nonsynonymous Indels alternations of coding exons 4 and 6 and the subsequent fixation of the meEM amino acid sequence in the eutherians. An additional outcome of our analyses is that two major Indel sites within the putative meEM motif need to be verified by physiological investigation of the expressed platypus SLC26A5 protein. We are currently creating a full coding cDNA construct and will use a combined approach of site-directed mutagenesis and chimeric engineered proteins with the platypus, chicken, frog, and gerbil prestin to recapitulate the evolution of the meEM motif. These latter studies would also be greatly assisted by the determination of the crystallographic structure of SLC26A5.
Supplementary Material
Acknowledgments
We wish to thank Lilian Calisto for her technical assistance and expertise in the associated BAC Southern blot hybridizations and sequence analysis of the platypus genome fragments. This investigation was supported in part by research grants from the USPHS National Institute on Deafness and Other Communication Disorders R01 DC05009 (K. W. B.), R01 DC05590 (B. F.), F32 DC08253 (M. W.), and from NSF/EPSCoR (R. H.).
REFERENCES
- Adam J, et al. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Adler HJ, Belyantseva IA, Merritt RC, Jr, Frolenkov GI, Dougherty GW, Kachar B. Expression of prestin, a membrane motor protein, in the mammalian auditory and vestibular periphery. Hear. Res. 2003;184:27–40. doi: 10.1016/s0378-5955(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Albert JT, et al. Voltage-sensitive prestin orthologue expressed in zebrafish hair cells. J. Physiol. 2007;580:451–461. doi: 10.1113/jphysiol.2007.127993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmore JF. A fast motile response in guinea-pig outer hair cells: the cellular basis of the cochlear amplifier. J. Physiol. 1987;388:323–347. doi: 10.1113/jphysiol.1987.sp016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhoff-Falk G. Hearing in Drosophila: development of Johnston’s organ and emerging parallels to vertebrate ear development. Dev. Dyn. 2005;232:550–558. doi: 10.1002/dvdy.20207. [DOI] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Brownell WE, Bader CR, Bertrand D, de Ribaupierre Y. Evoked mechanical responses of isolated cochlear outer hair cells. Science. 1985;227:194–196. doi: 10.1126/science.3966153. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Morrison HG, Pearson W. Exploring the relationship between sequence similarity and accurate phylogenetic trees. Mol. Biol. Evol. 2006;23:2090–2100. doi: 10.1093/molbev/msl080. [DOI] [PubMed] [Google Scholar]
- Chernova MN, et al. Functional comparison of mouse slc26a6 anion exchanger with human SLC26A6 polypeptide variants: differences in anion selectivity, regulation, and electrogenicity. J. Biol. Chem. 2005;280:8564–8580. doi: 10.1074/jbc.M411703200. [DOI] [PubMed] [Google Scholar]
- Choi IG, Kim SH. Evolution of protein structural classes and protein sequence families. Proc. Natl. Acad. Sci. USA. 2006;103:14056–14061. doi: 10.1073/pnas.0606239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallos P, Fakler B. Prestin, a new type of motor protein. Nat. Rev. Mol. Cell. Biol. 2002;3:104–111. doi: 10.1038/nrm730. [DOI] [PubMed] [Google Scholar]
- Dallos P, Zheng J, Cheatham MA. Prestin and the cochlear amplifier. J. Physiol. 2006;576(Pt 1):37–42. doi: 10.1113/jphysiol.2006.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danke J, et al. Genome resource for the Indonesian coelacanth, Latimeria menadoensis. J. Exp. Zool. A Comp. Exp. Biol. 2004;301:228–234. doi: 10.1002/jez.a.20024. [DOI] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 2007;134:2369–2378. doi: 10.1242/dev.001842. [DOI] [PubMed] [Google Scholar]
- Deak L, et al. Effects of cyclic nucleotides on the function of prestin. J. Physiol. 2005;563(Pt 2):483–496. doi: 10.1113/jphysiol.2004.078857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Brajkovic J, Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23:533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Durell SR, Hao Y, Guy HR. Structural models of the transmembrane region of voltage-gated and other K + channels in open, closed, and inactivated conformations. J. Struct. Biol. 1998;121:263–284. doi: 10.1006/jsbi.1998.3962. [DOI] [PubMed] [Google Scholar]
- Everett LA, et al. Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum. Mol. Genet. 2001;10:153–161. doi: 10.1093/hmg/10.2.153. [DOI] [PubMed] [Google Scholar]
- Everett LA, Morsli H, Wu DK, Green ED. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA. 1999;96:9727–9732. doi: 10.1073/pnas.96.17.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay RR, Popper AN. Comparative Hearing: Mammals. New York: Springer-Verlag; 1994. [Google Scholar]
- Fekete DM, Muthukumar S, Karagogeos D. Hair cells and supporting cells share a common progenitor in the avian inner ear. J. Neurosci. 1998;18:7811–7821. doi: 10.1523/JNEUROSCI.18-19-07811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Schmidt RS. The opossum ear and evolution of the coiled cochlea. J. Comp. Neurol. 1963;121:151–159. doi: 10.1002/cne.901210111. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Post-lethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornasari MS, Parisi G, Echave J. Quaternary structure constraints on evolutionary sequence divergence. Mol. Biol. Evol. 2007;24:349–351. doi: 10.1093/molbev/msl181. [DOI] [PubMed] [Google Scholar]
- Franchini LF, Elgoyhen AB. Adaptive evolution in mammalian proteins involved in cochlear outer hair cell electromotility. Mol. Phylogenet. Evol. 2006;41:622–635. doi: 10.1016/j.ympev.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. The amphibian octavo-lateralis system and its regressive and progressive evolution. Acta Biol. Hung. 1988;39:305–322. [PubMed] [Google Scholar]
- Fritzsch B. The ear of Latimeria chalumnae revisited. Zoology (Jena) 2003;106:243–248. doi: 10.1078/0944-2006-00120. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW. Evolution and development of the vertebrate ear. Brain Res. Bull. 2001;55:711–721. doi: 10.1016/s0361-9230(01)00558-5. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, et al. Development and evolution of inner ear sensory epithelia and their innervation. J. Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int. J. Dev. Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SB, Masterton RB. Hearing in primitive mammals: Monodelphis domestica and Marmosa elegans. Hear. Res. 1994;76:67–72. doi: 10.1016/0378-5955(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Furlong RF, Holland PW. Were vertebrates octoploid? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez J, Holland PW. Archetypal organization of the amphioxus hox gene cluster. Nature. 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebra-fish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–4644. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The Causes of Evolution. New York: Cornell University Press; 1932. [Google Scholar]
- Hallbook F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain Behav. Evol. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- He DZ, et al. Chick hair cells do not exhibit voltage-dependent somatic motility. J. Physiol. 2003;546(Pt 2):511–520. doi: 10.1113/jphysiol.2002.026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner RS. Primate hearing from a mammalian perspective. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2004;281:1111–1122. doi: 10.1002/ar.a.20117. [DOI] [PubMed] [Google Scholar]
- Hertzano R, et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum. Mol. Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, Wakefield MJ, Easteal S. Rates of genome evolution and branching order from whole genome analysis. Mol. Biol. Evol. 2007;24:1722–1730. doi: 10.1093/molbev/msm094. [DOI] [PubMed] [Google Scholar]
- Jiang Z, et al. Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nat. Genet. 2006;38:474–478. doi: 10.1038/ng1762. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Grichtchenko II, Boron WF, Aronson PS. Specificity of anion exchange mediated by mouse Slc26a6. J. Biol. Chem. 2002;277:33963–33967. doi: 10.1074/jbc.M202660200. [DOI] [PubMed] [Google Scholar]
- Jorgensen JM, Locket NA. The inner ear of the echidna Tachyglossus aculeatus: the vestibular sensory organs. Proc. Biol. Sci. 1995;260:183–189. doi: 10.1098/rspb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Karniski LP. Functional expression and cellular distribution of diastrophic dysplasia sulfate transporter (DTDST) gene mutations in HEK cells. Hum. Mol. Genet. 2004;13:2165–2171. doi: 10.1093/hmg/ddh242. [DOI] [PubMed] [Google Scholar]
- Keithley EM, Erkman L, Bennett T, Lou L, Ryan AF. Effects of a hair cell transcription factor, Brn-3.1, gene deletion on ho-mozygous and heterozygous mouse cochleas in adulthood and aging. Hear. Res. 1999;134:71–76. doi: 10.1016/s0378-5955(99)00070-2. [DOI] [PubMed] [Google Scholar]
- Kere J. Overview of the SLC26 family and associated diseases. Novartis Found. Symp. 2006;273:2–11. discussion 11–8, 261–4. [PubMed] [Google Scholar]
- King DC, et al. Finding cis-regulatory elements using comparative genomics: some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Carroll SB. A receptor tyrosine kinase from choano-flagellates: molecular insights into early animal evolution. Proc. Natl. Acad. Sci. USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladhams A, Pickles JO. Morphology of the monotreme organ of Corti and macula lagena. J. Comp. Neurol. 1996;366:335–347. doi: 10.1002/(SICI)1096-9861(19960304)366:2<335::AID-CNE11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Frantz GD, Carpenter DA, de Sauvage FJ, Gao WQ. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mech. Dev. 1998;78:159–163. doi: 10.1016/s0925-4773(98)00165-8. [DOI] [PubMed] [Google Scholar]
- Li M, Tian Y, Fritzsch B, Gao J, Wu X, Zuo J. Inner hair cell Cre-expressing transgenic mouse. Genesis. 2004;39:173–177. doi: 10.1002/gene.20040. [DOI] [PubMed] [Google Scholar]
- Li S, Price SM, Cahill H, Ryugo DK, Shen MM, Xiang M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development. 2002;129:3523–3532. doi: 10.1242/dev.129.14.3523. [DOI] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Liberman MC, Gao J, He DZ, Wu X, Jia S, Zuo J. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–304. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- Liu XZ, et al. Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum. Mol. Genet. 2003;12:1155–1162. doi: 10.1093/hmg/ddg127. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L, Griffiths CS. Protein structural influences in rhodopsin evolution. Mol. Biol. Evol. 2005;22:894–904. doi: 10.1093/molbev/msi081. [DOI] [PubMed] [Google Scholar]
- Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Zheng J, Du GG, Klocker N, Madison LD, Dallos P. N-linked glycosylation sites of the motor protein prestin: effects on membrane targeting and electrophysiological function. J. Neurochem. 2004;89:928–938. doi: 10.1111/j.1471-4159.2004.02377.x. [DOI] [PubMed] [Google Scholar]
- McGuire RM, Pereira FA, Raphael RM. Modulation of prestin function due to cysteine point mutation. 30th ARO Midwinter meeting; 2007. 41 Abs. [Google Scholar]
- Miyake T, Amemiya CT. BAC libraries and comparative genomics of aquatic chordate species. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;138:233–244. doi: 10.1016/j.cca.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- Muallem D, Ashmore J. An anion antiporter model of prestin, the outer hair cell motor protein. Biophys. J. 2006;90:4035–4045. doi: 10.1529/biophysj.105.073254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam D, Bai JP, Samaranayake H, Santos-Sacchi J. N-terminal-mediated homomultimerization of prestin, the outer hair cell motor protein. Biophys. J. 2005;89:3345–3352. doi: 10.1529/biophysj.105.068759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S, Montoya-Burgos JI, Margulies EH, Rougemont J, Nyffeler B, Antonarakis SE. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, et al. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- Ohno S. Patterns in genome evolution. Curr. Opin. Genet. Dev. 1993;3:911–914. doi: 10.1016/0959-437x(93)90013-f. [DOI] [PubMed] [Google Scholar]
- Ohta T. Pattern of nucleotide substitutions in growth hormone-prolactin gene family: a paradigm for evolution by gene duplication. Genetics. 1993;134:1271–1276. doi: 10.1093/genetics/134.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D, et al. Intracellular anions as the voltage sensor of prestin, the outer hair cell motor protein. Science. 2001;292:2340–2343. doi: 10.1126/science.1060939. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA. 2007;104 suppl. 1:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rajagopalan L, et al. Essential helix interactions in the anion transporter domain of prestin revealed by evolutionary trace analysis. J. Neurosci. 2006;26:12727–12734. doi: 10.1523/JNEUROSCI.2734-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer K. Hearing in the marsupial Monodelphis domestica as determined by auditory-evoked brainstem responses. Audiology. 1995;34:334–342. doi: 10.3109/00206099509071923. [DOI] [PubMed] [Google Scholar]
- Roth C, et al. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J. Exp. Zool. B Mol. Dev. Evol. 2007;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Santos-Sacchi J. Reversible inhibition of voltage-dependent outer hair cell motility and capacitance. J. Neurosci. 1991;11:3096–3110. doi: 10.1523/JNEUROSCI.11-10-03096.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Sacchi J, Dilger JP. Whole cell currents and mechanical responses of isolated outer hair cells. Hear. Res. 1988;35:143–150. doi: 10.1016/0378-5955(88)90113-x. [DOI] [PubMed] [Google Scholar]
- Santos-Sacchi J, Shen W, Zheng J, Dallos P. Effects of membrane potential and tension on prestin, the outer hair cell lateral membrane motor protein. J. Physiol. 2001;531(Pt 3):661–666. doi: 10.1111/j.1469-7793.2001.0661h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomoleculer graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Schaechinger TJ, Oliver D. Nonmammalian orthologs of prestin (SLC26A5) are electrogenic divalent/chloride anion exchangers. Proc. Natl. Acad. Sci. USA. 2007;104:7693–7698. doi: 10.1073/pnas.0608583104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinfest CW, et al. slc26a3 (dra)-deficient mice display chloride-losing diarrhea, enhanced colonic proliferation, and distinct up-regulation of ion transporters in the colon. J. Biol. Chem. 2006;281:37962–37971. doi: 10.1074/jbc.M607527200. [DOI] [PubMed] [Google Scholar]
- Scott DA, Karniski LP. Human pendrin expressed in Xenopus laevis oocytes mediates chloride/formate exchange. Am. J. Physiol. Cell. Physiol. 2000;278:C207–C211. doi: 10.1152/ajpcell.2000.278.1.C207. [DOI] [PubMed] [Google Scholar]
- Scott DA, Wang R, Kreman TM, Sheffield VC, Karniski LP. The Pendred syndrome gene encodes a chloride–iodide transport protein. Nat. Genet. 1999;21:440–443. doi: 10.1038/7783. [DOI] [PubMed] [Google Scholar]
- Shcheynikov N, et al. Coupling modes and stoichiometry of Cl−/ exchange by slc26a3 and slc26a6. J. Gen. Physiol. 2006;127:511–524. doi: 10.1085/jgp.200509392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon E, Pietrokovski S. Occurrence of protein structure elements in conserved sequence regions. BMC Struct. Biol. 2007;7:3. doi: 10.1186/1472-6807-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Hollich V. Scoredist: a simple and robust protein sequence distance estimator. BMC Bioinformatics. 2005;6:108. doi: 10.1186/1471-2105-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ota T, Fujiyama A, Kasahara M. Construction of a bacterial artificial chromosome library from the inshore hagfish, Eptatretus burgeri: a resource for the analysis of the agnathan genome. Genes Genet. Syst. 2004;79:251–253. doi: 10.1266/ggs.79.251. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Metcalfe RA, Watson PF, Weetman AP, Trembath RC. Mutations of the PDS gene, encoding pendrin, are associated with protein mislocalization and loss of iodide efflux: implications for thyroid dysfunction in Pendred syndrome. J. Clin. Endocrinol. Metab. 2002;87:1778–1784. doi: 10.1210/jcem.87.4.8435. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Regulation of polarized extension and planar cell polarity in the cochlea by the vertebrate PCP pathway. Nat. Genet. 2005a;37:980–985. doi: 10.1038/ng1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Renal and intestinal transport defects in Slc26a6-null mice. Am. J. Physiol. Cell. Physiol. 2005b;288:C957–C965. doi: 10.1152/ajpcell.00505.2004. [DOI] [PubMed] [Google Scholar]
- Weber T, et al. Expression of prestin-homologous solute carrier (SLC26) in auditory organs of nonmammalian vertebrates and insects. Proc. Natl. Acad. Sci. USA. 2003;100:7690–7695. doi: 10.1073/pnas.1330557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Currall B, Yamashita T, Parker LL, Hallworth R, Zuo J. Prestin–prestin and prestin–GLUT5 interactions in HEK293T cells. Dev. Neurobiol. 2007;67:483–497. doi: 10.1002/dneu.20357. [DOI] [PubMed] [Google Scholar]
- Yoshino T, et al. The immunohistochemical analysis of pendrin in the mouse inner ear. Hear. Res. 2004;195:9–16. doi: 10.1016/j.heares.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Zhang R, Qian F, Rajagopalan L, Pereira FA, Brownell WE, Anvari B. Prestin modulates mechanics and electromechanical force of the plasma membrane. Biophys. J. 2007;93:L07–L09. doi: 10.1529/biophysj.107.107573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, et al. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res. 2007;17:839–851. doi: 10.1101/gr.5586307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Du GG, Anderson CT, Keller JP, Orem A, Dallos P, Cheatham M. Analysis of the oligomeric structure of the motor protein prestin. J. Biol. Chem. 2006;281:19916–19924. doi: 10.1074/jbc.M513854200. [DOI] [PubMed] [Google Scholar]
- Zheng J, et al. The C-terminus of prestin influences nonlinear capacitance and plasma membrane targeting. J. Cell. Sci. 2005;118(Pt 13):2987–2996. doi: 10.1242/jcs.02431. [DOI] [PubMed] [Google Scholar]
- Zheng J, Long KB, Matsuda KB, Madison LD, Ryan AD, Dallos PD. Genomic characterization and expression of mouse prestin, the motor protein of outer hair cells. Mamm. Genome. 2003;14:87–96. doi: 10.1007/s00335-002-2227-y. [DOI] [PubMed] [Google Scholar]
- Zheng J, Long KB, Shen W, Madison LD, Dallos P. Prestin topology: localization of protein epitopes in relation to the plasma membrane. Neuroreport. 2001;12:1929–1935. doi: 10.1097/00001756-200107030-00032. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shen W, He DZ, Long KB, Madison LD, Dallos P. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- Zine A, et al. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.