Abstract
Pseudomonas aeruginosa secretes a 205 residue long hemophore (full-length HasAp) that is subsequently cleaved at the C’-terminal domain to produce mainly a 184 residue long truncated HasAp that scavenges heme [Letoffé, S., Redeker, V., and Wandersman, C. (1998) Mol. Microbiol. 28, 1223–1234]. HasAp has been characterized by X-ray crystallography and in solution by NMR spectroscopy. The X-ray crystal structure of truncated HasAp revealed a polypeptide αβ fold and a ferriheme coordinated axially by His32 and Tyr75, with the side chain of His83 poised to accept a hydrogen bond from the Tyr75 phenolic acid group. NMR investigations conducted with full-length HasAp showed that the carboxyl terminal tail (21 residues) is disordered and conformationally flexible. NMR spectroscopic investigations aimed at studying a complex between apo-HasAp and human met-hemoglobin were stymied by the rapid heme capture by the hemophore. In an effort to circumvent this problem NMR spectroscopy was used to monitor the titration of 15N-labeled holo-HasAp with hemoglobin. These studies allowed identification of a specific area on the surface of truncated HasAp, encompassing the axial ligand His32 loop that serves as a transient site of interaction with hemoglobin. These findings are discussed in the context of a putative encounter complex between apo-HasAp and hemoglobin that leads to efficient hemoglobin-heme capture by the hemophore. Similar experiments conducted with full-length 15N-labeled HasAp and hemoglobin revealed a transient interaction site in full-length HasAp similar to that observed in the truncated hemophore. The spectral perturbations observed while investigating these interactions, however, are weaker than those observed for the interactions between hemoglobin and truncated HasAp, suggesting that the disordered tail in the full-length HasAp must be proteolyzed in the extracellular milieu to make HasAp a more efficient hemophore.
The preferred aerobic metabolism of Pseudomonas aeruginosa requires respiratory enzymes that need iron or iron-containing cofactors for their function. The extremely low concentrations of free iron in mammalian hosts trigger a stress response in the opportunistic P. aeruginosa (and in many other pathogens) that involves the deployment of several iron-acquisition systems (1–4). The systems involved in the capture of iron typically fall in two categories, (i) excretion of low-molecular weight iron ligands (siderophores) and the receptors that take in iron-loaded siderophores through the outer membrane and (ii) direct binding of iron or iron-containing proteins at outer membrane receptors which internalize only the iron. Heme uptake and utilization of heme-iron is also a resource used by some pathogenic bacteria to overcome the highly restricted iron conditions encountered while colonizing a mammalian host. Hemoglobin (Hb) becomes available upon rupture of erythrocytes. Thus it is not surprising that under iron starvation conditions P. aeruginosa secretes very efficient hemolysins (5, 6) and cytotoxins (7) that rupture red blood cells and release Hb. There are two distinct heme uptake systems that enable P. aeruginosa to utilize heme-iron efficiently. One is the phu (Pseudomonas heme uptake) locus, consisting of a receptor gene (phuR) and the phuSTUVW operon encoding a typical ABC transporter (3). The second uptake system, has (heme acquisition system) consists of a heme receptor (hasR) and a protein that binds heme with high affinity, also known as the hemophore, HasAp (4). It is thought that in the early stages of infection, before tissue inflammation and damage ensues, the very low concentration of hemoglobin available to a pathogen is overcome by the secretion of hemophores, which can capture heme from hemoglobin for subsequent delivery to HasR for internalization. Once in the cytoplasm heme is delivered to a heme oxygenase (pa-HO) for its degradation to biliverdin, thus facilitating release of the heme-iron for subsequent metabolic use (8).
To date, the only structurally characterized hemophore is that of Serratia marcescens (HasAs), which captures heme and delivers it to the receptor HasR (9). In this organism HasR alone is able to take heme from hemoglobin but synergism with HasAs increases the efficiency of heme uptake from hemoglobin at least 100-fold (10, 11). A gene encoding a similar protein (HasAp) in P. aeruginosa is upregulated under iron-limiting conditions (12). HasAp, which shares approximately 50% identity with HasAs (Figure 1), is essential for P. aeruginosa uptake and utilization of hemoglobin iron (12). Like HasAs, HasAp is secreted to the extracellular milieu where it undergoes C’-terminal proteolytic cleavage that removes the last 15–21 amino acids, presumably by proteases also secreted by P. aeruginosa (13). In comparison, HasAs undergoes a single C’-terminal cleavage that removes the last 12 residues. It is therefore interesting that whereas the growth of HasAs mutants can be efficiently rescued by addition of full-length or cleaved forms of the S. marcescens HasAs to media where the only source of iron is hemoglobin, the growth of P. aeruginosa HasAp mutants can only be rescued by addition of truncated HasAp when hemoglobin is the sole source of iron (12). These observations suggest that proteolytic cleavage of HasAp is essential to the hemeuptake process.
Figure 1.
Alignment of amino acid sequences from HasAs and HasAp showing identical residues in bold face and residues involved in the coordination of the heme iron, including the “auxiliary” H83, which is thought to accept phenolic acid proton from Y75 (HasAp numbering), in red. The arrow indicates the length of truncated HasAp (full-length minus 21 amino acids) utilized in these investigations.
In this context, it is also important to note that expression of most virulence factors in P. aeruginosa is not constitutive but is regulated in a cell density-dependent manner (quorum sensing), in order to ensure that pathogenic characteristics are not expressed until the population has reached the critical density necessary to overwhelm the host defenses and establish an infection (14). It is therefore significant that a recent proteomics study revealed that among quorum-sensing regulated proteins in P. aeruginosa, several are involved in iron utilization, because it suggests a link between quorum sensing and the iron regulatory system (13). Moreover, two of the up-regulated proteins are the receptor phuR and the hemophore HasAp, which are components of each of the two heme uptake systems in P. aeruginosa (phu and has), therefore indicating that hemoglobin utilization is one of the phenotypes controlled through quorum sensing (13). Accordingly, while quorum sensing-impaired mutants show significantly reduced growth relative to wild type P. aeruginosa PAO1 in medium containing Hb as the sole source of iron, normal growth is rescued by supplementation with the quorum sensing signal molecule N-acyl homoserine lactone. The same authors reported that the most abundant form of secreted HasAp is the truncated form, wild type minus 21 amino acids (marked by an arrow in Figure 1), whereas the most abundant form of HasAp in quorum sensing-impaired mutants is the full-length protein (13). This observation is in agreement with the inability of full-length HasAp to rescue the growth of P. aeruginosa HasAp mutants in medium containing Hb as the sole source of iron (12) and underscores the importance of investigating HasAp in its full-length and truncated forms.
As part of an ongoing effort to understand the structure, function and dynamics of proteins involved in heme-iron metabolism by the opportunistic P. aeruginosa, we aim these studies at a molecular level understanding of hemoglobin-heme acquisition by the opportunistic P. aeruginosa. Results obtained from these investigations indicate that the hemophore fold, as well as the unusual axial heme coordination by His32 and Tyr75, first observed in the structure of HasAs (9) is likely a conserved motif in hemophores from different Gram-negative organisms. In addition, the present investigations revealed that the heme binding site of HasAp is conformationally disordered, a property that likely contributes to its high affinity for heme. Finally, NMR spectroscopic investigations revealed a specific complex between holo-HasAp and Hb that is discussed in the context of possible intermolecular interactions leading to hemoglobin-heme capture by hemophores.
EXPERIMENTAL
Cloning of the HasAp gene
A gene coding for full-length HasAp in Pseudomonas aeruginosa, strain PAO1 (PA3407), was synthesized and subcloned into the pET11a expression vector by Celtek Genes (Nashville, TN). The gene was engineered with silent mutations introducing codons favored by Escherichia coli (15). Restriction sites NdeI and BamHI were introduced at the 5' and 3' ends of the gene, respectively (Figure S1) to facilitate sublconing into the pET11a vector. The recombinant plasmid harboring the HasAp gene was then transformed into E. coli BL21-GOLD (DE3) host cell (Stratagene, La Jolla, CA) for subsequent expression. A gene coding for truncated HasAp was constructed from the recombinant pET11a plasmid harboring the gene coding for full-length HasAp using the QuickChange™ mutagenesis kit from Stratagene (La Jolla, CA). The oligonucleotides were synthesized by Integrated DNA Technologies, Inc. and used without further purification. The primers used are 5'-GCGACCCCGGCGGCGTAAGCGGCGGAAGTGGGC-3' and 5'- GCCCACTTCCGCCGCTTACGCCGCCGGGGTCGC-3'; the underlined codons represent mismatches designed to introduce a stop codon that prevents translation of the last 21 C’-terminal amino acids, as is indicated by the arrow in Figure 1. The mutation was confirmed by sequencing and the recombinant DNA plasmid transformed into E. coli BL21 (DE3) GOLD cells for subsequent protein expression.
Expression and Purification of Proteins
Full-length (205 residues) and truncated (184 residues) HasAp (see Figure 1) were expressed and purified as follows: A single colony of freshly transformed cells was cultured for 12 h in 10 mL of Luria-Bertani (LB) medium containing 200 µg/mL of ampicillin and used to inoculate 1.0 L of M9 minimal medium (200 µg/mL ampicillin). The resultant culture was incubated with continuous shaking at 225 rpm to an OD600 of 0.80 – 0.90, followed by centrifugation at 4000 rpm for 10 min. The cell pellet was resuspended in 1.0 L fresh minimal M9 medium containing ampicillin and cultured to an OD600 of 1.0 before addition of isopropyl- β-D-thiogalactopyranoside (IPTG; 1 mM final concentration), followed by culturing at 30 °C for 5 h and harvesting cells by centrifugation at 4000 rpm for 10 min. The cells were lysed by sonication as previously described (16) in 50 mM Tris-HCl (pH 7.8) containing 1 mM EDTA and 1 mM phenylmethysulfonyl fluoride (PMSF). The resultant suspension was centrifuged at 23,500 rpm for 2 h to separate the cell debris and the supernatant dialyzed against 20 mM Tris-HCl (pH 7.8) (3×4 L) at 4 °C. The protein was loaded into a Sepharose-Q Fast Flow column (2.6 cm i.d.×15 cm length) pre-equilibrated with 20 mM Tris-HCl (pH 7.6) and eluted with the same buffer with a linear gradient of NaCl (50–500 mM); the purity of the protein in the different fractions was determined by SDS-PAGE and the best fractions pooled. The apo-protein, which amounts to approximately 80% of total HasAp, was reconstituted with hemin dissolved in DMSO. To this end, heme was added gradually to the protein solution until the absorbance ratio (A280/A407) no longer changed. The resultant solution was incubated at 4 °C and concentrated to a final volume of 2–3 mL using Amicon ultracentrifuge filters and further purified by size-exclusion chromatography in a Sephacryl S-200 column (2.6 cm i.d.×90 cm length), equilibrated and eluted with 100 mM Tris-HCl containing 100 mM NaCl (pH 7.6). Fractions with an absorbance ratio (A280/A407) < 0.35 were pooled, dialyzed against phosphate buffer (µ = 0.10, pH 7.0), concentrated by ultrafiltration, and stored at −20 °C.
Expression and Purification of Isotopically Labeled Proteins
Uniformly (15N) and (15N, 13C) HasAp were expressed using the same procedure described above for unlabeled proteins with minor modifications: When the OD600 reached 0.80 – 0.90, the cells were centrifuged at 4000 rpm for 10 min. The cells were then resuspended and subcultured in 1.0 L of fresh M9 medium containing ampicillin and 1 g of 15NH4Cl and 1 g of D-glucose for producing [U-15N]-HasAp or 1 g of 15NH4Cl and 1 g 13C6-D glucose for producing [U-13C, 15N]-HasAp labeled samples. Polypeptide synthesis was induced by addition of IPTG to a final concentration of 1 mM. Cell lysis and protein purification were carried out as described above.
The expression of truncated HasAp labeled selectively with 15N-amino acids was carried out following protocols previously reported (17, 18) with minor changes. The M9 medium was supplemented with the following amino acids (g/L) prior to autoclaving, except for L-Trp (0.05), which was filter-sterilized: L-Ala (0.5), L-Arg (0.4), L-Asn (0.4), L-Asp (0.4), L-Cys (0.05), L-Gln (0.4), L-Glu (0.65), Gly (0.55), L-His (0.1), L-Ile (0.23), L-Leu (0.23), L-Lys hydrochloride (0.42), L-Met (0.25), L-Phe (0.13), L-Pro (0.1), L-Ser (2.10), L-Thr (0.23), L-Tyr (0.17), and L-Val (0.23). Supplemented M9 medium (1.0 L), which contained all amino acids except that to be labeled, was inoculated with 10 mL of an overnight LB culture. IPTG (1mM final concentration) and the appropriate 15N-labeled amino acid was added when the culture reached an OD600 ~ 1.0 and the protein was expressed and purified as described above. To prepare 15N-Val-HasAp, 0.23 g of 15N-L-Val was added. In the preparation of 15N-Leu-HasAp isotopic scrambling was minimized by preparing the M9 medium with 3 g/L of D-glucose and supplementing with 55 mg of 15N-L-Leu and additional 0.23 g of L-Ile and 0.23g L-Val immediately after the addition of IPTG. In the preparation of 15N-Thr-HasAp the M9 medium was also prepared with 3 g/L of D-glucose and supplemented with 55 mg of 15N-L-Thr and additional 0.55 g of L-Gly and 2.1 g of L-Ser. For the preparation of 15N-Tyr-HasAp the M9 medium contained 3 g/L of D-glucose and was supplemented with additional 170 mg of 15N-Tyr.
Electronic Absorption Spectroscopy
The electronic absorption spectra of full-length and truncated HasAp were recorded in phosphate buffer (µ = 0.1, pH 7.0) using an Ocean Optics UV-Vis spectrophotometer. Extinction coefficients of the proteins at 407 nm (ε407) were measured as described previously (19).
X-Ray Crystallography
Initial conditions for crystal growth were obtained from screening experiments conducted at the High Throughput Crystallization Screening Laboratory of the Hauptman-Woodward Medical Research Institute (Buffalo, NY) (20). For X-ray diffraction, crystals were grown using the hanging drop vapor diffusion method mixing 5 µL of a 120 mg/mL solution of truncated HasAp with an equal volume of reservoir solution, containing 100 mM MES buffer (pH 6.0), 100 mM NH4Cl, and 80% PEG 400, and incubating at 23 °C. Crystals grew reproducibly to a size of approximately 1mm per side in approximately 3 days. X-ray diffraction data were recorded at −180 °C in the Moffitt Cancer Center Structural Biology Core using the rotation method on a single flash-frozen crystal of HasAp [Detector: Rigaku HTC image plate; X-rays: CuKa, focused by mirror optics; Generator: Rigaku Micro-Max 007-HF (MSC, The Woodlands, TX)]. The data were reduced with XDS (21). The program package CNS (22) was employed for phasing and refinement. Model building was performed with O (23). The structure was solved by molecular replacement using HasAs (PDB code 1B2V), stripped of solvent molecules, ions and ligands as starting model. The asymmetric unit consists of two HasAp monomers. Refinement was performed using data to highest resolution with no sigma cut-off applied. Solvent molecules were added to the models at reasonable positions, and the heme molecules were modelled according to the clear electron density maps. Several rounds of minimization, simulated annealing (2500 °K starting temperature) and restrained individual B-factor refinement were carried out. Data collection and refinement statistics are summarized in Table 1.
Table 1.
Summary of data collection and structure refinement.
| Data set | HasAp – Heme complex |
|---|---|
| Space group | P31 |
| Unit cell dimensions (Å) | a=47.7 b=47.7 c=141.3 α=β=90° γ=120° |
| Protein atoms | 2 × 1327 |
| average main chain B-factor = 15.9 Å2] | |
| Ligands | Heme (2 ×43 atoms) |
| [average B-factor = 14.5 Å2] | |
| Solvent molecules | 250 |
| [average B-factor = 31.9 Å2] | |
| rmsda bonds (Å) | 0.011 |
| Rmsd angles (°) | 1.6 |
| Resolution range | 12-1.7 (1.8-1.7) |
| Unique reflections | 36833 (5430) |
| Completeness (%) | 93.1 (87.6) |
| I/σI | 22.2 (12.7) |
| Rmerge b(%) | 3.1 (6.3) |
| Rcryst c(%) | 16.6 |
| Rfree d (%) | 18.9 |
Values in parentheses refer to the highest resolution shell.
rmsd: root mean square deviation from ideal values.
Rmerge = 100 × Σ hΣi |Ihi - Ih| / ΣhiIhi where h are unique reflection indices.
Rcryst = 100 × Σ |Fobs-Fmodel | / ΣFobs where Fobs and Fmodel are observed and calculated structure factor amplitudes.
Rfree is Rcryst calculated for 1105 randomly chosen unique reflections, which were excluded from the refinement.
Resonance Raman and EPR spectroscopy
The resonance Raman spectra were obtained on room temperature samples using a custom McPherson 2061/207 spectrograph (set at 0.67 m with variable gratings) equipped with a Princeton Instruments liquid-N2-cooled CCD detector (LN-1100PB) and 90° geometry. A Kaiser Optical supernotch filter was used to attenuate Rayleigh scattering generated by the 413-nm excitation of an Innova 302 krypton laser (Coherent, Santa Clara CA). Frequencies were calibrated relative to aspirin, indene and CCl4 standards and are accurate to ±1 cm−1. EPR spectra were obtained on a Bruker E500 X-band EPR spectrometer equipped with a superX microwave bridge and a dual mode cavity equipped with a helium flow cryostat (ESR900, Oxford Instruments, Inc.).
NMR Spectroscopy
1H NMR spectra were collected on a Varian Unity Inova spectrometer equipped with a triple resonance probe operating at a 599.74 MHz 1H frequency. One-dimensional proton spectra were acquired with presaturation of the residual water peak over 40 k data points with a spectral width of 100 kHz, 200 ms acquisition time, 20 ms relaxation delay, and 1024 scans; spectra were referenced to a residual water peak at 4.80 ppm.
Protein samples (~ 2.5 mM) used for two- and three-dimensional NMR data collection were in phosphate buffer (µ = 0.1, 95% H2O, 5% D2O, pH 7.0). These experiments were performed at 32 °C unless otherwise noted. The following suite of 2-D and 3-D NMR experiments were carried out in a Bruker Avance 800 spectrometer equipped with a 5 mm TXI 1H-13C/15N/D xyz-gradient probe and a Varian Unity Inova 600 spectrometer equipped with a triple resonance probe, in order to perform backbone sequential assignments of truncated and full-length HasAp: 1H-15N-HSQC, HN(CO)CA, HNCA, CBCA(CO)NH, HNCACB, HNCO, (HCA)CO(CA)NH, NOESY-HSQC and 15N-separated NOESY-HSQC (110 ms mixing time). The data were processed using NMRPipe and analyzed with the program Sparky. The chemical shifts were referenced directly and indirectly using the proton frequency of the DSS resonance at 0.00 ppm.
Titration of HasAp with hemoglobin
A solution of [U-15N]-HasAp (2.5 mM) was titrated with incremental, 0.083 molar equivalents of human met-hemoglobin (met-Hb) added in small µL aliquots from a 3.5 mM stock solution, to a final molar ratio of 1:1.25 HasAp:Hb. The titration was monitored with the aid of 1H-15N HSQC NMR spectra. The HasAp and Hb solutions were prepared in phosphate buffer (µ = 0.1, pH 7.0). Prior to each experiment, lyophilized human Hb (Sigma) was dissolved in phosphate buffer (µ = 0.1, pH 7.0) and purified in a Sephacryl S-200 size exclusion column pre-equilibrated and eluted with phosphate buffer (µ = 0.1, pH 7.0). Fractions with purity ratio (A280/A406) < 0.25 were pooled and concentrated using Amicon ultracentrifuge filters to a final concentration of 3.5 mM.
RESULTS
Structural characterization of truncated HasAp
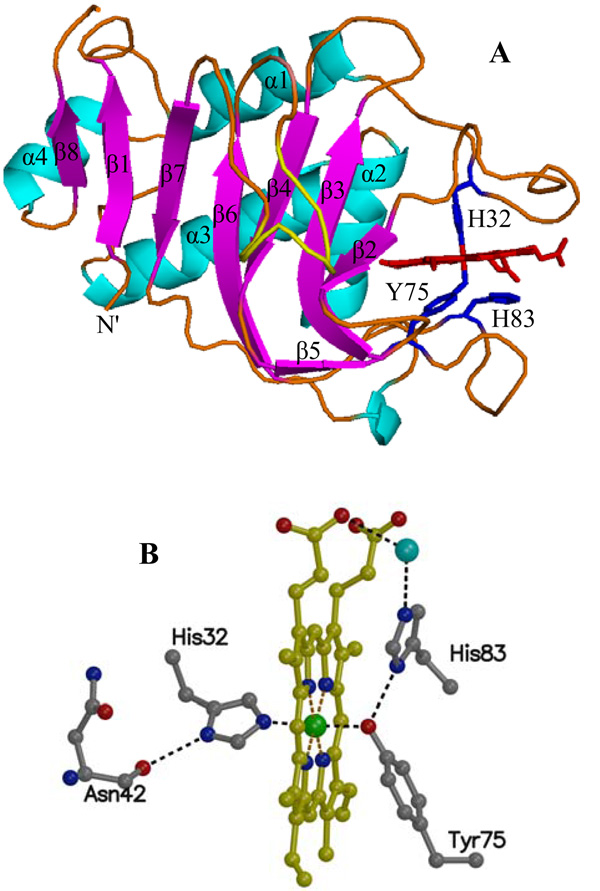
The polypeptide of truncated HasAp folds into the same αβ fold characteristic of HasAs (9), which consists of an extended “β-sheet wall” comprised of eight anti-parallel β-strands connected by hairpins (Figure 2-A). Four α-helices pack against the aformentioned β-sheet, thus forming an “α-helix wall” opposite to the “β-sheet wall”. The heme is contained within two extended loops: Loop 1 is located between α1 and β2 and contains one of the heme axial ligands, His32; hereafter this loop will be referred to as the His32 loop. A second extended loop is located between strands β5 and β6, and contains the other heme axial ligand, Tyr75. The Tyr75 loop continues onto the bulk of the “β-sheet wall”, thus the loops containing the heme can be thought of as flexible jaws hinged to the rigid β-sheet and α-helix walls. In the structure of HasAs electron density can be seen only to residue 174. In comparison, electron density in the structure of HasAp can be observed for all 184 residues, which reveals strand β-8 (residues 188–191) parallel to β-1 that extends the β-sheet wall by one strand relative to the structure of HasAs.
Figure 2.
(A) The fold of HasAp harbors four α-helices and eight anti-parallel β-strands. The axial ligands, His32 and Tyr75 are located in two extended loops above and below the heme. The side chain of His83 is located within hydrogen bonding distance of the Tyr75 phenolic acid proton. (B) Presentation of the heme and its coordination sphere in HasAp at 1.7 Å resolution depicting hydrogen bonding interactions (2.2 – 3.2 Å; black dotted lines).
As pointed out above, the heme is axially coordinated by His32 and Tyr75, a coordination motif that is identical to that seen in the structure of HasAs and in agreement with the conserved nature of these residues in hemophore sequences from distinct organisms (24). The hydrogen bonding network in the vicinity of the heme (Figure 2-B) is also very similar in the HasAs and HasAp structures. Notably, the orientation of the side chain of His83 places its Nδ within hydrogen bonding distance of the coordinated phenolate oxygen in Tyr75, a situation identical to that observed in the first crystal structure reported for HasAs (9), which was obtained from crystals grown at pH 4.6. It is worth noting that in a subsequent investigation (25) crystals of HasAs were grown at different pH values, and the structure obtained at pH 6.5 showed an alternative conformation of His 83 that is not hydrogen bonded to Try75. B factors for the heme in the latter structure were very high, an observation that was interpreted to suggest that loss of the hydrogen bond between His83 Nδ and the phenolate of Tyr75 may facilitate heme acquisition and/or release (25). In this context, it is interesting that the structure of HasAp, obtained from crystals grown at pH 6.0, shows low B-factors for the heme and confirms the presence of a hydrogen bond between the coordinated Tyr75 phenolate oxygen and Nδ of His83. The hemophore fold exposes a large portion of the heme to the aqueous environment. The heme propionates are exposed and are anchored to the protein via hydrogen bonding interactions with the NH of Gly35 and the guanidinium group of Arg29. The two heme edges contiguous to that harboring the heme propionates are also significantly exposed to the solvent with only a few hydrophobic contacts between the heme and the polypeptide: On one edge of the heme, His134 makes contact with heme-vinyl CBC and Val137 with heme-methyl CMC, whereas heme methyl CMD on the same edge and heme-methyl groups CMA, CMB and heme-vinyl CBB on the opposite edge are exposed, thus leaving only the back edge of the macrocycle with significant hydrophobic interactions with the polypeptide (see Figure S2). Given a similar degree of heme exposure in the crystal structure of HasAs (25), it is clear that the high affinity of the heme-hemophore complex is not achieved by steric confinement of the heme macrocycle, and despite the wealth of structural information, the determinants of high affinity remain to be defined.
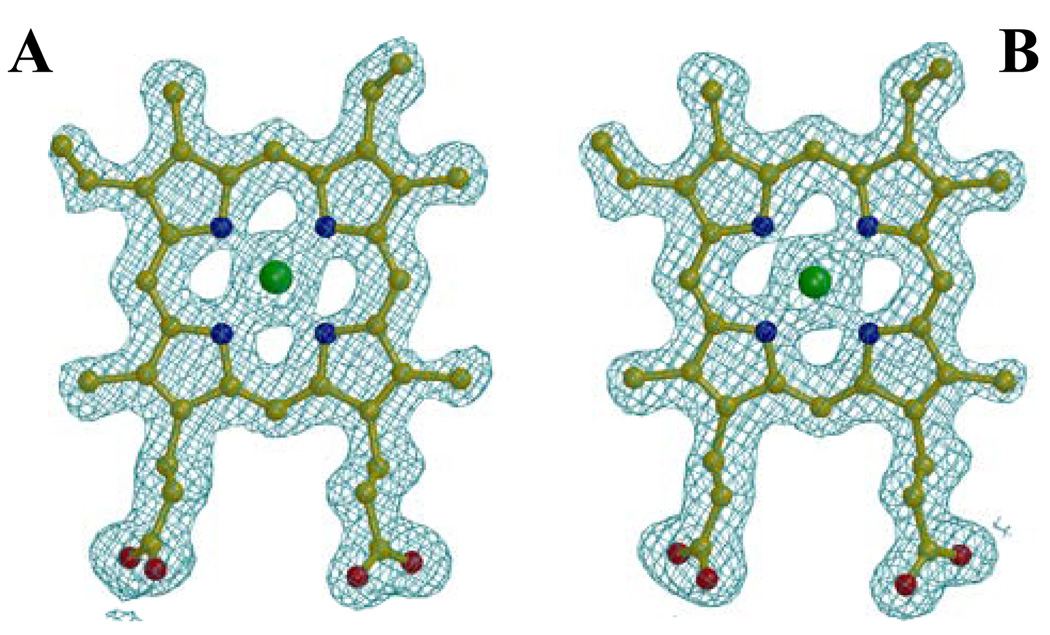
HasAp differs from HasAs in that the heme in HasAs binds to the polypeptide in two orientations that differ by a 180° rotation about the α-γ-meso axis, which is common in proteins where the heme is not bound covalently, i.e. myoglobin, cytochrome b5 and heme oxygenase (16, 26, 27). These two heme rotational isomers of HasAs, which exist with approximately similar abundance, have been observed in electron density maps of a 1.77 Å resolution structure (25) and in solution NMR spectroscopic studies (28). In comparison, analysis of heme electron density in HasAp indicates that the heme is bound in only one orientation, as can be seen in the electron density maps of the heme shown in Figure 3 and Figure S3 for each of the two monomers in the unit cell. Work performed with the cytochromes revealed that heme isomerism is a consequence of subtle packing interactions between amino acids lining the heme pocket and heme substituents, such as heme vinyl and heme methyl groups (29, 30). Analysis of the HasAp and HasAs structures does not reveal distinct packing interactions between heme substituents and the few residues making hydrophobic contact with the heme. The analysis, however, suggests that although the sequence PGPF51 in HasAp makes it possible to accommodate heme-methyl CMB without steric clash; the alternative heme orientation would place a bulkier heme-vinyl at the equivalent position, possibly creating unfavourable steric interactions with the side chain of Phe51. In comparison, the presence of two contiguous Gly residues and the replacement of Pro for Ser in the equivalent sequence of HasAs (GGSL50) likely make a more flexible environment that can accommodate a heme-vinyl or a heme-methyl, thus permitting heme to bind in the two possible orientations with almost equal abundance.
Figure 3.
Electron density for the heme in molecules A and B, contoured at 1σ, derived from a 2Fo-1Fc Fourier synthesis after the last refinement cycle. The electron density from a Fo-1Fc Fourier synthesis omitting the heme molecules is presented in the supplemental material (Figure S3). The data is consistent with heme binding in only one orientation in HasAp.
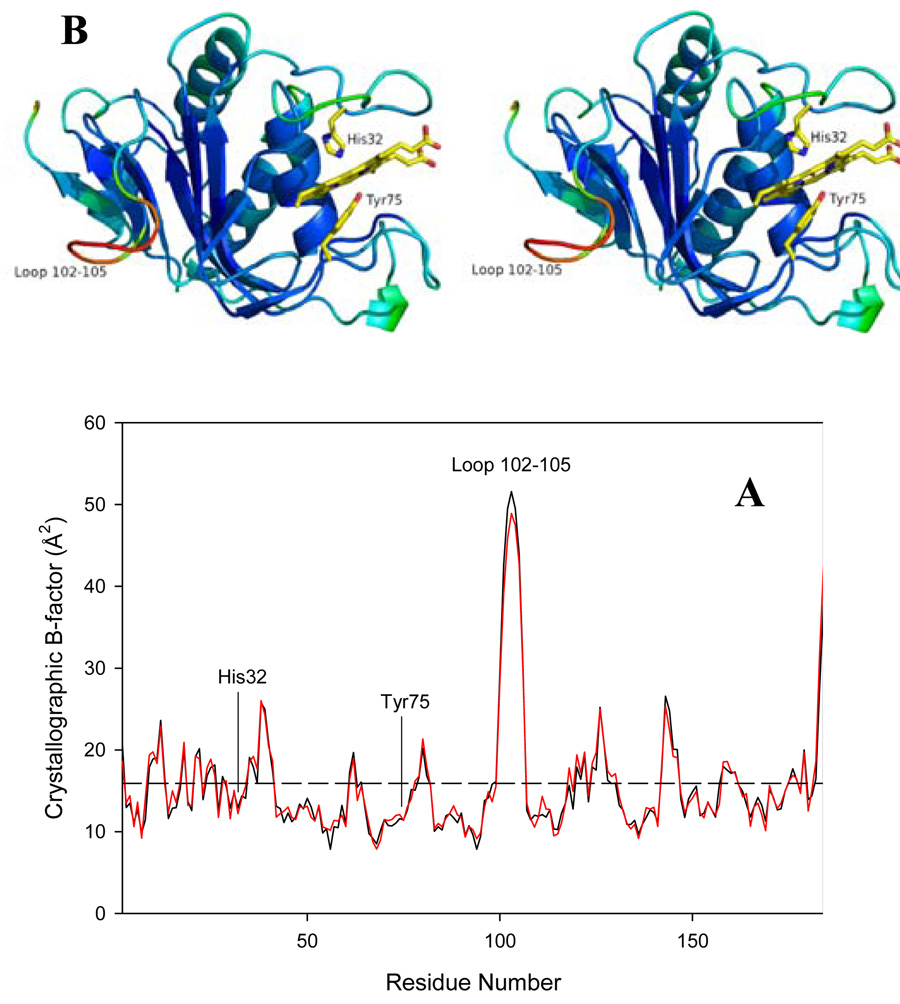
Analysis of thermal factors in the structure of HasAp is summarized in Figure 4: The plot shows per-residue temperature factors in the structures of each of the HasAp molecules, A (black line) and B (red line) in the unit cell; this information has been color-traced on the structure of molecule A, with temperature factors increasing from blue (8 Å2) to red (51 Å2) (Figure 3-B). Clearly, residues in the loop 102–105 exhibit high temperature factors, an observation that is in agreement with the poorly defined electron density of this section of the structure. A mutagenesis study of HasAs has implicated this loop as a putative surface of interaction with the receptor HasR (31). Hence, the high mobility of this loop may be important in the recognition and binding of the hemophore outer membrane receptor. The plot also reveals higher than average thermal factors for residues in helix α1, in sections of the His32 and Tyr75 loops and in the loop connecting α2 (the helix that forms the back wall of the heme) with α3. As will be discussed later, these sections of the protein may be involved in specific interactions between Hb and HasAp.
Figure 4.
(A) Per residue plot of B-factors of the Cα atoms in monomer A (black line) and monomer B (red line). The average B-factor of all main chain atoms is 15.9 Å2 for both monomers (dashed horizontal line). (B) Stereo representation of molecule A of the HasAp crystal structure color coded according to thermal factors, with blue representing 8 Å2 and red 51 Å2. The heme and its axial ligands His32 and Tyr75 are shown in yellow.
The spin state of the heme-iron in HasAp
The electronic absorbance spectra of full-length and truncated HasAp show a Soret maximum at 407 nm (ε407 = 77.7 mM−1 cm−1) with visible bands at 495, 540, 577, and 616 nm (Figure S4-A). The Soret and 616-nm bands are indicative of high-spin heme iron whereas the α/β bands at 577 and 540 nm suggest a low-spin species; hence, the Uv-vis spectrum suggests a mixture of low-spin (S = 1/2) and high-spin (S = 5/2) species (32). The 1H NMR spectra of full-length and truncated HasAp are identical (Figure S4-B) and show a single set of heme methyl resonances that support the presence of only one heme rotational isomer of HasAp in solution. The mean chemical shift of the four methyl groups of ferriheme is indicative of the heme-iron spin state (33, 34). This value in HasAp, 37 ppm, is between that of ferric high-spin and ferric low-spin species, thus further supporting a mixture of high-spin (S = 5/2) and low spin (S = 1/2) species in fast exchange relative to the NMR timescale. In agreement with this notion, the resonance Raman spectrum of HasAp obtained with Soret excitation shows a porphyrin skeletal ν4 at 1372 cm−1 characteristic of ferric hemes, and two sets of ν3, ν2, and ν10 frequencies representative of 6-coordinate high-spin (1477, 1561, and 1623 cm−1, respectively) and 6-coordinate low-spin (1504, 1579 and 1639 cm−1, respectively) species (Figure S4-C). These resonance Raman frequencies and the relative intensities of contributions from the high-spin and low-spin states are virtually identical to those observed earlier with HasAs (35). Finally, the EPR spectrum of HasAp (Figure S4-D) also reveals the presence of a mixture of high- and low-spin ferric heme, with a dominant S = 1/2 rhombic signal with g = 2.83, 2.20, and 1.71, and an S = 5/2 mostly axial signal with g = 6.09, 5.76, and 1.99. These EPR parameters and the relative intensities of the two signals are equivalent to those observed in HasAs (36).
Amide backbone resonance assignments
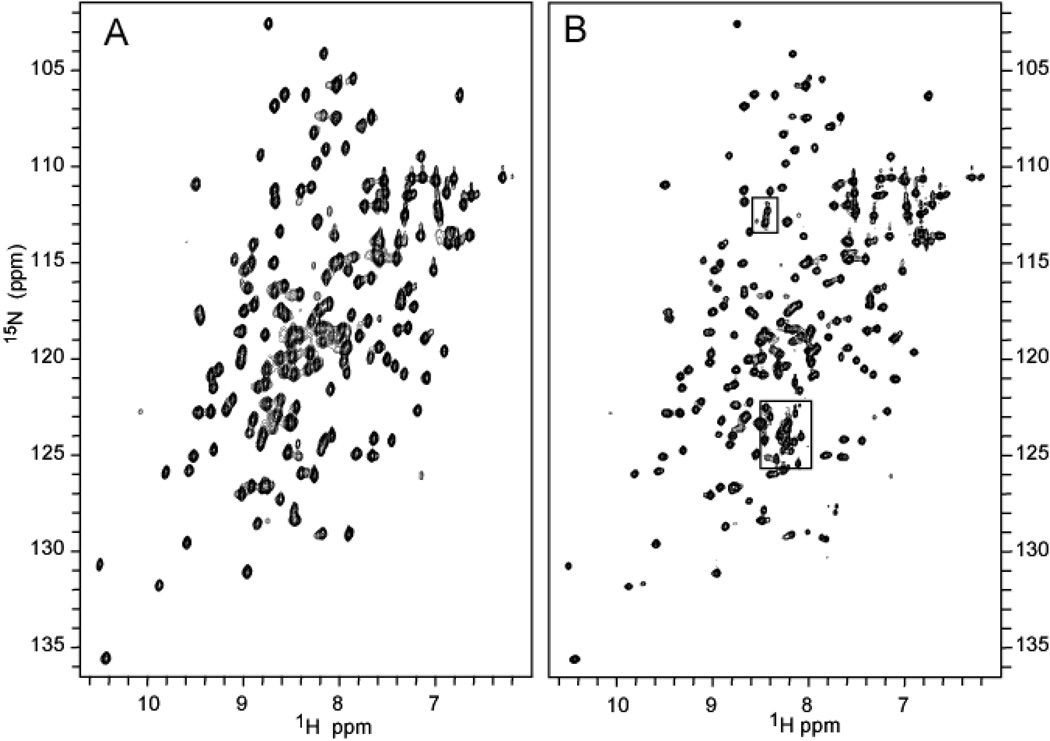
Figure 5 depicts a side-by-side comparison of the 1H-15N HSQC spectra of the full-length and truncated forms of HasAp. Cross-peaks highlighted in squares in the spectrum of the full-length form (Figure 5-B), which do not have equivalent peaks in the spectrum of the truncated form (Figure 5-A) correspond to residues in the C’-terminal tail. Not counting Pro198, 19 of the 21 C’-terminal residues exhibit cross-peaks. Sequential assignments were initially obtained for the truncated form using the strategy described below. These assignments and the similarity in the spectra of full-length and truncated forms of HasAp greatly simplified the sequential resonance assignment of the full-length form. The sequential backbone assignments for the truncated and full-length forms of HasAp are presented in Tables S1 and S2, respectively.
Figure 5.
1H-15N HSQC spectrum of (A) [U-15N]-truncated HasAp in phosphate buffer (µ = 0.1, pH 7.0 and 32 °C, obtained in a Varian Unity Inova 600 spectrometer. (B) [U-15N]-full-length HasAp under identical conditions. The cross-peaks within the boxes correspond to resonances originating from the C’-terminal tail in the full-length protein.
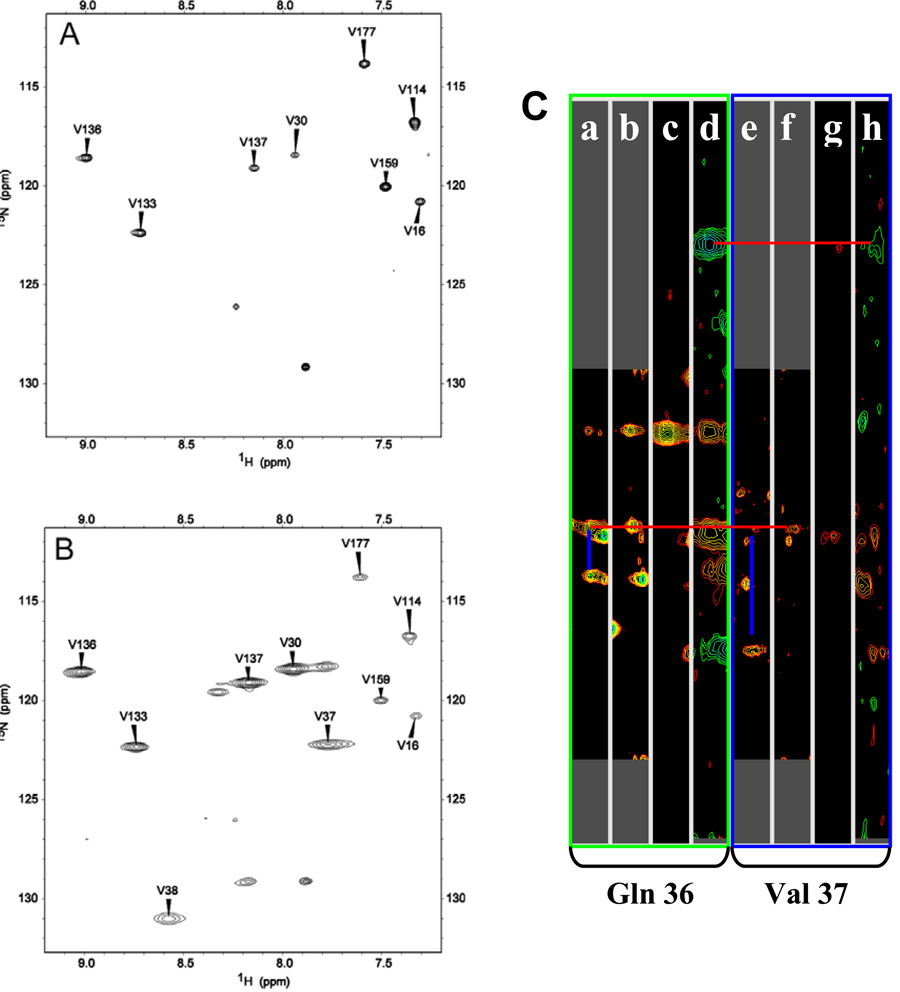
Conventional heteronuclear 2D and 3D NMR experiments performed with a sample of truncated [U-13C, U-15N]-HasAp allowed resonance assignment of a large majority of residues, with the exception of residues listed on Table 2. Not counting Ser2 and Ser103, these residues are located < 9 Å from the heme-iron, thus they are expected to be strongly influenced by the iron paramagnetism, which is manifested in weak or undetectable cross peaks in the HSQC spectrum. It is likely that cross-peaks corresponding to Ser2 and Ser103 cannot be detected due to conformational disorder because Ser2 becomes the amino terminal residue upon cleavage of the initiator Met during expression in E. coli and Ser 103 is located in the loop exhibiting the highest B-factors in the X-ray crystal structure. Resonances originating from several of the remaining residues listed in Table 2 were assigned with the aid of samples selectively labeled with 15N-Val, 15N-Thr, 15N-Leu, or 15N-Tyr and 2D 1H-15N HSQC experiments tailored to detect fast relaxing signals (37), in a manner similar to that used for the assignment of fast relaxing signals in heme oxygenase (18) and FPR (38) from Pseudomonas aeruginosa. For example, the HSQC spectrum of 15N-Val HasAp acquired with standard conditions (Figure 6-A) displays 8 cross-peaks corresponding to previously assigned Val residues. The isotopic label in 15N-Val is partially scrambled to Ala because the 1H-15N HSQC spectrum of 15N-Val HasAp acquired with standard conditions also shows cross-peaks with relative low intensity that correspond to all Ala residues in the sequence. The latter can be ascertained with confidence because cross-peaks corresponding to all Ala residues in the sequence had been previously assigned with the aid of conventional 3-D experiments. In comparison, the HSQC spectrum acquired with the same sample but with fast pulse repetition (Figure 6-B) exhibits 10 cross-peaks corresponding to Val residues; that is two additional cross-peaks are observed under these conditions relative to the spectrum acquired with standard conditions. The new cross-peak resonating at δ (1H) = 7.85 ppm and δ (15N) = 122.3 ppm corresponds to Val37 because it can be correlated to Gln36 with the aid of data from the suite of triple resonance experiments acquired with a sample of [U-13C, U-15N]-HasAp (Figure 6-C). Hence, the other cross-peak observed under fast repetition conditions, δ (1H) = 8.59 ppm and δ (15N) = 131.0 ppm can be safely assigned to the only Val residue left unassigned, that is, to Val38.
Table 2.
Distance from backbone NH to heme-iron for residues not assigned with the aid of conventional 2-D and 3-D NMR experiments.
| Residue | Distance from heme-iron (Å) |
|---|---|
| His32 | 7.70 |
| Arg33 | 6.92 |
| Val37a | 8.10 |
| Val38a | 9.29 |
| Thr43a | 7.36 |
| Thr76 | 8.01 |
| Leu77 | 6.71 |
| His83 | 6.52 |
| Leu85 | 7.92 |
Assigned with the aid of HSQC spectra acquired with fast repetition conditions.
Figure 6.
1H-15N HSQC spectra of [15N-Val]-truncated-HasAp acquired with standard conditions (A) 128 (t1)×1632 (t2) complex points; spectral width, 2.4 kHz (t1)×9.60 kHz (t2); acquisition time, 85 ms; recycle delay, 1 s; 32 scans per increment. (B) fast recycling conditions; 128 (t1)×1050 (t2) complex points; spectral width, 3.6 kHz (t1)×15 kHz (t2); acquisition time, 35 ms; recycle delay, 50 ms; 256 scans per increment. (C) Strip plots showing heteronuclear correlations of residues Gln36 and Val37 in: HNCA, a) and e); HN(CO)CA, b) and f); CBCA(CO)NH, c) and g); and HNCACB, d) and h) spectra obtained with [U-13C, U-15N]-truncated-HasAp. Blue lines are used to highlight HN to Ciα and Cα i-1 correlations in HNCA strips, whereas red lines illustrate correlations between HN of both Gln 36 and Val 37 to the Cα and Cβ cross-peaks of Gln 36.
Incorporation of 15N-Thr into HasAp using the conditions described above resulted in minimum isotopic scrambling, as can be seen in the 1H-15N-HSQC spectrum obtained from 15N-Thr- HasAp using standard acquisition conditions (Figure S5-A). The 1H-15N-HSQC spectrum acquired with fast pulse recycling (Figure S5-B) reveals the presence of only one additional cross-peak (δ (1H) = 6.87 ppm and δ (15N) = 105.7 ppm) relative to the spectrum obtained with standard conditions. This cross-peak was assigned to Thr43 on the basis of correlations to Asn42 and Gly44 in the suite of spectra acquired with [U-13C, U-15N]-HasAp (Figure S5-C).
Truncated HasAp was also labeled with 15N-Tyr in an effort to confirm the sequential assignment of the Tyr75 axial ligand. Relatively minor isotopic scrambling to Phe, Gln and Asp was observed under the present experimental conditions. Nevertheless, cross-peaks corresponding to Tyr residues are intense and allowed confirmation of the Tyr75 assignment. The spectrum obtained with 15N-Tyr-HasAp and fast repetition conditions (Figure S6-B) provided valuable unexpected information: It showed the presence of an additional low-intensity cross-peak (δ (1H) = 10.4 ppm and δ (15N) = 122.1 ppm) corresponding to Tyr138, which was sequentially assigned with the aid of 3-D experiments, as described above. In addition, the spectrum acquired with fast pulse recycling showed that Try75 also exhibits two cross-peaks, indicating that this residue is also conformationally disordered in HasAp.
The HSQC spectrum of [15N-Leu]-HasAp acquired with fast repetition conditions shows the presence of two additional cross-peaks, relative to those observed in the HSQC spectrum of the same sample acquired with standard conditions. Efforts to sequentially assign these cross-peaks, which correspond to Leu77 and Leu85, were unsuccessful most likely because the fast relaxation imparted by their proximity to the heme-iron prevents observation of correlations in the suite of 3-D spectra acquired with [U-13C, U-15N]-HasAp. Finally, it should be noted that cross-peaks originating from axial ligand His32 or from His83, which in the crystal structure of HasAs and HasAp forms a hydrogen bond to the phenolate oxygen of Tyr75, have not been identified. Selective labeling with His was not attempted due to cost considerations, thus it is not known whether the same strategy would have facilitated the identification and sequential assignment of cross-peaks originating from these residues.
Conformational disorder of residues lining the heme pocket of hasAp
As indicated above, HSQC spectra acquired with fast pulse recycling from amino acid-selective labeled samples revealed that Tyr75 and Tyr138 exhibit double cross-peaks. The possibility of finding additional residues exhibiting multiple cross-peaks was investigated by acquiring 1H-15N-HSQC spectra with fast pulse recycling using a sample of HasAp uniformly labeled with 15N. Analysis of this spectrum allowed identification of several residues that exhibit double cross-peaks; these are listed in Table S3 and highlighted in blue in the structure of HasAp shown in Figure 7. The large majority of residues exhibiting double cross-peaks are located in the extended loop containing His32, in the hairpin containing Tyr75 and in helix α2 and strand β2, both of which form the “back wall” of the heme pocket. It is important to underscore that the doubling of cross-peaks observed for these residues is not a consequence of heme rotational isomerism, as is sometimes observed with proteins binding heme b, because the heme in HasAp exists in only one orientation (see above). The doubling of cross-peaks is therefore a consequence of at least two backbone conformations associated with the corresponding residues that are in slow exchange relative to the chemical shift timescale. These observations indicate that the heme pocket or at least a significant component of it (residues in blue in Figure 7) is conformationally heterogeneous. In this context, it is interesting that similar sections of the structure exhibit larger than average thermal factors in the X-ray crystal structure of HasAp (see Figure 4). It is therefore conceivable that this trait may be important in the capture and delivery of heme, as well as in imparting the hemophore with a large affinity for heme.
Figure 7.
A view of the structure of HasAp where residues exhibiting double cross-peaks in the 1H-15N HSQC spectrum of [U-15N]-truncated HasAp acquired with fast repetition conditions have been highlighted in blue. The heme is shown in red.
Disorder in the carboxyl-terminal tail of full-length HasAp
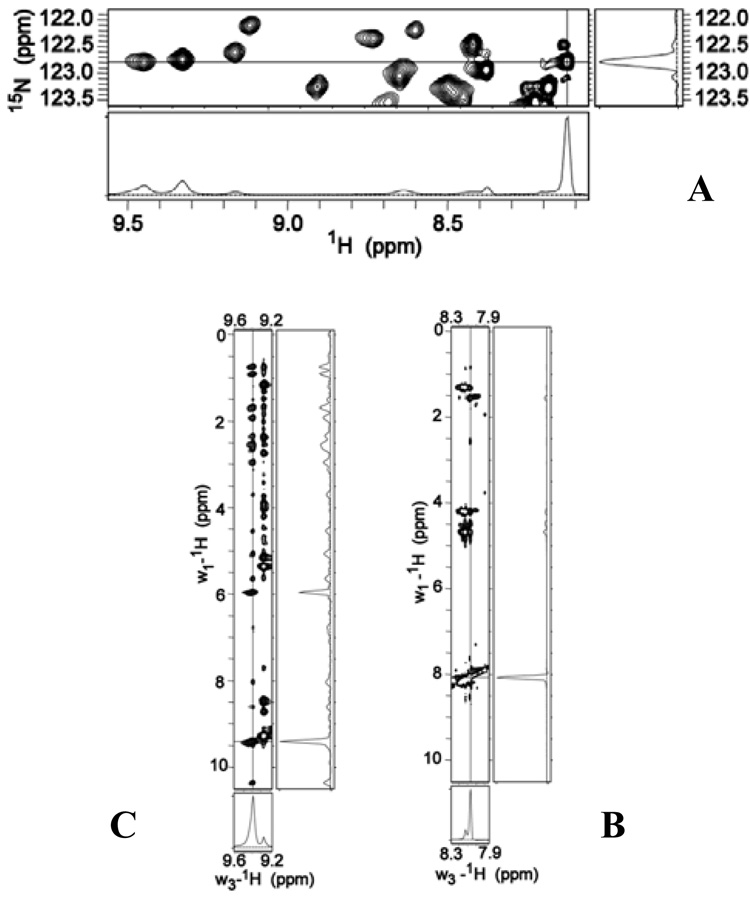
Close inspection of the 1H-15N HSQC spectrum of full-length HasAp shows that the cross-peaks corresponding to the 21 C’-terminal residues of the tail are more intense than cross-peaks originating from the remaining residues in the protein. This observation is illustrated in the 1-D slice taken at the cross-peak corresponding to Leu201, with coordinates δ(1H) = 8.13 ppm and δ(15N) = 122.8 ppm: The peak corresponding to Leu201 was chosen because it does not overlap with other cross-peaks and because it shares the same 15N frequency with two other cross-peaks exhibiting 1H frequencies at 9.34 and 9.46 ppm (see Figure 8-A). Comparison of these three 1H peaks in the slice makes it evident that the 1H-peak from Leu201 (8.13 ppm), which is located in the tail, is narrower and more intense than the other two 1H peaks (corresponding residues not in the tail) in the slice. Because the linewidth at half-height of NMR peaks is inversely proportional to transverse relaxation time T2 (Δν½ = 1/πT2 *), the sharper peaks corresponding to the C’-terminal tail strongly suggest that this section of holo-HasAp exhibits increased motional flexibility relative to the rest of the protein. Additional evidence suggesting that the C’-terminal tail of full-length HasAp does not pack against the structure of the protein can be seen in the 1H-1H 2D slice obtained at δ (15N) = 122.8 ppm of a 3D NOESY-HSQC. The diagonal cross-peak corresponding to Leu201 (δ(1H) = 8.13) exhibits only a few NOE correlations (Figure 8-B), most likely because the corresponding N-H is removed in space from other 1H nuclei in the protein or because a highly dynamic behaviour inhibits NOE build-up. Hence the few NOEs observed for the Leu201 N-H likely originate from intraresidue correlations or from correlations to residues adjacent in sequence. In contrast the diagonal cross-peak corresponding to Tyr56, at δ(1H) = 9.34 ppm (Figure 8-C) exhibits many NOE correlations, as is typical of 1H nuclei in structured proteins. Hence, the inefficient transverse relaxation and small number of NOE connectivities associated with N-H nuclei in the C’-terminal tail strongly suggest that this segment of full-length HasAp is highly mobile and unstructured.
Figure 8.
(A) A portion of the 1H-15N HSQC spectrum obtained from [U-15N]-full-length HasAp. The slices were obtained at the cross-peak corresponding to Leu201 (δ(1H) = 8.13 ppm and δ(15N) = 122.8 ppm). The intensity of this N-H hydrogen cross-peak is significantly larger than the intensity of two other N-H hydrogens attached to nitrogens resonating at the same 15N frequency. These are the protons at 9.34 and 9.46 ppm. (B) A 1H-1H 2D slice obtained from a 3D NOESY HSQC (tmix = 110 ms) spectrum of full-length HasAp. The slice was obtained at the 15N frequency of Leu201 and shows the few NOEs detected for the corresponding cross-peak at δ(1H) = 8.13 ppm. (C) Same as B, except that the cross-peak corresponding to Tyr56 at δ(1H) = 9.34 ppm shows several NOEs, as is typical of cross-peaks in structured proteins.
NMR Spectral Changes Upon Titration of holo-HasAp with Hemoglobin
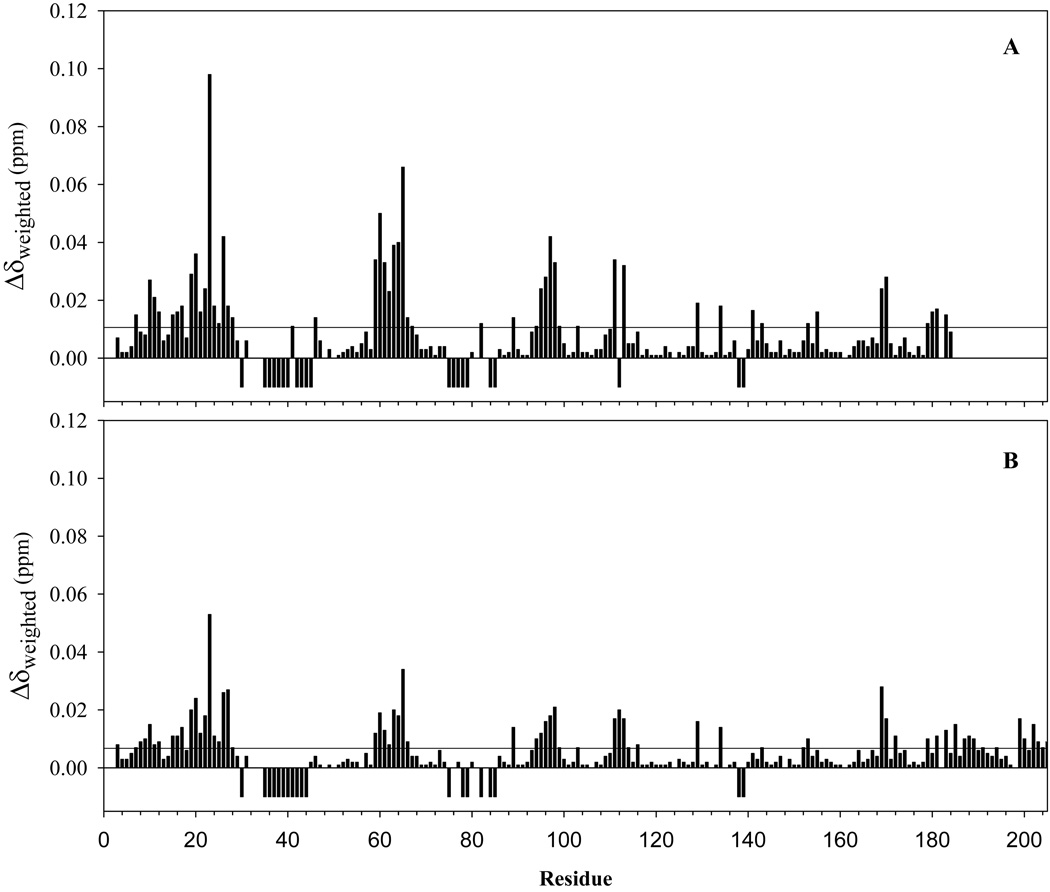
Attempts to study the interactions between apo-HasAp and met-Hb result in rapid heme uptake by the hemophore and precludes most experimental approaches aimed at mapping the surface of interaction between these proteins. In an effort to circumvent these limitations and gain insight into potential protein-protein interactions leading to heme capture we titrated truncated U-15N-holo-HasAp with met-Hb, while monitoring the chemical shift perturbations of backbone N-H resonances with the aid of 1H-15N HSQC spectra. Cross-peaks affected by the titration can be categorized in three groups: (1) Cross-peaks that do not show appreciable chemical shift perturbation (Δδweighed), (2) cross-peaks that shift as the titration progresses; the perturbations in this category are in fast exchange, i.e., Δδmax < kex , and (3) cross-peaks that decrease in intensity and disappear as the titration progresses; the perturbations in this category are in the intermediate exchange regime, where Δδmax ~ kex. These results are summarized in Table 3 and in the per-residue plot of average chemical shift perturbations depicted in Figure 9-A, where residues in categories 1 and 2 are represented by positive bars and residues in category 3 by negative bars; absence of a bar for a particular residue indicates either a Pro, lack of assignment, or peak overlap that prevent its analysis.
Table 3.
Residues exhibiting cross-peak perturbations upon titration of holo-15N-HasAp with Hb
| Residues affected | in holo-HasApa | Equivalent residues | in apo-HasAsa |
|---|---|---|---|
| Category 2 | Category 3 | Category 2 | Category 3 |
| T10 | D29 | S10 | D29 |
| Y19 | N31 | Y19 | N31 |
| L20 | G35 | L20 | G35 |
| D22 | Q36 | Q22 | N36 |
| W23 | V37 | W23 | V37 |
| Y26 | V38 | T26 | T38 |
| K59 | D39 | S58 | D39 |
| S60 | G40 | S59 | A40 |
| T61 | N42 | T60 | N41 |
| S63 | T43 | N62 | S43 |
| D64 | G45 | Q63 | G44 |
| A65 | Y75 | V64 | Y75 |
| G95 | T76 | G95 | T76 |
| D96 | L77 | D96 | L77 |
| T97 | F78 | G97 | F78 |
| L98 | S79 | L98 | N79 |
| S111 | T84 | V110 | T84 |
| E113 | L85 | D112 | L85 |
| D170 | Q112 | D167 | P111 |
| Y138 | Y137 | ||
| G139 | G138 |
Sequence numbering as in Figure 1
Figure 9.
Per residue plot of cross-peak perturbations associated with the titration of (A) truncated- [U-15N]-HasAp and (B) full-length-[U-15N]-HasAp with met-Hb. The weighted values of chemical shift perturbations were obtained from . The average of all chemical shift perturbations is 0.011 in (A) and 0.0067 in (B) The two plots are presented with the same vertical scale two illustrate that the chemical shift perturbations are larger in the case of truncated HasAP. The negative peaks correspond to residues whose resonances disappear in the course of the titration.
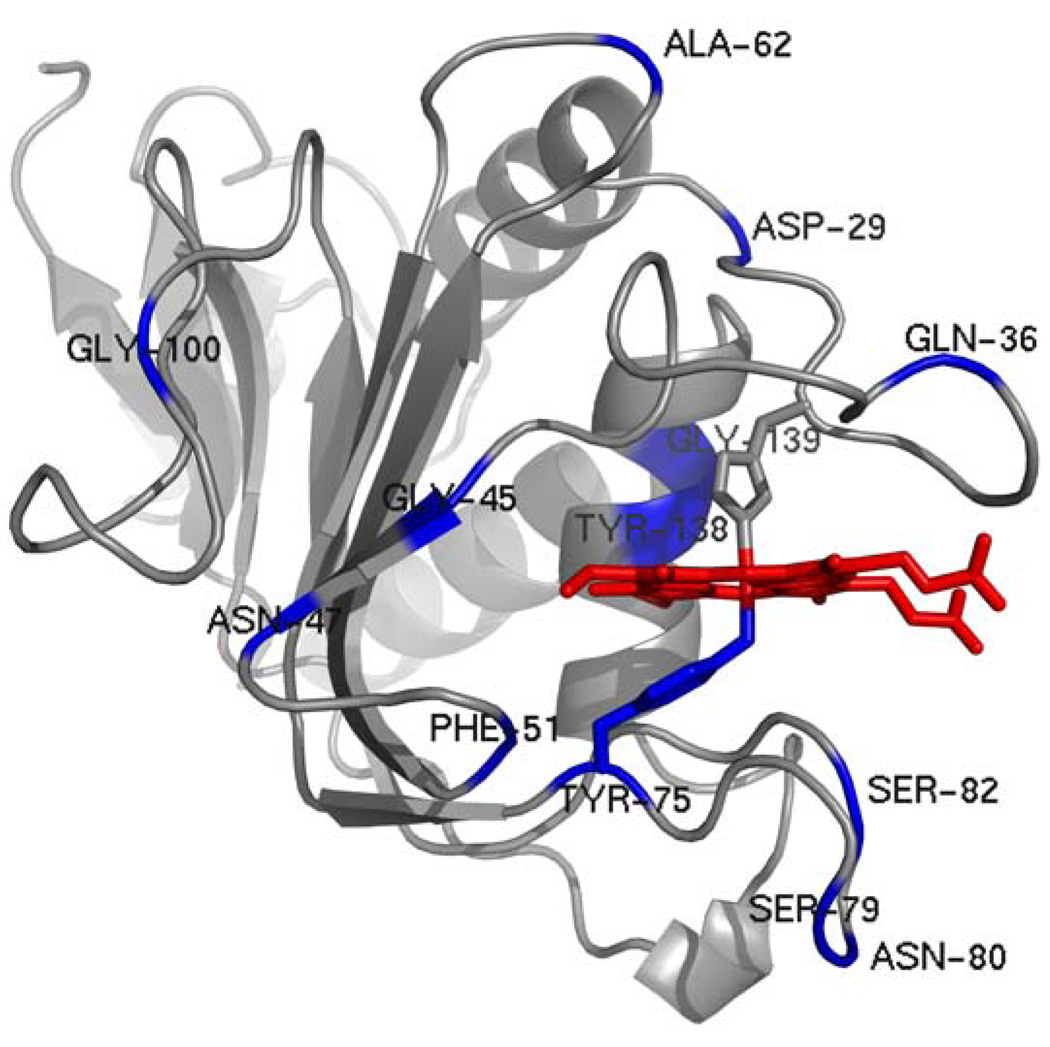
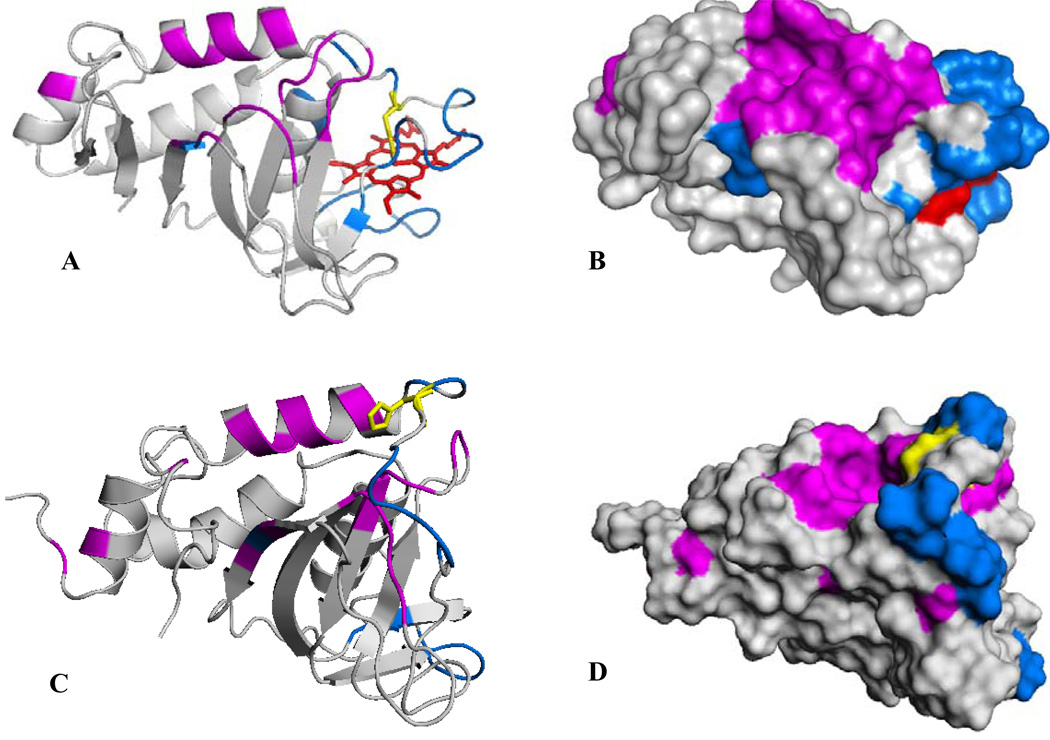
Residues exhibiting chemical shift perturbations (category 2) with Δδavg > 0.03 (3 times the average of all Δδweighed) have been highlighted magenta on the structure of holo-HasAp and resonances whose corresponding cross-peaks disappear as the titration progresses (category 3) have been highlighted in blue (Figure 10-A). It can be observed that the portions of HasAp affected by the presence of met-Hb are concentrated on the loops containing both axial ligands, helix α2 (the back wall of the heme pocket), helix α1 and on the hairpins connecting β3 to β4 and β6 to β7. A surface rendering representation of the perturbations shown in Figure 10-A, with identical view and color coding, is presented in Figure 10-B. The surface representation shows that residues in categories 2 and 3 are nearly perfectly segregated into distinct parts of a nearly contiguous surface; residues exhibiting cross-peaks in category 3 (blue) are localized mainly on the loop containing His32 and in portions of the loop harboring Tyr75, and residues exhibiting cross-peaks in category 2 (magenta) are localized on helix α2 and on the hairpins connecting β3 to β4 and β6 to β7. The occurrence of two surfaces that differ in the timescale of their corresponding cross-peaks (i.e. categories 2 and 3) can be interpreted as (a) surfaces that interact with met-Hb with distinct binding affinities or (b) a single binding event in which the binding of the surface exhibiting cross-peaks affected by chemical shift perturbations (magenta in Figure 10-B) triggers the acceleration of backbone motions in residues comprising the axial ligand-containing loops (blue in Figure 10-B).
Figure 10.
(A) A view of the structure of truncated holo-HasAp where residues whose cross-peaks exhibit weighted chemical shift perturbations larger than three times the average of all chemical shifts (0.011) upon titration with met-Hb are highlighted in magenta and residues whose corresponding cross-peaks disappear upon addition of met-Hb are highlighted in blue. His32 is shown in yellow. (B) Surface representation of the view shown in (A). (C) A view of the structure of apo-HasAs (PDB ID 1YBJ) (45), where residues highlighted in magenta are equivalent to residues in holo-HasAp whose corresponding cross-peaks are affected by chemical shift perturbations and residues highlighted in blue are equivalent to those in holo- HasAp whose cross-peaks disappear upon titration with met-Hb. (D) Surface representation of the view shown in (C).
A similar experiment was carried out by titrating met-Hb into a solution of full-length holo HasAp labeled uniformly with 15N. The results were very similar to those obtained with truncated holo-HasAp. Indeed, the per-residue plot obtained from titration of full-length holo-HasAp with met-Hb (Figure 9-B) shows a pattern of chemical shift perturbations and peaks that disappear during the titration nearly identical those observed upon titration of truncated holo-HasAp with met-Hb (Figure 9-A). However, the magnitude of the chemical shift perturbations corresponding to residues in category 2 is uniformly smaller in full-length HasAp, with Δδavg of 0.067, compared to 0.011 in truncated HasAp. These observations suggest that the disordered tail in full-length HasAp partially interferes with its binding to met-Hb, thus giving rise to the smaller chemical shift perturbations. It is therefore possible that cleavage of the tail by proteolysis in the extracellular milieu makes HasAp more efficient at sequestering heme from hemoglobin.
DISCUSSION
The two structurally characterized hemophores, HasAs (9) and HasAp share ~ 50% sequence identity (see Figure 1). Nevertheless their three-dimensional structures are nearly identical, indicating that the αβ fold observed first in HasAs is characteristic of hemophores and that the axial coordination of the heme by His32 and Tyr75 is also typical of this family of proteins. The structure of HasAp also shows the imidazole side chain of His83 within hydrogen bonding distance of the phenolic group in Tyr75, and is consistent with the notion that the side chain of His83 functions as a hydrogen bond acceptor that stabilizes a coordinative tyrosinate state of the Tyr75 axial ligand (28). The triad His32, Tyr75 and His83 may become a recurring structural motif in hemophores, despite the fact that His/Tyr axial coordination has been rarely observed thus far. NMR and resonance Raman spectroscopic investigations of HasAs in solution led to the conclusion that the heme-iron is in fast exchange between 6-coordinate high- and low-spin states (28, 36). The spectroscopic studies presented above also establish a six-coordinate high-spin/low-spin equilibrium in HasAp that may be linked to tautomers of the hydrogen bond interaction between the coordinating Tyr75 phenolate group and the Nδ of His83.
The multiplicity of HSQC cross-peaks corresponding to the backbone of several residues lining the heme suggests that the heme binding site of HasAp is inhomogeneous and dynamic. In this context, it is noteworthy that recent investigations have shown that protein conformational heterogeneity and dynamics constitute conformational entropy in a ligand-protein complex, which contributes significantly to the high-affinity of the complex (39). In other words, conformational disorder in a protein-ligand complex makes the normally unfavorable entropic contribution to binding, less unfavorable, thus increasing binding affinity. This idea has been used to introduce the concept of induced disorder in drug design, whereby a boost of conformational entropy has been engineered into the Bcr-Abl kinase-drug complex that significantly increases its stability (40). In this frame, it is not unreasonable to postulate that the conformational disorder of the heme active site in HasAp contributes to the very high binding affinities exhibited by the heme-hemophore complex (41), despite the fact that the heme is largely exposed to the aqueous environment. Clearly, additional investigations aimed at correlating plasticity in the heme binding site of HasAp and mutants with corresponding heme binding constants are needed to test this notion and to investigate the nature of the high binding affinity of the heme-hemophore complex.
Hemoglobin HasAp interactions
Attempts to study possible protein interactions between apo-HasAp and met-Hb by NMR spectroscopy were stymied by the relatively rapid transfer of heme to the hemophore, which results in the formation of significant concentrations holo-HasAp in the dead time of the experiment (approximately 15 min). It was therefore decided to monitor interactions between holo-HasAp and met-Hb in an effort to gain some insights regarding potential proteinprotein interactions leading to heme capture by hemophores. Observations made while performing these experiments demonstrate selective interactions between met-Hb and holo-HasAp (see Figure 9 and Figure 10). These findings are surprising and significant because previous studies carried out with the aid of analytical centrifugation and co-immunoprecipitation techniques showed that HasAs does not form a stable complex with Hb. This observation led to the assumption that heme is transferred passively from Hb to the hemophore, via a mechanism where heme released naturally by hemoglobin is trapped by the hemophore due to its higher heme binding affinity, rather than being transferred in a hemophore-Hb complex (10, 42). Although analytical centrifugation and immunoprecipitation are very useful experimental approaches to study long-lived complexes such as that formed between HasAs and its outer-membrane receptor (HasR) (10), these techniques are not well suited to detect the formation of transient complexes. NMR spectroscopy, on the other hand, is commonly used to investigate transient inter-protein complexes (43) and the results obtained from the NMR spectroscopic studies reported herein indicate that holo-HasAp undergoes transient protein-protein interactions with met-Hb. Analysis of the titration data revealed cross-peaks that display chemical shift perturbations in fast exchange that originate from residues highlighted in magenta in Figure 10-B, and cross peaks that broaden and disappear, which correspond to residues highlighted in blue. It is striking that residues in each of these surfaces, whose resonances are affected differently by addition of Hb, are segregated into two surfaces. Consequently, it is of interest to consider the distinct relaxation properties of cross-peaks from residues highlighted in magenta and blue in the context of a complex between holo-HasAp and met-Hb. One possible interpretation suggests that binding of holo-HasAp to met-Hb occurs mainly via the surface highlighted in magenta, which triggers the chemical shift perturbations affecting the cross-peaks corresponding to these residues. The binding process, in turn, induces enhanced conformational dynamics in the His-32 loop, therefore giving rise to the line broadening and concomitant disappearance of cross-peaks corresponding to residues highlighted in blue. The dynamic process giving rise to the enhanced relaxation of cross-peaks corresponding to these residues could be a consequence of frustrated attempts at changing the conformation of the His32 loop from “closed” to “open” upon sensing the Hb surface. These conformational rearrangements are restrained (frustrated) by the His32 to iron coordination bond. This view, which implicates His32 in the capture of heme from met-Hb is in agreement with recent observations in which CO has been shown to displace the His32 ligand, giving rise to a Tyr-Fe-CO axial coordination (44), because the displacement of His32 and not the axial Tyr by CO is in agreement with a relatively labile His-32 to iron coordination bond in the hemophores. An alternative interpretation of the titration data would suggest that holo-HasAp interacts with Hb using two distinct surfaces, each interacting with a different binding affinity. It is interesting that this interpretation of the titration data also highlights selective areas on the surface of holo-HasAp used to interact with met-Hb, which is also in agreement with the notion that His32 and the loop harboring it may play an important role in an encounter complex between apo-HasAp and met-Hb that leads to heme transfer.
Although the relevance of the interactions observed with holo-HasAp to the complex between apo-HasAp and met-Hb is not straightforward, it is possible to gain potentially important insight by considering the titration data obtained with holo-HasAp in the context of the recently determined structure of apo-HasAs (45). The structure of apo-HasAs (Figure 10-C) is very similar to that of the holo-protein (Figure 10-A), except for the loop containing His32, which undergoes a large conformational change. Given that the structures of holo-HasAp and holo-HasAs are nearly identical, it seems reasonable to assume that the structure of apo-HasAp will undergo similar conformational rearrangement in its His32-containing loop, while maintaining the rest of the structure unchanged. Within this frame it is interesting to map the chemical shift perturbations and the disappearance of cross-peaks observed in the titration of holo-HasAp with met-Hb on the structure of apo-HasAs. The results of doing this are depicted in Figure 10-C and in surface rendering mode in Figure 10-D; the affected residues are listed in Table 3. As in the case of the holo-protein, residues in apo-HasAs equivalent to those experiencing chemical shift perturbations in holo-HasAp upon addition of met-Hb are highlighted in magenta and residues equivalent to those whose resonances disappear during the titration are shown in blue. The comparison is informative because it shows how the surface of the protein changes as the His32 loop relocates from a “closed” conformation in the holo-protein (Figure 10-B) to an “open” conformation in apo-HasAs (Figure 10-D). The “open” conformation of the His32-containing loop places the surface highlighted in blue close to the surface labeled in magenta and positions His32 (yellow) such that it becomes part of the putative interaction surface in the apo-hemophore. Consequently, it is tempting to hypothesize that the conformation adopted by the His32 loop in the apoprotein plays an important role in heme capture, either by actively utilizing His32 as the initial contact with heme, or alternatively, as a constituent of the recognition surface in an encounter complex between apo-HasAp and Hb. Clearly the notions discussed here must be investigated further in order to establish the formation of a transient complex between apo-HasAp and Hb and to determine the role played by the His32 loop in binding met-Hb and in “stealing” its heme.
Supplementary Material
DNA sequence of HasAp engineered with silent mutations to include E. coli-favored codons, schematic representation of the heme binding site in HasAp, UV-vis, 1-D 1H NMR, resonance Raman and EPR spectra of HasAp, HSQC spectra of 15N-Thr-HasAp and 15N-Tyr-HasAp acquired with standard and fast repetition conditions and tables of backbone NMR assignments for truncated and full-length HasAp. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENT
We thank Dr. James Whitaker for the use of his helium cryostat during EPR experiments.
Footnotes
This work was supported by grants from the National Institute of Health, GM-50503 (M.R.), NSF-MCB-0818488 (M.R.) and NSF-MCB-0811888 (P.M.L.).
Coordinates and crystallographic structure factors for HasAp have been deposited in the protein data bank under accession code 3ELL. Backbone resonance assignments for truncated and full-length holo-HasAp have been deposited in the BMRB under access code 15962 and 15963, respectively.
REFERENCES
- 1.Wandersman C, Stojiljkovic I. Bacterial Heme Sources: The Role of Heme, Hemoprotein Receptors and Hemophores. Curr. Oppin. Microbiology. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 2.Genco CA, Dixon DW. Emerging Strategies in Microbial Haem Capture. Mol. Microbiol. 2001;39:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 3.Ochsner UA, Johnson Z, Vasil AI. Genetics and Regulation of Two Distinct Haem-Uptake Systems, phu and has in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 4.Wandersman C, Delepelaire P. Bacterial Iron Sources: From Siderophores to Hemophores. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 5.Berka RM, Vasil ML. Phospholipase C (Heat-Labile Hemolysin) of Pseudomonas aeruginosa: Purification and Preliminary Characterization. J. Bacteriol. 1982;152:239–245. doi: 10.1128/jb.152.1.239-245.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard A, Vasil ML. Nucleotide Sequence and Expression of a Phosphate-Regulated Gene Encoding a Secreted Hemolysin of Pseudomonas aeruginosa. J. Bacteriol. 1986;167:291–298. doi: 10.1128/jb.167.1.291-298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasil ML, Kabat D, Iglewski BH. Structure-Activity Relationships of an Exotoxin of Pseudomonas aeruginosa. Infect. Immun. 1977;16:353–361. doi: 10.1128/iai.16.1.353-361.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. Homologues of Neisserial Heme Oxygenase in Gram-Negative Bacteria: Degradation of Heme by the Product of the pigA Gene of Pseudomonas aeruginosa. J. Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjek M. The Crystal Structure of HasA, a Hemophore Secreted by Serratia marcescens. Nature Structural Biology. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 10.Létoffé S, Nato F, Goldberg ME, Wandersman C. Interactions of HasA, a Bacterial Haemophore, with Haemoglobin and with its Outer Membrane eceptor HasR. Mol. Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 11.Ghigo JM, Léttofé S, Wandersman C. A New Type of Hemophore-Dependent Heme Acquisition System of Serratia Marcescens Reconstituted in Escherichia coli. J. Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Létoffé S, Redeker V, Wandersman C. Isolation and Characterization of an Extracellular Haem-Binding Protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia Marcescens HasA Haemophore. Mol. Microbiol. 1998;28:1223–1224. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 13.Arevalo-Ferro C, Hentzer M, Reil G, Gorg A, Kjelleberg S, Givskov M, Riedel K, Eberl L. Identification of Quorum-Sensing Regulated Proteins in the Opportunistic Pathogen Pseudomonas aeruginosa by Proteomics. Environ. Microbiol. 2003;5:1350–1369. doi: 10.1046/j.1462-2920.2003.00532.x. [DOI] [PubMed] [Google Scholar]
- 14.de Kievit TR, Iglewski BH. Bacterial Quorum Sensing in Pathogenic Relationships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikemura T. Codon Usage and tRNA Content in Unicellular and Multicellular Organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 16.Caignan GA, Deshmukh R, Wilks A, Zeng Y, Huang H, Moënne-Loccoz P, Bunce RA, Eastman MA, Rivera M. Oxidation of Heme to β- and δ-biliverdin by Pseudomonas aeruginosa Heme Oxygenase as a Consequence of an Unusual Seating of the Heme. J. Am. Chem. Soc. 2002;124:14879–14892. doi: 10.1021/ja0274960. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Westler WM, Xia B, Oh B-H, Markley JL. Protein Expression, Selective Isotopic Labeling, and Analysis of Hyperfine-Shifted NMR Signals of Anabaena 7120 Vegetative [2Fe-2S]Ferredoxin. Arch. Biochem. Biophys. 1995;316:619–634. doi: 10.1006/abbi.1995.1082. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez JC, Wilks A, Rivera M. Backbone NMR Assignments and H/D Exchange Studies on the Ferric Azide- and Cyanide-Inhibited Forms of Pseudomonas aeruginosa Heme Oxygenase. Biochemistry. 2006;45:4578–4592. doi: 10.1021/bi0600188. [DOI] [PubMed] [Google Scholar]
- 19.Eakanunkul S, Lukat-Rodgers GS, Sumithran S, Ghosh A, Rodgers KR, Dawson JH, Wilks A. Characterization of the Periplasmic Heme-Binding Protein ShuT from the Heme Uptake System of Shigella dysenteriae. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 20.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A Deliberate Approach to Screening for Initial Crystallization Conditions of Biological Macromolecules. J. Struct. Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constraints. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Jones TA, Zhou JY. Improved Methods for Binding Protein Models in Electron Density Maps and the Location of Errors in These Models. Acta Crystallogr Section A: Foundations of Crystallography. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Rosi MS, Fetherston JD, Létoffé S, Perry RD, Ghigo JM. Identification and Characterization of the Hemophore-Dependent Heme Acquisition System of Yersinia Pestis. Infect. Immun. 2001;69:6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnoux P, Haser R, Izadi-Pruneyre N, Lecroisey A, Czjek M. Functional Aspecs of the Heme Bound Hemophore HasA By Structural Analysis of Various Crystal Forms. Proteins: Struct. Funct. Genet. 2000;41:202–210. doi: 10.1002/1097-0134(20001101)41:2<202::aid-prot50>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Rivera M, Barillas-Mury C, Christensen KA, Little JW, Wells MA, Walker FA. Gene Synthesis, Bacterial Expression, and 1H NMR Spectroscopic Studies of the Rat Outer Mitochondrial Membrane Cytochrome b5. Biochemistry. 1992;31:12233–12240. doi: 10.1021/bi00163a037. [DOI] [PubMed] [Google Scholar]
- 27.Lee KB, Jun E, La Mar GN, Rezzano IN, Ravindra KP, Smith KM, Walker FA, Buttlaire DH. Influence of Heme Vinyl- and Carboxylate-Protein Contacts on Structure and Redox Properties of Bovine Cytochrome b5. J. Am. Chem. Soc. 1991;113:3576. [Google Scholar]
- 28.Caillet-Saguy C, Delepierre M, Lecroisey A, Bertini I, Piccioli M, Turano P. Direct-Detected 13C NMR to Investigate the Iron(III) Hemophore HasA. J. Am. Chem. Soc. 2006;128:150–158. doi: 10.1021/ja054902h. [DOI] [PubMed] [Google Scholar]
- 29.Altuve A, Silchenko S, Lee KH, Kuczera K, Terzyan S, Zhang X, Benson DR, Rivera M. Probing the Differences Between Rat Liver Outer Mitochondrial Membrane Cytochrome b5 and Microsomal Cytochromes b5. Biochemistry. 2001;40:9469–9483. doi: 10.1021/bi010636i. [DOI] [PubMed] [Google Scholar]
- 30.Cowley AB, Altuve A, Kuchment O, Terzyan S, Zhang X, Rivera M, Benson DR. Toward Engineering the Stability and Hemin-Binding Properties of Microsomal Cytochromes b5 int Rat Mitochondrial Membrane Cytochrome b5: Examining the Influence of Residues 25 and 71. Biochemistry. 2002;41:11566–11581. doi: 10.1021/bi026005l. [DOI] [PubMed] [Google Scholar]
- 31.Barjon C, Wecker K, Izadi-Pruneyre N, Delepelaire P. Mutagenesis and Molecular Modeling Reveal Three Key Extracellular Loops of the Membrane Receptor HasR that are Involved in Hemophore HasA Binding. J. Bacteriol. 2007;189:5379–5382. doi: 10.1128/JB.00251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izuka T, Yonetani T. Spin Changes in Hemoproteins. Advan. Biophys. 1970;1:157–182. [PubMed] [Google Scholar]
- 33.Walker FA, Simonis U. Proton NMR Spectroscopy of Model Hemes. In: Berliner LJ, Reuben J, editors. Biological Magnetic Resonance. New York: Plenum Press; 1993. pp. 133–274. [Google Scholar]
- 34.La Mar GN, Satterlee JD, De Ropp JS. Nuclear Magnetic Resonance of Hemoproteins. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. Academic Press; 2000. pp. 185–297. [Google Scholar]
- 35.Izadi-Pruneyre N, Huché F, Lukat-Rodgers GS, Lecroisey A, Gilli R, Rodgers KR, Wandersman C, Delepelaire P. The Heme Transfer from the Soluble HasA Hemophore to Its Membrane-bound Receptor HasR is Driven by Protein-Protein Interactions from a High to a Lower Affinity Binding Site. J. Biol. Chem. 2006;281:25541–25550. doi: 10.1074/jbc.M603698200. [DOI] [PubMed] [Google Scholar]
- 36.Caillet-Saguy C, Turano P, Piccioli M, Lukat-Rodgers GS, Czjzek M, Guigliarelli B, Izadi-Pruneyre N, Rodgers KR, Delepierre M, Lecroisey A. Deciphering the Structural Role of Histidine 83 for Heme Binding in Hemophore HasA. J. Biol. Chem. 2008;283:5960–5970. doi: 10.1074/jbc.M703795200. [DOI] [PubMed] [Google Scholar]
- 37.Machonkin TE, Westler WM, Markley JL. Strategy for the Study of Paramagnetic Proteins with Slow Electronic Relaxation Rates by NMR Spectroscopy: Application to Oxidized Human [2Fe-2S] Ferredoxin. J. Am. Chem. Soc. 2004;126:5413–5426. doi: 10.1021/ja037077i. [DOI] [PubMed] [Google Scholar]
- 38.Wang A, Rodríguez JC, Han H, Schönbrunn E, Rivera M. X-Ray Crystallographic and Solution State Nuclear Magnetic Resonance Spectroscopic Investigations of NADP+ Binding to Ferredoxin NADP Reductase from Pseudomonas aeruginosa. Biochemistry. 2008;47:8080–8093. doi: 10.1021/bi8007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational Entropy in Molecular Recognition by Proteins. Nature. 2007;448:325–330. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crespo A, Fernández A. Induced Disorder in Protein-Ligan Complexes as a Drug-Design Strategy. Mol. Pharmaceutics. 2008;5:430–437. doi: 10.1021/mp700148h. [DOI] [PubMed] [Google Scholar]
- 41.Deniau C, Gilli R, Izadi-Pruneyre N, Lettofe S, Delepierre M, Wandersman C, Briand C, Lecroisey A. Thermodynamics of Heme Binding to the HasASM Hemophore: Effect of Mutations at the Three Key Residues for Heme Uptake. Biochemistry. 2003;42:10627–10633. doi: 10.1021/bi030015k. [DOI] [PubMed] [Google Scholar]
- 42.Cescau S, Cwerman H, Léttofé S, Delepelaire P, Wandersman C, Biville F. Heme Acquisition by Hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 43.Zuiderweg ERP. Mapping Protein-Protein Interactions in Solution by NMR Spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 44.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel Heme Ligand Displacement by CO in the Soluble Hemophore HasA and Its Proximal Ligand Mutants: Implications for Heme Uptake and Release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 45.Wolff N, Izadi-Pruneyre N, Couprie J, Habeck M, Linge J, Rieping W, Wandersman C, Nilges M, Delepelaire P, Lecroisey A. Comparative Analysis of Structural and Dynamic Properties of the Loaded and Unloaded Hemophore HasA: Functional Implications. J. Mol. Biol. 2008;376:517–525. doi: 10.1016/j.jmb.2007.11.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequence of HasAp engineered with silent mutations to include E. coli-favored codons, schematic representation of the heme binding site in HasAp, UV-vis, 1-D 1H NMR, resonance Raman and EPR spectra of HasAp, HSQC spectra of 15N-Thr-HasAp and 15N-Tyr-HasAp acquired with standard and fast repetition conditions and tables of backbone NMR assignments for truncated and full-length HasAp. This material is available free of charge via the Internet at http://pubs.acs.org.