Abstract
Background
IL-13, a critical cytokine in allergy, is regulated by as-yet-elusive mechanisms.
Objective
We investigated IL-13 posttranscriptional regulation by HuR, a protein associating with adenylate-uridylate-rich elements in the 3′ untranslated regions (UTRs) of mRNA, promoting mRNA stability and translation.
Methods
IL-13 mRNA decay was monitored in human TH2-skewed cells by using the transcriptional inhibitor actinomycin D. The IL-13 3′UTR was subcloned into an inducible β-globin reporter transiently expressed in H2 cells in the absence or presence of overexpressed HuR. Association of HuR with IL-13 mRNA was detected by means of immunoprecipitation of ribonucleoprotein complexes and a biotin pull-down assay. The effects of HuR transient overexpression and silencing on IL-13 expression were investigated.
Results
IL-13 mRNA half-life increased significantly in restimulated TH2-skewed cells compared with baseline values. Decay of β-globin mRNA was significantly faster in H2 cells transfected with the IL-13 3′UTR-containing plasmid than in those carrying a control vector. HuR overexpression increased the β-globin IL-13 3′UTR reporter half-life. Significant enrichment of IL-13 mRNA was produced by means of immunoprecipitation of Jurkat cell ribonucleoprotein complexes with anti-HuR. HuR binding to the IL-13 3′UTR was confirmed by means of pull-down assay of biotin-labeled RNA probes spanning the IL-13 3′UTR. Two-dimensional Western blot analysis showed stimulus-induced posttranslational modification of HuR. In Jurkat cells mitogen-induced IL-13 mRNA was significantly affected by HuR overexpression and silencing.
Conclusions
Mitogen-induced IL-13 expression involves changes in transcript turnover and a change in phosphorylation of HuR and its association with the mRNA 3′UTR.
Keywords: Asthma, inflammation, mRNA turnover, posttranscriptional gene regulation, RNA-binding proteins, IL-13, TH2 cytokines, T cells
Clinical and experimental studies have clearly established that IL-13 is a key player in the pathophysiology of allergic diseases, in particular of asthma.1
IL-13 is primarily secreted on antigen-mediated stimulation of TH2 lymphocytes and controls a vast array of pathogenic immune and inflammatory processes, such as IgE production, tissue eosinophil recruitment, mucus secretion, airway hyperre-activity, and airway wall remodeling, among others. Therefore identification of the molecular pathways leading to IL-13 overexpression might uncover primary immunomodulatory therapeutic targets.
Studies on the regulation of IL-13 expression in T cells have thus far focused on transcriptional activation and the associated signaling.2,3 However, growing knowledge indicates a key role of posttranscriptional gene regulation after T-cell activation. Essential T-cell costimulatory signals, mediated by CD28 and leukocyte function-associated antigen-1 (LFA-1), critically modulate T cell–derived cytokine expression through changes in mRNA stability.4,5 Earlier genome-wide studies in human T cells indicated that rapid and significant changes in mRNA stability occur for hundreds of early-response genes after mitogenic stimulation.6 A recent array-based comparative analysis of nuclear gene transcription versus total mRNA expression revealed that mRNA turnover accounts for as much as 50% of the mitogen-induced changes in gene expression in the Jurkat T-cell line.7
Despite increasing knowledge of the molecular regulation of mRNA stability of other T-cell products, such as IL-2, IL-4, and CD40 ligand,8-11 almost no information is available on posttranscriptional regulation of IL-13 expression. Butler et al12 report that in activated T cells from the TH2-biased mouse strain DBA/2, increased expression of IL-4 and IL-13 over that seen in the TH1-biased C56BL/6 strain was coupled with increased stabilization of their transcripts. These data indicate that the regulation of TH2 cytokine mRNA stability could also act as an important novel mechanism contributing to the relative prevalence of a TH1- or TH2-driven immune response.
Messenger RNA decay and translation are regulated through binding to elements present in the 5′ and 3′ untranslated regions (UTRs), as well as in the coding regions, of mature cytoplasmic mRNA. The adenylate-uridylate–rich elements (AREs) present in the 3′UTR of many early-response and inflammatory genes are the most conserved and best characterized sequences regulating mRNA turnover and translation. Several proteins exert posttranscriptional control through high-affinity binding to such regions in the context of specific RNA secondary structures.13
HuR is the ubiquitous member of the Hu protein family of RNA-binding proteins.14 It is functionally characterized as a positive regulator of mRNA stability, translation, or both, acting in balance with ARE-binding factors that mediate acceleration of mRNA decay, such as tristetraprolin, adenylate-urydilate–binding factor 1, and KH-type splicing regulatory protein,15 although its activity is pleiotropic.16 Involvement of HuR is increasingly recognized in T-cell gene regulation.9,17,18 T-cell activation triggers shuttling of HuR to the cytoplasm, an event deemed necessary for its regulatory functions.9,18-20 HuR binds to ARE in mRNAs encoding key regulators of proliferation and stress responsiveness,21,22 transcription factors,23 and a growing number of immune and inflammatory genes, such as IL-2, TNF-α, IL-3, and many more.18,19,24-29
The aim of this study was to investigate the contribution of mRNA turnover to IL-13 expression in activated human T cells and to examine the involvement of HuR in this process. Our data indicate that in human T cells mitogen-induced increase in steady-state levels is paralleled by significant stabilization of IL-13 mRNA. We demonstrate that HuR associates with the 3′UTR of IL-13 mRNA and present evidence of a functional role for HuR in regulating IL-13 mRNA levels in this cell type.
Methods
Cell cultures, plasmid constructs, small interfering RNA, and transfection protocols
Blood was drawn from healthy subjects under a protocol approved by a Johns Hopkins Medicine Institutional Review Board. T cells were cultured under TH2-skewing conditions, as described in the Methods section of the Online Repository, which is available at www.jacionline.org.30 Intracellular staining for IL-13, IL-4, and IFN-γ in these cultures30 is described in the Methods section of the Online Repository, and the results are shown therein as Fig E1 (available at www.jacionline.org).
The human Jurkat T-cell line was cultured in RPMI, 10% FBS, 2 mmol/L l-glutamine (Invitrogen, Carlsbad, Calif), and 100 μg/mL gentamicin (Quality Biological, Gaithersburg, Md). The H2 cell line is a clone of the human cell line H1299, a non–small cell lung carcinoma stably transfected with the pTet-Off plasmid (Clontech, Mountain View, Calif),31 and was cultured in Dulbecco's modified Eagle's medium (Invitrogen), 10% FBS, and penicillin (100 U/mL)/streptomycin (100 μg/mL).
Description of the plasmid constructs, small interfering RNA (siRNA), and the protocol for transient transfections can be found in the Methods section of the Online Repository.
RNA isolation and analysis
Total RNA was extracted with TRIzol (Invitrogen).32 Cytoplasmic RNA was isolated and DNase treated with the RNAeasy kit (Qiagen, Valencia, Calif). RNA was reverse-transcribed with the Gene Amp Kit (Perkin Elmer Life Sciences, Boston, Mass) and PCR amplified with the SYBR Green reagent Kit (Perkin Elmer). Northern blot analysis was carried out as previously described.29 The DNA probe was a cDNA encompassing the full-length coding region of rabbit β-globin.33 In the Methods section of the Online Repository are listed the primers for IL-13, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), enhanced green fluorescent protein (EGFP), and β-globin, as well as the methods for the amplification, data collection, and analysis of the PCR results29,34 and the protocol for Northern blot analysis.
In vitro biotin pull-down assay
Biotinylated transcripts were generated by means of RT-PCR of Jurkat RNA by using specific primers (listed in the Methods section of the Online Repository). PCR products were purified from agarose gels, as previously described,21 and used as templates for the synthesis of biotinylated RNAs by using T7 RNA polymerase and biotin-conjugated cytidine 5′-triphosphate. After an established protocol,35 cytoplasmic fractions of unstimulated Jurkat cells (40 μg) were incubated for 1 hour at room temperature with 1 μg of biotinylated transcripts, and then ribonucleoprotein complexes were isolated with streptavidin-conjugated Dynabeads (Invitrogen). The presence of HuR in the pull-down pellet was verified by means of Western blot analysis with the mouse monoclonal anti-HuR antibody 3A2 (Santa Cruz Biotechnology, Santa Cruz, Calif)36 and the enhanced chemiluminescence protocol (GE Healthcare, Piscataway, NJ), as previously described.29
Immunoprecipitation of endogenous messenger ribonucleoprotein complexes
We used a modification29 of an established protocol37 described in detail in the Methods section of the Online Repository for immunoprecipitation (IP) of endogenous messenger ribonucleoprotein complexes (mRNPs).
Two-dimensional Western blot analysis
This assay was performed by using IPG strips (11 cm, pH 7-10; Bio-Rad, Hercules, Calif) for isoelectric focusing (IEF) in the first dimension followed by electrophoresis in SDS-containing gels (4% to 15% acrylamide) in the second dimension, as previously reported38 and described in detail in the Methods section of the Online Repository.
Statistical analysis
Data were analyzed by using the repeated-measures ANOVA test with post-hoc analysis.
Results
To test the hypothesis that IL-13 expression can be regulated at the level of mRNA turnover, we studied the decay of IL-13 mRNA after T-cell activation. IL-4–primed primary CD4+ T cells, henceforth referred to as TH2-skewed cells, were left untreated or restimulated with phorbol 12-myristate 13-acetate (PMA; 50 ng/mL) and the Ca2+ ionophore A23187 (250 ng/mL) alone or in combination for 3 hours. Cells were then either harvested (as time 0) without any additional treatment or further cultured in the presence of the transcriptional inhibitor actinomycin D (3 μg/mL) for 1, 3, or 5 hours. Steady-state levels of IL-13 mRNA were detectable by means of real-time PCR at baseline (cycle threshold [CT] = 27.4 ± 0.4) and increased significantly after cell challenge with PMA (fold over control, 46.04 ± 6.8), the Ca2+ ionophore A23187 (17.1 ± 2.3), and their combination (234.1 ± 67), as expected (Fig 1, A). Treatment with actinomycin D revealed that in unstimulated T cells IL-13 mRNA has a relatively fast turnover; in fact, only 22% of IL-13 mRNAwas detectable within 1 hour from transcriptional termination (Fig 1, B). In contrast, more than 50% of the IL-13 mRNA was still detectable at the same time point after treatment with PMA or A23187. More strikingly, in cells treated with both stimuli, the levels of IL-13 mRNA after 1 hour were still virtually unchanged compared with those at time 0, indicating a rapid and significant stimulus-dependent stabilization of the IL-13 mRNA (P < .002, ANOVA). Accordingly, analysis of the decay curve revealed that half-life of IL-13 mRNA, calculated for each condition as the time (in hours) required for the transcript to decrease to 50% of its initial abundance, was 1.6 hours in unstimulated cells, became longer after either stimulation (3.7 hours after PMA and 3.0 hours after A23187), and was significantly prolonged after combined stimulation (6.3 hours) over unstimulated cells.
FIG 1.
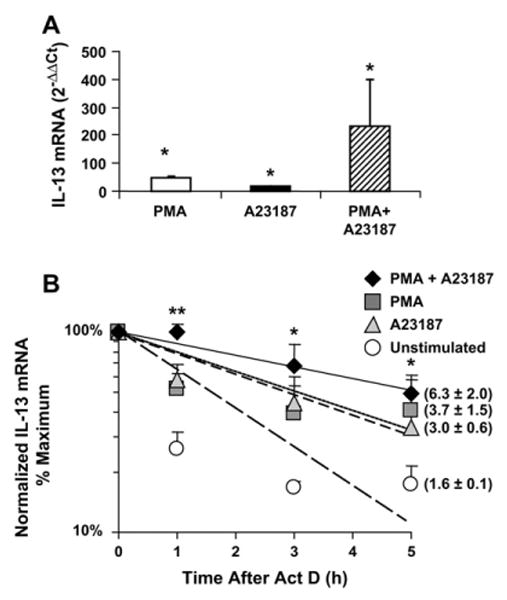
Increase of IL-13 steady-state levels and mRNA stabilization in restimulated T cells. IL-13 mRNA detected by means of real-time PCR in TH2-skewed cultures (n = 3) at time 0 after the indicated treatment (A; *P < .05 vs unstimulated cells) or after treatment with actinomycin D (Act D; B) is shown. Shown in parentheses in Figs 1 to 3 is the mRNA half-life (in hours). **P < .001 and *P < .05 versus unstimulated cells.
The 3′UTR of IL-13 mRNA displays AREs with a class 1 configuration,39 with 4 scattered AUUUA sequences embedded in an A- and U-rich milieu, indicating potential regulation of mRNA turnover by ARE-binding proteins (Fig 2, A). We used an established model31 for the isolated study of the cis-element (the ARE) and the transfactor (HuR) potentially involved in IL-13 mRNA turnover. We generated the construct pTet-BBB-IL13UTR by subcloning the full-length IL-13 3′UTR in pTetBBB, which expresses the ARE-less rabbit β-globin reporter under the control of a Tet-off promoter.40 The 2 constructs were transiently transfected in parallel in H2 cells, together with a green fluorescent protein (GFP) expression vector for normalization (Fig 2, B). H2 cells31 were used in place of Tet-off Jurkat cells because the latter lack the adenovirus receptor. In Fig 2, C, densitometric analysis of the Northern blot, performed with cytoplasmic RNA extracted from H2 after transcriptional shutoff, shows that the insertion of the IL-13 3′UTR significantly accelerated the slow turnover of the ARE-less chimeric β-globin mRNA,40 displaying a half-life of 2.1 hours compared with 11.4 hours in cells transfected with pTet-BBB (n = 6). The fast turnover of the chimeric transcript resembles the decay of the IL-13 mRNA in unstimulated TH2-skewed cells, suggesting that the IL-13 3′UTR does contain functional regulatory elements of mRNA turnover.
FIG 2.
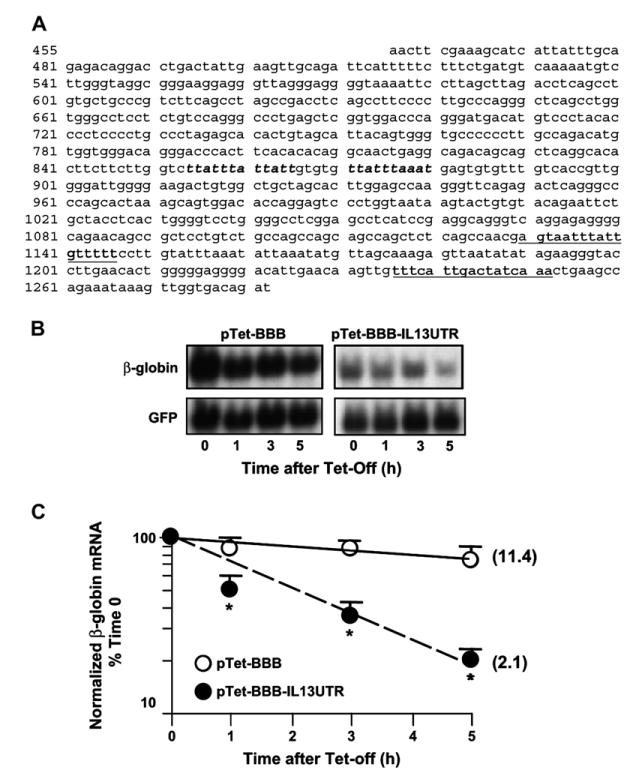
The 3′UTR of IL-13 mRNA mediates changes in the stability of a β-globin mRNA reporter. A, The 3′UTR of the human IL-13 mRNA. AU-rich elements are bolded. Underlined are additional putative HuR recognition sites. Representative Northern blot (B) and densitometric analysis (C; means ± SEMs of n = 6) of β-globin mRNA expression in H2 cells transfected with the indicated plasmids is shown. *P < .01 compared with pTet-BBB-transfectants.
When HuR was transiently overexpressed in H2 cells (Fig 3, C), it induced a profound stabilization of the labile chimeric β-globin mRNA only in cells cotransfected with pTet-BBB-IL13UTR (Fig 3, A, right). In these cells this can be appreciated by comparing cells transfected with the empty vector with those transfected with HuR, especially at the 3- and 5-hour data points (P < .02, ANOVA). Densitometric analysis indicates a 10-fold increase in reporter mRNA half-life over mock-transduced cells (22.7 vs 2.8 hours, respectively; Fig 3, B). These results, confirmed by means of densitometry of Northern blot analysis (see Fig E2 in the Online Repository at www.jacionline.org), suggest that HuR is involved in transcript stabilization mediated by the 3′UTR of IL-13 mRNA.
FIG 3.
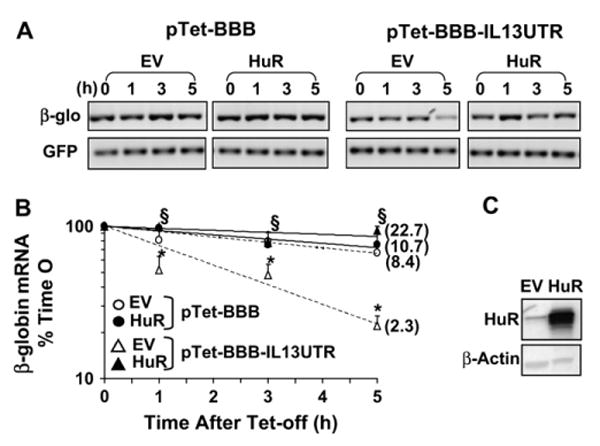
Overexpression of HuR increases the stability of the IL-13 3′UTR-bearing β-globin reporter. Representative end point PCR (A) and densitometric analysis (B; means ± SEMs of n = 6) of β-globin (upper panels) and GFP (lower panels) mRNA in H2 cells transfected with the indicated plasmids after transduction with Ad-HuR (HuR) or Ad-empty vector (EV) is shown. *P < .001, β-globin mRNA in pTet-BBB-IL13UTR-transfectants compared with pTet-BBB-transfectants; §P < .05, pTet-BBB-IL13UTR plus Ad-HuR cotransfectants compared with Ad-EV cotransfectants. C, Western Blot of HuR and β-actin levels in transfected H2.
To verify the occurrence of a stimulus-dependent association of HuR with the endogenous IL-13 mRNA and to validate in a physiologically relevant model the mechanisms identified in the reporter system, we performed IP of mRNPs obtained from Jurkat cells stimulated with or without PMA and A23187 for 3 hours using a monoclonal anti-HuR (3A2) or an isotype-matched antibody. The specificity of the IP with anti-HuR was confirmed by Western blotting to monitor HuR presence in the mRNPs (data not shown). The immunoprecipitated mRNA pools were then assayed for the presence of IL-13 mRNA by using real-time PCR. IL-13 mRNA was barely detectable in the IP samples of unstimulated cells (data not shown). Conversely, after cell activation, we found a consistent enrichment of IL-13 mRNA in the samples obtained by means of IP with anti-HuR over the background mRNA levels in the control antibody immunoprecipitates. In Jurkat cells the difference of 6 cycles in the CT value for IL-13 detection between the HuR and the mock IP (ΔCT) indicated a 2-(-6) = 64-fold enrichment in IL-13 mRNA in the HuR IP. Importantly, selective association of IL-13 mRNA with HuR was also confirmed in TH2-skewed cells (Fig 4, A, right panel), where the ΔCT for IL-13 (6.6 cycles) indicated a 2-(-6.6) = 97.7-fold enrichment in IL-13 mRNA in the HuR IP. In both cases detection of GAPDH mRNA, used as a negative control because it is known not to be a target of HuR, showed no difference between the 2 IP conditions, indicating a lack of specific enrichment despite the more abundant message, as indicated by the lower CT number.
FIG 4.
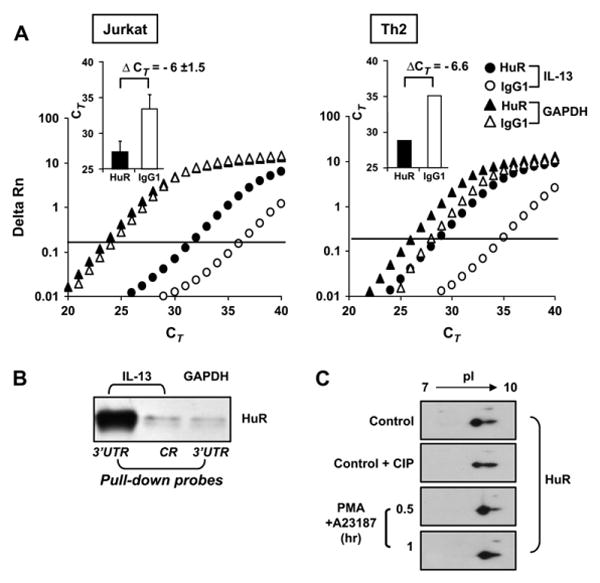
Association of HuR with IL-13 mRNA and posttranslational changes of HuR. A, Representative real-time PCR amplification plot of fluorescence intensity over background (Delta Rn) against PCR cycle (CT) of IL-13 and GAPDH mRNA in stimulated Jurkat (n = 3) and TH2-skewed cells (n = 2) subjected to IP with the anti-HuR antibody (solid symbols) or with control antibody (open symbols). In the bar graph inserts the IL-13 datasets are expressed as the means ± SEMs CT of the IP with anti-HuR (black bars) or the control antibody (white bars). B, Western blot of HuR after biotin pull-down assay (representative of n = 3). C, Detection of HuR by means of 2-dimensional Western blot analysis (representative of n = 4) in Jurkat cells unstimulated (Ctrl) or treated for the indicated times. Lysates from unstimulated cells were run in the absence or presence of treatment with calf intestinal phosphatase (CIP) before separation in the first dimension.
We then confirmed the interaction of HuR with the 3′UTR of IL-13 mRNA by using the biotin pull-down assay (Fig 4, B). Cytoplasmic proteins from Jurkat cells were incubated with biotinylated transcripts spanning the coding region or the full-length 3′UTR of IL-13 or the full-length 3′UTR of GAPDH as a negative control. The presence of HuR in the pulled-down pellets isolated with streptavidin-coated beads was detected by means of Western blotting. Only residual binding was detected in the IP samples pulled down with the transcript encompassing the coding region, which was comparable with that obtained by using the ARE-less 3′UTR of GAPDH. In contrast, substantial amounts of HuR were associated with the IL-13 3′UTR, which is in line with the mRNA-stabilizing effects of overexpressed HuR in our IL-13 3′UTR-bearing β-globin reporter model (Fig 3).
Levels of endogenous HuR did not change after cell stimulation (data not shown). Therefore to assess whether mitogens trigger posttranslational modification of HuR, 2-dimensional Western blot analysis of HuR was performed by using lysates from Jurkat cells unstimulated or treated with PMA and A23187. After stimulation up to 1 hour, the HuR signal shifted rightward, indicating a loss in negative charge (n = 4; Fig 4, C). Treatment with calf intestinal phosphatase of unstimulated samples also produced a rightward shift, although less marked than that observed after mitogen treatment, suggesting that the latter posttranslational change of HuR could be due, at least in part, to a loss of phosphorylation.
We then explored the functional role for HuR association by transiently modulating HuR levels in Jurkat cells. The level and specificity of transient overexpression and silencing of HuR was verified by means of Western blotting (Fig 5, left panels). Transfected cells were stimulated with or without PMA and ionomycin and harvested for analysis of IL-13 mRNA. Results were normalized to GAPDH mRNA expression, the negative control also used for the experiments of IP and the biotin pull-down assay, which was not altered by changes in HuR levels (data not shown). Mitogen-induced levels of IL-13 mRNA were significantly increased in cells overexpressing HuR (12.5 ± 3.3–fold over mock-transfected cells, P < .05). Accordingly, IL-13 levels decreased by 41% when HuR expression was suppressed with a specific siRNA (Fig 5, left panels).
FIG 5.
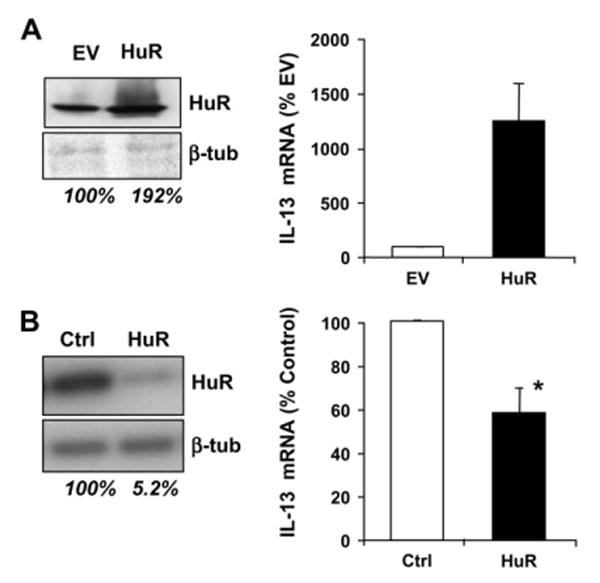
Changes in HuR levels affect IL-13 mRNA expression. Left panels, Western blot of HuR and β-tubulin after transfection of Jurkat cells with an HuR expression vector or an empty vector (EV;A) and HuR-specific siRNA or a scrambled siRNA (Ctrl;B). Numbers below the blots represent the normalized densitometric readings. Right panels, Mitogen-induced IL-13 mRNA in transfected Jurkat cells (Fig 5, A: n = 3; Fig 5, B, n = 5; P < .05).
Discussion
Our studies provide evidence for a thus far undescribed regulatory component of IL-13 expression in T cells that involves modulation of mRNA turnover. In resting human primary T cells, the low steady-state levels of IL-13 mRNA displayed a relatively short half-life. After cell restimulation, the expected increase in IL-13 mRNA levels was paralleled by a profound prolongation of the transcript's half-life. These findings demonstrate that on cell activation, mRNA stabilization occurs in conjunction with the increase in IL-13 mRNA steady-state levels, which is in agreement with previous studies recognizing the process of stabilization of cytokine mRNAs as a central regulatory mechanism of T-cell activation.5,6 Signaling cascades activated by proinflammatory stimuli that regulate transcription also control downstream regulatory events at the level of mRNA transport, stability, and translation.41 Therefore the observed changes in mRNA turnover are likely to be integrated with transcriptional events to produce the robust increase in steady-state levels of IL-13 that occurs after T-cell activation in vitro.
Our data indicate that the 3′UTR plays an important role in determining the turnover rate of the IL-13 mRNA. Subcloning of this region significantly accelerated the slow decay of the ARE-less β-globin mRNA but also mediated transcript stabilization after HuR overexpression. Changes in mRNA turnover rates and translation of ARE-bearing genes are modulated by complex remodeling of ribonucleoprotein complexes, which provides a tight control of the timing and magnitude of their expression through the promotion of sequential transient increases in mRNA stabilization and accelerated decay. It is possible that in resting conditions the more rapid decay seen in both the endogenous IL-13 mRNA and in the chimeric reporter could be under the control of decay-promoting RNA-binding proteins, such as tristetraprolin, as shown for the control of IL-2 mRNA decay in human T cells.42 In activated cells such a pattern of rapid decay could be changed by the activation of HuR, which would determine an increase in IL-13 levels by increasing mRNA stabilization. In support of this view, in addition to mRNA stabilization, we also show that mitogen stimulation promotes association of HuR with the endogenous IL-13 mRNA. A functional role for this ribonucleoprotein complex in IL-13 transcript turnover is further supported by the strong stabilization of the chimeric mRNA bearing the IL-13 3′UTR after overexpression of HuR and by the significant changes in IL-13 mRNA levels after overexpression or silencing of HuR.
The putative AREs that bind HuR show a high degree of heterogeneity in both human subjects and mice.35 Recently, novel binding regions for HuR have been described within the coding regions of IL-4 and CD83.9,10 In addition to class 1 AREs, the IL-13 3′UTR contains 2 additional putative HuR-binding sites identified through computational analysis.35 In our study we demonstrate, by means of mRNP IP and biotin pull-down assay, that HuR is bound to the 3′UTR and not to the coding region of the IL-13 mRNA. Further deletional and mutational analysis of the IL-13 3′UTR will clarify the role of the discrete sequences controlling the observed changes in mRNA turnover and will help identify more specific recognition sites for HuR.
HuR-mediated functions are dependent on the increase in its cytoplasmic levels after cell activation. Increased nucleocytoplasmic shuttling of HuR has been documented in human and mouse T cells after activation through the T-cell receptor by costimulatory molecules (eg, CD28 and LFA-1), by IL-4, and by PMA.5,9,18,20,43 This process is increasingly viewed as a central feature of T-cell activation, granting an important mechanistic insight into the regulation of T-cell responses. Because only a small percentage of HuR shuttles to the cytoplasm, this process can be best appreciated in the thin cytoplasm of activated T cells by using confocal microscopy.5,18 Although because of technical limitations we did not document a stimulus-induced increase in cytoplasmic levels of HuR in our system, we observed that the total levels of HuR were not changed by cell treatment in Jurkat and TH2-skewed cells (data not shown), suggesting that activation of HuR, rather than increased synthesis, is involved in IL-13 mRNA turnover. Posttranslational modifications of HuR, such as methylation or phosphorylation, have been shown to be mediated by multiple pathways38-45 and have been linked to either stimulus-dependent nucleocytoplasmic shuttling or to the mRNA-stabilizing function of HuR. The loss of negative charge induced in HuR by mitogen treatment is reproduced, at least in part, by phosphatase treatment of unstimulated samples. These results suggest that a loss in phosphorylation might be involved in mitogen-mediated posttranslational changes of HuR, either through inhibition of kinases38,45 or activation of as-yet-unidentified phosphatases. Further studies are warranted to characterize these pathways and their role in the effect of HuR on IL-13 mRNA stability.
The functions of HuR in T cells appear to be pleiotropic, accounting for either increased stabilization, cytoplasmic accumulation, or translation of T-cell transcripts.9,10,17,18,25,46 In our study the significant changes in IL-13 mRNA levels subsequent to transient changes in HuR levels provide additional support for the proposed role of HuR as a positive regulator of IL-13 mRNA stability. Also in support of this hypothesis, IL-13 and IL-4 mRNA displayed increased stability, along with higher levels of expression, in activated T cells in a TH2-biased mouse strain in comparison with those observed in cells from a TH1-biased background.12 Importantly, HuR has been recently identified as a mediator of IL-4 mRNA stabilization in mouse CD4+ T cells.9
Strikingly, HuR has been thus far reported to mediate mRNA stabilization of the majority of the cytokine genes clustered on chromosome 5q: IL-3, IL-4, GM-CSF, and, in the present study, IL-13.9,19,25,47,48 On this basis, we propose that in T cells HuR could be exerting a critical role by regulating posttranscriptionally the coordinate expression of this cytokine cluster. This is in line with the current view of the control of cytoplasmic mRNA by RNA-binding proteins as a key regulatory mechanism for the coordinate expression of functionally related genes.49 It is also interesting to note that HuR is encoded in chromosome 19p13.2,50 a region known to include candidate genes in asthma and other atopic disorders.51
Taken together, our data indicate that posttranscriptional regulation critically affects the expression of IL-13 in T cells and support the hypothesis that HuR is an important amplifier of inflammatory and allergic responses. By binding to ARE-bearing and functionally related transcripts after T-cell activation, HuR could coordinate their cytoplasmic fate and ultimately sustain their increased production in response to inflammatory stimuli. Identification of the molecular pathways of HuR activation and function will further our knowledge of how inflammation works and will likely identify valuable targets for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Dr Ann-Bin Shyu for kindly providing the pTet-BBB construct.
Supported by National Institutes of Health grants R01 AI060990 (CS) and R01 AI041463 (VC).
Abbreviations used
- ARE
Adenylate-uridylate–rich element
- CT
Cycle threshold
- DTT
Dithiothreitol
- EGFP
Enhanced green fluorescent protein
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GFP
Green fluorescent protein
- IEF
Isoelectric focusing
- IP
Immunoprecipitation
- mRNP
Messenger ribonucleoprotein complex
- PMA
Phorbol 12-myristate 13-acetate
- siRNA
Small interfering RNA
- Tet
Tetracycline
- UTR
Untranslated region
Footnotes
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
Clinical implications: Posttranscriptional events are critical in T-cell gene regulation and potentially targetable for therapy.
References
- 1.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–90. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 2.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol. 2001;167:4414–20. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 3.Keen JC, Cianferoni A, Florio G, Guo J, Chen R, Roman J, et al. Characterization of a novel PMA-inducible pathway of interleukin-13 gene expression in T cells. Immunology. 2006;117:29–37. doi: 10.1111/j.1365-2567.2005.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–43. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 5.Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, et al. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J Immunol. 2006;176:2105–13. doi: 10.4049/jimmunol.176.4.2105. [DOI] [PubMed] [Google Scholar]
- 6.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, et al. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–38. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–48. [PMC free article] [PubMed] [Google Scholar]
- 9.Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol. 2006;177:4426–35. doi: 10.4049/jimmunol.177.7.4426. [DOI] [PubMed] [Google Scholar]
- 10.Prechtel AT, Chemnitz J, Schirmer S, Ehlers C, Langbein-Detsch I, Stulke J, et al. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J Biol Chem. 2006;281:10912–25. doi: 10.1074/jbc.M510306200. [DOI] [PubMed] [Google Scholar]
- 11.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J Immunol. 2004;173:976–85. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 12.Butler NS, Monick MM, Yarovinsky TO, Powers LS, Hunninghake GW. Altered IL-4 mRNA stability correlates with Th1 and Th2 bias and susceptibility to hypersensitivity pneumonitis in two inbred strains of mice. J Immunol. 2002;169:3700–9. doi: 10.4049/jimmunol.169.7.3700. [DOI] [PubMed] [Google Scholar]
- 13.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–7. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–27. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–89. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J Biol Chem. 2001;276:47958–65. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 18.Seko Y, Azmi H, Fariss R, Ragheb JA. Selective cytoplasmic translocation of HuR and site-specific binding to the interleukin-2 mRNA are not sufficient for CD28-mediated stabilization of the mRNA. J Biol Chem. 2004;279:33359–67. doi: 10.1074/jbc.M312306200. [DOI] [PubMed] [Google Scholar]
- 19.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–51. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 20.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111(suppl):3145–56. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–50. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–9. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–78. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–30. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ming XF, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–89. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg-Cohen I, Furneauxb H, Levy AP. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem. 2002;277:13635–40. doi: 10.1074/jbc.M108703200. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, Forstermann U, et al. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein. HuR J Biol Chem. 2000;275:26040–9. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 29.Atasoy U, Curry SL, López de Silanes I, Shyu AB, Casolaro V, Gorospe M, et al. Regulation of eotaxin gene expression by TNF and IL-4 through messenger RNA stabilization: involvement of the RNA-binding protein HuR. J Immunol. 2003;171:4369–78. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 30.De Fanis U, Mori F, Kurnat RJ, Lee WK, Bova M, Adkinson NF, et al. GATA3 up-regulation associated with surface expression of CD294/CRTH2: a unique feature of human Th cells. Blood. 2007;109:4343–50. doi: 10.1182/blood-2006-05-025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, et al. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–13. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 33.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–31. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 34.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 35.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–92. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–8. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–8. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 38.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–57. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–46. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 40.Xu N, Loflin P, Chen CY, Shyu AB. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 1998;26:558–65. doi: 10.1093/nar/26.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kracht M, Saklatvala J. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine. 2002;20:91–106. doi: 10.1006/cyto.2002.0895. [DOI] [PubMed] [Google Scholar]
- 42.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–61. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 43.Xu YZ, Di Marco S, Gallouzi I, Rola-Pleszczynski M, Radzioch D. RNA-binding protein HuR is required for stabilization of SLC11A1 mRNA and SLC11A1 protein expression. Mol Cell Biol. 2005;25:8139–49. doi: 10.1128/MCB.25.18.8139-8149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, et al. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase J Biol Chem. 2002;277:44623–30. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 45.Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell. 2007;18:2137–48. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millard SS, Vidal A, Markus M, Koff A. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol Cell Biol. 2000;20:5947–59. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esnault S, Malter JS. Granulocyte macrophage-colony-stimulating factor mRNA is stabilized in airway eosinophils and peripheral blood eosinophils activated by TNF-alpha plus fibronectin. J Immunol. 2001;166:4658–63. doi: 10.4049/jimmunol.166.7.4658. [DOI] [PubMed] [Google Scholar]
- 48.Winzen R, Gowrishankar G, Bollig F, Redich N, Resch K, Holtmann H. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol. 2004;24:4835–47. doi: 10.1128/MCB.24.11.4835-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–7. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 50.Ma WJ, Furneaux H. Localization of the human HuR gene to chromosome 19p13.2. Hum Genet. 1997;99:32–3. doi: 10.1007/s004390050305. [DOI] [PubMed] [Google Scholar]
- 51.Hamshere M, Cross S, Daniels M, Lennon G, Brook JD. A transcript map of a 10-Mb region of chromosome 19: a source of genes for human disorders, including candidates for genes involved in asthma, heart defects, and eye development. Genomics. 2000;63:425–9. doi: 10.1006/geno.1999.6075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


