Abstract
Clock genes are known to be the molecular core of biological clocks of vertebrates. They are expressed not only in those tissues considered central pacemakers, but also in peripheral tissues. In the present study, partial cDNAs for six of the principal clock genes (Period 1-3 and Cryptochrome 1-3) were cloned from a teleost fish, the goldfish (Carassius auratus). These genes showed high homology (approximately 90%) with the respective cDNAs of zebrafish (Danio rerio), the only other teleost from which clock genes have been cloned. The daily expression pattern of each gene in retina, gut and liver of goldfish was investigated using quantitative RT-PCR and cosinor analysis. All clock genes analyzed in the retina showed circadian rhythmicity; however, only Per 2-3 and Cry 2-3 were rhythmic in goldfish liver and gut. The amplitude and phase of the expression in liver and gut were different from those found in goldfish retina. Such differences suggest that other cues, such as feeding time, may contribute to the entrainment of oscillators in goldfish liver and gut. Our results support the use of goldfish as a teleost model to investigate the location and functioning of the circadian oscillators.
Keywords: goldfish, Period gene, Cryptochrome gene, circadian oscillators, retina, nonphotic entrainment
INTRODUCTION
Living organisms exhibit daily rhythms in physiology, behaviour, and gene expression due to the existence of endogenous circadian clocks. These clocks synchronize biological processes to the 24-h light/dark cycle, allowing the organisms to anticipate changes in their environment. In metazoans, the generation of circadian rhythmicity appears to be consequence of specialised tissues known as “master clocks”, which have different locations among species, i.e. lateral neurons in Drosophila (Stanewsky, 2002), the pineal gland and retina in fish (Falcon, 1999), pineal gland, retina and hypothalamus in birds (Underwood et al., 2001), and the hypothalamic suprachiasmatic nuclei (SCN) in mammals (Klein et al., 1991).
The core oscillator of the circadian clock consists of transcriptional-translational feedback loops that involve a highly conserved set of “clock genes”. The mechanism of the endogenous clocks is known to be conserved along the phylogenetic scale, from unicellular organisms and plants to insects and mammals (Panda et al., 2002). In mammals, these loops are formed by positive elements, CLOCK and BMAL1, which heterodimerize and enhance the transcription of the negative components Period (Per1, Per2 and Per3) and Cryptochrome (Cry1 and Cry2) genes. The PER and CRY proteins form complexes that inhibit their own transcription by binding to the CLOCK:BMAL1 complex and blocking its function. This negative loop allows a daily rhythm in expression of Per and Cry transcripts and protein products (Iuvone et al., 2005; Okamura et al., 2002). The molecular basis of circadian clocks in teleost fish has been studied in zebrafish, where the homologues of the mammalian clock genes have been cloned (Cahill, 2002). The main differences with respect to the mammalian system may be the regulation of Per2 and the number of Cry genes; up to six Cry genes have been found, all of them rhythmically expressed (Kobayashi et al., 2000).
However, the expression of clock genes is not restricted to the master clocks. The mRNA products of these genes have been found in peripheral tissues such as liver, heart, muscle, kidney, pancreas, adipose tissue and lung from mammals (Balsalobre, 2002; Muhlbauer et al., 2004; Peirson et al., 2006; Zvonic et al. 2006), heart, spleen and gall bladder in zebrafish (Kaneko et al., 2006), and lung, heart, liver, muscle and testis in lizard (Della Ragione et al., 2005; Vallone et al., 2007). Although these circadian oscillators may need an external messenger to sustain their rhythmicity, they can be entrained to other cues besides the light/dark cycle, such as temperature, the reproductive cycle, or feeding time (Brown et al., 2002; Mendoza, 2007; Vallone et al., 2007). Regarding feeding time, some studies have shown it to be a potent zeitgeber that is able to entrain the master circadian clock in the absence of photic cues in mammals (Castillo et al., 2004) and in fish (Boujard and Leatherland, 1992), where feeding time also influences the daily changes of metabolic parameters such as cortisol, glucose and triglyceride plasma levels (Polakof et al., 2007). These facts, together with the well-studied increase in locomotor activity prior to food intake (food anticipatory activity, FAA), have led investigators to propose the existence of a food-entrainable oscillator (FEO), whose location is still unknown (Mendoza, 2007).
The goldfish is a well studied species, with a robust circadian system and a demonstrated capacity of entrainment by feeding cues, as shown by its pattern of FAA (Aranda et al., 2001; Vera et al., 2007). However, there is no data available on clock gene expression in this species.
The aim of the present study was to investigate some of the main components of the molecular clock in the goldfish (Carassius auratus). First, six cDNA transcripts from the Period and Cryptochrome genes were cloned in this teleost species. Second, their expression pattern in central and peripheral tissues was characterized. Lastly, possible daily rhythms of expression of their mRNA were analyzed.
MATERIALS AND METHODS
1. Animals and tissue collection
All animal experiments were conducted in accord with the NIH Guide for the Care and Use of Laboratory Animals and complied with the Spanish legal requirements. For the cloning and tissue expression of the genes, goldfish (Carassius auratus) with a body weight of 9–12 g were used. Fish were kept in 100 l tanks with filtered and aerated water, and were fed once a day at zeitgeber time (ZT) 2 on a commercial pellet diet (1% bw, Sera Biogran). Animals were maintained under 12L:12D photoperiod, with lights on at ZT0. For the tissue distribution study, fish were killed at ZT2. Brain, neural retina, heart, liver and gut were removed and immediately placed in RNAlater (Sigma, Saint Louis, MO, USA) for tissue preservation. For the study of daily variations in gene expression, we used 30 goldfish of 10–15 g, kept under the same housing conditions. Fish were sacrificed at ZT2, ZT8, ZT14, ZT20 and ZT2 of the following day. Neural retina, liver and gut were excised and rapidly immersed in RNAlater.
2. RNA isolation and first-strand cDNA synthesis
Tissues removed from RNAlater were homogenized in RLT buffer (Qiagen Inc., Valencia, CA, USA) by a rotor homogenizer (Polytron, Brinkmann Instruments, Westbury, NY). Total RNA was extracted by a silica-based filter-binding RNeasy mini kit (Qiagen Inc.) and treated with RNase-free DNase I (1 U) according to the manufacturer’s instructions (Qiagen Inc.). The yield and quality of RNA were quantified by the 260/280 nm absorbance ratio (SmartSpec3000, Bio-Rad Laboratories, Hercules, CA, USA) and by electrophoresis under non-denaturing conditions on 1.5% agarose gel.
An aliquot of 200 ng of total RNA from tissues was reverse transcribed (RT) in a 20 μl reaction volume using oligo-dT or random primers, RNase inhibitor and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The reaction was carried out for 50 min at 55°C, followed by 15 min at 70°C to inactivate the reverse transcriptase. In the case of random primers, there was a 5-min step at 25°C prior to the inactivation of the enzyme.
3. RT-PCR cloning of goldfish Period and Cryptochrome cDNAs
To amplify the cDNAs of the clock genes, PCR was performed by using specific primers (Table 1) for the homologous Per1, Cry1, Cry2 and Cry3 of zebrafish (Danio rerio) transcripts. For Per2 and Per3, degenerate primers were designed using the CLUSTALW algorithm from the sequences of zebrafish, Siganus guttatus and Xenopus tropicalis.
Table 1.
Sequences of primers used for real-time PCR.
| PRIMER NAME | TARGE T GENE | SEQUENCE (from 5′ to 3′) | PCR product (bp) | ACCESSION NUMBER | SPECIES | |
|---|---|---|---|---|---|---|
| For gene cloning: | ||||||
| zP1F-1377 | Forward | Per1 | 5′-TGTACCGCAGCTGAATGAACTGA-3′ | 531 | NM001030183 | Danio rerio |
| zP1R-1974 | Reverse | 5′-TCAAAGGTCGTTCCTGCAGCAC-3′ | ||||
| zCry1F831 | Forward | Cry1a | 5′-AAAGAACAGCTCACCTCCTCTG-3′ | 396 | NM131789 | Danio rerio |
| zCry1R1311 | Reverse | 5′-GCGATATCTGCCAGTCTTAAGG-3′ | ||||
| zCry2F830 | Forward | Cry2a | 5′-GAAAACCAGCACTCCTCCGCT-3′ | 492 | NM131791 | Danio rerio |
| zCry2R1322 | Reverse | 5′-CTATTCTCCGAGGTTTCCCTGC-3′ | ||||
| zCry3F711 | Forward | Cry3 | 5′-AGGACTCAGGGTGACAGCTTACA-3′ | 712 | NM131786 | Danio rerio |
| zCry3R1423 | Reverse | 5′-GAGGTACATCCCAAAGTTAAAGG-3′ | ||||
| g18SF571 | Forward | 18s rRNA | 5′-ATGATTAAGAGGGACGGCCGG-3′ | 509 | EF189737 | Carassius auratus |
| g18SR1080 | Reverse | 5′-AATAGTTACGCGGCCCCGTG-3′ | ||||
| Degenerate primers for gene cloning: | ||||||
| dPer2F-2 | Forward | Per2 | 5′-AGCTTYRTCAATCCCTGGAGY-3′ | 849 | NM182857 | Danio rerio |
| dPer2R-2 | Reverse | 5′-CAGYTACAGCAGCACMATTGT-3′ | EF208027 | Siganus guttatus | ||
| AF199499 | Xenopus laevis | |||||
| dPer3F | Forward | Per3 | 5′-GCCTGTTCATAATAATGGATCCAG-3′ | 460 | NM131584 | Danio rerio |
| dPer3R | Reverse | 5′-CAGCCTGAGTCMGAAGTCACT-3′ | NM001079228 | Xenopus tropicalis | ||
|
| ||||||
| For qRT-PCR: | ||||||
| gP1F56 | Forward | Per1 | 5′-AGCGCCACTTCCTCCTCTGA-3′ | 130 | EF690698 | Carassius auratus |
| gP1R186 | Reverse | 5′-CCAACGGACAGCAGGTCTTCA-3′ | ||||
| gP2F4 | Forward | Per2 | 5′-TTTGTCAATCCCTGGAGCCGC-3′ | 116 | EF690697 | Carassius auratus |
| gP2R119 | Reverse | 5′-CGTGGCTGAGGGCAAATCCTT-3′ | ||||
| gP3F26 | Forward | Per3 | 5′-GGCTATGGCAGTCTGGCTAGTAA-3′ | 130 | EF690699 | Carassius auratus |
| gP3R155 | Reverse | 5′-GACATTGCAGCGGTTTTGTGCTG-3′ | ||||
| gC1F11 | Forward | Cry1 | 5′-TACCGGCTGCCACCAACAAC-3′ | 106 | EF690700 | Carassius auratus |
| gC1R117 | Reverse | 5′-TTTGGCCAAGTGGGCTGAGG-3′ | ||||
| gC2F16 | Forward | Cry2 | 5′-CGCTCTCCCTGTATGGTCAAC-3′ | 102 | EF 690701 | Carassius auratus |
| gC2R118 | Reverse | 5′-AAGGCAACCCGATCTGTGTGC-3′ | ||||
| gC3F38 | Forward | Cry3 | 5′-GGTGAGACAGAAGCCCTGGAA-3′ | 102 | EF690702 | Carassius auratus |
| gC3R140 | Reverse | 5′-CGCTCAATCACTGTTCGCAAGC-3′ | ||||
| g18SF571 | Forward | 18s rRNA | 5′-ATGATTAAGAGGGACGGCCGG-3′ | 143 | ||
| g18SR714 | Reverse | 5′-GTCGGAGGTTCGAAGACGATCA-3′ | ||||
The reaction mixture contained cDNA, 10 pmol each of forward and reverse primers (see Table 1), 1 U Taq DNA polymerase (Invitrogen), 5 μl of 10× PCR buffer, 1 μl of 10 mM dNTPs (200nM final), and 2 μl of 50 mM MgCl2 (2 mM final). PCRs were performed in a total volume of 50 μl. PCR conditions were set at 95 °C for 2 min, followed by 40 cycles of denaturation, annealing and extension, each one at 95 °C, 54 °C and 72 °C for 30 s. The amplified products were gel purified using QIAE’s gel extraction kit (Qiagen). Purified PCR products (426–849 bp) were ligated into pGEM® T-vector (Promega, Madison, WI, USA). Ligated products were transformed into Escherichia coli JM109 cells. Positive clones were obtained and plasmid DNA extraction was performed using the QIAprep Spin miniprep kit (Qiagen). Nucleotide sequences were determined by sequencing with Agencourt Bioscience Corporation (Beverly, MA) with two T-vector-specific primers (T7 and SP6).
4. Quantitative Real-time RT-PCR (qRT-PCR)
Primers (Table 1) for qRT-PCR were designed based on the sequences of the cloned cDNAs and on the available sequence for goldfish 18s rRNA. Amplification reactions were performed in an iCycler (Bio-Rad) using cDNA (2μl from each sample), 1x SYBR Green PCR Master mix (Biorad) and 400nM gene-specific forward and reverse primers, up to a volume of 25μl. Samples were incubated at 95°C for 3 min, followed by 45 cycles of denaturation, annealing and extension, each one at temperatures of 95°C, 54°C and 72°C for 30 seconds. All samples were assayed in duplicates. PCR products were checked by agarose electrophoresis, and quantification was carried out by comparing the threshold cycle for amplification of the unknown product with those of six concentrations of standard cDNA for each gene (standard curves were generated using from 1 fg to 100 pg of cDNA). The gene expression levels were calculated and normalized by dividing the calculated values for the mRNA samples by that of 18S rRNA.
5. Statistical analysis
To test for variation in mRNA levels among time points, statistical analyses were carried out with one-way ANOVA. To evaluate rhythmicity in gene expression, cosinor analysis was performed by fitting periodic sinusoidal functions to the expression values for the genes across the five time points using the formula f(t)=M+Acos(tπ/12−ϕ), where f(t) was the gene expression level in a given time, the mesor (M) is the mean value, A is the sinusoidal amplitude of oscillation, t is time in hours and ϕ is the acrophase (time of peak expression). Non-linear regression allows the estimation of M, A, and ϕ, and their standard error (SE) (Delgado et al, 1993). Significance of cosinor analysis was defined by the noise/signal of amplitude calculated from the ratio SE(A)/A. Expression was considered to display a daily rhythm if it had both P<0.05 by ANOVA and SE(A)/A <0.3 by cosinor analysis.
RESULTS
Cloning of goldfish Period and Cryptochrome genes
PCR-generated partial cDNAs of Per1, 2, and 3 and Cry1, 2, and 3 of goldfish were cloned into a pGEM®-T easy vector (Promega, Madison, WI) and sequenced. The sequences were compared to the homologous zebrafish sequences. The size of the PCR products, their homology with corresponding zebrafish genes, and the GenBank accession numbers of the cloned goldfish clock genes are presented in Table 2. More than 80% and 90% sequence identity was observed between goldfish Periods and Cryptochromes cDNA products and their corresponding zebrafish sequences, respectively (Table 2).
Table 2.
Characteristics of the partial cDNA sequences for each gene.
Expression of Period and Cryptochrome genes in goldfish
The mRNA expression pattern of Per1, Per2, Per3, Cry1, Cry2 and Cry3 genes was examined in goldfish retina, brain, heart, liver, and gut, dissected at ZT2 (Figure 1). The expression of Cryptochromes was higher than that of Periods in all the tissues examined. Per 1 and Per3 were more highly expressed in the retina and liver compared to the other tissues. Per2 expression appeared to be relatively low in all tissues. The relative tissue distribution of Periods and Cryptochromes is presented in Table 3.
Fig 1. Tissue distribution of goldfish clock genes.
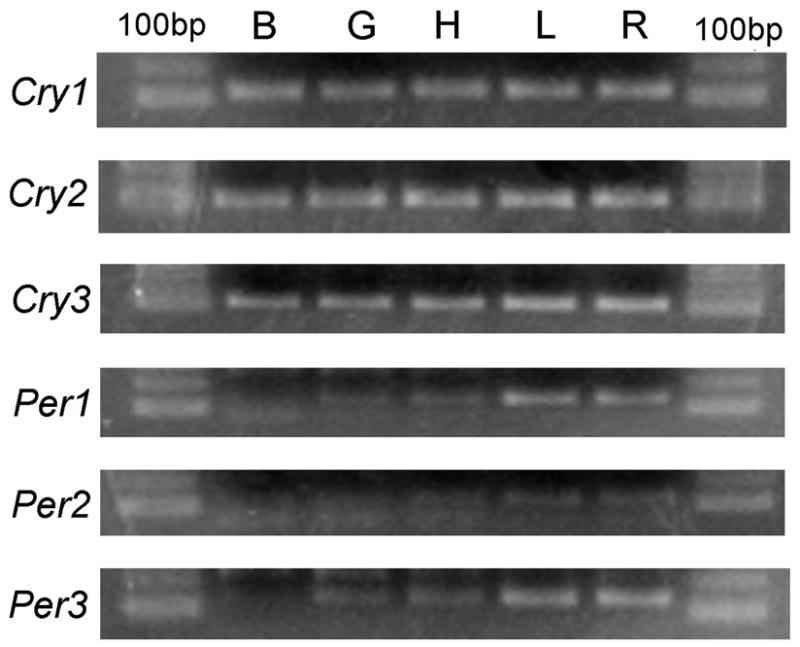
RT-PCR analysis of Cry1-3 and Per1-3 transcripts in different tissues collected at ZT2, as shown in a 1.5% agarose electrophoresis gel with ethidium bromide and 100bp molecular marker. Abbreviations: B, brain; G, gut; H, heart; L, liver; R, retina.
Table 3.
Relative tissue expression pattern of Period and Cryptochrome transcripts in the goldfish.
| GENE | TISSUE EXPRESSION PATTERN |
|
|---|---|---|
| High expression | Low expression | |
| Per1 | Liver, retina | Gut, brain, heart |
| Per2 | Retina, gut, liver, heart, brain | |
| Per3 | Liver, retina | Gut, brain, liver, heart |
| Cry1 | Brain, gut, heart, liver, retina | |
| Cry2 | Gut, brain, retina, heart, liver | |
| Cry3 | Gut, heart, brain, retina, liver | |
Rhythmic expression of circadian clock genes
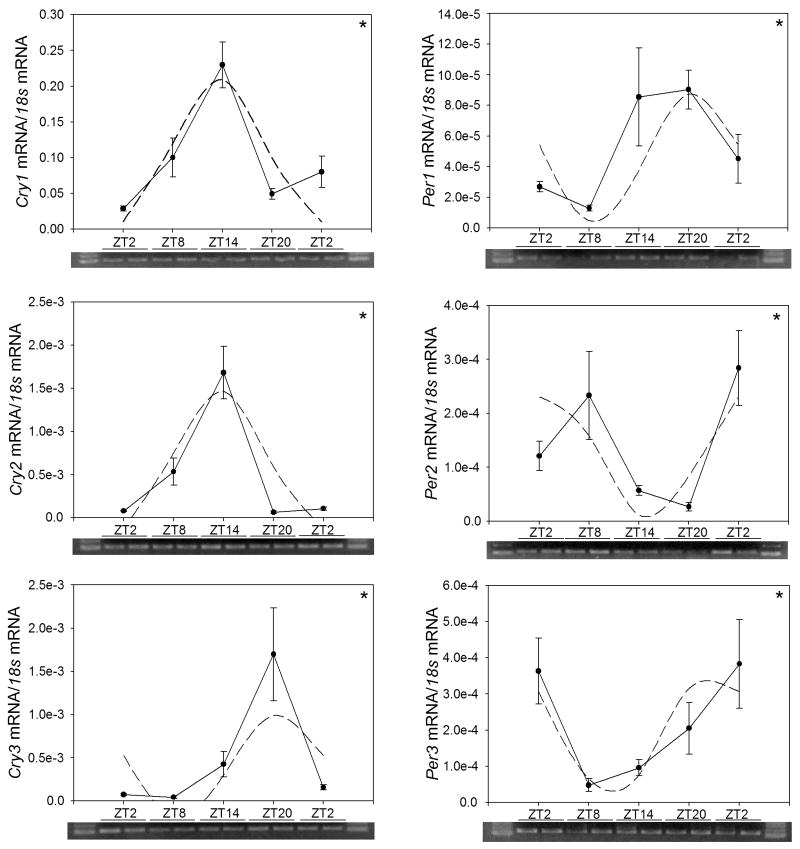
The daily rhythms of Per1, Per2, Per3 and Cry1, Cry2, and Cry3 transcript levels were examined by qRT-PCR in the goldfish retina, liver, and gut during a 12: 12h light/dark cycle (Figs. 2–4). Statistical analysis, including cosinor and ANOVA, showed that mRNAs for the six clock genes in retina displayed significant cyclic oscillations as a function of a 24-h cycle. The data presented in Table 4 show that the mean expression (mesor) of Cryptochrome genes in goldfish retina was higher than that of Periods, and that their peak expression (acrophase) occurred at the day-night transition (Cry1 and 2) or mid-night (Cry3). Peak expression of Per1 also occurred at night. In contrast, peak expression of Per2 and Per3 transcripts occurred during the daytime.
Fig 2.
Expression of clock genes in the retina of goldfish (Carassius auratus) during the daily light-dark cycle. Relative mRNA levels were quantified by qRT-PCR. Each data point represents the mRNA amount of the corresponding clock gene normalized to 18s rRNA, expressed as mean ± S.E.M. (n=6 retinas per time point). The dashed lines in the graphs represent the periodic sinusoidal functions determined by the cosinor analysis. The asterisks indicate a significant rhythm (SE(A)/A<0.3). ANOVA: Per1 P=0.01; Per2 P=0.004; Per3 P=0.02; Cry1 P=0.03; Cry2 P<0.001; Cry3 P=0.008.
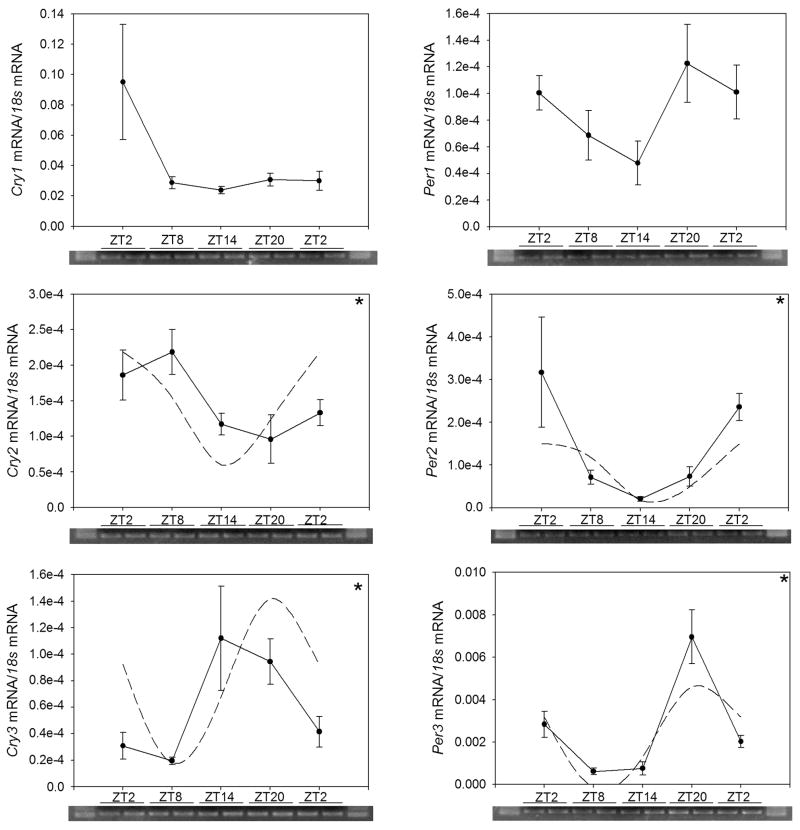
Fig 4.
Expression levels of clock genes in the liver of goldfish (Carassius auratus) during the daily light-dark cycle. Relative mRNA levels were quantified by qPCR. Each data point represents the mRNA amount of the correspondent clock gene normalized to 18s rRNA, and are expressed as mean ± S.E.M. (n=6 livers per time point). The dashed line in the graphs represents the periodic sinusoidal functions determined by the cosinor analysis. The asterisks indicate a significant rhythm (SE(A)/A<0.3). ANOVA: Per1 P=0.103; Per2 P=0.211; Per3 P=0.001; Cry1 P=0.365; Cry2 P=0.002; Cry3 P<0.001.
Table 4.
Parameters defining the gene expression rhythms in the retina of goldfish (Carassius auratus).
| Mesor ± SE (mRNA/18s mRNA) | Amplitude ± SE (mRNA/18s mRNA) | Acrophase ± SE (hours) | |
|---|---|---|---|
| Per1 | (0.46 ± 0.04)·10−4 | (0.42 ± 0.06)·10−4 | 18.55 ± 0.50 |
| Per2 | (1.21 ± 0.21)·10−4 | (1.16 ± 0.31)·10−4 | 6.15 ± 0.92 |
| Per3 | (1.91 ± 0.37)·10−4 | (1.70 ± 0.49)·10−4 | 0.30 ± 1.25 |
| Cry1 | 0.13 ± 0.01 | 0.11 ± 0.01 | 12.14 ± 0.43 |
| Cry2 | (0.67 ± 0.09)·10−3 | (0.80 ± 0.12)·10−3 | 12.87 ± 0.61 |
| Cry3 | (0.41 ± 0.06)·10−3 | (0.59 ± 0.09)·10−3 | 18.67 ± 0.47 |
The cosinor analysis was used to obtain the rhythmic parameters as defined by a sinusoidal function. Statistical significance was assumed when the ratio SE(A)/A was below 0.3
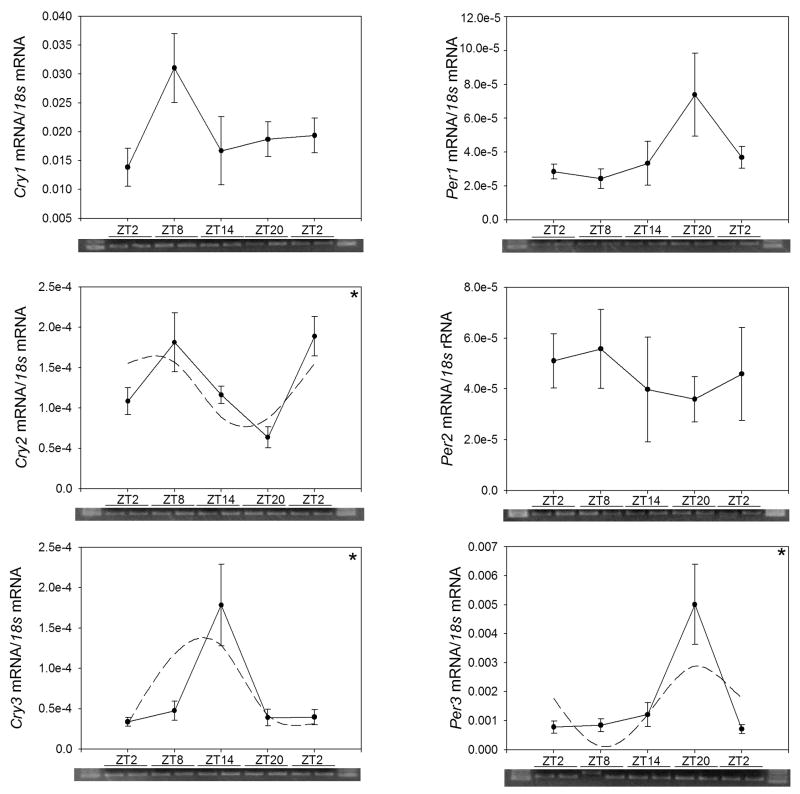
In gut, Per2 and Per3, but not Per1, transcript levels showed a significant daily variation by ANOVA and cosinor analysis (Figure 3; Table 5). Similarly, Cry2 and Cry3 showed significant daily rhythms, but Cry1 did not. The rhythms in Per2, Per3, Cry2 and Cry3 showed differences in amplitude and phase (Fig. 3, Table 5). Thus, Per2 and Cry2 peaked in the early and mid-daytime, respectively, while Per3 and Cry3 had their acrophase at near midnight. Per1 and Cry1 did not show significant rhythms.
Fig 3.
Expression levels of clock genes in the gut of goldfish (Carassius auratus) during the daily light-dark cycle. Relative mRNA levels were quantified by qPCR. Each data point represents the mRNA amount of the correspondent clock gene normalized to 18s rRNA, and are expressed as mean ± S.E.M. (n=6 intestines per time point). The dashed line in the graphs represents the periodic sinusoidal functions determined by the cosinor analysis. The asterisks indicate a significant rhythm (SE(A)/A<0.3). ANOVA: Per1 P=0.138; Per2 P=0.005; Per3 P=0.006; Cry1 P<0.001; Cry2 P=0.033; Cry3 P=0.018.
Table 5.
Parameters defining the statistically significant gene expression rhythms of peripheral tissues of the goldfish (Carassius auratus).
| Mesor ± SE (mRNA/18s mRNA) | Amplitude± SE(mRNA/18s mRNA) | Acrophase± SE(hours) | |
|---|---|---|---|
| LIVER | |||
| Per3 | (1.50 ± 0.19)·10−3 | (1.41 ± 0.29)·10−3 | 18.88 ± 0.65 |
| Cry2 | (1.34 ± 0.09)·10−4 | (0.79 ± 0.14)·10−4 | 7.49 ± 0.59 |
| Cry3 | (8.00 ± 1.24)·10−5 | (6.19 ± 1.60)·10−5 | 13.73 ±1.17 |
|
| |||
| GUT | |||
| Per2 | (8.38 ± 1.35)·10−5 | (7.44 ± 1.76)·10−5 | 1.94 ± 1.05 |
| Per3 | (2.23 ± 0.24)·10−3 | (2.55 ± 0.37)·10−3 | 20.94 ± 0.46 |
| Cry2 | (1.39 ± 0.12)·10−4 | (0.81 ± 0.18)·10−4 | 6.99 ± 0.72 |
| Cry3 | (1.30 ± 0.14)·10−4 | (1.13 ± 0.20)·10−4 | 18.34 ± 0.63 |
The cosinor analysis was used to obtain the rhythmic parameters as defined by a sinusoidal function. Statistical significance was assumed when the ratio SE(A)/A was below 0.3. Parameters are shown only for transcripts displaying significant rhythms.
Significant daily rhythms of expression of Cry2, and Cry3 and Per3 transcripts were observed in the liver, as assessed by both ANOVA and cosinor analysis, although with different temporal profiles of expression (Fig. 4; Table 5). Peak expression Cry3 mRNA occurred near the day-to-night transition, while Cry2 peaked in the middle of the daytime. Per3 mRNA levels were highest in the middle of the night. The liver Per1, Per2, and Cry1 transcript levels did not display significant rhythms.
Comparing the significant rhythms in the three tissues, Cry3 peaked earlier in the liver (~ ZT14) than in gut and retina (~ ZT18). The levels of mRNA were similar for both peripheral tissues, but approximately 10 times more abundant in retina. In liver and gut, Cry2 had a similar acrophase at the middle of the day, while the peak in retina was delayed until the beginning of the dark phase. As occurs with Cry3 expression, mRNA expression for Cry2 was approximately an order of magnitude higher in retina than in gut and liver. The maximum values for Per3 expression in liver and gut were acquired late in the scotophase (ZT21 for gut and ZT19 for liver), whereas it occurred at the beginning of the subjective day in retina. The amount of Per3 mRNA gene was an order of magnitude lower in retina than in peripheral tissues.
DISCUSSION
In the present study, we have cloned, for the first time in the goldfish, partial cDNAs of some of the principal clock genes, which exhibit robust oscillations of mRNA expression through the light-dark cycle in retina as well as in liver and gut.
The partial cDNAs of the goldfish clock genes have a high degree of homology with zebrafish clock genes. Although alignment is not shown, goldfish Per1, Per2, and Per3 cDNA sequences are 84–88% identical with those of zebrafish, which are true homologs of the three mammalian Period genes (Cahill, 2002; Delaunay et al., 2003), supporting the role played by these genes in the molecular clock machinery of the goldfish. Regarding to Cryptochrome transcripts, the cDNA sequences are 91–93% identical with those of zebrafish. While there are only two Cryptochrome genes and proteins characterized in mammals (CRY1 and CRY2) (Kobayashi et al., 1998), six rhythmically expressed Crytochrome genes have been reported in zebrafish (Kobayashi et al., 2000). Both being teleosts, zebrafish and goldfish are expected to have two paralogs of most mammalian genes due to genome duplication (Postlethwait et al., 1998), but this fact is not sufficient to explain the six zebrafish Cryptochromes. In the present study we obtained the goldfish cDNAs from primers designed upon sequences of zebrafish Cry1a and Cry2a (both most similar to mCry1) and zebrafish Cry3, whose product does not inhibit the transcription mediated by the CLOCK:BMAL1 dimer (Kobayashi et al., 2000). The high homology of the goldfish cDNA fragments with their zebrafish homologs, and therefore with mammals, highlights their function in goldfish circadian system. Moreover, the presence of a probable zCry3 homolog in goldfish is a first approach to delve into the existence and role of Cryptochrome genes in the teleost lineage.
Analysis of goldfish clock gene expression in retina reveals that, under LD conditions, all transcripts are rhythmic. Regarding the Period genes, Per3 expression is highest at dawn, as occurs in the retina of zebrafish or Japanese quail (Delaunay et al., 2000; Yoshimura et al., 2000). Moreover, Per1 and Per2 also exhibit rhythms but with opposite phase relationships: while Per1 peaks during midnight, Per2 shows its maximum at midday. These patterns of rhythmicity are the inverse from that previously reported for Xenopus photoreceptors (Zhuang et al., 2000). However, goldfish Per2 parallels the rhythm described for quail, chicken and mammalian retina, with higher levels during the daytime (Chaurasia et al., 2006; Kamphuis et al., 2005; Yoshimura et al., 2000). The high nocturnal expression of cryptochrome transcripts in goldfish retina is similar to the expression profile of Cry2 in zebrafish. In contrast, zCry1a and zCry3 are expressed during daytime (Kobayashi et al., 2000) as occurs with Cry1 in the quail (Fu et al., 2002) and chicken retina (Chaurasia et al., 2006; Haque et al., 2002). These findings support the key role of Period and Cryptochrome genes in the molecular clock of goldfish retina, although further experiments may be necessary to unravel the implication of each component.
Our results demonstrate the expression of all three Cryptochrome and Period genes in peripheral organs of the goldfish, such as liver or intestine. Rhythmic clock gene expression in such tissues is already reported in organisms like zebrafish (Kaneko et al., 2006; Whitmore et al., 1998), lizard (Della Ragione et al., 2005), chicken (Chong et al., 2003), Japanese quail (Fu et al., 2002), sheep (Andersson et al., 2005), mice (Peirson et al., 2006) and humans (Pardini et al., 2005). This rhythmic expression of clock genes in mammalian peripheral tissues is self-sustained (Balsalobre, 2002; Yoo et al., 2004), temperature compensated (Reyes et al., 2008), and can synchronize to cues different from the light/dark cycle, such as temperature (Brown et al., 2002) or feeding (Damiola et al., 2000; Stokkan et al., 2001). Characteristics of peripheral clock gene expression in fish have been studied most in zebrafish, where photo-entrainable peripheral oscillators have been found in tissues such as heart, kidney or spleen, suggesting a circadian system made of a number of widely distributed pacemakers synchronized by light, perhaps due to its semi-transparent body (Kaneko et al., 2006; Whitmore et al., 1998). Thus, our results of rhythmic circadian clock gene expression in goldfish peripheral tissues, together with their robust FAA both along the light/dark cycle and with restricted feeding (Aranda et al., 2001; Vera et al., 2007), support the use of goldfish as a model to study the existence of a food entrainable oscillator (FEO) in fish.
The clock genes studied have been implicated in entrainment by feeding cues, and therefore affecting food anticipatory activity (FAA), principal output of the so-called FEO (Stephan et al., 1979). Studies with Clock mutant mice showed the FAA persisted even in constant darkness, suggesting that Clock gene is not part of the FEO clockwork (Pitts et al., 2003), although it might be necessary in maintaining rhythmicity in peripheral oscillators (DeBruyne et al., 2007). The genes considered a part of the FEO due to its involvement in food anticipatory behaviours are the Cryptochromes, which appear to be necessary for the development of the FAA (Iijima et al., 2005), and Period genes. Specifically, Per2 mutant mice have significant impairment of the FEO output (Feillet et al., 2006; Mendoza et al. 2005).
Clock gene expression in goldfish liver and gut has revealed rhythms of both Period and Cryptochrome genes under LD conditions. Per2 is rhythmic only in goldfish gut, with a maximum at ZT2, preceding the peak in retina and occurring just prior to the feeding time. To our knowledge, circadian expression of clock genes in the gastrointestinal tract has been studied only in mice, where Per2 exhibits its maximum expression at the beginning of the night with ad libitum feeding; this peak shifts to midday when food availability is restricted (Hoogerwerf et al., 2007). Rhythmic expression of Per3 occurs in goldfish gut and liver with similar acrophase late in the night, different from the peak in retina. In the case of mice, Per3 in the gastrointestinal tract shows an expression pattern parallel to Per2, peaking early in the night when fed ad libitum and shifting to early day in timed feeding (Hoogerwerf et al., 2007). Moreover, Per3 expression in mouse liver reaches its maximum at the beginning of the dark phase with ad libitum food access (Peirson et al., 2006), but shifts to midnight when food is restricted to their activity period (Damiola et al., 2000).
Concerning Cryptochrome genes, our study shows that Cry3 is rhythmically expressed in liver and gut of goldfish with a maximum during midnight in gut (similar to retina) and early in the night in liver. There is no data available to compare these results, as this is the first time that the expression rhythm of this gene is characterized in a teleost species, and Cry3 is not expressed in mammals. In both goldfish peripheral tissues, Cry2 exhibits a maximum in the middle of the light phase, while in retina it occurs early in the night. This could be due to an entrainment of the peripheral oscillator of the goldfish to restricted feeding (feeding time in our experiment was at ZT2), resulting in an uncoupling from the central pacemaker. Our results agree with studies in the colon of mice subjected to restricted feeding (Hoogerwerf et al., 2007). In liver, ad libitum-fed mice present Cry2 maximums at the beginning of the dark phase (Peirson et al., 2006) but early in the light phase with restricted feeding (Damiola et al., 2000). Thus, feeding time for the goldfish may be essential to synchronize its peripheral oscillators, as occurs in mammals, supporting the existence of a FEO in this species.
In summary, Period and Cryptochrome genes are expressed in a circadian manner in retina, liver and gut of the goldfish, with differences in phase and amplitude, suggesting entrainment of goldfish peripheral tissues to cues different from light and, therefore, highlighting a probable role of feeding. Thus, the goldfish can be considered a model to delve into the mechanisms of peripheral oscillators, as well as to unravel the location and physiology of the food entrainable clock.
Acknowledgments
This study was supported by the Spanish Ministerio de Educación y Ciencia (MEC) project AGL2007-65744-C03-03 and the National Institutes of Health (PMI) R01EY04864 and P30EY006360. E. Velarde and C. Azpeleta are recipients of predoctoral fellowships from the Spanish MEC.
References
- Andersson H, Johnston JP, Messager S, Hazlerigg D, Lincoln G. Photoperiod regulates clock gene rhythms in the ovine liver. Gen Comp Endocrinol. 2005;142:357–363. doi: 10.1016/j.ygcen.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Aranda A, Madrid JA, Sánchez-Vázquez FJ. Influence of light on feeding anticipatory activity in goldfish. J Biol Rhythms. 2001;16:50–57. doi: 10.1177/074873040101600106. [DOI] [PubMed] [Google Scholar]
- Balsalobre A. Clock genes in mammalian peripheral tissues. Cell Tissue Res. 2002;309:193–199. doi: 10.1007/s00441-002-0585-0. [DOI] [PubMed] [Google Scholar]
- Boujard T, Leatherland JF. Circadian rhythms and feeding time in fishes. Environ Biol Fish. 1992;35:109–131. [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Cahill GM. Clock mechanisms in zebrafish. Cell Tissue Res. 2002;309:27–34. doi: 10.1007/s00441-002-0570-7. [DOI] [PubMed] [Google Scholar]
- Castillo MR, Hochstetler KJ, Tavernier RJ, Jr, Greene DM, Bult-Ito A. Entrainment of the master circadian clock by scheduled feeding. Am J Physiol Regulatory Integrative Comp Physiol. 2004;287:R551–R555. doi: 10.1152/ajpregu.00247.2004. [DOI] [PubMed] [Google Scholar]
- Chaurasia SS, Pozdeyev N, Haque R, Visser A, Ivanova TN, Iuvone PM. Circadian clockwork machinery in neural retina: evidence for the presence of functional clock components in photoreceptor-enriched chick retinal cell cultures. Mol Vis. 2006;12:215–223. [PubMed] [Google Scholar]
- Chong NW, Chaurasia SS, Haque R, Klein DC, Iuvone PM. Temporal.-spatial characterization of chicken clock genes: circadian expression in retina, pineal gland and peripheral tissues. J Neurochem. 2003;85:851–860. doi: 10.1046/j.1471-4159.2003.01723.x. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Gene Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–R539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B. An inherited functional circadian clock in zebrafish embryos. Science. 2000;289:297–300. doi: 10.1126/science.289.5477.297. [DOI] [PubMed] [Google Scholar]
- Delaunay F, Thisse C, Thisse B, Laudet V. Differential regulation of Period 2 and Period 3 expression during development of the zebrafish circadian clock. Gene Expr Patterns. 2003;3:319–324. doi: 10.1016/s1567-133x(03)00050-4. [DOI] [PubMed] [Google Scholar]
- Delgado MJ, Alonso-Gómez AL, Gancedo B, De Pedro N, Valenciano AI, Alonso-Bedate M. Serotonin N-acetyltransferase (NAT) activity and melatonin levels in the frog retina are not correlated during the seasonal cycle. Gen Comp Endocrinol. 1993;92:143–150. doi: 10.1006/gcen.1993.1151. [DOI] [PubMed] [Google Scholar]
- Della Ragione F, Comitato R, Angelini F, D’Esposito M, Cardone A. Molecular cloning and characterization of the clock gene period2 in the testis of lizard Podarcis sicula and its expression during seasonal reproductive cycle. Gene. 2005;363:105–112. doi: 10.1016/j.gene.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Falcon J. Cellular circadian clocks in the pineal. Prog Neurobiol. 1999;58:121–162. doi: 10.1016/s0301-0082(98)00078-1. [DOI] [PubMed] [Google Scholar]
- Feillet CA, Ripperger JA, Magnone MC, Dulloo A, Albrecht U, Challet E. Lack of food anticipation in Per2 mutant mice. Curr Biol. 2006;16:2016–2022. doi: 10.1016/j.cub.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Fu Z, Inaba M, Noguchi T, Kato H. Molecular cloning and circadian regulation of Cryptochrome genes in Japanese quail (Coturnix coturnix japonica) J Biol Rhythms. 2002;17:14–27. doi: 10.1177/074873002129002302. [DOI] [PubMed] [Google Scholar]
- Haque R, Chaurasia SS, Wessel JH, III, Iuvone PM. Dual regulation of cryptochrome 1 mRNA expression in chicken retina by light and circadian oscillators. Neuroreport. 2002;13:2247–2251. doi: 10.1097/00001756-200212030-00016. [DOI] [PubMed] [Google Scholar]
- Hoogerwerf WA, Hellmich HL, Cornelissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone V. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Iijima M, Yamaguchi S, van der Horst GTJ, Bonnefont X, Okamura H, Shibata S. Altered food-anticipatory activity rhythm in Cryptochrome-deficient mice. Neurosci Res. 2005;52:166–173. doi: 10.1016/j.neures.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, Chaurasia SS. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24:433–456. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hernandez-Borsetti N, Cahill GM. Diversity of zebrafish peripheral oscillators revealed by luciferase reporting. Proc Natl Acad Sci USA. 2006;103:14614–14619. doi: 10.1073/pnas.0606563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Commun. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- Klein DC, Moore RY, Reppert SM. Suprachiasmatic nucleus: the mind’s clock. Oxford University Press; New York: 1991. [Google Scholar]
- Kobayashi K, Kanno S, Smit B, van der Host GT, Takao M, Yasui A. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res. 1998;26:5086–5092. doi: 10.1093/nar/26.22.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Toh H, Fukuda I, Tsujimura T, Terada N, Kamei Y, Yuba S, Iwai S, Todo T. Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells. 2000;5:725–738. doi: 10.1046/j.1365-2443.2000.00364.x. [DOI] [PubMed] [Google Scholar]
- Mendoza J. Circadian clocks: setting time by food. J Neuroendocrinol. 2007;19:127–137. doi: 10.1111/j.1365-2826.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- Mendoza J, Graff C, Dardente H, Pevet P, Challet E. Feeding cues alter clock gene oscillation and photic responses in the suprachiasmatic nuclei of mice exposed to a light/dark cycle. J Neurosci. 2005;25:1514–1522. doi: 10.1523/JNEUROSCI.4397-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlbauer E, Wolgast S, Finckh U, Peschke D, Peschke E. Indication of circadian oscillations in the rat pancreas. FEBS Lett. 2004;564:91–96. doi: 10.1016/S0014-5793(04)00322-9. [DOI] [PubMed] [Google Scholar]
- Okamura H, Yamaguchi S, Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res. 2002;309:47–56. doi: 10.1007/s00441-002-0572-5. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Pardini L, Kaeffer B, Trubuil A, Bourreille A, Galmiche JP. Human intestinal circadian clock: expression of clock genes in colonocytes lining the crypt. Chronobiol Int. 2005;22:951–961. doi: 10.1080/07420520500395011. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Duffield GE, Takher S, Sharma P, Foster RG. Comparison of clock gene expression in SCN, retina, heart, and liver of mice. Biochem Biophys Res Comm. 2006;351:800–807. doi: 10.1016/j.bbrc.2006.10.118. [DOI] [PubMed] [Google Scholar]
- Pitts S, Perone E, Silver R. Food-entrained circadian rhythms are sustained in arrhythmic Clk/Clk mutant mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R57–R67. doi: 10.1152/ajpregu.00023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakof S, Ceinos RM, Fernández-Durán B, Míguez JM, Soengas JL. Daily changes in parameters of energy metabolism in liver, white muscle, and gills of rainbow trout: dependence on feeding. Comp Biochem Physiol. 2007;146:265–273. doi: 10.1016/j.cbpa.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates M, Horne S, Amores A, Brownlie A, Donovan A, Egan E, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw P, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish genome map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Reyes BA, Pendergast JS, Yamazaki S. Mammalian circadian peripheral oscillators are temperature compensated. J Biol Rhythms. 2008;23:95–98. doi: 10.1177/0748730407311855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R. Clock mechanisms in Drosophila. Cell Tissue Res. 2002;309:11–26. doi: 10.1007/s00441-002-0569-0. [DOI] [PubMed] [Google Scholar]
- Stephan FK, Swann JM, Sisk CL. Entrainment of circadian rhythms by feeding schedules in rats with suprachiasmatic lesions. Behav Neural Biol. 1979;25:545–554. doi: 10.1016/s0163-1047(79)90332-7. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Underwood H, Steele CT, Zivokovic B. Circadian organization and the role of the pineal in birds. Microsc Res Techn. 2001;53:48–62. doi: 10.1002/jemt.1068. [DOI] [PubMed] [Google Scholar]
- Vallone D, Frigato E, Vernesi C, Foa A, Foulkes NS, Bertolucci C. Hypothermia modulates circadian clock gene expression in lizard peripheral tissues. Am J Physiol Regul Integr Comp Physiol. 2007;292:R160–R166. doi: 10.1152/ajpregu.00370.2006. [DOI] [PubMed] [Google Scholar]
- Vera LM, De Pedro N, Gómez-Milán E, Delgado MJ, Sánchez-Muros MJ, Madrid JA, Sánchez-Vázquez FJ. Feeding entrainment of locomotor activity rhythms, digestive enzymes and neuroendocrine factors in goldfish. Physiol Behav. 2007;90:518–524. doi: 10.1016/j.physbeh.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Burh ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Suzuki Y, Makino E, Suzuki T, Kuroiwa A, Matsuda Y, Namikawa T, Ebihara S. Molecular analysis of avian circadian clock genes. Brain Res Mol Brain Res. 2000;78:207–215. doi: 10.1016/s0169-328x(00)00091-7. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Wang Y, Steenhard BM, Besharse JC. Differential regulation of two period genes in the Xenopus eye. Brain Res Mol Brain Res. 2000;82:52–64. doi: 10.1016/s0169-328x(00)00177-7. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]