Abstract
The filoviruses, Ebola and Marburg, cause severe hemorrhagic fever in humans and nonhuman primates, with high mortality rates. Although the filovirus replication pathway is now understood in considerable detail, no antiviral drugs have yet been developed that directly inhibit steps in the replication cycle. One potential target is the filovirus VP40 matrix protein, the key viral protein that drives the budding process, in part by mediating specific virus-host interactions to facilitate the efficient release of virions from the infected cell. This review will summarize current knowledge of key structural and functional domains of VP40 believed to be necessary for efficient budding of virions and virus-like particles. A better understanding of the structure and function of these key regions of VP40 will be crucial, as they may represent novel and rational targets for inhibitors of filovirus egress.
Keywords: filovirus, Ebola virus, Marburg virus, viral budding, VP40 matrix protein, virus-like particles, antiviral therapy
Introduction
Ebola (EBOV) and Marburg (MARV) viruses are the sole members of the Filoviridae family and are important pathogens of humans and nonhuman primates (Ascenzi et al., 2008; Bray and Murphy, 2007; Casillas et al., 2003; Feldmann, Klenk, and Sanchez, 1993; Peters and Khan, 1999). EBOV and MARV have been the cause of sporadic and deadly outbreaks of hemorrhagic fever in many countries since their initial outbreaks in 1976 and 1967, respectively (Ascenzi et al., 2008; Bray and Murphy, 2007; Feldmann, Klenk, and Sanchez, 1993; Peters and Khan, 1999). Depending on the virus strain initiating the outbreak, the mortality rate is variable and can be as high as 90%. The filoviruses have been classified by the CDC as Category A bioterrorism agent, and a Category A NIAID priority pathogen (Bray, 2003). Currently, there are no approved vaccines, nor antiviral drugs available to prevent or treat filovirus infections (Bausch et al., 2008; Bray and Paragas, 2002).
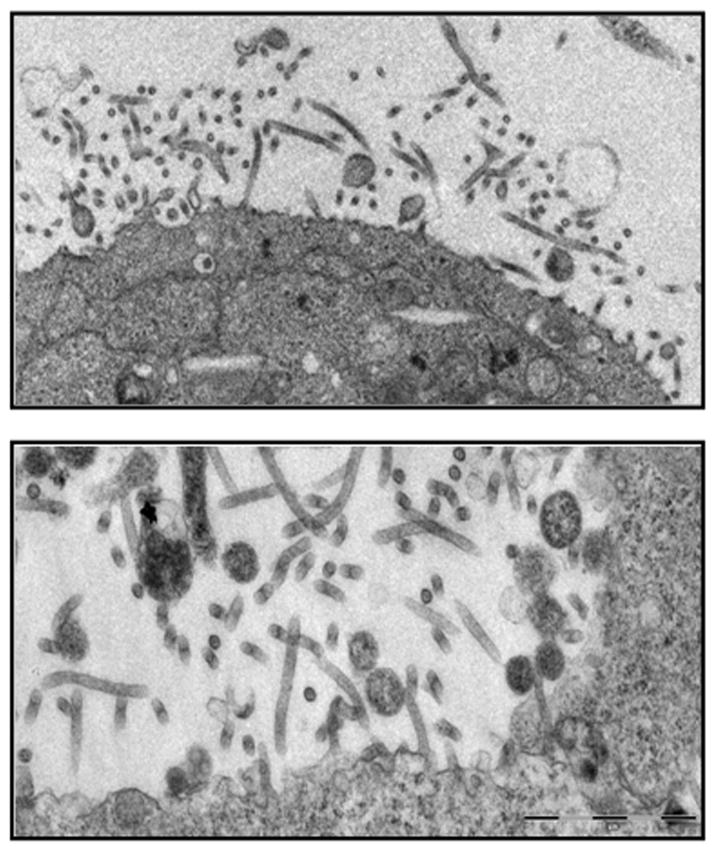
One of the major obstacles toward development of filovirus vaccines and therapeutics is that live virus experiments can be conducted only under Biosafety Level-4 (BSL-4) conditions. Nevertheless, much progress has been made toward our understanding of the molecular aspects of filovirus replication by investigating the structure and function of the viral proteins independently under less stringent conditions. A better understanding of the molecular events that govern filovirus replication will be essential for future development of vaccines and/or therapeutics. For example, our understanding of the budding process and identification of important virus-host interactions that contribute to efficient virus egress has progressed rapidly over the last decade (Chen and Lamb, 2008; Hartlieb and Weissenhorn, 2006; Jasenosky and Kawaoka, 2004; Schmitt and Lamb, 2004). One of the key approaches that has helped provide us with an abundance of valuable insight into filovirus budding has been the use of virus-like particle (VLP) budding assays, which are relatively straightforward to perform under BSL-2 conditions and accurately mimic the budding process of authentic, infectious virus. For example, human 293T cells are transfected with a plasmid encoding the filovirus VP40 matrix protein, and both cell lysates and cell culture media are harvested 24–48 hours post-transfection. The media sample is then layered onto a 20% sucrose cushion, and the VLPs are pelleted through the cushion by high speed centrifugation. VLPs can be purified further by floatation gradient centrifugation. The amount of VP40 present in the VLPs can be quantitated by immunoprecipitation and SDS-PAGE analyses, and the budding VLPs can also be visualized by electron microscopy (Fig. 1) (Johnson et al., 2006; Noda et al., 2002). Co-expression of additional filovirus proteins (e.g. GP and NP) along with VP40 results in their incorporation into budding VLPs and enhances the release of VLPs over that observed by expressing VP40 alone (Licata et al., 2004). Thus, this late stage of filovirus replication represents a viable and promising target for development of novel antivirals as our fundamental understanding of the budding process grows.
Figure 1.
Electron micrographs of EBOV VP40 VLPs budding from the surface of human 293T cells. Ultrathin sections were examined with a Philips CM-100 transmission electron microscope equipped with a KeenView digital camera system.
There are precedents for targeting late stages of virus assembly, maturation, and release with antiviral drugs. One example is Bevirimat, a novel anti-HIV-1 drug currently in clinical trials and designed to inhibit virion maturation (Salzwedel, Martin, and Sakalian, 2007). A second example includes the family of neuraminidase inhibitors of influenza viruses (Tambyah, 2008). These drugs were designed to block neuraminidase activity, which is required for efficient release and spread of influenza viruses. Antivirals targeting filovirus budding would be predicted to dampen or slow down virus budding and spread in an infected host, thus allowing more time for the individual’s immune system to respond and control the infection. The filovirus VP40 L-domain/host interaction represents a particularly attractive target since many additional human pathogens (e.g. HIV-1, Lassa fever virus, and Nipah/Hendra viruses) utilize L-domains for efficient budding, and thus inhibitors of this process could potentially have broad-spectrum activity and application.
Functional domains of viral matrix proteins
Early studies on retroviral Gag proteins paved the way for identification of functional protein domains required for virus budding. Pioneering work from Wills and Craven as well as others helped to identify three modular domains within the Gag proteins of Rous sarcoma virus and HIV-1 that were crucial for the budding process (Accola, Strack, and Gottlinger, 2000; Craven and Parent, 1996; Gottlinger et al., 1991; Patnaik and Wills, 2002). The M (membrane-binding), I (interaction), and L (late) domains were determined to be the minimal essential components of Gag required for budding (Patnaik and Wills, 2002). The M domains of RSV and HIV-1 Gag mapped to their respective N-termini, the I domains mapped to the region of the Gag polyprotein of RSV and HIV-1 that is involved in nucleocapsid (NC) formation, and the L-domains mapped to the N-terminal p2b region of RSV Gag and the C-terminal P6 region of HIV-1 Gag (Patnaik and Wills, 2002). The working model was that Gag localized and bound to the plasma membrane (M domain), began to self-interact or oligomerize (I domain), and then budded or “pinched off” (L domain) from the cell surface (Patnaik and Wills, 2002). Results from subsequent studies supported this model of budding, not only for retroviruses, but also for other RNA viruses (Craven et al., 1999; Harty et al., 1999; Licata et al., 2003; Martin-Serrano, Zang, and Bieniasz, 2001; Noda et al., 2002; Schmitt and Lamb, 2004; Timmins et al., 2001). Thus, it is fairly well accepted that RNA viral matrix proteins (e.g. filovirus VP40) that are functional homologues of Gag and that can bud independently as VLPs must possess domains equivalent to M, I, and L to promote efficient budding of VLPs and mature virions.
VP40 matrix protein
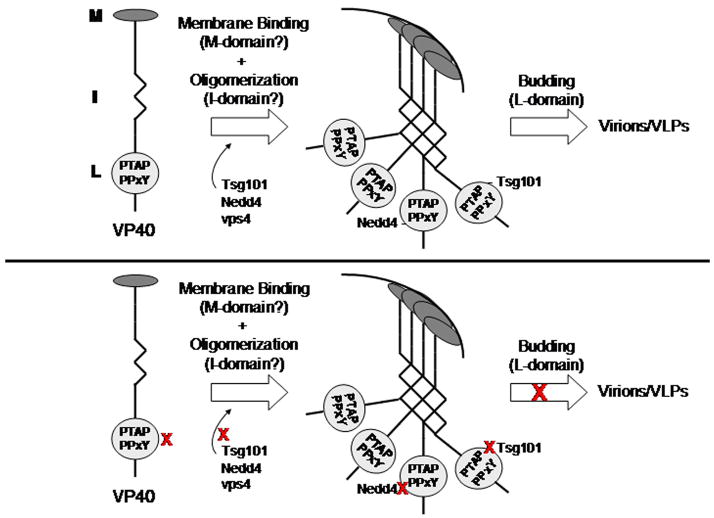
VP40 is the most abundant protein in mature filovirus virions and is the key building block for virion maturation and subsequent egress (Feldmann, Klenk, and Sanchez, 1993). Functional homologs of filovirus VP40 include the Gag polyprotein of retroviruses, and the M proteins of rhabdoviruses and paramyxoviruses. Like the Gag proteins of Rous sarcoma virus and HIV-1 and the M protein of vesicular stomatitis virus (VSV), Ebola VP40 was found to bud from mammalian cells in the absence of other viral proteins (Harty et al., 2000). VP40 is believed to possess at least three domains essential for efficient budding: the M, I, and L-domains (Fig. 2). While the L-domain region of Ebola VP40 has been characterized extensively, precise identification of the more complex M and I domains of VP40 remains to be determined. However, recent studies have provided new insights into these structural and functional regions of filovirus VP40.
Figure 2.
Top - Diagram depicting a monomer of EBOV VP40 with the M, I, and L domains highlighted. The locations of the M and I domains are for illustrative purposes and have yet to be defined precisely. In the absence of inhibitors, the M domain will mediate membrane binding, the I domain will mediate homo-oligomerization, and the L-domains will mediate interactions with host proteins Tsg101 and Nedd4 leading to efficient budding of virions or VLPs. Bottom – The red Xs represent potential target sites for inhibitors of budding. Inhibitors of L-domain function would prevent VP40 interactions with Tsg101 or Nedd4, and inhibitors of vps4 would disrupt the ESCRT machinery to impair virus egress.
Oligomerization of VP40
Homo-oligomerization is a key feature of viral matrix proteins thought to be required for efficient budding of VLPs and virions. Unlike the concise, well characterized L-domains (discussed below), the precise identity and location of the oligomerization (I) domain(s) of EBOV and MARV VP40 remains to be determined. Since the I domains are likely to be more complex than the L-domains, targeting the oligomerization function of VP40 with small molecular inhibitors will represent a significant challenge. To date, there are no existing strategies to target this activity of VP40; however, as our understanding of the oligomerization and self-assembly functions of VP40 grows, the potential to target this process with small molecule inhibitors may be feasible.
Results from a number of studies have shown that the oligomerization process of EBOV VP40 is complex and critical for several stages of virus replication. These investigations have provided fundamental information on the structure of VP40 and the regions involved in the formation of VP40 multimers. For example, elegant studies pioneered primarily by the Weissenhorn lab have demonstrated that homo-oligomerization of VP40 is important for both RNA binding and virion assembly/budding (Gomis-Ruth et al., 2003; Panchal et al., 2003; Ruigrok et al., 2000; Scianimanico et al., 2000; Timmins, Ruigrok, and Weissenhorn, 2004; Timmins et al., 2003a). Ruigrok et al. determined that the N-terminal region of Ebola VP40 mediated the formation of hexamers which were suggested to be important for virus assembly (Ruigrok et al., 2000). Timmins et al. went on to use a plethora of experimental approaches to demonstrate further that the N-terminal region of Ebola VP40 oligomerizes in vitro into both hexameric and octameric ringlike structures, with octamer formation depending primarily on interactions with nucleic acids (Timmins et al., 2003a). These results suggested that the different oligomeric forms of VP40 were essential for specific and separate stages during filovirus replication (Timmins et al., 2003a), and perhaps regulation of these conformational changes of VP40 may in turn regulate the various stages of the virus life cycle (Hoenen et al., 2005; Panchal et al., 2003; Scianimanico et al., 2000). Lastly, the role of the N-terminal portion of VP40 in oligomerization appeared to be conserved, since this region of Marburg VP40 was also found to form ringlike structures, which were observed to polymerize into rods comprised of stacked rings (Timmins et al., 2003a).
More recently, McCarthy et al. targeted amino acids 212KLR214 of Ebola VP40 for mutagenesis to determine whether these residues were important for VP40 VLP budding (McCarthy et al., 2007). This region of VP40 comprises a loop connecting two beta sheets in the C-terminal region of VP40 and was predicted to be important for VP40 structure/oligomerization based on the reported crystal structure (Dessen et al., 2000a; Dessen et al., 2000b). Interestingly, several of the KLR mutants of VP40 were defective in their ability to bud as VLPs compared to that of wild type VP40 (McCarthy et al., 2007). Addressing the mechanism of this budding defect, several of the KLR mutants displayed intracellular localization and oligomerization patterns that were altered from those observed for wild type VP40 (McCarthy et al., 2007). Although further experimentation is needed to define precisely the role of the 212KLR214 residues of VP40 in budding, these findings suggest that the 212KLR214 residues may be important for ordered assembly/oligomerization of VP40 and subsequent budding of VLPs. The authors suggested that the KLR mutants may aggregate in a disordered manner primarily in the cytoplasm, rather than oligomerize in an ordered fashion at the site of budding at the plasma membrane (McCarthy et al., 2007).
Membrane-binding domains of VP40
The second property of VP40 deemed critical for efficient budding of VLPs and virions is its ability to interact directly with lipid membranes and/or host proteins that may chaperone VP40 to the site of budding (Fig. 2). The plasma membrane is considered to be the primary site of budding for Ebola virus, whereas studies on Marburg virus demonstrate that budding can occur on basolateral membranes in polarized cells (Kolesnikova et al., 2007b; Sanger et al., 2001) as well as on internal MVB membranes (Kolesnikova et al., 2004; Kolesnikova et al., 2002). While the precise region of Ebola VP40 required to mediate interactions with lipid bilayers remains to be elucidated, studies by Ruigrok et al were the first to suggest that the C-terminal region of Ebola VP40 was important for association with membranes (Ruigrok et al., 2000). These investigators isolated full-length and a C-terminally truncated (114 amino acids) form of EBOV VP40 from Escherichia coli and demonstrated that the full-length monomeric VP40 associated efficiently with negatively-charged lipid membranes, whereas the C-terminally truncated hexameric VP40 associated inefficiently with membranes (Ruigrok et al., 2000).
Scianimanico et al. demonstrated that deletion of the C-terminal seven amino acids of VP40 resulted in destabilization of VP40 monomers and induction of VP40 hexamers (Scianimanico et al., 2000). Importantly, these investigators found that membrane association of wild type VP40 induced a conformational change from monomeric to hexameric forms of VP40 that were postulated to form the ordered building blocks necessary for efficient virus assembly/budding at the membrane (Dessen et al., 2000a; Dessen et al., 2000b; Ruigrok et al., 2000; Scianimanico et al., 2000). Thus, the hexameric form of VP40 was postulated to be critical for efficient assembly and budding. As such, Nguyen et al. went on to determine the molecular structure of hexameric EBOV VP40 by using three-dimensional EM (Nguyen et al., 2005). This study provided key insights into the mechanism of VP40 monomer-hexamer transition and its biological relevance to the late stages of filovirus replication (Nguyen et al., 2005). In sum, oligomerization and membrane localization/binding of VP40 required for efficient budding appear to be intricate and tightly regulated, and thus the feasibility of targeting these activities of VP40 with small molecular inhibitors could prove problematic.
Lastly, many viruses have been shown to assemble and bud from specialized domains within the plasma membrane known as lipid rafts (Suzuki and Suzuki, 2006). These specific microdomains are thought to aid in concentrating viral proteins, and excluding host proteins, in the membrane for efficient assembly, budding, and in some cases entry of different viruses (Suzuki and Suzuki, 2006). Important observations made by Bavari et al. and later by Panchal et al. demonstrated that these cholesterol-enriched, detergent-resistant domains were biologically significant for filovirus budding (Bavari et al., 2002; Panchal et al., 2003). Association of VP40 with lipid raft domains and efficient release of VLPs were directed by key C-terminal sequences of VP40 including amino acids 309-317 and proline residues 283 and 286 (Panchal et al., 2003). These findings provide further evidence that the C-terminal region of VP40 plays a role in binding to membranes, particularly to specialized lipid raft microdomains.
Intracellular trafficking of VP40
The mechanism by which VP40 traverses through the host cell to the site of budding has remained an important and active area of investigation. The process of intracellular trafficking of VP40 is of particular interest since it likely involves host interactors serving as chaperones and/or scaffolding matrices to mediate efficient assembly and budding. A better understanding of this process could reveal novel targets for inhibitors of filovirus assembly and budding pathways.
Some recent findings have begun to shed some light on potential host proteins involved in intracellular transport and localization of VP40. Not too surprisingly, results from several studies suggest that host cytoskeletal proteins (e.g. actin and microtubules) associate with both Ebola and Marburg VP40 and may be important for VLP formation and egress (Han and Harty, 2005; Kolesnikova et al., 2007a; Ruthel et al., 2005). More recently, a region centered around proline-53 of EBOV VP40 has been implicated in modulating intracellular transport of VP40 leading to efficient production of VLPs (Yamayoshi and Kawaoka, 2007). Introduction of alanine substitutions between amino acids 51–56 of VP40 led to reduced levels of VLP production. Importantly, several of these VP40 mutants were deemed defective in localization to the site of budding at the plasma membrane, but were not defective in homo-oligomerization (Yamayoshi and Kawaoka, 2007). Although the mechanism by which this region of VP40 functions in budding remains to be defined, Yamayoshi and Kawaoka note that the 51-LRPIA-55 of VP40 is conserved in other viral matrix proteins (e.g. Gag protein of HIV-1 and Z protein of Lassa fever virus) and may represent a conserved domain for mediating interactions with novel host factors/chaperones (Yamayoshi and Kawaoka, 2007).
In a related study, Yamayoshi et al. used co-immunoprecipitation and mass spectrographic analyses to identify the host protein Sec24C, a component of the COPII vesicular transport system, as an interactor with Ebola VP40 (Yamayoshi et al., 2008). More specifically, amino acids 303–307 of Ebola VP40 were shown to interact with host Sec24C, which is thought to facilitate intracellular transport of VP40 (Yamayoshi et al., 2008). While the COPII system typically involves transport of proteins from the ER to the Golgi, it will be of interest to determine whether this machinery of the cell is usurped by the virus for the purpose of escorting VP40 to the site of budding. Adding to the potential significance of this interaction was the finding that host Sec24C also interacted with Marburg VP40. Whether Sec24C or the COPII system plays a similar role in directing both EBOV and MARV VP40 to the site of budding will be of interest since localization of Marburg VP40 has been observed in both internal membranes of MVBs and in filopodia-like projections at the plasma membrane (Dolnik, Kolesnikova, and Becker, 2008; Kolesnikova et al., 2004).
L-domains of VP40
The third domain that is highly conserved among many viral matrix proteins and has been shown to be important for efficient VLP production and virus budding is the L-domain. The L-domain motifs conserved within Ebola VP40 were shown to be important for efficient release of both VLPs (Fig. 1) and infectious virus in cell culture (for review see, Chen and Lamb, 2008; Dolnik, Kolesnikova, and Becker, 2008; Hartlieb and Weissenhorn, 2006; Jasenosky and Kawaoka, 2004; Pornillos, Garrus, and Sundquist, 2002; Schmitt and Lamb, 2004). The nature of the L-domain region of EBOV VP40 was found to be unique in that it was composed of two overlapping core motifs; PTAP and PPEY (Licata et al., 2003), whereas the putative L-domain of Marburg VP40 is composed only of a PPPY motif (Urata et al., 2007). Licata et al. were able to demonstrate that individually, each L-domain motif was able to promote budding of Ebola VP40 VLPs (Licata et al., 2003). In contrast, mutations that disrupted both L-domain motifs resulted in a significant decrease in Ebola VP40 VLP budding (Licata et al., 2003).
These findings were corroborated by Irie et al., who successfully recovered VSV recombinants using reverse-genetics in which the L-domain region of VSV M was removed and replaced with that from Ebola VP40. Thus, the recovered VSV recombinants had either wild type or mutated Ebola L-domains and flanking residues inserted into the M protein of VSV (Irie, Licata, and Harty, 2005). The inserted Ebola VP40 L-domains were found to be functional in the context of a VSV infection, such that recombinant virus titers were virtually equivalent to those of wild type VSV (Irie, Licata, and Harty, 2005). Only the VSV/Ebola recombinant containing mutations in both VP40 L-domain motifs was found to be impaired in budding (Irie, Licata, and Harty, 2005). In addition, efficient budding of VSV recombinants carrying the foreign PTAP-type L-domain from Ebola VP40 was now dependent on expression of host Tsg101, a PTAP L-domain interactor (see below).
The biological relevance of VP40 L-domains was assessed in the context of an actual Ebola virus infection, in which Neumann et al. recovered Ebola virus recombinants containing L-domain mutations (Neumann et al., 2005). The results indicated that the VP40 L-domains were not essential for Ebola virus replication in cell culture, but L-domain activity was important for efficient release of virions from infected cells. Indeed, titers of Ebola virus recombinants lacking L-domain activity were reduced by one order of magnitude compared to those achieved by wild type virus infection in cell culture (Neumann et al., 2005). The functional importance of filovirus L-domains during infection and budding in vivo, in promoting efficient budding in a cell type-dependent manner in vivo, and in serving as a useful and effective target for budding inhibitors in vivo, remain to be determined.
L-domain/host protein interactions
Viral L-domains mediate interactions with specific host proteins to facilitate VLP or virus egress. Most of the key host interactors identified to date (e.g. Tsg101, Nedd4, and Alix) are associated with the cellular ESCRT (Endosomal Sorting Complex Required for Transport) machinery. The function and relevance of the mammalian ESCRT machinery to L-domain interactions and virus budding has been reviewed extensively elsewhere, and thus will not be discussed in detail (Chen and Lamb, 2008; Dolnik, Kolesnikova, and Becker, 2008; Hartlieb and Weissenhorn, 2006; Jasenosky and Kawaoka, 2004; Martin-Serrano, Zang, and Bieniasz, 2003; Pornillos, Garrus, and Sundquist, 2002; von Schwedler et al., 2003; Welsch, Muller, and Krausslich, 2007). The ESCRT pathway is highly conserved in eukaryotes, and is essential for protein sorting on endosomal membranes (Hurley and Emr, 2006; Raiborg, Rusten, and Stenmark, 2003; Saksena et al., 2007; Slagsvold et al., 2006). The multivesicular body (MVB) is an integral part of the ESCRT pathway, and it is the inward invagination and ultimate scission of vesicles into the lumen of the MVB that topologically mimics the process of virus budding from the plasma-membrane. Thus, it follows that hijacking and usurping of host proteins involved in MVB formation and biogenesis by VP40 would likely be beneficial for the process of virus budding (Welsch, Muller, and Krausslich, 2007).
At least three host proteins linked to the ESCRT machinery of the cell, (Tsg101, Nedd4, and vps4) have been implicated in numerous reports in facilitating efficient budding of Ebola and Marburg VP40 (Han and Harty, 2007; Harty et al., 2000; Irie, Licata, and Harty, 2005; Irie et al., 2004; Licata et al., 2003; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Martin-Serrano, Zang, and Bieniasz, 2001; Okumura, Pitha, and Harty, 2008; Panchal et al., 2003; Pornillos et al., 2002b; Silvestri et al., 2007; Timmins et al., 2003b; Urata et al., 2007; Yasuda et al., 2003) (Fig. 2). Although the focus of the discussion is centered around host proteins Tsg101, Nedd4, and vps4, it should be noted that evidence implicating additional components of the cellular VPS pathway in playing a role in virus budding continues to accumulate.
Interactions with host proteins Tsg101 and Nedd4
Human Tsg101 was first implicated in playing a role in virus budding by Garrus (2001) et al., who demonstrated that the UEV domain of Tsg101 interacted with the PTAP L-domain motif of HIV-1 Gag to facilitate efficient budding (Garrus et al., 2001). Elegant structural studies have revealed insights into the physical interactions between the PTAP peptide and the UEV binding pocket of Tsg101, and a series of biochemical approaches have been employed to demonstrate that Tsg101 expression is important for overall budding efficiency (Garrus et al., 2001; Pornillos et al., 2002a, 2002b). A number of subsequent reports have demonstrated that Tsg101 functions in a similar manner by interacting physically and functionally with the PTAP L-domain motif within Ebola VP40, as well as in other viral matrix proteins, to promote efficient budding (Irie, Licata, and Harty, 2005; Licata et al., 2003; Martin-Serrano, Zang, and Bieniasz, 2001; Silvestri et al., 2007; Timmins et al., 2003b; Urata et al., 2007; Yasuda et al., 2003).
Although the precise role of Tsg101 in filovirus budding remains unclear, one thought is that Tsg101 may represent an early upstream entry point for VP40 to access and/or hijack the complete ESCRT machinery of the cell. It has also been suggested that Tsg101 may act in concert with additional host factors linked to both ESCRT function and the ubiquitination machinery of the cell to promote virus budding (Blot et al., 2004; Gottwein et al., 2003; Licata et al., 2003; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Medina et al., 2008; Medina et al., 2005; Pincetic et al., 2008; Timmins et al., 2003b; Urbe, 2005; Usami et al., 2008). Indeed, a role for the cellular ubiquitination machinery in the process of EBOV budding has been described (Harty et al., 2000; Licata et al., 2003; Malakhova and Zhang, 2008; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Okumura, Pitha, and Harty, 2008; Timmins et al., 2003b; Yasuda et al., 2003). Viral matrix proteins like EBOV VP40 are thought to be monoubiquitinated, rather than polyubiquitinated, by the host. Whereas polyubiquitination often targets proteins for degradation in the lysosome, monoubiquitination is thought to represent a tag necessary for engagement of, or entry into, the ESCRT pathway. Ebola VP40 has been shown to be ubiquitinated in vitro and in vivo, and disruption of the cellular ubiquitination machinery resulted in a decrease in overall efficiency of budding (Harty et al., 2000; Licata et al., 2003; Malakhova and Zhang, 2008; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Okumura, Pitha, and Harty, 2008; Timmins et al., 2003b; Yasuda et al., 2003).
The ubiquitination process in the cell represents a series of steps involving E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes, and E3 ubiquitin ligases (Bonifacino and Weissman, 1998; Urbe, 2005). The E3 ubiquitin ligase is typically responsible for target recognition and modification by enzymatically catalyzing the transfer of a ubiquitin moiety onto the target protein. The host E3 ligase implicated by many studies in playing a role in filovirus VP40 budding is Nedd4 (Harty et al., 2000; Irie et al., 2004; Licata et al., 2003; Malakhova and Zhang, 2008; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Okumura, Pitha, and Harty, 2008; Timmins et al., 2003b; Yamayoshi and Kawaoka, 2007; Yasuda et al., 2003). Nedd4 is a member of the HECT (Homologous to the E6-AP Carboxyl Terminus) family of E3 ubiquitin ligases (Bernassola et al., 2008; Chen and Matesic, 2007; Harvey and Kumar, 1999; Ingham, Gish, and Pawson, 2004; Rotin, Staub, and Haguenauer-Tsapis, 2000; Shearwin-Whyatt et al., 2006). We and others have shown that ubiquitination of EBOV VP40 by host Nedd4 E3 ligase is dependent on an interaction between select WW-domains of Nedd4 and the PPxY L-domain of VP40 (Harty et al., 2000; Irie et al., 2004; Licata et al., 2003; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Okumura, Pitha, and Harty, 2008; Yasuda et al., 2003). Interestingly, the PPPY motif of Marburg VP40 was reported to be necessary for mediating interactions with host Tsg101 (Urata et al., 2007). Whether that motif is able to mediate interactions with WW-domains of host Nedd4, leading to ubiquitination, remains to be determined.
Finally, host protein vps4 (isoforms vps4A and vps4B) is an AAA-ATPase and downstream component of the ESCRT machinery. The catalytic activity of Vps4 is thought to be necessary for removal and recycling of the ESCRT components from the membrane, thus promoting membrane fission and vesicularization in MVBs and possibly the final pinching-off stage of virus budding. Numerous studies employing dominant negative mutants of vps4 have demonstrated the importance of this enzymatic activity for efficient egress of VLP and virus budding (Garrus et al., 2001; Gottwein et al., 2003; Irie et al., 2004; Langelier et al., 2006; Licata et al., 2003; Martin-Serrano, Zang, and Bieniasz, 2003; Medina et al., 2005; Schmitt et al., 2005; Shehu-Xhilaga et al., 2004; Silvestri et al., 2007; Urata et al., 2006). Strategies to target both L-domain/host interactions as well as vps4 function to inhibit budding of filoviruses will be discussed below.
VP40 in the bull’s-eye
As the driving force behind filovirus budding, VP40 represents a logical target for antiviral drugs designed to impair filovirus budding and replication. If these antivirals can inhibit the egress and spread of virus, then the host may be afforded more time to mount a robust and protective immune response. As with any attempt to inhibit virus replication, there may be both advantages and disadvantages. For example, the advantages of targeting L-domain/host protein interactions include a detailed and fundamental understanding of how these short peptides interact with host proteins, and the potential broad-based implications of an antiviral that could inhibit L-domain mediated budding of many emerging human pathogens. On the other hand, the disadvantages include potentially deleterious side-effects associated with the disruption of normal host protein function and uncertainty regarding the robustness of inhibition in vivo afforded by such inhibitors.
Strategies to inhibit filovirus budding
Inhibitors capable of disrupting VP40 L-domain interactions with either Tsg101 or Nedd4 would be predicted to impair filovirus budding. By understanding the detailed atomic structure of a protein-protein interaction, a rational approach can be undertaken to screen a large database of small molecules with predicted drug-like properties to identify specific inhibitors of this protein-protein interaction (McInnes, 2007; Stoermer, 2006). Indeed, elegant work by the Sundquist lab revealed the solution structure of the domain of Tsg101 bound to the PTAP peptide that constitutes one active L-domain core motif of Ebola VP40 (Pornillos et al., 2002a, 2002b). Pornillos et al. demonstrated that the PTAP peptide lies in a bifurcated groove above the vestigial enzyme active site of tsg101 (Pornillos et al., 2002a).
Using this information as a starting point, Michael Lee and Mark Olson (USAMRIID, Ft. Detrick, MD) devised a strategy to utilize the ZINC database (Irwin and Shoichet, 2005) which currently lists 5 million commercially-available compounds with drug-like chemical properties, and screen for those that could potentially inhibit binding of the PTAP peptide to the pocket within Tsg101 (Fig. 3). Each molecule was flexibly docked into a rigid binding pocket of tsg101 using the Autodock4 program (Huey et al., 2007) and automated by the DOVIS pipeline (Zhang et al., 2008). The top scoring compounds were re-evaluated by flexible optimization of the protein atoms using a molecular dynamics package and re-scored using an empirically-fitted binding free energy scoring function [e.g., Ligscore (Krammer et al., 2005) and Glide XP (Friesner et al., 2006)]. Finally, the top ranking compounds from this re-scoring process were filtered by qualitative visual evaluation and quantitative criteria such as the number of hydrogen bonds between protein and ligand. The net result of this approach yielded a greatly reduced set of molecules (N ~ 50) that are amenable to further screening (Fig. 3).
Figure 3.
Protocol to identify candidate inhibitors of an EBOV VP40 PTAP-Tsg101 interaction.
In collaboration with the Lee and Olson laboratories, we are currently in the process of screening the top candidate drugs identified by this approach using our Ebola VP40 VLP budding assay. As an initial method of validation, each compound will be assessed for its ability to disrupt a PTAP-dependent VP40-Tsg101 interaction and inhibit release of VP40 VLPs in a dose-dependent manner. In addition to using the traditional VLP budding assay to determine the efficient of VP40 VLP egress, we are also attempting to convert our VLP budding assay into one that is less-cumbersome, more rapid, and amenable to a high-throughput format (McCarthy, Licata, and Harty, 2006)Liu and Harty, unpublished data). Toward this end, we have created a plasmid expressing a luciferase-VP40 fusion protein, with the intent of converting luciferase into a budding protein that is L-domain dependent (Liu and Harty, unpublished data). Such an assay would allow us to rapidly and quantitatively detect luciferase-VP40 in the cell supernatant and be useful for screening and validation of inhibitors of filovirus budding. Indeed, Capul and de la Torre have successfully utilized this approach to generate a luciferase-based budding assay for the Z protein of Lassa fever virus (Capul and de la Torre, 2008).
A second strategy, similar to that described above, is to identify small-molecule inhibitors of a VP40-Nedd4 interaction mediated by the PPxY L-domain of VP40 and the WW-domains of Nedd4. Although not yet proven, it is likely that the PPPY motif of Marburg VP40 will also interact with WW-domains of Nedd4 or those of Nedd4 family members. A number of reports have yielded insight into detailed structural interactions between WW-domains of Nedd4 and PPxY ligands (Harty et al., 1999; Henry et al., 2003; Hu et al., 2004; Kanelis et al., 2006; Kanelis et al., 2000; Kanelis et al., 1998; Kanelis, Rotin, and Forman-Kay, 2001; Sudol et al., 1995), which will help provide the foundation for developing a strategy to identify budding inhibitors that target VP40-Nedd4 interactions. For example, the solution structures of WW domain 3 from human and Drosophila Nedd4 complexed to their respective PPxY ligands have been solved (Kanelis et al., 2006; Kanelis et al., 2001). Importantly, results from both of these studies suggest that, in addition to the core PPxY motif, amino acids flanking the PPxY motif contribute significantly to the high-affinity interaction with the corresponding WW domains of Nedd4. Interestingly, several studies have reported that activity of viral L-domain motifs are context-dependent, in that amino acids flanking the core L-domain motifs are crucial for efficient L-domain function (Irie and Harty, 2005; Li et al., 2002; Martin-Serrano, Perez-Caballero, and Bieniasz, 2004; Zhai et al., 2008). Indeed, if efficient budding of filoviruses is dependent on ubiquitination of VP40 by host E3 ligases, then small molecules designed to interfere with binding of the ligase to its substrate (Oberst et al., 2007) would be predicted to interfere with virus budding.
A third potentially promising strategy to inhibit filovirus budding involves disruption of vps4 catalytic activity. This L-domain independent approach would impair the overall function of the cellular ESCRT machinery, and thus prevent the virus from usurping this machinery for efficient virion release. Indeed, Silvestri et al. recently reported that using phosphorodiamidite morpholino antisense oligonucleotides (PMOs) to block expression of vps4 protected mice from lethal EBOV infection (Silvestri et al., 2007). Mice receiving the vps4A PMO had a delayed time to death compared to the control group, and 70% of the mice survived an otherwise uniformly lethal challenge. Importantly, this approach would be predicted to inhibit budding of both Ebola and Marburg, as well as other viruses that tap into the VPS pathway.
Conclusion
Assembly and budding are essential late events in the replication of Ebola and Marburg viruses. The study of these virus-host interactions continues to progress rapidly, and a plethora of recent studies summarized here have revealed novel and important insights into the structure and function of both viral and host proteins involved in filovirus egress. With this fundamental knowledge of the budding process serving as the foundation, we are now poised to develop novel therapeutics to inhibit filovirus budding by targeting specific functions of VP40 and/or specific VP40-host interactions. Although the identification and validation of small molecules targeting VP40 L-domain/host-protein interactions, as well as host VPS proteins, will be challenging, they should be of high priority. These targets are particularly attractive, since these drug candidates may possess broad-spectrum activity against many emerging human pathogens that utilize similar mechanism for efficient budding. Ideally, an antiviral cocktail targeting multiple steps in the budding pathway (e.g. membrane binding and oligomerization of VP40) would likely provide the most benefit.
Acknowledgments
This work was supported in part by NIH grants to R.N.H.. I would like to thank past and present members of my lab who contributed to this work, including: Jillian Licata, Takashi Irie, Ziying Han, Reed Johnson, Sarah McCarthy, and Atsushi Okumura. I also thank Drs. Michael Lee and Mark Olson for allowing me to share their unpublished findings, and Peter Bell for assistance with electron microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accola MA, Strack B, Gottlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi P, Bocedi A, Heptonstall J, Capobianchi MR, Di Caro A, Mastrangelo E, Bolognesi M, Ippolito G. Ebolavirus and Marburgvirus: insight the Filoviridae family. Mol Aspects Med. 2008;29:151–85. doi: 10.1016/j.mam.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bausch DG, Sprecher AG, Jeffs B, Boumandouki P. Treatment of Marburg and Ebola hemorrhagic fevers: a strategy for testing new drugs and vaccines under outbreak conditions. Antiviral Res. 2008;78:150–61. doi: 10.1016/j.antiviral.2008.01.152. [DOI] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F, Abriel H, Staub O, Dokhelar MC, Pique C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117:2357–67. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. Defense against filoviruses used as biological weapons. Antiviral Res. 2003;57:53–60. doi: 10.1016/s0166-3542(02)00200-0. [DOI] [PubMed] [Google Scholar]
- Bray M, Murphy FA. Filovirus research: knowledge expands to meet a growing threat. J Infect Dis. 2007;196(Suppl 2):S438–43. doi: 10.1086/520552. [DOI] [PubMed] [Google Scholar]
- Bray M, Paragas J. Experimental therapy of filovirus infections. Antiviral Res. 2002;54:1–17. doi: 10.1016/s0166-3542(02)00005-0. [DOI] [PubMed] [Google Scholar]
- Capul AA, de la Torre JC. A cell-based luciferase assay amenable to high-throughput screening of inhibitors of arenavirus budding. Virology. 2008;382:107–14. doi: 10.1016/j.virol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas AM, Nyamathi AM, Sosa A, Wilder CL, Sands H. A current review of Ebola virus: pathogenesis, clinical presentation, and diagnostic assessment. Biol Res Nurs. 2003;4:268–75. doi: 10.1177/1099800403252603. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–32. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- Craven RC, Harty RN, Paragas J, Palese P, Wills JW. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–65. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RC, Parent LJ. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- Dessen A, Forest E, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystallization and preliminary X-ray analysis of the matrix protein from Ebola virus. Acta Crystallogr D Biol Crystallogr. 2000a;56:758–60. doi: 10.1107/s0907444900004388. [DOI] [PubMed] [Google Scholar]
- Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. Embo J. 2000b;19:4228–36. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnik O, Kolesnikova L, Becker S. Filoviruses: Interactions with the host cell. Cell Mol Life Sci. 2008;65:756–76. doi: 10.1007/s00018-007-7406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Klenk HD, Sanchez A. Molecular biology and evolution of filoviruses. Arch Virol Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–96. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Dessen A, Timmins J, Bracher A, Kolesnikowa L, Becker S, Klenk HD, Weissenhorn W. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11:423–33. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88:3195–9. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol. 2003;77:9474–85. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Harty RN. Packaging of actin into Ebola virus VLPs. Virol J. 2005;2:92. doi: 10.1186/1743-422X-2-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Harty RN. Influence of calcium/calmodulin on budding of Ebola VLPs: implications for the involvement of the Ras/Raf/MEK/ERK pathway. Virus Genes. 2007;35:511–20. doi: 10.1007/s11262-007-0125-9. [DOI] [PubMed] [Google Scholar]
- Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A. 2000;97:13871–6. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–9. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–9. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- Henry PC, Kanelis V, O’Brien MC, Kim B, Gautschi I, Forman-Kay J, Schild L, Rotin D. Affinity and specificity of interactions between Nedd4 isoforms and the epithelial Na+ channel. J Biol Chem. 2003;278:20019–28. doi: 10.1074/jbc.M211153200. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Volchkov V, Kolesnikova L, Mittler E, Timmins J, Ottmann M, Reynard O, Becker S, Weissenhorn W. VP40 octamers are essential for Ebola virus replication. J Virol. 2005;79:1898–905. doi: 10.1128/JVI.79.3.1898-1905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ. A map of WW domain family interactions. Proteomics. 2004;4:643–55. doi: 10.1002/pmic.200300632. [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–52. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–98. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–84. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- Irie T, Harty RN. L-domain flanking sequences are important for host interactions and efficient budding of vesicular stomatitis virus recombinants. J Virol. 2005;79:12617–22. doi: 10.1128/JVI.79.20.12617-12622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, Harty RN. Functional characterization of Ebola virus L-domains using VSV recombinants. Virology. 2005;336:291–8. doi: 10.1016/j.virol.2005.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J Virol. 2004;78:2657–65. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–82. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky LD, Kawaoka Y. Filovirus budding. Virus Res. 2004;106:181–8. doi: 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Johnson RF, Bell P, Harty RN. Effect of Ebola virus proteins GP, NP and VP35 on VP40 VLP morphology. Virol J. 2006;3:31. doi: 10.1186/1743-422X-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanelis V, Bruce MC, Skrynnikov NR, Rotin D, Forman-Kay JD. Structural determinants for high-affinity binding in a Nedd4 WW3* domain-Comm PY motif complex. Structure. 2006;14:543–53. doi: 10.1016/j.str.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Kanelis V, Donaldson L, Muhandiram DR, Rotin D, Forman-Kay JD, Kay LE. Sequential assignment of proline-rich regions in proteins: application to modular binding domain complexes. J Biomol NMR. 2000;16:253–9. doi: 10.1023/a:1008355012528. [DOI] [PubMed] [Google Scholar]
- Kanelis V, Farrow NA, Kay LE, Rotin D, Forman-Kay JD. NMR studies of tandem WW domains of Nedd4 in complex with a PY motif-containing region of the epithelial sodium channel. Biochem Cell Biol. 1998;76:341–50. doi: 10.1139/bcb-76-2-3-341. [DOI] [PubMed] [Google Scholar]
- Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat Struct Biol. 2001;8:407–12. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L, Berghofer B, Bamberg S, Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol. 2004;78:12277–87. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007a;9:939–51. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L, Bugany H, Klenk HD, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol. 2002;76:1825–38. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L, Ryabchikova E, Shestopalov A, Becker S. Basolateral budding of Marburg virus: VP40 retargets viral glycoprotein GP to the basolateral surface. J Infect Dis. 2007b;196(Suppl 2):S232–6. doi: 10.1086/520584. [DOI] [PubMed] [Google Scholar]
- Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M. LigScore: a novel scoring function for predicting binding affinities. J Mol Graph Model. 2005;23:395–407. doi: 10.1016/j.jmgm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80:9465–80. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen C, Puffer BA, Montelaro RC. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J Virol. 2002;76:1569–77. doi: 10.1128/JVI.76.4.1569-1577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata JM, Johnson RF, Han Z, Harty RN. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol. 2004;78:7344–51. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77:1812–9. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–7. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. J Virol. 2004;78:5554–63. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–9. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Johnson RF, Zhang YA, Sunyer JO, Harty RN. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J Virol. 2007;81:11452–60. doi: 10.1128/JVI.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Licata JM, Harty RN. A luciferase-based budding assay for Ebola virus. J Virol Methods. 2006;137:115–9. doi: 10.1016/j.jviromet.2006.06.007. [DOI] [PubMed] [Google Scholar]
- McInnes C. Virtual screening strategies in drug discovery. Curr Opin Chem Biol. 2007;11:494–502. doi: 10.1016/j.cbpa.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Medina G, Pincetic A, Ehrlich LS, Zhang Y, Tang Y, Leis J, Carter CA. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology. 2008;377:30–8. doi: 10.1016/j.virol.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina G, Zhang Y, Tang Y, Gottwein E, Vana ML, Bouamr F, Leis J, Carter CA. The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic. 2005;6:880–94. doi: 10.1111/j.1600-0854.2005.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Ebihara H, Takada A, Noda T, Kobasa D, Jasenosky LD, Watanabe S, Kim JH, Feldmann H, Kawaoka Y. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol. 2005;79:10300–7. doi: 10.1128/JVI.79.16.10300-10307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TL, Schoehn G, Weissenhorn W, Hermone AR, Burnett JC, Panchal RG, McGrath C, Zaharevitz DW, Aman MJ, Gussio R, Bavari S. An all-atom model of the pore-like structure of hexameric VP40 from Ebola: structural insights into the monomer-hexamer transition. J Struct Biol. 2005;151:30–40. doi: 10.1016/j.jsb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–65. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Malatesta M, Aqeilan RI, Rossi M, Salomoni P, Murillas R, Sharma P, Kuehn MR, Oren M, Croce CM, Bernassola F, Melino G. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci U S A. 2007;104:11280–5. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105:3974–9. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, Aman MJ. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc Natl Acad Sci U S A. 2003;100:15936–41. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik A, Wills JW. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J Virol. 2002;76:2789–95. doi: 10.1128/JVI.76.6.2789-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters CJ, Khan AS. Filovirus diseases. Curr Top Microbiol Immunol. 1999;235:85–95. doi: 10.1007/978-3-642-59949-1_6. [DOI] [PubMed] [Google Scholar]
- Pincetic A, Medina G, Carter C, Leis J. Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRTproteins to facilitate the budding process. J Biol Chem. 2008 doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002a;9:812–7. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. Embo J. 2002b;21:2397–406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002c;12:569–79. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–55. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Schoehn G, Dessen A, Forest E, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J Mol Biol. 2000;300:103–12. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Demmin GL, Kallstrom G, Javid MP, Badie SS, Will AB, Nelle T, Schokman R, Nguyen TL, Carra JH, Bavari S, Aman MJ. Association of ebola virus matrix protein VP40 with microtubules. J Virol. 2005;79:4709–19. doi: 10.1128/JVI.79.8.4709-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–73. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Salzwedel K, Martin DE, Sakalian M. Maturation inhibitors: a new therapeutic class targets the virus structure. AIDS Rev. 2007;9:162–72. [PubMed] [Google Scholar]
- Sanger C, Muhlberger E, Ryabchikova E, Kolesnikova L, Klenk HD, Becker S. Sorting of Marburg virus surface protein and virus release take place at opposite surfaces of infected polarized epithelial cells. J Virol. 2001;75:1274–83. doi: 10.1128/JVI.75.3.1274-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:145–96. doi: 10.1007/978-3-662-06099-5_5. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–97. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, Weissenhorn W. Membrane association induces a conformational change in the Ebola virus matrix protein. Embo J. 2000;19:6732–41. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617–28. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO. Late domain-dependent inhibition of equine infectious anemia virus budding. J Virol. 2004;78:724–32. doi: 10.1128/JVI.78.2.724-732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri LS, Ruthel G, Kallstrom G, Warfield KL, Swenson DL, Nelle T, Iversen PL, Bavari S, Aman MJ. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J Infect Dis 196 Suppl. 2007;2:S264–70. doi: 10.1086/520610. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–26. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stoermer MJ. Current status of virtual screening as analysed by target class. Med Chem. 2006;2:89–112. doi: 10.2174/157340606775197750. [DOI] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Suzuki Y. Virus infection and lipid rafts. Biol Pharm Bull. 2006;29:1538–41. doi: 10.1248/bpb.29.1538. [DOI] [PubMed] [Google Scholar]
- Tambyah PA. Update on influenza anti-virals. Respirology. 2008;13(Suppl 1):S19–21. doi: 10.1111/j.1440-1843.2008.01249.x. [DOI] [PubMed] [Google Scholar]
- Timmins J, Ruigrok RW, Weissenhorn W. Structural studies on the Ebola virus matrix protein VP40 indicate that matrix proteins of enveloped RNA viruses are analogues but not homologues. FEMS Microbiol Lett. 2004;233:179–86. doi: 10.1111/j.1574-6968.2004.tb09480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003a;312:359–68. doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- Timmins J, Schoehn G, Ricard-Blum S, Scianimanico S, Vernet T, Ruigrok RW, Weissenhorn W. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol. 2003b;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol. 2007;81:4895–9. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. Cellular factors required for Lassa virus budding. J Virol. 2006;80:4191–5. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbe S. Ubiquitin and endocytic protein sorting. Essays Biochem. 2005;41:81–98. doi: 10.1042/EB0410081. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–13. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Welsch S, Muller B, Krausslich HG. More than one door - Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–97. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Kawaoka Y. Mapping of a region of Ebola virus VP40 that is important in the production of virus-like particles. J Infect Dis. 2007;196(Suppl 2):S291–5. doi: 10.1086/520595. [DOI] [PubMed] [Google Scholar]
- Yamayoshi S, Noda T, Ebihara H, Goto H, Morikawa Y, Lukashevich IS, Neumann G, Feldmann H, Kawaoka Y. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe. 2008;3:168–77. doi: 10.1016/j.chom.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77:9987–92. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–9. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhang S, Kumar K, Jiang X, Wallqvist A, Reifman J. DOVIS: an implementation for high-throughput virtual screening using AutoDock. BMC Bioinformatics. 2008;9:126. doi: 10.1186/1471-2105-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]