Abstract
Apolipoprotein B (apoB) is the most abundant protein in low density lipoproteins and plays key roles in cholesterol homeostasis. The co-translational degradation of apoB is controlled by fatty acid levels in the endoplasmic reticulum (ER) and is mediated by the proteasome. To define the mechanism of apoB degradation, we employed a cell-free system in which proteasome-dependent degradation is recapitulated with yeast cytosol, and we developed an apoB yeast expression system. We discovered that a yeast Hsp110, Sse1p, associates with and stabilizes apoB, which contrasts with data indicating that select Hsp70s and Hsp90s facilitate apoB degradation. However, the Ssb Hsp70 chaperones have no effect on apoB turnover. To determine whether our results are relevant in mammalian cells, Hsp110 was overexpressed in hepatocytes, and enhanced apoB secretion was observed. This study indicates that chaperones within distinct complexes can play unique roles during ER-associated degradation (ERAD), establishes a role for Sse1/Hsp110 in ERAD, and identifies Hsp110 as a target to lower cholesterol.
Apolipoprotein B (apoB)3 is a ~540-kDa protein in chylomicrons and very low (VLDL) and low density (LDL) lipoproteins, the atherogenic particles secreted from the liver and small intestine. With the aid of microsomal triglyceride transfer protein (MTP), lipids are added co-translationally to apoB in the endoplasmic reticulum (ER), which results in the formation of a primordial lipoprotein (1, 2). These primordial particles mature upon the acquisition of additional lipids in the secretory pathway and are ultimately transported to the plasma to deliver cholesterol, cholesteryl esters, and triglycerides to peripheral tissues. While much attention has been given to mutations of the LDL receptor causing hypercholesterolemia and accelerated atherosclerosis, mutations in apoB that interfere with LDL endocytosis also lead to hypercholesterolemia (3). Targeted down-regulation of hepatic apoB production lowers the levels of circulating cholesterol in rodents and non-human primates and may thereby reduce atherosclerosis (4–6). Thus, any means to lower apoB levels has the potential for a therapeutic benefit.
ApoB synthesis, and consequently the formation of LDL and VLDL particles, is under tight metabolic control, particularly during its co-translational translocation into the ER (2). When lipids are limiting or if MTP is absent or its function is blocked, apoB translocation is slowed but continued apoB translation exposes large cytoplasmic domains. The resulting translocon-associated apoB species is bound by the cytosolic Hsp70 and Hsp90 molecular chaperones and the polypeptide is polyubiquitinated and degraded by the proteasome. This process ensures that lipoprotein particles, which require apoB for assembly, form only if cellular lipids are abundant and prevents the self-aggregation of hydrophobic domains.
The Hsp70- and Hsp90-catalyzed degradation of apoB is in accordance with the roles that these chaperones play during the targeting of aberrant proteins for proteasomal degradation. Although Hsp70s and Hsp90s facilitate the folding and assembly of multi-protein complexes, these chaperones also recruit components in the ubiquitin proteasome pathway (UPP), most notably E3 ubiquitin ligases, if a polypeptide substrate is unable to achieve its native conformation (7). In its most general form, chaperone-mediated protein “triage” appears to be conserved from yeast to man. For example, we found that in vitro translated apoB was degraded when apoB-containing microsomes were incubated with cytosol prepared from either hepatic cells or yeast; however, degradation was significantly compromised if cytosol was obtained from yeast harboring mutant alleles in the genes encoding either a cytosolic Hsp70 (Ssa1p) or Hsp90 (Hsp82p) (8). These results were consistent with the Hsp70- and Hsp90-mediated enhancement of apoB degradation observed in mammalian cells (8–10).
The proteasomal destruction of ER-resident and membrane-bound proteins has been termed ER-associated degradation (ERAD) (11), and even though molecular chaperones such as Hsp70 and Hsp90 participate to varying extents in the degradation of almost every ERAD substrate examined, the mechanism by which the turnover of apoB occurs may be unique. Not only is apoB degradation metabolically regulated, but it is robust only in select cell types and occurs co-translationally (9, 10, 12–16). These studies suggest that apoB, in addition to being subject to the general ERAD machinery, will also require a unique set of factors that facilitate its biogenesis and degradation. Consistent with this hypothesis, apoB is one of the few ERAD substrates whose degradation requires P58IPK, a chaperone/TPR domain-containing protein (17).
Based on its role in preventing protein aggregation, we hypothesized that Hsp110 may impact the biogenesis of this hydrophobic, aggregation-prone protein. Hsp110s are abundant, cytoplasmic heat shock proteins that maintain the solubility of denatured proteins and possess an N-terminal ATP-binding domain that is homologous to the Hsp70 N-terminal domain (18). Both mammalian Hsp110 and the yeast Hsp110s, Sse1p and Sse2p, interact with and act as nucleotide exchange factors for cytosolic Hsp70s (19–22), and Sse1p function is required to support several Hsp90-dependent activities in yeast (23, 24). Importantly, the degradation of the von Hippel-Lindau (VHL) tumor suppressor is abrogated in yeast lacking Sse1p, indicating that Hsp110 contributes to the turnover of a client of the Hsp90 complex (25). Moreover, Sse1p associates with Ssb1p, an Hsp70 in the ribosome-associated complex (RAC), and sse1 mutants are hypersensitive to translation inhibitors (19–22, 26). These results suggested to us that Hsp110/Sse1p may facilitate the Hsp70/Hsp90-dependent degradation of apoB co-translationally.
We report here that Hsp110/Sse1p, rather than promoting apoB ERAD, is required to stabilize apoB. Our initial results were obtained from cell-free assays that utilize components isolated from wild type and mutant yeast strains. To confirm and extend these data, we also established a yeast apoB expression system and then examined the effects of Hsp110 overexpression in mammalian cells. These results not only identify Hsp110 as a target to reduce cholesterol, but implicate apoB as an Hsp110 client.
EXPERIMENTAL PROCEDURES
Yeast Strains, Molecular Methods, and Antisera
Yeast strains (Table 1) were grown under standard conditions at 30 °C unless otherwise noted, and established media and manipulations were used (27). Antibodies used in this study are listed in supplemental Table S2.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| W3031b | MATα, ade2, his3, leu2, trp1, ura3, can1-100 | 59 |
| SSE1 | MATα, his3, leu2, trp1, ura3 | 23 |
| sse1 Δ (JG014a) | MATα, his3, leu2, trp1, sse1::URA3 | 23 |
| SSE1 (RAY3Aa) | MATα, his3, leu2, ura3 | 59 |
| sse1 Δ (E0020) | MATα, his3, leu2, ura3, sse1::HIS3 | 59 |
| sse1 Δ (W303) | MATα, ade2-1, can1-100, his2-11,15, leu2-3,112, trp1-1, ura2-1, SSE1::Kan | 45 |
| SSB1SSB2 | MATa, his3, leu2, lys2, trp1, ura3 | 60 |
| ssb1 Δ ssb2 Δ | MATa his3, leu2, lys2, trp1, ura3, ssb1-1, ssb2-1 | 60 |
| CIM3 | MATa, ura3-52, his3Δ200, leu2-Δ1 | 38 |
| cim3-1 | MATa, cim3-1, ura3-52, his3Δ200, leu2-Δ1 | 38 |
| PEP4 | MATa, can1-100, leu2-3,112, his3-11, trp1-1, ura3-1, ade2-1 | 42 |
| pep4 Δ | MATa, PEP4::TRP1, leu2-3,112, his3-11, trp1-1, ura3-1, ade2-1 | 42 |
| KAR2 | MATa, ura3-52, leu2-3,112, ade2-101 | 62 |
| kar2-1 | MATa, ura3-52, leu2-3,112, ade2-101, kar2-1 | 62 |
| UBC6UBC7 | MATα, lys2-801, his3Δ200, ura3-52, trp1-1, leu2-3,112 | 63 |
| ubc6 Δ ubc7 Δ | MATα, lys2-801, his3Δ200, ura3-52, trp1-1, leu2-3,112, ubc6::HIS3, ubc7::LEU2, | 63 |
| UFD1 | MATa, his4-519, ura3-52, ade1-100, leu2-3,112 | 64 |
| ufd1-1 | MATa, his4-519, ura3-52, ade1-100, leu2-3,112, ufd1-1 | 64 |
| DOA10HRD1 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1 | 65 |
| doa10 Δ hrd1 Δ | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, doa10:: HIS3, hrd1:: LEU2 | 65 |
| CDC48 | MATa, lys2-801 leu2-3,112 ura3-52 | 55 |
| cdc48-10 | MATa, lys2-801 leu2-3,112 ura3-52 cdc48-10ts | 55 |
| UBC7 | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1 | 63 |
| ubc7 Δ | MATα, his3-Δ200, leu2-3,112, ura3-52, lys2-801, trp1-1, ubc7::LEU2 | 63 |
| sti1 Δ (W303) | MATα, ade2, his3, leu2, trp1, ura3, can1-100, sti1::HIS3 | 61 |
| sba1 Δ (W303) | MATα, ade2, his3, leu2, trp1, ura3, can1-100, sba1::URA3 | 61 |
To express apoB29 in yeast, plasmid pSLW1 was first constructed using pJJB20 (kindly provided by Dr. R. Fuller, University of Michigan), which contains a pBM258 backbone with amino acids 1–100 from the yeast mating factor alpha 1 locus inserted into the BamHI and SalI restriction sites (28). Next, a triple hemagglutinin (HA) tag (YPYDVPDYA) was PCR-amplified with a 5′ XbaI, an internal ClaI, a 3′ SalI restriction site, and tandem stop codons (5′-TAA TGA-3′), and was inserted into pJJB20 at the XbaI and SalI sites to form plasmid pSLW1 (supplemental Table S1). ApoB29 was then amplified from the SP6-apoB48 plasmid (8) by PCR with the following primers: (forward) 5′-ATT GCC AGC ATT GCT AAA GAA GAA GGG GTA TCA CTA CTC AAG AGG AAA ATG TCA GCC TGG TCG TC-3′ and (reverse) 5′-GGG ATA GCC CGC ATA CTC AGG AAC ATC GTA TGG GTA ATC GAT ACT GTA GGA GGC GGA CCA GTT GCT-3′. Finally, ClaI- and XbaI-digested pSLW1 and the apoB29 PCR product were co-transformed into yeast strain W3031b, and colonies containing the gap-repaired pSLW1 plasmid with the apoB29 insert were selected by growth on synthetic complete medium lacking uracil. Plasmid DNA from yeast harboring the pSLW1-B29 plasmid (Fig. 2 and supplemental Table S1) was prepared, and the integrity of the plasmid was confirmed by sequence analysis. All other plasmids used in this study are listed in supplemental Table S1.
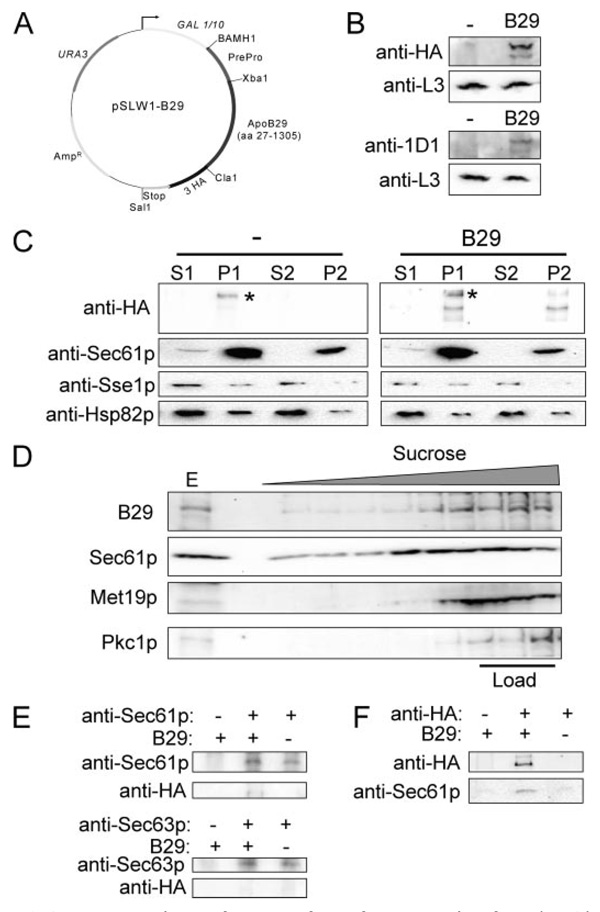
FIGURE 2. ApoB29 is membrane- and translocon-associated.
A, plasmid map of pSLW1-B29, a galactose-inducible, multicopy yeast apoB29 expression vector. B, both anti-HA and anti-apoB (1D1) antibodies detect apoB29, a ~160-kDa protein, expressed in yeast. Blots containing extracts from control (−) or apoB29-expressing (B29) cells were also probed for a ribosomal protein, L3, as a loading control. C, apoB29 fractionates with membranes after differential centrifugation of lysates from apoB29-expressing (B29) or control cells (−). Note the slower migrating band that nonspecifically cross-reacts with the HA antibody (marked with *). S1, 16,000 × g supernatant; P1, 16,000 × g pellet; S2, 150,000 × g supernatant; P2, 150,000 × g pellet. Protein levels were normalized by SDS-PAGE and Coomassie Brilliant Blue staining prior to Western blot analysis. D, apoB29 and the ER membrane-associated protein, Sec61p, migrate to a lower sucrose density when extracts from apoB-expressing cells are layered in a sucrose gradient. The soluble cytosolic proteins glucose-6-phosphate dehydrogenase (Met19p) and protein kinase C (Pkc1p) remain in the fractions where the extracts were loaded into the gradient. The lane marked E indicates 1% of the input (Load). A similar flotation pattern was observed for Sec61p, Met19p, and Pkc1p in cells transformed with the empty vector control (data not shown). E, apoB29 co-precipitates with Sec61p. Cell extracts were prepared from cells transformed with a vector control or pSLW1-B29 (B29) and were mock-treated or were incubated with anti-Sec61p or anti-Sec63p and protein A-Sepharose. The total proteins in the precipitates were resolved by SDS-PAGE and were immunoblotted with the indicated antisera. F, in the reciprocal experiment, Sec61p co-precipitates with apoB29. Cell extracts were prepared and incubated with anti-HA resin (+) or Sepharose 6B beads (−). The precipitates were immunoblotted with the indicated antisera.
In Vitro and in Vivo Methods to Quantify ApoB Degradation
ApoB48 degradation was assessed in vitro as previously described (8), except that time points were taken at 5, 15, and 30 min. The cytosol used in the reactions was prepared as previously described (11) and was diluted to a final concentration of 5–10 µg/µl and preincubated with either 250 µm MG132 (Peptides International) or an equivalent volume of Me2SO for 15 min on ice. The reactions were quenched by adding an equal volume of 125mm Tris, pH 6.8, 4% SDS, 6 m urea, 1mm EDTA, 10 mm dithiothreitol, 250 mm β-mercaptoethanol, 20% glycerol, 0.05% bromphenol blue, and the samples were heated at 96 °C for 4 min prior to SDS-PAGE. Phosphorimager data were analyzed using Image Gauge software (Fuji Film Science Laboratory). To examine the effect of purified Sse1p on apoB stabilization, hexahistidine-tagged Sse1p was purified as previously described (23), dialyzed into 20 mm HEPES, pH7.4, 110 mm KCl, 5 mm MgCl2, and added into the cytosol at a final concentration of 3% of the total protein. The reaction was incubated on ice for 15 min before the degradation reaction commenced.
To measure apoB29 ERAD in yeast, cells transformed with pSLW1-B29 (see above) were grown to logarithmic phase (A600 = 0.4–1.0) overnight at 26 °C in synthetic complete medium lacking uracil but supplemented with glucose to a final concentration of 2%. To obtain maximal expression of apoB29, the cells were harvested and resuspended to an initial concentration of 0.5 A600/ml in complete medium (Yeast Extract-Peptone) containing galactose at a final concentration of 2% and were grown at 30 °C for 5 h. To monitor apoB degradation, protein synthesis was stopped by the addition of cycloheximide to a final concentration of 50 µg/ml, 2 absorbance units of cells were harvested at indicated time points, and total protein was precipitated as previously described (29). The Supersignal West Pico Chemiluminescent Substrate (Pierce) was utilized for anti-L3 immunoblots and the Supersignal West Femto Maximum Sensitivity Substrate (Pierce) was used for anti-HA and anti-apoB immunoblots. The signals were quantified using a Kodak 440CF Image Station and the associated Kodak 1D (v3.6) software (Rochester, NY). The cycloheximide chase analysis of CFTR-HA and Ste6p*-HA and the pulse chase analysis of ppαfΔG-HA and CPY*-HA were performed as previously described (29–32). Previous studies on ERAD in yeast have failed to uncover a difference in chaperone dependence on the degradation of ER-associated proteins when either cycloheximide or metabolic labeling followed by a methionine chase was employed.4 For all cycloheximide chase analyses, each time point was normalized to the L3 loading control, and the relative amount of apoB was calculated by dividing the signal at each time by the value at t = 0.
Biochemical and Immunological Methods
ApoB29 expression was induced as above and a total of 100 ODs of cells were harvested and fractionated as previously published (33). To assess whether apoB29 was carbonate-extractable, ~40 µg of yeast lysate prepared as described for the fractionation analysis were treated with 100 mm NaCO3, pH 11.5 (33). The apoB sucrose gradient flotation assay was performed as published (29).
For apoB interaction studies, cell extracts were prepared by glass bead lysis from 100 absorbance units of yeast grown as described above, and immunoprecipitations were conducted in the presence of 20 mm NaMoO4 (20) using anti-Sse1p, anti-Ssa1p, anti-Ssb1p, anti-Sec61p, or anti-Sec63p antiserum (see above). For apoB immunoprecipitation experiments with the anti-HA resin (Roche Applied Science), cell extracts from 100 absorbance units of yeast were prepared by 3 5-min agitations with glass beads in modified Roche lysis buffer (50 mm Tris, pH 7.5, 25 mm NaCl, 0.1% Nonidet P40, 20 mm NaMoO4) with protease inhibitors (0.25 mm MG132, 1 mm phenylmethylsulfonyl fluoride, 1 µg/ml leupeptin, 0.5 µg/ml pepstatin A). Next, 1 mg of cell extract was incubated overnight at 4 °C with anti-HA resin or Sepharose 6B beads that were used as a negative control (Sigma). The beads were washed twice with Roche lysis buffer containing 150 mm NaCl and 300 mm NaCl, respectively. The isolated protein precipitates were resolved by SDS-PAGE, and the relevant proteins were identified by immunoblot analysis as described above.
Hsp110 Overexpression in Rat Hepatoma Cells
Rat hepatoma McA-RH7777 cells were cultured and transfected with either the pcDNA3.1 vector (control) or the pcDNA3.1-Hsp110 expression vector (provided by Dr. J. Subjeck, Roswell Park Cancer Institute), and 48 h after transfection a pulse-chase analysis was performed as previously described (8). Experiments to which either oleic acid or the MTP inhibitor was added (at a final concentration of 0.625 µm and 0.1 nm respectively) measured the amount of apoB-precipitable material from vector versus Hsp110-overexpressing cells after a 15-min pulse and 60-min chase.
RESULTS
Degradation of Microsomal ApoB48 Is Enhanced in Yeast Cytosol Lacking Sse1p
To determine if Sse1p contributes to the regulated degradation of apoB, we utilized cytosols prepared from sse1Δ yeast and an isogenic wild-type strain in a previously established in vitro ERAD assay (8). In this assay, an isoform of apoB (“apoB48”) is translated in the presence of dog pancreas microsomes and 35S-labeled methionine, and after reisolation of the apoB-containing vesicles, the degradation of the radiolabeled substrate is measured in the presence of yeast cell lysates. ApoB48, which is ~48% of the size of full-length apoB (260 kDa), is expressed endogenously and secreted from rodent hepatic and rodent and human intestinal cells, and undergoes MTP and lipid-dependent maturation (34).
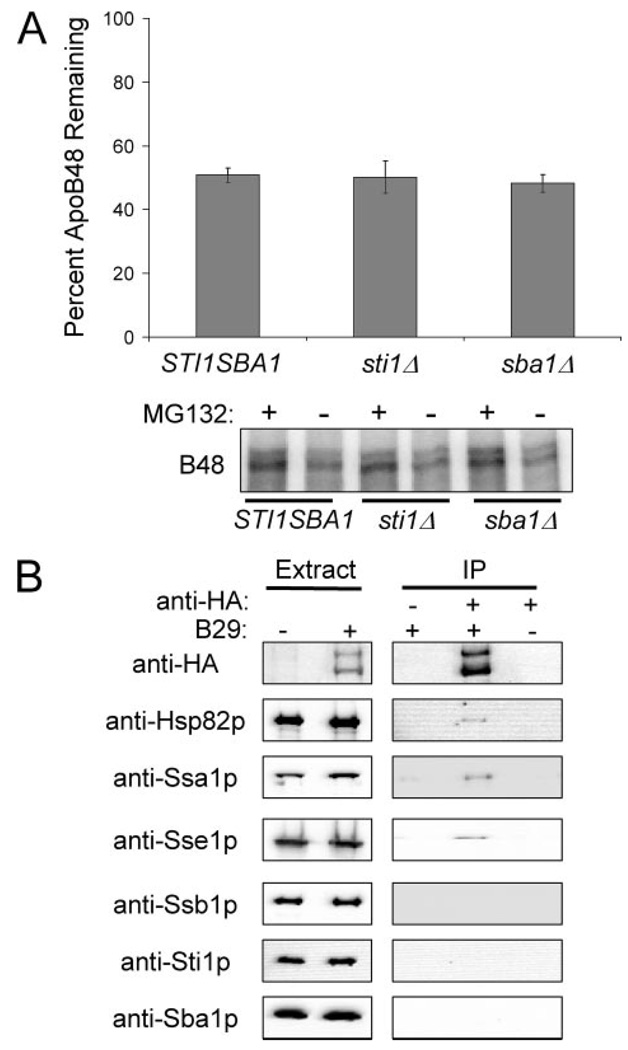
As published previously (8), ~70% of the apoB remained after 30 min in a reaction supplemented with cytosol from a wild-type yeast strain. The observed degradation was also MG132-inhibitable, indicating that it was proteasome-dependent (Fig. 1, A and B). In contrast, when apoB degradation was examined in the presence of cytosol from the sse1Δ strain, only ~50% of the translated apoB remained following a 30-min incubation.
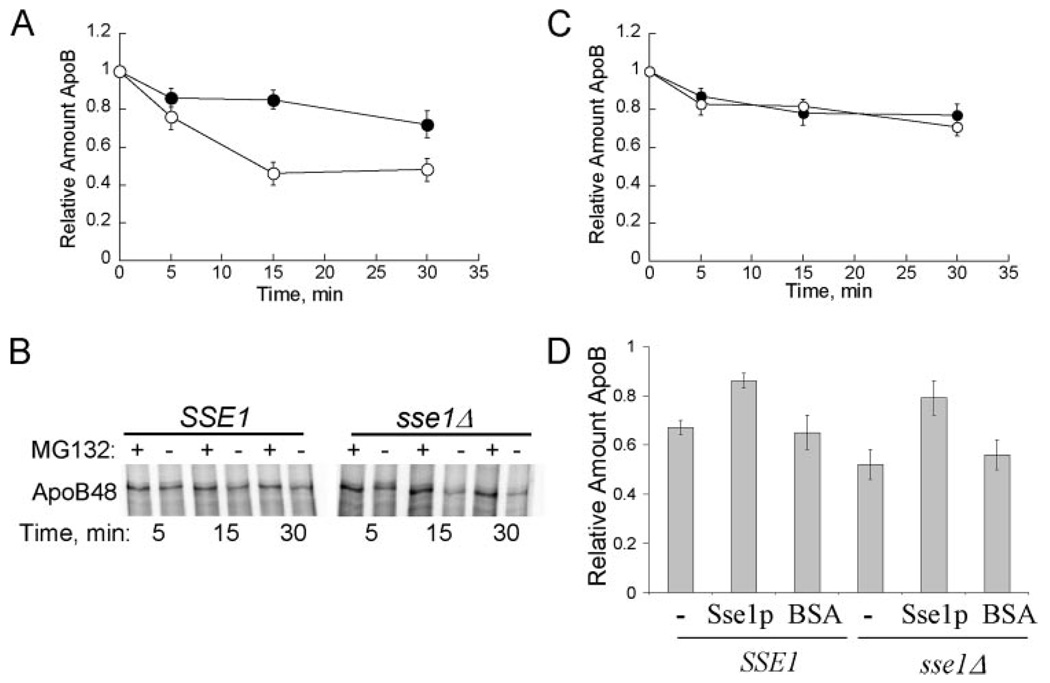
FIGURE 1. Sse1p contributes to apoB48 stabilization in vitro.
A, apoB48 degradation was assessed in vitro at 37 °C using cytosol (5 µg/µl) from SSE1 (●) or sse1Δ (JG014a) (○) yeast. The percentage of apoB remaining was calculated by normalizing the amount of apoB in the Me2SO-treated samples to those samples treated with MG132, thus reflecting only the degree of proteasome-mediated degradation. Data represent the means from six independent experiments ± S.E. of the means: 15 min, p < 0.0002; 30 min, p < 0.0005. B, phosphorimage of 35S-labeled apoB48 during a representative degradation assay in cytosol from SSE1 or sse1Δ (JG014a) yeast. Reactions were treated with the proteasome inhibitor MG132 (250 µm) or Me2SO (−) as indicated. C, apoB48 degradation is similar in cytosol from ssb1Δssb2Δ cells and the isogenic wild-type cells. ApoB48 degradation was assessed in vitro at 37 °C using cytosol (5 µg/µl) from SSB1SSB2 (●) or ssb1Δssb2Δ (○) yeast, and quantified as above. Data represent the means from six independent experiments ± S.E. of the means. D, degradation reactions containing the indicated source of cytosol at a final concentration of 5 µg/µl were supplemented with Sse1p at a final concentration of 3% of the total protein or an equal amount of bovine serum albumin (BSA). Reactions were incubated at 37 °C for 15 min. Data represent the means of 3–9 experiments ± S.E. of the mean. The activity of purified Sse1p was confirmed in steady state ATPase assays: 0.5 nmol of ATP hydrolyzed min−1 mg protein−1. SSE1, cytosol ± purified Sse1p, p < 0.02; sse1Δ, (JG014a) cytosol ± purified Sse1p, p < 0.002.
Sse1p is reported to interact with and modulate the ATPase activity of two cytosolic Hsp70s in yeast, Ssa1p and Ssb1p (20–22). Even though Ssa1p facilitates apoB degradation in vitro (8), the contribution of Ssb1p, an Hsp70 in the RAC, on apoB degradation had not been previously tested. To this end, cytosols were prepared from a strain deleted for SSB1 and the SSB2 homologue and from an isogenic wild-type strain. Next, apoB degradation was assessed in vitro (Fig. 1C). In contrast to the pro-degradative effect of Ssa1p on apoB and the stabilizing effect of Sse1p on apoB, we found that the levels of apoB were unaltered when the Ssb chaperones were absent. It should be noted that the extent of degradation observed was lower than in Fig. 1A, which is consistent with the previously observed difference in apoB ERAD when different yeast strain backgrounds were utilized (8). Nevertheless, these data suggest that individual Hsp70s and Hsp70 facilitators can exert unique effects during the biogenesis of a given polypeptide, and point to the complexity with which these chaperones and co-chaperones act.
To determine if the enhanced degradation of apoB in cytosols lacking Sse1p was specific and not the result of secondary consequences, purified Sse1p was supplemented into the degradation reactions. As shown in Fig. 1D, Sse1p stabilized apoB regardless of whether it was added to cytosols prepared from sse1Δ cells or the isogenic wild-type strain. The addition of an irrelevant protein, bovine serum albumin, had no impact on apoB degradation.
ApoB29 Is Membrane- and Translocon-associated in Yeast
To confirm and extend these results (and to create a genetic system in which other components required for apoB biogenesis can be isolated or examined) a yeast apoB expression system was developed. For this purpose, a galactose-inducible yeast expression vector was constructed to produce an HA-tagged apoB isoform that is ~29% of the size of full-length apoB (Fig. 2A, pSLW1-B29). In mammalian cells, apoB29 forms a lipoprotein particle and is the shortest form of apoB that matures and traffics normally through mammalian cells (35). Because of yeast codon bias (data not shown), the apoB signal sequence was replaced with the signal sequence and pro-region from yeast pre-pro-alpha factor. This pre-pro sequence was chosen because it was used previously to express the β-amyloid precursor protein (APP) in yeast (28).
Under induction conditions (see “Experimental Procedures”), the pSLW1-B29 plasmid (Fig. 2A) directed the expression of a protein of the correct molecular mass and that was detected by both anti-HA and anti-apoB antibodies (Fig. 2B). The localization of the expressed protein was then examined by subcellular fractionation. As shown in Fig. 2C, apoB29 was present in the pellet fractions (P1 and P2), as was Sec61p, a component of the ER translocon, suggesting membrane association. In contrast, both Sse1p and Hsp82p, which loosely associate with the ER, were detected primarily in the second supernatant (S2). To confirm that apoB29 associates with membranes in yeast, cell extracts were mixed with a dense sucrose solution and overlaid with sucrose solutions of lower density. Following high-speed centrifugation, much of the apoB29 was found in fractions of lower sucrose density with Sec61p (Fig. 2D). Note that the cytoplasmic proteins, glucose-6-phosphate dehydrogenase (Met19p) and protein kinase C (Pkc1p) remained in the load fractions. Finally, an intimate membrane association of apoB29 in yeast was established by carbonate extraction (see supplemental Fig. S1).
To assess whether the apoB isoform expressed in yeast interacted with the translocon, as observed in mammalian cells (12, 13, 36), we employed non-denaturing conditions to immunoprecipitate Sec61p, and probed the precipitate to determine if apoB29 was present. ApoB29 was detected in the precipitate with Sec61p, suggesting that the protein associates with the translocon (Fig. 2E). Correspondingly, when apoB29 was immunoprecipitated with anti-HA resin, Sec61p was found in the precipitate (Fig. 2F). In contrast, apoB29 was absent from the precipitate when Sec63p, another integral membrane protein in the ER, was immunoprecipitated (Fig. 2E). These data indicate that apoB29 is targeted to the ER and interacts with the translocon during its biogenesis in yeast.
The Degradation of ApoB29 in Yeast Requires Components of the Ubiquitin Proteasome Pathway
ApoB degradation depends upon the UPP both in vitro and in mammalian cells (see Introduction). To determine whether the degradation of apoB29 in yeast similarly required components of the UPP, cycloheximide chase analyses were performed to evaluate apoB29 degradation in strains with mutations in UPP-encoding genes.
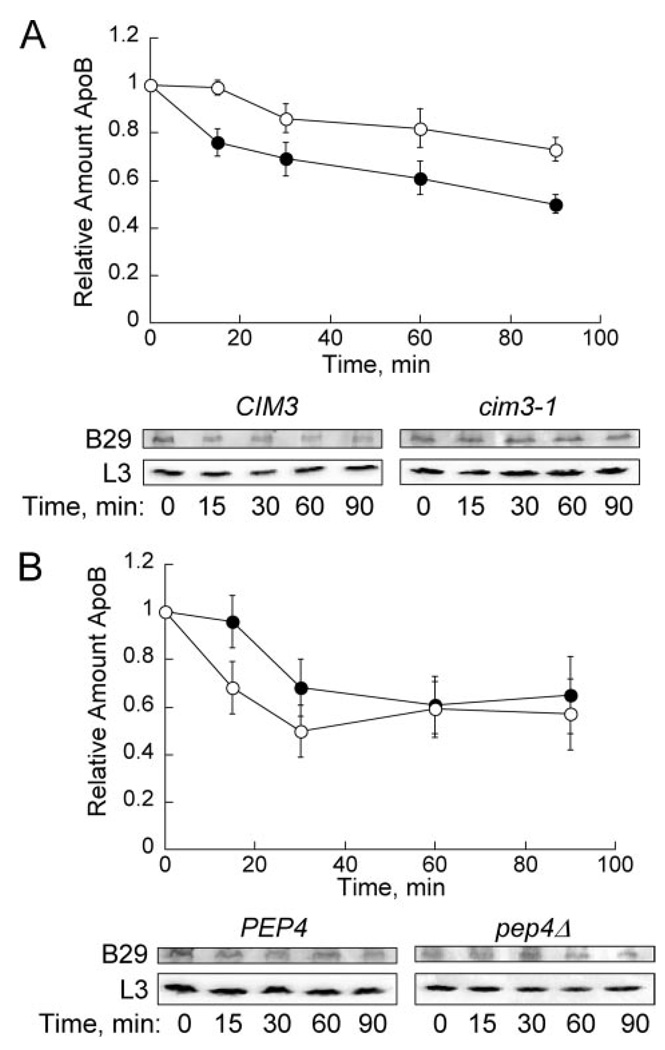
We first tested the contribution of CIM3, which encodes one of six AAA ATPases in the 19S regulatory particle of the 26S proteasome (37). In cim3-1 cells, ubiquitinated substrates are stabilized (38), and the ERAD of a mutant form of carboxypeptidase Y (CPY*) and pro-alpha factor are slowed (39, 40). As displayed in Fig. 3A, apoB29 was significantly stabilized in the cim3-1 strain. ApoB29 ERAD was also abrogated in strains containing mutations in KAR2, DOA10HRD1, UFD1, UBC6UBC7, and UBC7 each of which are known to compromise proteasome activity and/or ERAD to varying extents (supplemental Fig. S2) (41). In contrast, apoB turnover was robust in yeast lacking UBC6 (an ER-associated E2; data not shown) or in cells deleted for PEP4, which encodes a vacuolar protease; loss of Pep4p abrogates nearly all vacuolar protease activity (42) (Fig. 3B). Together, these data establish apoB29 as an ERAD substrate in yeast.
FIGURE 3. ApoB29 is degraded by the proteasome in yeast.
A, cycloheximide chase was performed in CIM3 (●) and cim3-1 (○) cells transformed with pSLW1-B29. ApoB29 was detected using anti-HA antibody, and L3 was used as a loading control. For all cycloheximide chase analyses, each time point was normalized to the L3 loading control, and the relative amount of apoB was calculated by dividing the signal at each time by the value at t = 0. Data represent the means from nine independent experiments ± S.E. of the means: 60 min, p < 0.02; 90 min, p < 0.002. B, cycloheximide chase in PEP4 cells (●) and pep4Δ cells (○) transformed with pSLW1-B29 was performed. Data represent the means from six independent experiments ± S.E. of the means.
Sse1p Stabilizes and Associates with ApoB29 in Yeast
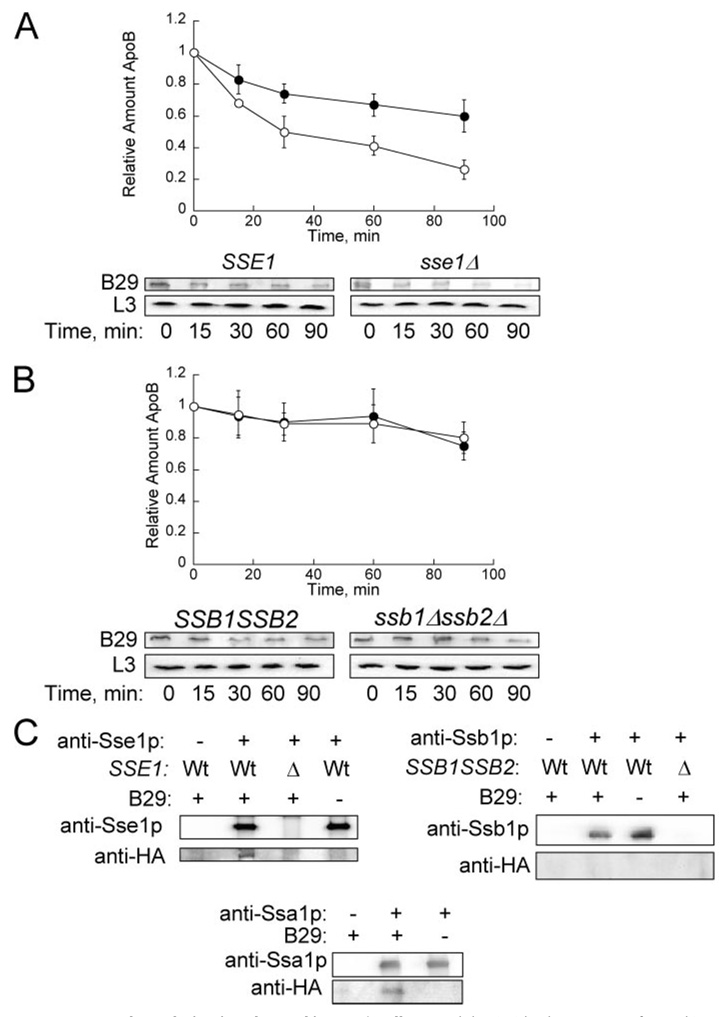
To ascertain whether the results obtained using yeast cytosols could be recapitulated in vivo, cycloheximide chase analyses were performed to evaluate the degradation of apoB29 in sse1Δ, in ssb1Δssb2Δ, and in the respective isogenic wild-type strains. As in the cell-free studies, we found that the proteolysis of apoB29 was enhanced in sse1Δ cells but that degradation was unchanged in cells lacking Ssb1p and Ssb2p (Fig. 4, A and B). To determine whether Sse1p and apoB co-precipitate, extracts were prepared from SSE1 and sse1Δ yeast transformed with either pSLW1-B29 or with the vector control, and a specific anti-Sse1p antiserum (23) was used to immunoprecipitate the chaperone. We found that apoB29 only precipitated from strains that expressed Sse1p and apoB29 (Fig. 4C).
FIGURE 4. ApoB29 degradation is enhanced in sse1Δ cells.
A, cycloheximide chase was performed in SSE1 (●) and sse1Δ (E0020) cells (○) transformed with pSLW1-B29. ApoB29 was detected using anti-HA. L3 was used as a loading control. Data represent the means from eight independent experiments ± S.E. of the means. 60 min, p < 0.01; 90 min, p < 0.004. B, cycloheximide chase was performed in SSB1SSB2 (●) and ssb1Δssb2Δ cells (○) transformed with pSLW1-B29. Data represent the means from five independent experiments ± S.E. of the means. C, apoB29 co-precipitates with Sse1p and Ssa1p, but not with Ssb1p. Extracts were prepared from cells transformed with a vector control or with pSLW1-B29 (B29) and were treated with anti-Sse1p, anti-Ssa1p, or anti-Ssb1p, and protein A-Sepharose. The proteins in the precipitates were resolved by SDS-PAGE and were immunoblotted with the indicated antisera.
Because all of the Sse1p in the cell was reported to be in heterodimeric complexes with either Ssb1p or Ssa1p (19), we wanted to assess if apoB interacted with Ssb1p even though deletion of this Hsp70 did not impact the rate of apoB degradation. However, apoB29 failed to co-precipitate with Ssb1p, implying that the Sse1p-apoB interaction occurs independently of this Hsp70. In contrast, when we immunoprecipitated Ssa1p, the Hsp70 that facilitates apoB degradation in yeast (8), apoB29 resided in the precipitate. Therefore, while both Sse1p and Ssa1p are proposed to be in a heterodimeric complex and we can detect the interaction of these chaperones with apoB, they affect the ERAD of this substrate differently.
We next addressed whether the enhanced degradation of apoB in the sse1Δ strain is substrate-specific. We found that there was no difference in the rate of degradation of CFTR, CPY*, and pαf in the SSE1 and sse1Δ strains. In contrast, Ste6p* stabilization was observed in the sse1Δ strain (supplemental Fig. S3). Others have reported that there is no difference in the rate of degradation of CPY* lacking its signal sequence in sse1Δ and wild-type cells, but reduced degradation of VHL was noted in strains lacking Sse1p (25, 43). Although these collective data indicate diverse effects on the turnover of ERAD substrates in Sse1p-deficient strains, none of the substrates tested exhibited the enhanced degradation we observed for apoB29 in the sse1Δ strain. Overall, Sse1p appears to play a unique role during the ERAD of apoB.
Other Co-factors in the Hsp90 Complex Do Not Contribute to ApoB ERAD
Hsp90 assists in the targeting of apoB for ERAD and elevated Hsp90 levels result in increased apoB degradation (8). Furthermore, Sse1p is a component of the Hsp90 complex and is involved in the folding of substrates associated with the Hsp90 complex (23, 24). Therefore, to determine if other components of the Hsp90 complex contribute to apoB degradation we examined apoB disappearance in cytosols from sti1Δ and sba1Δ cells, which are deleted for the HOP and p23 homologs, respectively. As shown in Fig. 5A, apoB ERAD is similar in cytosols from the wild type and deletion strains. In addition, apoB degradation was tested in cytosol from sse1Δsti1Δ cells and the extent of degradation was similar to levels observed in the cytosol from the sse1Δ cells (data not shown). We also investigated the contributions of the functionally redundant Hsp40s, Ydj1p, and Hlj1p, during apoB degradation because of the reported genetic interaction between Sse1p and Ydj1p and the functional association of these Hsp40s with Ssa1p (23, 44). When examined by cycloheximide chase analysis, the degradation of apoB was slowed by 23% (p < 0.02) in a yeast strain containing the temperature-sensitive ydj1-151 allele and that was deleted for HLJ1, suggesting that these Hsp40s may participate in the targeting of apoB for degradation.5
FIGURE 5. The Hsp90 co-chaperones, Sba1p and Sti1p, do not contribute to ApoB ERAD.
A, apoB48 degradation was assessed in vitro at 37 °C for 30 min using cytosol (5 µg/µl) from STI1SBA1, sti1Δ, or sba1Δ yeast. The percentage of apoB remaining was calculated by normalizing the amount of apoB in the Me2SO-treated samples to those samples treated with MG132, thus reflecting only the degree of proteasome-mediated degradation. Data represent the means from six independent experiments ± S.E. of the means. The lower panel contains a representative phosphorimage of 35S-labeled apoB48 during the degradation assay in cytosol from STI1SBA1, sti1Δ, or sba1Δ yeast. Reactions were treated with the proteasome inhibitor MG132 (250 µm) or Me2SO (−), as indicated. B, apoB29 co-precipitates with Hsp82p, Ssa1p, and Sse1p, but not with Ssb1p, Sti1p, or Sba1p. Extracts were prepared from cells transformed with a vector control or with pSLW1-B29 (B29) and were treated with anti-HA resin or an unconjugated Sepharose resin control. The proteins in the precipitates were resolved by SDS-PAGE and were immunoblotted with the indicated antisera.
Does apoB interact with all of the components of the Hsp90 complex? To answer this question, the protein was again immunoprecipitated using non-denaturing conditions and Hsp82p, Ssa1p, and Sse1p were detected in the precipitate by Western blot. However, we failed to observe Ssb1p, Sti1p, or Sba1p co-precipitating with apoB (Fig. 5). Therefore, chaperones that impact apoB ERAD (i.e. Sse1p, Ssa1p, Hsp82p) associate with apoB, whereas factors that have no effect on the degradation of apoB fail to associate with the lipoprotein (i.e. Ssb1p, Sti1p, Sba1p).
ATP Binding Is Required for Sse1p-mediated Stabilization of ApoB
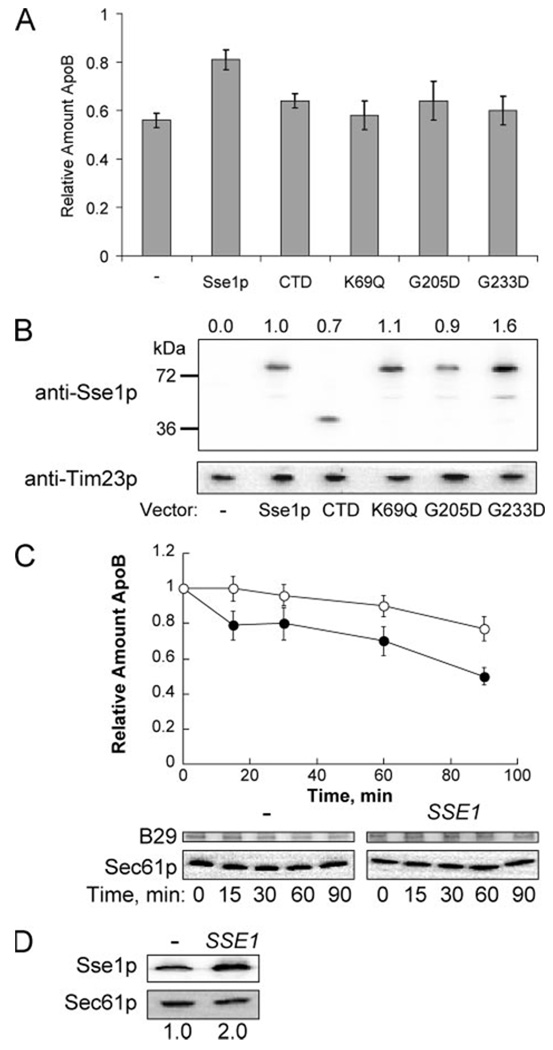
Unlike the Hsp70 chaperones, Sse1p is a “holdase” that retains proteins in solution but is unable to catalyze folding, and the N-terminal ATP-binding domain is dispensable for holdase activity (23). However, the ability of Sse1p to support viability in some strain backgrounds may require ATP binding, but not ATPase activity (45). To determine which features of Sse1p help stabilize apoB, we assessed apoB29 degradation in an sse1Δ strain transformed with Sse1p expression constructs that encode wild type or mutant forms of the protein (45). We first observed that cells expressing full-length Sse1p (from the introduced plasmid) stabilized apoB29 compared with cells transformed with the vector control (Fig. 6A), as anticipated based on the data presented above. However, cells expressing a K69Q mutant (that binds ATP but is unable to hydrolyze nucleotide), Sse1p-G205D (that is unable to bind ATP), Sse1p-G233D (that cannot bind Ssa1p and lacks ATPase activity), and the C-terminal peptide-binding domain of Sse1p that maintains some holdase activity (CTD, Ref. 23) did not significantly stabilize apoB, even though the proteins were expressed to similar or even higher levels than the wild-type control (Fig. 6, A and B).
FIGURE 6. The Sse1p ATP-binding domain is required for apoB29 stabilization.
A, apoB29 degradation was assessed by cycloheximide chase analysis in sse1Δ (W303) cells transformed with pSLW1-B29 and the indicated Sse1p expression constructs (see supplemental Table S1). Data correspond to the means of the 90-min time point from six independent experiments ± S.E. of the means: p < 0.004 for cells transformed with the control vector versus cells transformed with the wild-type expression vector. B, relative levels of Sse1p expression in each strain were examined and compared with the amount of full-length Sse1p in the wild-type control. Values above the blot correspond to the means for four independent blots. The mitochondrial protein, Tim23p, was used as a loading control. C, apoB29 degradation was assessed by cycloheximide chase analysis in SSE1 (RAY3Aa) cells transformed with pSLW1-B29 and the p414TEF-SSE1 expression vector (○) or an empty vector control (●). Data represent the means from six independent experiments ± S.E. of the means. 60 min, p < 0.02; 90 min, p < 0.005. D, Sse1p steady state levels increased 2-fold in the cells transformed with p414TEF-SSE1 compared with the empty vector control. Sse1p was detected using anti-Sse1p. Sec61p was used as a loading control. Data represent the means from three independent experiments.
Because purified Sse1p stabilized apoB in vitro (Fig. 1D), we asked whether increased amounts of Sse1p would also stabilize apoB29 in yeast. Sse1p was overexpressed from an introduced plasmid about 2-fold and enhanced stabilization of apoB29 was noted when compared with control cells (Fig. 6, C and D). Interestingly, these conditions were unable to lead to detectable apoB secretion (data not shown). This phenomenon was also evident when apoB100 was examined in HepG2 cells that had been treated with proteasome inhibitors, i.e. an increase in intracellular apoB did not lead to an increase in secreted material (data not shown) (13). In any event, these data do indicate that a modest increase in Sse1p protects apoB from ERAD.
Hsp110 Overexpression in Hepatic Cells Enhances ApoB Secretion
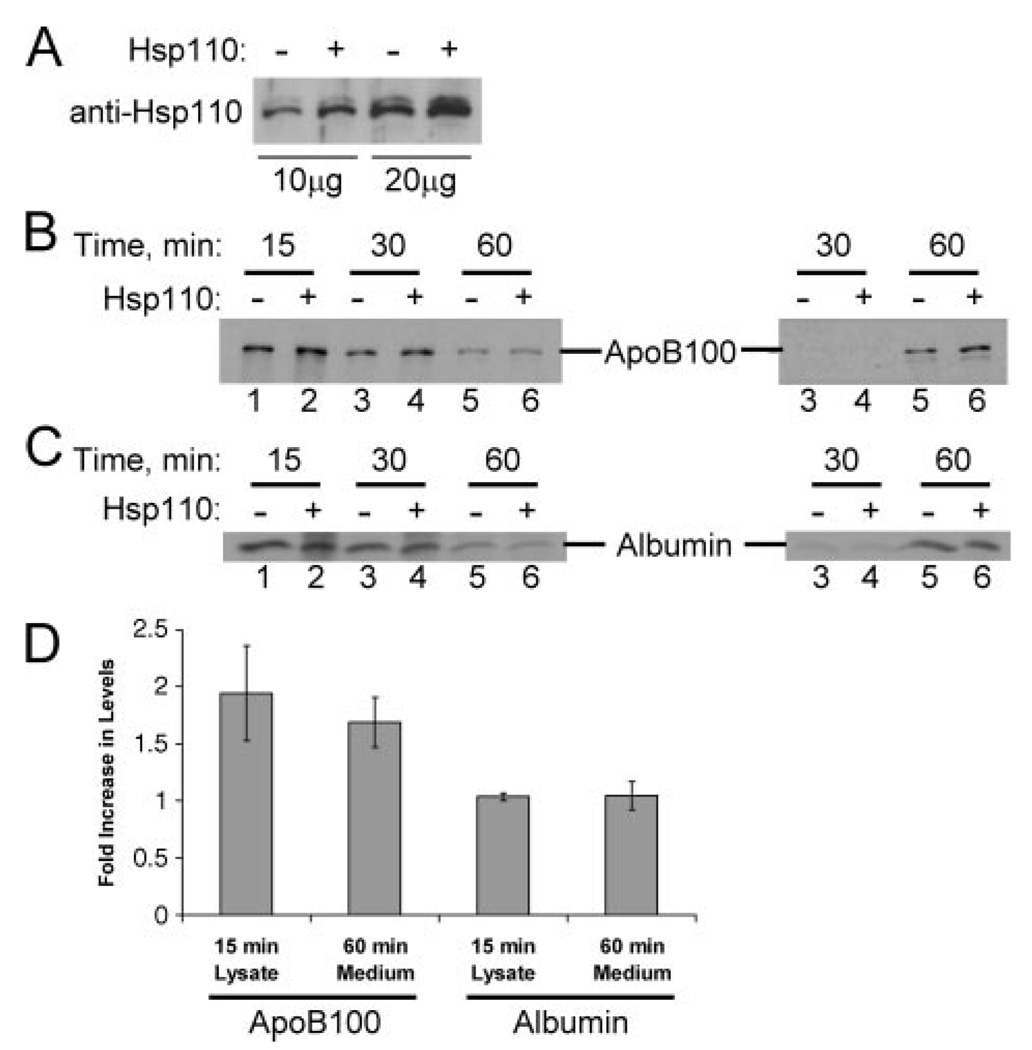
ApoB is normally synthesized in hepatic cells and is secreted in mature VLDL particles, which are then converted to LDL particles. Although mammalian Hsp110 failed to improve the slow growth phenotype of sse1 yeast,6 we were curious whether Hsp110 might also be involved in apoB biogenesis in hepatic cells. To test this hypothesis, we first showed that Hsp110 was present in McArdle-RH7777 cells (see below), which are a rat hepatoma cell line and an established model of apoB metabolism, and in liver extracts (data not shown). In addition, the cells secrete apoB100, the form that is produced by human liver cells. Next, the cells were transfected with an Hsp110 overexpression plasmid or with a control vector and a pulse chase analysis was performed. We initially observed that Hsp110 expression increased only 1.8-fold in cells transfected with the Hsp110 expression vector (Fig. 7A), suggesting that the effect on apoB, if any, would be subtle. Nevertheless, we observed 1.9-fold more apoB intracellularly at the 15 min time point, an effect that translated into an 1.7-fold increase in apoB secretion after 60 min (Fig. 7, B and D). The lack of apoB in the conditioned medium samples at the 30-min time point (Fig. 7B) was expected based on previous data that it takes ~40 min for newly synthesized apoB to be secreted from hepatic cells (46). As a control for this experiment, we found that the levels and secretion of albumin, another protein synthesized in hepatic cells, was unaffected regardless of whether Hsp110 was overexpressed (Fig. 7, C and D). When we examined apoB recovery in Hsp110 overexpressing cells and control cells in the presence of oleic acid, a compound that stimulates apoB secretion, we observed an increase in the percent of apoB recovered in the lysate and the medium fractions from 49.6% (control) to 78% (upon Hsp110 overexpression) (data not shown). Furthermore, we found that the overexpression of Hsp110 in these cells in the presence of an MTP inhibitor (BMS-200150) (47) increased the percent of apoB recovered in the lysate and medium fractions from 14 to 28% (data not shown).
FIGURE 7. Hsp110 overexpression protects apoB and increases its secretion from rat hepatoma cells.
A, equal amounts of cell lysates from McArdle-RH7777 cells transfected with pcDNA3.1 or a pcDNA3.1-Hsp110 expression plasmid were analyzed by Western blotting with an anti-Hsp110 antibody. B and C, McArdle-RH7777 cells were transfected either with pcDNA3.1 (lanes 1, 3, and 5) or the pcDNA3.1-Hsp110 expression plasmid (lanes 2, 4, and 6), and 48 h after transfection cells were pulse-labeled with [35S]methionine/cysteine for 10 min and chased in isotope-free medium for 15 (lanes 1 and 2), 30 (lanes 3 and 4), or 60 min (lanes 5 and 6). Cell lysates (left), and conditioned media samples (right) containing an equal number of trichloroacetic acid-insoluble counts per mg of total protein were immunoprecipitated with antibodies against apoB (B) or albumin (C), and the immunoprecipitates were analyzed by SDS-PAGE and fluorography. Note that the form of apoB displayed is apoB100, the species secreted by a normal human liver. Data were reproducible in five complete experiments. D, fold increase in apoB100 and albumin levels in McArdle-RH7777 cells transfected with pcDNA3.1-Hsp110 compared with control cells transfected with pcDNA3.1. Data represent the means from five independent experiments ± S.E. of the means. 15 min, p < 0.04; 60 min, p < 0.02.
DISCUSSION
In this work, we identify apoB as a substrate for Hsp110 and find that the chaperone stabilizes apoB in vitro and in both yeast and mammalian cells. This result was unexpected given that the yeast Hsp110 homologues, Sse1p and Sse2p, interact with an Hsp70 chaperone that enhances apoB degradation. Equally surprising was our discovery that other Sse1p-interacting Hsp70s, Ssb1p, and Ssb2p, have no effect on apoB biogenesis. Because all of the Sse1p in the cell may be associated with either Ssa1p or the Ssbs (19), these data suggest that related chaperones or that chaperones within a single complex can act uniquely during the “decision” to protect or degrade a newly synthesized polypeptide at the ER membrane.
How might Sse1p/Hsp110 protect apoB from proteasome-mediated degradation? Because Sse1p and apoB co-immunoprecipitate, and because apoB is targeted for ERAD co-translationally (9, 10, 15–17, 48), we suggest that the chaperone binds to and shields the substrate as it is being synthesized. This hypothesis is consistent with the reported ribosome-association of Sse1p and the sensitivity of sse1Δ mutants to translation poisons (26). This hypothesis is also consistent with the fact that large, hydrophobic loops of untranslocated apoB are exposed to the cytosol if MTP activity is absent (10, 12, 13). Such hydrophobic tracts are prime binding sites for chaperones, which may be required to retain apoB in solution and/or target it to the UPP. However, Sse1p is not simply protecting apoB by virtue of its “holdase” activity, which only requires the C-terminal domain in vitro (23). Instead, we found that a functional ATP-binding domain is required for Sse1p to stabilize apoB. Consistent with this observation we previously reported that the Sse1p ATP-binding domain is required to rescue the temperature-sensitive growth defect of yeast expressing a mutant form of an ER-associated Hsp40 chaperone, Ydj1p (23). Therefore, the ATP binding and hydrolytic activities of Sse1p are critical for substrate protection or to recruit other cytoplasmic factors that may protect newly synthesized apoB from the UPP. It is tempting to speculate that these events require the recently described nucleotide exchange activity of Sselp (19, 21, 22). To begin to address this hypothesis, we examined apoB ERAD in a strain deleted for two other known Hsp70 exchange factors, SNL1 and FES1, but failed to note any change in apoB stability (data not shown). How the NEF activity of Sse1p is coupled to substrate/co-factor recruitment awaits further study.
We also describe in this report the first yeast expression system for an apoB isoform, and by co-opting this system we discovered that the ERAD of apoB requires both cytosolic and lumenal factors. The importance of cytoplasmic and lumenal factors, especially chaperones, is consistent with the apoB bitopic orientation across the ER membrane before it is degraded (2). For example, we found that the ERAD of apoB was attenuated in yeast containing a thermosensitive mutation in the gene encoding BiP (supplemental Fig. S2), a lumenal Hsp70 chaperone that associates with apoB (49–52). BiP facilitates the degradation of some ERAD substrates because it prevents the aggregation of ER lumenal polypeptides prior to their retro-translocation (33). Given the large, lipophilic N-terminal domains of apoB that reside initially in the ER it is not surprising that BiP may augment the retro-translocation competence of apoB. Consistent with this view it was reported that overexpression of BiP resulted in accelerated apoB turnover (53). We also found that apoB degradation was attenuated in yeast containing a mutation in the gene encoding Ufd1p. Ufd1p and Npl4p are adaptors for Cdc48p, which is the yeast homologue of p97/VCP. Cdc48p is a member of the AAA ATPase family that when coupled with the Npl4p and Ufd1p adaptors is thought to drive the extraction of polyubiquitinated ERAD substrates from the ER membrane and may function as a protein “disaggregase” (54). In addition, we observed reduced apoB degradation in a strain containing the cdc48-10 mutant allele (55) and in vitro when cytosols were prepared from yeast containing a ufd1 mutant allele.7 To date, it is not clear whether Cdc48p/p97 directly impacts the degradation of apoB in mammalian cells, but our data suggest that this complex may function similarly during the disposal of apoB.
Do members of the Hsp110 family play a role in the biogenesis of other proteins at the ER membrane? We previously reported that the degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) in yeast was unaffected in cells lacking Sse1p (44), and have found that Hsp110 overexpression has no effect on CFTR biogenesis in HEK293 cells.8 However, the degradation of a cytoplasmic, human protein, VHL, was slowed in yeast deleted for SSE1 (25). In addition we found that the degradation of a yeast membrane protein, Ste6p*, was compromised in sse1Δ yeast. We again confirmed that there was no impact on the degradation of another membrane protein, CFTR, in yeast. There was no difference in the degradation of the soluble substrates, CPY* and pαF in cells lacking Sse1p. At present, then, there is no way to predict beforehand what effect the Hsp110 chaperones may have on a cellular substrate.
Based on yeast proteomic and genomic analyses, a large number of chaperones and co-chaperones are known to interact with Sse1p. With the exception of Cdc37p, which is involved primarily in kinase maturation (56), we examined each of these factors (Sti1p, Ssb1p, Ssa1p, Hsp82p, and Ydj1p) for their effects on apoB degradation. We also observed interactions between apoB and each of the chaperones that impact apoB ERAD (Hsp82p, Sse1p, and Ssa1p). In contrast, we were unable to establish interactions between those chaperones and co-factors that do not contribute to the degradation of apoB (Sti1p, Sba1p, Ssb1p). Similarly, proteomic analyses in mammalian cells indicate that the degradation of wild-type CFTR and the disease-causing ΔF508-CFTR have distinct co-chaperone interactions and requirements (57). These data suggest that each ERAD substrate possesses specific, but likely overlapping chaperone requirements during folding and/or degradation.
The decision to either degrade or stabilize apoB must be finely balanced because VLDL particles are rapidly assembled and secreted when lipids are abundant; therefore, factors that slow apoB degradation are vital to regulate the transition between ERAD and VLDL assembly (58). One protein, P58IPK (17), was recently reported to enhance apoB degradation but not the turnover of most other ERAD substrates, and here we report on a factor that instead stabilizes apoB. Although disabling P58IPK function is predicted to increase VLDL and LDL production, and thus circulating cholesterol levels, Hsp110 inhibition would have the opposite, and desirable, effect. Future research efforts will seek to identify additional factors involved in regulating apoB biogenesis and lipoprotein production.
Supplementary Material
Acknowledgments
We thank Drs. Robert Fuller, John Subjeck, Ross Milne, Elizabeth Craig, Jon Warner, Avrom Caplan, Toshi Endo, Mark Hochstrasser, Davis Ng, and Kevin Morano for reagents, and Sheara Fewell, Ursula Andreo, Anthony Petruso, Jacob McBride, and Donald Schleicher for technical assistance. We also thank Christine Wright and Shruthi Vembar for critical review of the manuscript.
Footnotes
This work was supported in part by National Institutes of Health Grants HL058541 (to E. A. F. and J. L. B.) and GM75061 (to J. L. B.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
Supported in part by Predoctoral Grant 0515316U from the American Heart Association.
The abbreviations used are: apoB, apolipoprotein B; ERAD, endoplasmic reticulum-associated degradation; ER, endoplasmic reticulum; LDL, low density lipoprotein; MTP, microsomal triglyceride transfer protein; HA, hemagglutinin; UPP, ubiquitin proteasome pathway; VLDL, very low density lipoprotein; RAC, ribosome-associated complex.
J. L. Brodsky, unpublished data.
J. McBride, S. L. Hrizo, and J. L. Brodsky, data not shown.
K. Morano, personal communication.
D. Schleicher, S. L. Hrizo, and J. L. Brodsky, data not shown.
A. Ahner and J. L. Brodsky, unpublished data.
REFERENCES
- 1.Hussain MM, Shi J, Dreizen P. J. Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 2.Fisher EA, Ginsberg HN. J. Biol. Chem. 2002;277:17377–17380. doi: 10.1074/jbc.R100068200. [DOI] [PubMed] [Google Scholar]
- 3.Whitfield AJ, Barrett PH, van Bockxmeer FM, Burnett JR. Clin. Chem. 2004;50:1725–1732. doi: 10.1373/clinchem.2004.038026. [DOI] [PubMed] [Google Scholar]
- 4.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 6.Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Cyr DM, Hohfeld J, Patterson C. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 8.Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. J. Biol. Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- 9.Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. J. Biol. Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- 10.Zhou M, Fisher EA, Ginsberg HN. J. Biol. Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 11.McCracken AA, Brodsky JL. J. Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pariyarath R, Wang H, Aitchison JD, Ginsberg HN, Welch WJ, Johnson AE, Fisher EA. J. Biol. Chem. 2001;276:541–550. doi: 10.1074/jbc.M007944200. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell DM, Zhou M, Pariyarath R, Wang H, Aitchison JD, Ginsberg HN, Fisher EA. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14733–14738. doi: 10.1073/pnas.95.25.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeung SJ, Chen SH, Chan L. Biochemistry. 1996;35:13843–13848. doi: 10.1021/bi9618777. [DOI] [PubMed] [Google Scholar]
- 15.Liao W, Yeung SC, Chan L. J. Biol. Chem. 1998;273:27225–27230. doi: 10.1074/jbc.273.42.27225. [DOI] [PubMed] [Google Scholar]
- 16.Benoist F, Grand-Perret T. J. Biol. Chem. 1997;272:20435–20442. doi: 10.1074/jbc.272.33.20435. [DOI] [PubMed] [Google Scholar]
- 17.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, Taunton J, Katze MG, Ron D. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 18.Easton DP, Kaneko Y, Subjeck JR. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaner L, Wegele H, Buchner J, Morano KA. J. Biol. Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 20.Yam AY, Albanese V, Lin HT, Frydman J. J. Biol. Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
- 21.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Mol. Biol. Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu XD, Morano KA, Thiele DJ. J. Biol. Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- 25.McClellan AJ, Scott MD, Frydman J. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 27.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 28.Zhang H, Komano H, Fuller RS, Gandy SE, Frail DE. J. Biol. Chem. 1994;269:27799–27802. [PubMed] [Google Scholar]
- 29.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W, Spear ED, Ng DT. Mol. Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Ng DT, Spear ED, Walter P. J. Cell Biol. 2000;150:77–88. doi: 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loayza D, Tam A, Schmidt WK, Michaelis S. Mol. Biol. Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu AL, Windmueller HG. J. Biol. Chem. 1981;256:3615–3618. [PubMed] [Google Scholar]
- 35.Segrest JP, Jones MK, De Loof H, Dashti N. J. Lipid Res. 2001;42:1346–1367. [PubMed] [Google Scholar]
- 36.Chen Y, Le Caherec F, Chuck SL. J. Biol. Chem. 1998;273:11887–11894. doi: 10.1074/jbc.273.19.11887. [DOI] [PubMed] [Google Scholar]
- 37.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghislain M, Udvardy A, Mann C. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 39.Lee RJ, Liu CW, Harty C, McCracken AA, Latterich M, Romisch K, DeMartino GN, Thomas PJ, Brodsky JL. EMBO J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH. J. Biol. Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa S, Brodsky JL, Nakatsukasa K. J. Biochem. (Tokyo) 2005;137:551–555. doi: 10.1093/jb/mvi068. [DOI] [PubMed] [Google Scholar]
- 42.Jones EW, Zubenko GS, Parker RR. Genetics. 1982;102:665–677. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. Mol. Biol. Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Mol. Biol. Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. J. Biol. Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- 46.Borchardt RA, Davis RA. J. Biol. Chem. 1987;262:16394–16402. [PubMed] [Google Scholar]
- 47.Jamil H, Gordon DA, Eustice DC, Brooks CM, Dickson JK, Jr, Chen Y, Ricci B, Chu C-H, Harrity TW, Ciosek CP, Jr, Biller SA, Gregg RE, Wetterau JR. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11991–11995. doi: 10.1073/pnas.93.21.11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du EZ, Fleming JF, Wang SL, Spitsen GM, Davis RA. J. Biol. Chem. 1999;274:1856–1862. doi: 10.1074/jbc.274.3.1856. [DOI] [PubMed] [Google Scholar]
- 49.Linnik KM, Herscovitz H. J. Biol. Chem. 1998;273:21368–21373. doi: 10.1074/jbc.273.33.21368. [DOI] [PubMed] [Google Scholar]
- 50.Rashid KA, Hevi S, Chen Y, Le Caherec F, Chuck SL. J. Biol. Chem. 2002;277:22010–22017. doi: 10.1074/jbc.M112448200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Herscovitz H. J. Biol. Chem. 2003;278:7459–7468. doi: 10.1074/jbc.M207976200. [DOI] [PubMed] [Google Scholar]
- 52.Adeli K, Macri J, Mohammadi A, Kito M, Urade R, Cavallo D. J. Biol. Chem. 1997;272:22489–22494. doi: 10.1074/jbc.272.36.22489. [DOI] [PubMed] [Google Scholar]
- 53.Qiu W, Kohen-Avramoglu R, Mhapsekar S, Tsai J, Austin RC, Adeli K. Arterioscler. Thromb. Vasc. Biol. 2005;25:571–577. doi: 10.1161/01.ATV.0000154142.61859.94. [DOI] [PubMed] [Google Scholar]
- 54.Jentsch S, Rumpf S. Trends Biochem. Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandal AK, Lee P, Chen JA, Nillegoda N, Heller A, Distasio S, Oen H, Victor J, Nair DM, Brodsky JL, Caplan AJ. J. Cell Biol. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, Riordan JR, Kelly JW, Yates JR, 3rd, Balch WE. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 58.Tietge UJ, Bakillah A, Maugeais C, Tsukamoto K, Hussain M, Rader DJ. J. Lipid Res. 1999;40:2134–2139. [PubMed] [Google Scholar]
- 59.Shirayama M, Kawakami K, Matsui Y, Tanaka K, Toh-e A. Mol. Gen. Genet. 1993;240:323–332. doi: 10.1007/BF00280382. [DOI] [PubMed] [Google Scholar]
- 60.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 61.Fang Y, Fliss AE, Rao J, Caplan AJ. Mol. Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- 63.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 64.Ye Y, Meyer HH, Rapoport TA. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 65.Swanson R, Locher M, Hochstrasser M. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.