Abstract
Molecular imprinting is a generic technology that allows for the introduction of sites of specific molecular affinity into otherwise homogeneous polymeric matrices. Commonly this technique has been shown to be effective when targeting small molecules of molecular weight <1500, while extending the technique to larger molecules such as proteins has proven difficult. A number of key inherent problems in protein imprinting have been identified, including permanent entrapment, poor mass transfer, denaturation, and heterogeneity in binding pocket affinity, which have been addressed using a variety of approaches. This review focuses on protein imprinting in its various forms, ranging from conventional bulk techniques to novel thin film and monolayer surface imprinting approaches.
1. Introduction
Specific molecular recognition is the fundamental process governing control of both biological form and function. This impressive and ubiquitous biological phenomenon is mediated, in the main, by proteins. Proteins are complex macromolecule assemblies possessing sites that can controllably and specifically interact with and bind target molecules. The binding process arises as a result of attractive forces that exist between complimentary loci on the protein (or host) and ligand (or guest molecule). Binding of the host to the guest arises when these forces result in a host-guest complex leading to a decrease in the free energy of the system. Therefore binding specificity is simply an indirect measure of the energy of the system. These forces result in either entropy- or enthalpy-driven reduction in the free energy of the system. The binding process depends on the appropriate geometric organization of functional groups, including hydrophilic domains, within the host molecule that match or fit reciprocal functionality on the guest. The conceptionally simple “lock and key” hypothesis of protein-guest interaction is perhaps the most useful illustration of the principle. Moreover, biological recognition in macromolecules, in particular antibodies, has led to development of highly useful and adaptable laboratory tools. Extending upon the natural systems, the production of artificial recognition elements (e.g., “plastic antibodies”) that mimic these highly selective agents is also a highly desirable research objective that may afford unique advantages over the biological counterparts.
A number of divergent strategies, both natural and synthetic, have been described that operate as molecular structures and act as “host” sites for a ligand or “guest” moiety. Crown ethers (1), cyclophanes (2), phage display generated peptides (3), cyclodextrins (4), and dendrimers (5) are examples of these. Methods to synthesize these materials rely upon either “design and synthesis” or “coincidental fit” approach. One drawback to the host-guest route is the lack of a generic process, meaning that each recognition problem requires a novel solution. Approaches that utilize naturally occurring macromolecules to act as selective recognition sites are well documented (6).
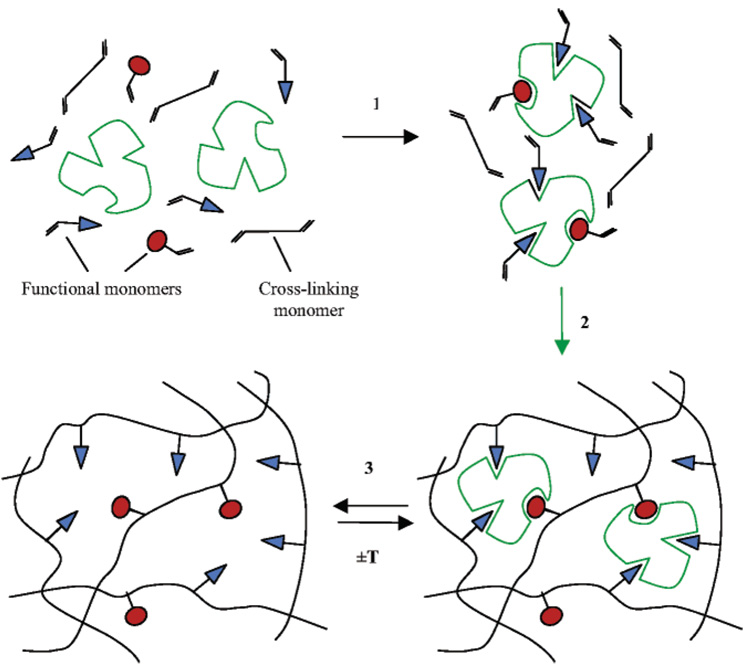
An alternative strategy is to prearrange the recognition site around the target molecule or template. This strategy is the basis for the process of molecular imprinting. Compounds with functional groups reciprocal to those of a target molecule or template are selected and used to form a scaffold around the chosen template. This complex is then preserved within a matrix to form an imprint that is chemically and sterically complementary to the template (Figure 1). Molecular imprinting is now a well-established technology in the field of synthetic molecular recognition, offering a generic, robust, and cost-effective alternative to existing techniques such as monoclonal antibodies (7). The nature of molecular imprinting lends itself to the generation of 3D imprint systems where binding sites are formed throughout a bulk material. However, molecular imprinting on surfaces (2D imprinting) is gaining momentum especially when forming imprints against large macromolecules such as proteins and in sensor design (8).
Figure 1.
Schematic of molecular imprinting process adapted from Byrne (9). 1: formation of pre-polymerization complex; 2: polymerization; 3: template removal/rebinding.
Molecular imprinting has been extensively reviewed in both the periodic press (7, 10–13) and in a series of books (14–17), and the number of original articles and patents have continued to increase (18). However, despite the current good health of molecular imprinting in general, the commercially important subdiscipline of protein imprinting remains underdeveloped and remains largely in its infancy.
Commonly, small (MW ⋦1000) multifunctional template molecules have been shown to give rise to high affinity and specificity MIPs (19); however, an increase in molecule size and functionality on the template leads to increased binding site heterogeneity seen in respective polymers (20). Commonly these systems have utilized polar noncovalent interactions to establish polymer-template reciprocal functionality on which MIP function relies. Examples of this type of strategy includes amino acids (21), drugs such as theophylline and diazepam (22), nucleotides (23), and pesticides (24). An alternative approach relies initially on covalent interactions to establish reciprocal functionality, while subsequent rebinding has been demonstrated through either the re-establishment of covalent linkages or via a noncovalent mechanism (25). Examples of the type of templates addressed using this approach are sugars (26), monoalcohols (27), and steroids (28).
1.1. Protein Imprinting
A challenging area that is currently receiving a great deal of attention is the molecular imprinting of bio-macromolecules and in particular proteins. In the area of selective protein binding current laboratory practice is almost entirely dependent on antibodies to achieve specific protein entrapment in assays and in isolation, extraction, and biosensors (29). However, such systems are notoriously expensive and often suitable only for single use due to the fragility and labile nature of the protein recognition element involved. Hence there is a strong demand for inexpensive, robust, and reusable alternatives to these biological moieties (30) that afford levels of selectivity and specificity similar to those of their biological counterparts, as has been shown with small molecule imprinting (12, 31, 32).
The potential application of such materials could also extend well beyond the laboratory. From the biological or cellular perspective, biologically functional proteins rarely act independently, and protein–protein interactions influence all aspects of cell regulation and control. Therefore it is possible that protein-selective MIPs could be developed as useful tools through which it would be possible to influence control over biological systems (29).
1.2. Obstacles Faced when Imprinting Proteins
Despite the obvious advantages in the development of these systems, there is a paucity of literature on the development of MIPs for the recognition of biological macromolecules (29). This suggests intrinsic limitations in the scalability of traditional molecular imprinting techniques from small bioactives to macromolecules.
The historical dominance of small molecule imprinting means that, to a great extent, both the covalent and the noncovalent approaches have focused on the development of materials that favor the smaller templates, in terms of polymer composition and structure. In particular the arguably key technique of preparing dense (meth)acrylate polymer monoliths in a nonpolar solvent is wholly unsuited for protein imprinting. When imprinting proteins there are a number of key issues to address that are largely absent when targeting small molecules, and these are related to the molecular size, complexity, conformational flexibility, and solubility.
1.2.1. Size
Traditional polymer monoliths tend to be relatively dense, leading to difficulty in a macromolecular template reaching (or leaving) any formed binding site. Such poor mass transport and permanent entrapment result in inadequate recognition properties. While a porogen may be added during imprinting to better control the MIP surface area (7), the pervasiveness of these problems often necessitates shattering and grinding of the monolith in an effort to expose binding sites. This in turn can damage a formed binding site.
1.2.2. Complexity
Unlike smaller templates, proteins present a large number of potential recognition sites over a relatively large surface area. Different regions of a protein exhibit distinct physiochemical properties and hence problems can arise with recognition and cross reactivity. It is accepted that MIPs exhibit greatest selectivity when the number of points of interaction is small but the interactions themselves are strong. Multiple weak interactions are less likely to produce a highly specific MIP and tend to favor nonspecific binding (14).
1.2.3. Conformational Flexibility
Polymerization conditions can also be problematic when applying traditional imprinting procedures to proteins. The nonphysiological conditions often employed during polymerization may denature proteins or force them into conformations or aggregates that are not suitable for imprinting. This flexibility could affect selectivity as a protein might alter conformation to fit into an imprint formed for an alternate template. This could be especially true with families of proteins or those that have been selectively modified (by site-directed mutagenesis for example). It can be theorized that this effect could work to our advantage as imprints could be used as a method of forcing a protein into a known conformation, hence obtaining a form of catalysis.
1.2.4. Solubility
Perhaps the greatest challenge of all is the limited choice of solvent. The majority of molecular imprinting takes place in apolar, organic solvents in an attempt to maximize electrostatic interactions, such as hydrogen bonding, upon which many MIPs rely for recognition. However, the poor stability and solubility of most proteins in apolar solvents limits their use. This also limits the choice of monomers available for selection as many popular monomers are insoluble or partially soluble in water. Water is far from the ideal solvent for conventional molecular imprinting because it will compete for and potentially disrupt any hydrogen-bonding sites on the template and monomers (13).
It is clear that the size and complexity of proteins and their behavior in solution will play an important part in an attempt to imprint them. However the major factor limiting this work is the effects that the organic solvents, normally used for imprinting, have upon the structure and surface properties of protein. Solubility still limits the choice of polymer composition and suitable monomers as it is vital to maintain the conformation of the template upon polymerization. This makes imprinting in aqueous solutions a necessity, as specific imprints of proteins must be tailored to the native structure to be of any use in an assay or sensor system. The influence of water competing for binding sites is an important factor that has to be considered, but it has been shown that this effect is more pronounced with small molecule binding, and thus using a macromolecular template with increased surface interactions can lower or remove this effect (16). Many groups in the past have studied imprinting within aqueous solution, with a variety of templates. We therefore look to these examples to obtain clues to best imprint our target macromolecular templates in these conditions.
1.3. Imprinting in Aqueous Environments
It is theorized that most imprints work best in the solvent that was used for polymerization (7); therefore the use of water as a solvent has been studied for several targets with the aim of using the resultant imprinted polymers in aqueous systems. Hydrophobic interactions, caused by the exclusion of water, are frequently employed for imprinting in aqueous systems, and hydrophobically driven molecular imprints have proven successful with a variety of small molecules in a range of polymer matrices.
Kugiyama relied on hydrophobic interactions between pyridine monomers and the chosen template sialic acid to produce a MIP capable of selective recognition in aqueous media (33). Likewise, Haupt imprinted the hydrophobic herbicide 2,4,5-trichlorophenoxyacetic acid, in a mixture of methanol and water (34). This polar medium elicited hydrophobic binding sites within a 4-vinylpyridine (4-VP) functionalized imprinted polymer.
Perez-Moral produced core-shell particles with surface imprints that used shape-specific hydrophobic interactions to recognize the hydrophobic molecule cholesterol (35, 36). Similarly, Carter formed core–shell particles, prepared in water using emulsion polymerization. These particles were noncovalently imprinted with caffeine and theophylline, using both hydrophobic and electrostatic interactions (37, 38). The caffeine imprinted system displayed selective molecular recognition, although this method did not produce a system selective for theophylline over caffeine. In a different approach to the same templates, Han demonstrated a novel method of aqueous imprinting using an oil/water emulsion (39). The selected monomer and template sequestered into water droplets within the oil and formed complexes. These complexes were then transferred to a microporous membrane substrate and photopolymerized. Using this method, selectivity between theophylline and caffeine was demonstrated with a concentration-dependent separation factor of 4.9 ± 0.8. This patented method offers a possible scale-up to macromolecules as mass transfer problems are reduced by the thin film imprints.
Dirion used a high-throughput synthesis and evaluation of MIP sorbents to optimize a MIP for the local anaesthetic bupivacaine in aqueous systems (40). The resultant MIPs showed high imprinting factors in water, attributed to reduced nonspecific binding to the imprinted polymer. Batches of these sorbents were successfully employed for extraction of the template from blood plasma samples.
In proteins, most hydrophobic residues are generally sequestered in the core of the native structure, though patches of hydrophobicity often exist on the protein surface and could thus be targets for molecular recognition. Similarly, polar residues presented on the surface of a protein are more suited to hydrogen bonding and electrostatic interactions. Proteins use these types of interactions to maintain conformation and folding. A reciprocal hydrogen bond acceptor or donor group positioned within a polymer matrix, corresponding to a suitable region on the surface of a protein, would offer excellent binding, using the directional properties of a hydrogen bond to its advantage.
Mathew developed a polymer capable of recognizing adenine over other nucleotide bases, in an aqueous medium, using methacrylic acid (MAA) and 4(5)-vinylimidazole (41). This polymer also demonstrated pH-dependent binding of adenosine 5′-triphosphate (ATP). They interpreted this in terms of hydrogen bonding and ionic interactions between ATP and corresponding ligands within the polymer.
The group of Shea presented an approach toward the development of oligonucleotide receptors (42). A 2′-deoxyadenosine dimer was imprinted in aqueous based solvents using the conventional noncovalent imprinting compounds MAA and ethylene glycol dimethacrylate (EGDMA). The developed MIPs demonstrated selectivity for adenine over guanine. The same group also reported a new procedure for imprinting peptides, utilizing the affinity of Ni(II) for N-terminal histidine (His) residues (43). A complex between Ni(II) linked with a polymerizable ligand and an N-terminal His peptide was prepared in water. This complex was then copolymerized with other hydrophilic monomers, including the water-soluble cross-linker N,N′-ethylenebis(acrylamide) (BisA) and optimized. The resultant material used the His-Ni(II) interaction, combined with secondary interactions between peptide and polymer, to distinguish between a series of simple peptides.
Sol–gel systems are highly compatible with aqueous environments (44). Typically, the formation of this type of polymer requires mild conditions (temperature, pH), with the functionalized interactions between template and monomer favoring weak intermolecular forms (hydrogen bonding, van der Waals). Relying on these forces between template and monomer, Zhang demonstrated stereoselectivity within an imprinted sol-gel thin film in aqueous solution (45). Selectivity was demonstrated, using a quartz crystal microbalance (QCM), for several amino acids and l and d isomers of histidine. Similarly, Makote produced a selective imprint for dopamine in a sol-gel hybrid film using cyclic voltammetry and UV–vis adsorption spectroscopy for detection (46).
It is clear that forming imprints and using them for selective rebinding in aqueous systems is achievable in a variety of matrices using different forms of polymer/template interactions. Hydrophobic interactions have been shown to work, but this type of bond is non-directional. This may lead to lower specificity, especially when working with macromolecules with similar structure, as well as problems with solubility and conformation due to apolar effects. Polar interactions on the other hand are directed and therefore more favorable in obtaining specificity. By using these residues on the surface as targets we can position interaction sites that will conform to complementary sites on our protein. Therefore, unlike the simple, small molecules for which the majority of MIPs have been demonstrated, the size and complex structural makeup of any target protein provide numerous angles for building multivalent binding sites, which may be singular in recognition nature or a collage of hydrophobic, hydrogen bonding, and other electrostatic interactions as discussed next.
2. Molecular Imprinting of Proteins
The use of the term “protein imprinting” can cause some confusion because it is used to describe two separate, although related, areas of research The two areas can be distinguished according to the role of the protein, specifically, whether the protein is employed as the imprintable matrix, a process often referred to as bioimprinting, or as the template molecule (47). This review will focus on the latter case where the protein is used as a template to form binding sites in synthetic materials with template-specific binding characteristics. A current overview of research in protein imprinting is provided in the following sections and summarized in Table 1.
Table 1.
Examples of Protein Imprinting Research
| refs | imprint platform | separation/ detection system | protein(s) | matrix | solvent | affinity/ detection | specificity (imprinting factor)b | comments |
|---|---|---|---|---|---|---|---|---|
| 48, 49 | amphoteric polymer for direct protein separation by HPLC | chromatographic phase | bovine serum albumin, lysozyme | methacrylic acid, N-[3-(dimethylamino) propyl]-methacrylamide | water + CaCO3 (porogen) | lysozyme imprint shows enhanced rebinding of initial template | slight cross-reactivity between two systems | acid wash to remove CaCO3 followed by 48 h pronase incubation and phosphate buffer wash to remove remnants of protein; lysozyme shown to enter but not interact with BSA imprint through chromatographic elution |
| 50 | recognition of a DNA protein using DNA as backbone of formed polymer | solution depletion measured by enzyme activity, via gel electrophoresis | EcoR1 | DNA, psoralenterminated polyisopropyl-acrylamide | series of buffers during polymerization step | demon-started between active and denatured EcoR1 | non-imprinted DNA blocks nearly all protein/DNA interactions | highly complex system with specifically synthesized monomer; requires specific DNA chain for protein recognition; possible uses for separation of DNA-binding proteins |
| 51 | combination of affinity separation and molecular imprinting for specific protein separation | solution depletion | trypsin chymotrypsin | acrylamide/N,N-ethylenebis(acrylamide) gel with additional polymerizable inhibitor | water/DMF | capacity of ~0.7 mg/g of polymer | 2.92 for Trp imprinted/non-imprinted. 1.92 for Trp/CTrpon the Trp specific polymer | polymer dried and washed with acetone/chloroform; well-studied system used for polymerization incorporating a specific monomer; potential protein entrapment in gels as shown by Ou (52) |
| 53–55 | polyacrylamide gel based polymers for chromatographic separation of various proteins | chromatographic phase | hemoglobin, cytochrome c, RNase, human growth hormone, transferrin | acrylamide/N,N-ethylenebis(acrylamide) | aqueous buffers and water | various | various | well-studied system used for polymerization and demonstrated with several proteins; different wash steps performed for each protein; binding attributed to weak interactions; possible difficulties seen with cross-reactivity and protein entrapment |
| 52, 56 | polyacrylamide gel based polymers, designed with specific electrostatic groups | solution depletion | lysozyme | methacrylic acid, N,N-diethylamino ethyl methacrylate | aqueous Tris buffer | 12.5–43.8% w/wa | 1.34–3.38 | gentle salt wash; good imprinting efficiency; nonspecific binding seen; ~27% of lysozyme trapped in system |
| 57–59 | peptide chain selectivity for an angiotensin II octapeptide; “epitope approach” | chromatographic phase | angiotensin II octapeptide; attempts to bind the whole corresponding protein | sodium acrylate/poly(ethylene glycol) diacrylate | water | 0.4 µg/mL detection limit by HPLC for octapeptide | 19.2–2.10 depending on environment | uses target epitope as an anchor for imprint, lowering cross reactivity due to template size; peptide “anchor” imprint recognized but failed to with the whole protein; small point of interaction not giving strong enough affinity between polymer and protein |
| 60, 61 | traditionally imprinted sol-gel monolith | solution depletion | urease, bovine serum albumin | 3-amino-propyltriethoxysilane, tetraethylorthosilicate. | potassium phosphate buffer pH 7, 0.1 M | 60–90% rebinding | preferential binding factor of 1.5 seen for both protein specific MIPs | ~25% protein entrapment seen in crushed polymer after 160 h pronase digestion; repeated protocol using hemoglobin and myoglobin showed no selectivity |
| 62 | direct imprint of protein onto silane modifiedsilica particles | chromatographic phase | transferrin | borate-silane complex | potassium phosphate buffer pH 7, 0.1 M | not shown | relative retention of transferrin over BSA of 2.16 | uses a specific functional monomer; gentle salt wash to remove protein; no evidence of template entrapment. |
| 63, 64 | macroporous chitosan beads with acrylamide | HPLC, solution depletion | hemoglobin | acrylamide | phosphate buffer pH 6.8, 10 mM | adsorption capacity ~12 mg/g | KD: 42.7 (Hb)1.41 (BSA)c | mechanically stable materials used in two forms of analysis; high selectivity demonstrated; time-consuming protocol to obtain equibrium due to mass transfer |
| 65, 66 | thins films formed around proteins coated with disaccharide layers | competitive adsorption using radiolabel target protein | bovine serum albumin, immunoglobulin G, fibrinogen, lysozyme, RNase | disaccharide-coated hexa-fluoropropylene (C3F6), on fixed support | phosphate buffered saline | not directly measured | 5–26 depending on substrate | demonstrated for a variety of templates; however complexity limits large scale applications; protein recognition only shown in competitive assays |
| 67 | acrylate beads by inverse-phase suspension polymerization | solution depletion | bovine serum albumin | acrylamide/N,N-ethylenebis(acrylamide), methacrylic acid | phosphate buffer pH 3.7 | not shown | defined imprinting effect compared to non-imprinted beads; specificity between BSA and ovalbumin demonstrated | simple protocol with clear optimization; cross reactivity data shown for 1 protein; stability issues of gels not discussed; protocol requires demonstration in chromatographic conditions |
| 68–71 | modification of silica beads with acrylate-based polymer | batch binding/enzyme activity test/ QCM | glucose oxidase, lysozyme | N,N-1,2 dihydroxy-ethylenebis(acrylamide), N,N’-methylenebisacrylamide, acrylamide, acrylic acid | range of phosphate buffers | 0.557 mg/g of polymer, for GO imprint. 0.8 mg/mL by QCM for lysozyme | not shown for GO system. demonstrated between Hb and Lzy on Lzy system | simple protocol, demonstrated in two formats and for two proteins; gentle salt wash used for template removal; lack of cross reactivity data; nonspecific binding seen for both proteins with QCM |
| 72–74 | supported polymers grafted to wells of polystyrene microplate; grafted layers to glass surfaces (microcalorimetry); grafted layers onto gold QCM crystal | solution depletion, QCM, microcalorimetry | horseradish peroxidase, hemoglobin, microperoxidase, lactoperoxidase, lysozyme, cytochrome c | 3-aminophenylboronic acid | water | various for different polymers | various between different polymers and templates | simple method, using only one polymer component; rebinding subject to environmental conditions; further demonstration with QC by Rick and Chou showing “dual imprint” |
| 75 | modified silica surfaces | solution depletion | hemogloblin | 3-amino-propyl-trimethoxysilane, trimethoxypropyl silane | MOPS pH 7, 10mM | not directly measured | demonstrated between hemoglobin and a range of competing proteins | complex protocol for formation of highly selective imprints; template not completely removed from material; indirect detection |
| direct detection of protein using film formed on electrode | pulsed amperometric detection | bovine leukaemia virus glycoprotein | polypyrrole | KCI 100 mM | 10 ng/mL detection shown | not shown | direct detection; regeneration problems, matrix not suited for all proteins; high nonspecificity seen | |
| 77 | specific monomer used to coat silica particles | stationary chromatography phase | ribonuclease A, lysozyme | metal chelating monomer, N-(4-vinyl)-benzyl iminodiacetic acid | 70/30 water/DMF rebinding in 70/30 HEPES buffer/DMF | capacity factor of 5.79 compared to 2.68 on ref | 2.35 between ribonuclease A and lysozyme | good recognition of target template; however, this technique is specific for His- bearing proteins; requires presence of Cu2+ ions |
| 78 | micro-contact approach with thin films (protein stamping) | competitive binding by ELISA | C-reactive protein, lysozyme, human serum albumin | O-(4-nitrophenylphosphoryl)-choline/PEG400 dimethacrylate | water + CaCO3 1 mM | 3.78 ng/cm2 for CRP 2.78 µg/cm2 for HSA | 0.08 ng/cm2 for HAS 0.27 µg/cm2 for HSA | low cross reactivity, demonstrated for two templates; microscale application; stability and reusability issues due to fragility of material |
| 79–82 | imprinted Langmuir monolayer formed at the air/water interface, measured in situ or transferred to hydrophobic support | SPR, QCM | ferritin | methyl stearate, dioctadecyldimethyl-ammonium bromide, poly(ethylene glycol) bearing phospholipids in various ratios | water | demonstrated between varying ratios of monolayers components | 6 on PEG:SME:DOMA 6:3:1 layers by QCM, 3.8 on 20:9:1 layer (molar ratios) | generic protocol, with imprinting coming though complementary patterning of monolayers; stability issues with formed layers; specificity not demonstrated |
Protein absorbed/total protein initial × 100.
Imprinted activity/nonimprinted activity.
Distribution coefficient KD = Cp/Cs where Cp = concentration of protein in beads (mg/g) and Cs = concentration of protein in solution (mg/mL).
Strategies for imprinting protein can conveniently be subdivided into either 2D or 3D platforms. 2D imprinting refers to those systems that limit the template protein to the surface of the imprint material, whereas 3D imprinting refers to those methods that bring about an imprinting effect by trapping the template within the bulk of the matrix.
2.1. 3D Imprinting of Proteins: Acrylates, Hydrogels, Sol-Gels, and Hybrids
a Key issue when developing 3D protein imprinting strategies is the limitations that the imprinted polymer imparts on the movement of the protein. A fundamental requirement of any system is that the template protein can freely be removed and reintroduced into the imprinted site. Control of porosity and pore size is a major design criteria, and a number of different matrices have been used.
2.1.1. 3D Acrylate Imprinting of Proteins
A number of studies have used conventional (meth)acrylate chemistries to prepare protein MIPs (7). However, a major limitation of this approach is that conventionally this method is carried out using solvents in which proteins are largely insoluble. Additionally, the dissolution or dispersion of protein in nonaqueous media commonly results in the protein adopting a conformation that is significantly different from what it adopts in water. This may have implications for the efficacy of the resulting MIP in aqueous environments. Several groups have therefore used water-soluble acrylic monomers to prepare bulk hydrogel MIPs with compositions analogous to electrophoretic gels (83).
Vaidya et al. used AAm and BisA to molecularly imprint trypsin (51). A complex of trypsin and its polymerizable inhibitor N-acryloyl p-aminobenzamidine (acting as a specific monomer) was polymerized in a mixture of DMF and water, and the protein was extracted from the resultant gel using acetone. The cross-linker concentration was optimized to ensure minimum swelling while maintaining imprint integrity and suitable pore size to allow maximum transport of protein. Solution depletion experiments demonstrated trypsin binding and the original template was favored in a competitive binding assay mixture of trypsin and chymotrypsin. The Scatchard analysis of this system showed that the imprinted polymers worked like a true receptor (straight line suggesting selective binding), differing from the non-imprinted, which exhibit the properties of an affinity chromatography system (exhibiting nonlinear characteristics and hence nonselective binding).
Similarly the group of Hjerten polymerized AAm and N,N′-methylenebisacrylamide (MBisA) in the presence of protein to form a gel (53–55). The gel was pressed through a sieve, and the resultant particles were packed into a column for chromatography. The protein was removed from the gel using a solution of acetic acid (HOAc) and sodium dodecyl sulfate (SDS).When a variety of proteins including the template were run on the column, the template was selectively adsorbed. This technique was demonstrated for bovine hemoglobin (Hb), cytochrome C (CyC), and transferrin (Tf) (54) and later for human growth hormone (HgH), RNase, and horse myoglobin (Mb) (53). It was hypothesized that a large number of weak electrostatic bonds formed between the gel and the protein, giving an overall strong interaction and hence the success of the imprint. This is in opposition to traditional imprinting theory that suggests that fewer strong bonds are better than numerous weak ones. In part Hjerten’s theory is supported by the demonstration of imprints of small proteins binding specifically, but with less adsorption volume than larger proteins to the same gel (larger proteins having greater surface area and therefore more interactions with the gel).
Methacrylic acid based gel beads were imprinted with bovine serum albumin (BSA) by Pang, using an inverse-phase suspension method (67). They obtained good quality spherical beads exhibiting excellent macroporous structure, which is important in facilitating the movement of proteins through the material. The produced beads demonstrated higher adsorption over their non-imprinted counterparts, and more importantly, a relatively high separation factor (α = 4.71) was obtained between the BSA (MW 67 kD, pI 4.8) and a challenging protein (ovalbumin, MW 44 kD, pI 4.5). The authors attributed this success to steric factors and multiple point electrostatic interactions, similar to that suggested by Hjerten.
Acrylamide gel was used by Guo as an entrapment material for the imprinting of Hb (64).Macroporous cross-linked chitosan beads were used as a support in which the acrylamide and protein were allowed to diffuse into the pores before polymerization. The protein was then removed by washing with an acetic acid/SDS solution. The imprinted chitosan beads were shown to have greater capacity and higher selectivity for the template protein than the non-imprinted chitosan/acrylamide beads and pure polyacrylamide beads. This work was then carried further by the use of these beads in the chromatographic phase, where selectivity of Hb and BSA was demonstrated (63). The addition of the chitosan as a surface to support the imprints was demonstrated to improve the mechanical stability of the beads.
Huang et al. developed an amphoteric imprinted polymer for application in protein separation (49). They imprinted BSA and hen egg white lysozyme (Lzy) in a suspension of two functional monomers (MAA and N-[3-(dimethylamino) propyl]methacrylamide), the crosslinker (BisA), and CaCO3 as a pore-forming agent in a phosphate buffer. The resultant gels were then treated with acid to remove the CaCO3 and then with a pronase to digest the templates. These polymers were applied in chromatographic stationary phases to assess capacity and selectivity. The protein Lzy showed preferential binding to its equivalent polymer but not the BSA imprinted system. This was attributed to cooperative multiple electrostatic interactions.
A further method of imprinting Lzy was reported by Ou et al., using MAA and AAm as functional monomers and 2-(dimethylamino)ethyl methacrylate as cross-linker (52). The resultant polymer was wet sieved and then washed with deionized water, NaCl solution, and then water again to remove the template (this mild elution condition is contrasted with the acidic wash step used by Hjerten (53) and Huang (49)). The gels were lyophilized and subjected to solution depletion assays. Selected imprinted polymers gave imprinting factors (binding of protein to imprinted polymer/ binding of protein to nonimprinted polymer) of 1.83–3.38. However, rebinding was not strictly template-specific as the polymer also bound albumin (~13.3% w/w from protein solution). It was also noted that >25% of the original template remained in the polymer.
Imprinting of proteins using acrylate chemistry in aqueous solution relies on forming gels with a low degree of cross-linking to ensure mobility of large molecules. Some success was gained with the methods described above; however, it is known that low cross-linked materials lose their imprinted properties rapidly and are in general less stable with respect to changes in environment (84). The acrylate imprinting protocols are also somewhat limited in terms of suitable monomers, thus leading to exploration of other novel materials.
2.1.2. Hydrogels
In recent years a class of novel swellable polymers known as hydrogels has been synthesized. These stimuli-responsive gels can undergo a reversible volume transition between the swollen and collapsed phases, which can be triggered by such external stimuli as temperature, solvent composition, pH, ionic strength, light, and specific chemicals. Such gels are often termed “smart” gels and are commonly used as controlled release systems, actuators, and sensors, as well as for the demonstrations of artificial muscles (29). These responses would be of value in controlling specific binding of proteins and various efforts have been made at combining hydrogel and molecular imprinting technologies to prepare hydrogels exhibiting enzyme-like behavior.
Karmalkar reported a metal ligand mediated approach, positioning imidazole, carboxyl, and hydroxyl groups within a hydrogel matrix to mimic the serine protease class of enzymes (e.g., α-chymotrypsin) (85). The template molecule, 2-([(isobutyrylamino) caproyl]-l-phenylalanyl)2-aminopyridine, was removed, and the catalytic behavior of the polymer was tested by monitoring the hydrolysis of nitrophenyl esters. The imprinted polymer catalyzed the hydrolysis at a rate comparable to that of α-chymotrypsin, whereas the non-imprinted hydrogel exhibited only modest activity.
Kofinas’ group has reported noncovalent molecular imprinting of poly(allylamine hydrochloride) with glucose phosphate, producing a MIP hydrogel capable of quantitative, isomer specific glucose recognition (86, 87). A pre-polymerization complex relying on ionic interactions is formed and then cross-linked with epichlorohydrin. MIP hydrogels templated with glucose were selective in rebinding the template over fructose.
Watanabe synthesized temperature sensitive hydrogels by copolymerizing N-isopropylacrylamide and acrylic acid (AA) with a cross-linker in the presence of various template molecules, including norephedrine (88). After template removal the hydrogel exhibited characteristic swelling at temperatures below the lower critical solution temperature (LCST) with complete collapse seen at higher temperatures. The swollen gel at a low temperature showed no change in swelling upon re-exposure to the template. However, the collapsed high-temperature hydrogel exhibited increased swelling upon increased template concentration, implying that the imprinted hydrogel in the collapsed state could memorize the template, whereas the swollen state could not. Specificity for norephedrine was inferred from swelling with increased ephedrine concentration and no change upon exposure to adrenaline.
Imprintable hydrogels, although only demonstrated for small molecules, offer a possible solution to the problem of protein entrapment as their ability to swell would allow for controlled protein removal (89). This is further supported by several papers reporting the use of these materials as protein delivery agents, with protein encapsulation a major exponent of these systems (90, 91). This encapsulation and entrapment of proteins while maintaining biological activity has been demonstrated in a wide range of materials (92, 93). Sol–gel chemistry has in particular proved to be an ideal solution for producing these doped polymers (94). Gill and Ballesteros developed highly porous sol-gels for encapsulation of macromolecular compounds. These polymers, based on a poly(glyceryl) silicate, have demonstrated high stability compared to more traditional sol-gels. This stability combined with unusually high porosity could be used for the benefit of imprinting (95).
2.1.3. 3D Sol-Gel Protein Imprinting
Sol-gels have proved able to specifically interact with a variety of proteins for protein encapsulation (92, 93). This has been adapted to protein imprinting utilizing the benefits of working with sol-gels (mild polymerization conditions (pH, ionic strength) and that they favor aqueous solutions).
Venton prepared imprints for urease and BSA in polysiloxane copolymers, using 3-aminopropyltriethoxysilane and tetraethylorthosilicate (60, 61). The polymers were formed as monoliths and then crushed, and the entrapped proteins were removed by a pronase digestion. Increased affinity for each template was demonstrated for its corresponding polymer (+30% for urease and +3% for BSA, when compared to opposing protein), and selectivity was shown between the two proteins. However, a similar study by the same group using Hb and Mb failed to show any imprinting effect for either MIP (60). Rebinding experiments using labeled proteins demonstrated that the bound proteins did not equilibrate with labeled proteins in solution, suggesting strong association between polymer and template, possibly hiding specificity. Experiments measuring the release of urea in the presence of the specific MIP and its template (Hb and Mb) suggested that specific imprints were formed, as the amount of urea released was less for the specific template association than for the other protein. No mention of the influence of porosity is made, but this work shows that ~25% of protein still remains trapped in the polymer matrix.
As discussed above, the sol-gel reaction is highly suitable for imprinting proteins (mild polymerization conditions, water compatible). However, the bulk imprinting of proteins seems to be limited to the above example. This is possibly due to the dense structures of sol-gel formed and the requirement of grinding to expose binding sites. This may greatly diminish specificity for the macromolecules, as larger binding sites are destroyed by the mechanical process of grinding, unlike those of smaller molecules.
2.1.4. 3D Hybrid-DNA Protein Imprinting
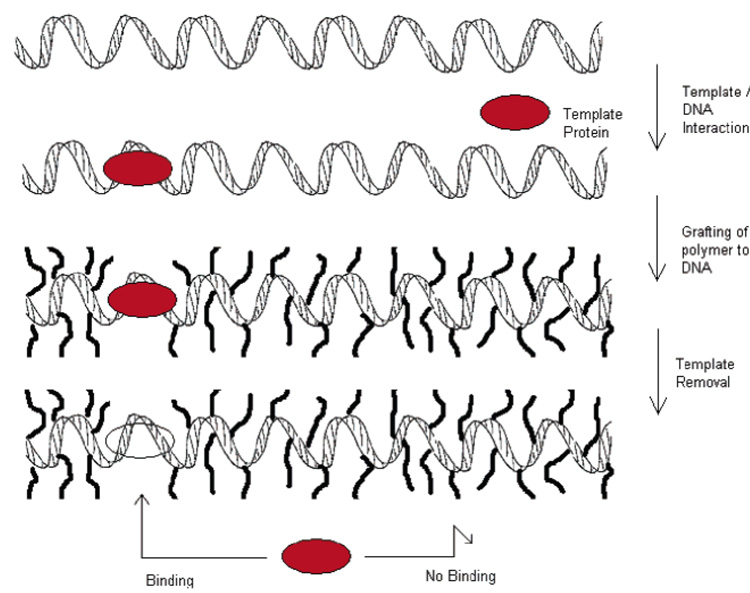
The interactions between a protein and DNA are known to be highly specific. A novel method of protein imprinting has been demonstrated by Umeno who used a polymer-coated DNA strand for protein recognition (50). A temperature-responsive monomer designed to interact with DNA was synthesized (poly(N-isopropyl acrylamide terminated with psoralen) and combined with a DNA strand to form a conjugate (photoinduced reaction between DNA and psoralen end group). The conjugated polymer coating the strand elicited a blocking effect to proteins trying to interact with the DNA. This blocking effect was then used to imprint the binding site of EcoR1 (a restriction endonuclease) by incubating the DNA with the endonuclease before conjugation with the polymer. A binding pocket, specific for the binding site of EcoR1, was formed and preserved (Figure 2). This novel technique demonstrates the selectivity that can be obtained when building upon an existing target site, which is then singled out by essentially occupying (and rendering inaccessible) all other potential binding sites. Although the practical use of this system is clearly limited (possible uses for separation of DNA-binding proteins), this represents a unique biomimetic approach toward MIPs.
Figure 2.
Protein imprinting on DNA backbone using protein/DNA recognition for protein immobilization adapted from Umeno (50).
These varied approaches toward protein imprinting illustrate the potential of applying traditional bulk and hybrid molecular imprinting technologies for specific protein entrapment and separation. However, the stability of such systems for use as robust biosensors has yet to be determined. The requirement of high porosity for protein transport through these materials has led to favoring the formation of gels. These lack the high degree of cross-linking required to maintain stability over long periods, compared to traditional monolith imprinted materials. Environmental changes also affect the abilities of these gels and in some situations can lead to pore collapse and permanent loss of function. Additional requirements of harsh washing steps (acids, pronases) to remove templates will also affect the function of these materials. A more favorable situation for macromolecule templates, such as proteins, is that of surface imprinting.
2.2. 2D Protein Imprinting: Designer Surfaces and Interfaces
The compatibility of bulk imprinted materials for use with large templates is limited. An alternative approach has been to produce polymers in thin films or fixed to a supporting surface (8). This 2D or in-plane approach allows free access to binding sites for larger molecules and allows specific areas of proteins to be a target for imprints (anchor points). However, these 2D approaches may not offer as high a specificity as only a small portion of the surface of the protein will be recognized. Binding site heterogeneity for such systems may also be high. These limitations may be outweighed by the potential advantages afforded by high mass transfer, integration with sensor platforms, and robustness gained by having a support. Due to these benefits and the increased interest in surface chemistry (a large and rapidly expanding field in its own right), several novel methods of imprinting at a surface have been reported.
2.2.1. 2D Sol-Gel Imprinting
The first reported successful protein imprint was by the group of Mosbach (62). They developed a method of coating an imprinted polymer for the glycoprotein transferrin (Tf) onto the surface of porous silica beads. A mixture of silanes was polymerized on the surface of porous silica particles in aqueous solution, including a new boronate-silane functional monomer designed to interact with the carbohydrate portion of Tf in the mixture. Using HPLC they showed the Tf-imprinted polymer to have a higher affinity for its template than for BSA (relative retention of 2.16). These coated beads were compared to a bulk material produced using the same mixture and ground. The beads were found to have superior enzymatic properties (40% for the imprinted siloxane on beads compared with 5% for the siloxane only ground material). This was attributed to superior protein entrapment compared to that of the bulk polymer.
Shiomi imprinted porous silica with hemoglobin (Hb) by covalently immobilizing it to the silica using imine bonds (75). After imprinting the protein was removed by an oxalic acid wash, to break the covalent attachments. These imprints were observed to be superior to those formed using free (in solution) Hb when studied for rebinding of hemoglobin and competing proteins. The results showed binding affinity to be linked to isoelectric point (pI) of the protein and the size of the protein as well as the characteristics of the binding site itself (shape, hydrophobicity).
Despite numerous sol-gel examples of protein interaction and entrapment (93) and integration with sensors and electrochemistry (96), examples of sol-gel protein imprinting (2D or 3D) are few.
2.2.2. 2D Metal Coordination
Metal ions and their role in coordinating specific binding in imprinted polymers has been reviewed elsewhere (84). By combining favored groups into polymeric structures these interactions have been used with protein residues to gain specificity toward protein templates.
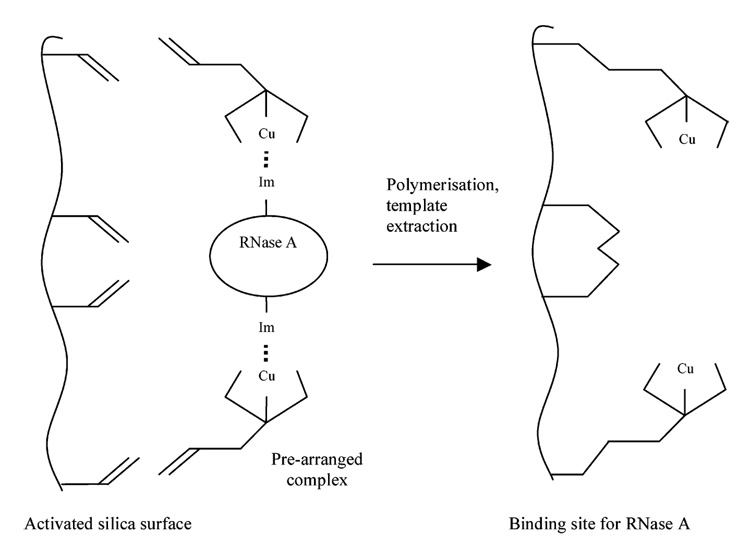
Mallik et al. used metal ions to coordinate His residues via imidazole functionality (30). A series of bis-imidazole protein analogues (two imidazoles linked with organic spacers) were investigated, with a view to producing a synthetic receptor capable of coordinating proteins. Using a similar approach, Kempe reported a surface-imprinting procedure for the preparation of selective adsorbents for HPLC separation of proteins (77). They employed a metal chelating monomer, N-(4-vinyl)-benzyl iminodiacetic acid (VBIDA), with methacrylate-derivatized silica particles, in the presence of the protein ribonuclease A (RNase A) and metal ions. Reference polymers were made with BSA. Imidazole groups on the surface exposed His residues, which combined with the monomer VBIDA to coordinate Cu2+ ions. When polymerized the metal binding ligands are positioned on the silica surface at precise distances as to enable selective rebinding of the protein in the presence of the Cu2+ ions (Figure 3). RNase A was recognized over Lzy using this method. However, this method is limited to proteins with exposed His residues and as such is not universally applicable for protein imprinting.
Figure 3.
Creation of RNase A binding site on surface of modified silica. Adapted from Kempe et al. (77).
2.2.3. 2D Acrylate Chemistry
Burow imprinted glucose oxidase (GO) using acrylates on functionalized silica bead supports (68, 69). A mixture of two cross-linking monomers (MBisA and N,N′-1,2-dihydroxyethylenebisacrylamide), and two functional monomers (AAm and AA) was polymerized in the presence of the protein and surface activated silica beads, leading to the polymer forming a thin layer, or coat, around the silica. Rebinding assays showed selectivity for the imprinted protein GO over BSA. The recognition effect was attributed to both weak electrostatic interactions and a shape conformation between template protein and polymer (69). Hirayama, using the same technique, also reported a Lzy-imprinted polymer that modest exhibited selectivity for Lzy over Hb, although nonspecific binding of both proteins did occur (71).
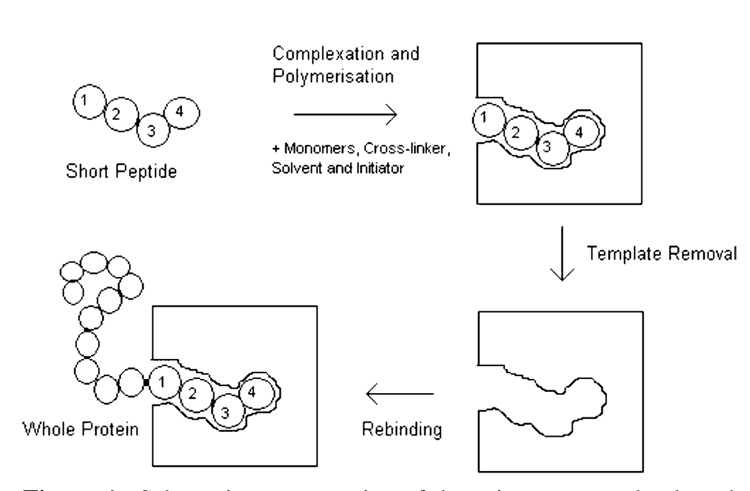
The methods discussed above target several sites upon the surface of a protein. By forming numerous interaction points they attempt to obtain specificity through steric properties as well as complementary chemistries. An alternative to this entitled the “epitope” approach has been developed by Rachkov, where the template used for polymer development is not the whole protein but a short peptide that represents part of a larger polypeptide or protein, as an epitope represents an antigen in immunology (Figure 4) (57–59). This is technically not imprinting of proteins in the traditional sense but is included as the ultimate goal is to obtain recognition of the parent molecule.
Figure 4.
Schematic representation of the epitope approach adapted from Rachkov and Minoura (59).
The imprinting procedure used was akin to more conventional methods (MAA and EGDMA were the functional monomer and cross-linker respectively in an organic solvent). Selective recognition of oxytocin, a nonapeptide, was demonstrated by a polymer imprinted with a tetrapeptide, YPLG. Both compounds possessed the same three amino acid sequence at the C-terminal. This allows for specific, strong interactions with part of the protein to be formed, rather than relying on numerous weak interactions all over the molecule, thus (theoretically) increasing specificity and affinity. This also allows for traditional imprinting methods in organic solvents to be used, as the small peptides are not subject to the major conformational and degradation problems seen with whole proteins. The results demonstrated selectivity and high affinity for the template molecule. However, when this was repeated using an angiotensin II octapeptide in aqueous solvent, the formed polymer retained for the octapeptide, while its parent protein angiotensin II was not bound at all to the polymer (57).
A commercial product based upon a concept similar to Rachkov’s epitope approach has been recently launched. Aspira Biosystems has designed ProteinPrint technology (48, 97). A synthetic peptide whose sequence corresponds to a small portion of a protein is used as a template molecule to form cavities on the surface of polymer arrays or beads, as shown by Hirayama (71). Aspira plans to market films for single proteins, or arrays of different proteins, for separation and analysis (97). Such films are planar polymer surfaces containing sequence-specific locations, each of which contains multiple binding sites. Their patented technology claims high affinity films can be made to capture a single protein across an entire film area, or a microarray of imprints can be constructed to capture a family of proteins (97).
Though acrylate chemistry is considered the general and most popular technique for imprinting, we have shown above that it is not ideal for working with proteins. A number of other materials exist and several groups have demonstrated the use of non-acrylate polymerizable matrices with regards to imprinting.
2.2.4. Other 2D Methods for Protein Imprinting
Ramanaviciene prepared a polypyrrole-based MIP for the direct detection of the bovine leukaemia virus (BLV) glycoprotein gp51 (gp51) (76). The polymer was prepared as a surface coating onto a platinum electrode in a potassium chloride solution. Using pulsed amperometric detection (PAD), they demonstrated binding, washing, and rebinding of the protein over several cycles; however, the recognition and affinity degraded with each repeat.
Bossi used 3-aminophenylboronic acid (APBA) oxidized by ammonium persulfate to form imprints against several proteins (HRP, Hb, Microperoxidase, Lactoperoxidase) (72). During the polymerization process, the resultant polymers are grafted to the polystyrene surface of a microplate. This grafting procedure was demonstrated to stabilize and aid the imprinting process and hence recognition of the target proteins. For all proteins used, good imprinting factors were obtained with respective low cross reactivity. The effects of the washing steps on rebinding of template supported formation of specific binding sites. A steady increase in reabsorbed protein per washing step is seen, in contrast to the blank, which decreases.
Rick and Chou used a similar APBA protocol to form films specific for Lzy and CyC (74). Using microcalorimetry, they demonstrated successful imprints shown by the significant differences in the enthalpy observed upon binding when comparing template with a competing protein. The enthalpy of binding produced by the template protein (that used to form the imprint) was less than that created by the addition of a foreign protein. An interesting discovery was made when they performed a dual imprint with both proteins present. It was found that the maximum enthalpy change was obtained when the two proteins were present in an equimolar ratio (73). Also, solutions of the component proteins in different ratios offered the best recognition when present at the same ratio used for the imprint. Neither of the component proteins offered any recognition when exposed on their own to the “dual” imprint. Therefore they concluded that a new motif had been formed from the two proteins and that recognition comes from a protein–protein complex (Figure 5). The work also demonstrated the ease of integration of the APBA protocol with a known sensor technique (in this case QCM). The APBA chemistry is a fairly simple protocol that conforms to the environmental constraints of protein imprinting and one that can be integrated into sensor systems, but as Bossi showed, the interactions are highly dependent on environment with respect to rebinding (72).
Figure 5.
Representation of protein–protein interaction as in Rick and Chou (73). The imprint (A) is formed between two proteins interacting with one another. The two constituent proteins (Lzy and CyC) are unable to bind on their own (B) and only when placed in their original ratio will form the complex which is the specific motif for the imprint (C).
The same group developed another method of imprinting using a polymerizable analogue of a proteins natural ligand. In this case the pentameric C-reactive protein (CRP), known to be a marker and pathway activator in pneumococcal pneumonia, was imprinted. A series of imprinted polymers and their respective blanks (differing by cross-linking degree) were produced by a microcontact or “protein stamping” approach. A layer of the protein/polymerizable ligand was placed on a glass coverslip, sandwiched between the slip and glass slide, and then cured by photopolymerization. The cover slip was then removed, leaving the thin film imprint on the glass support. Competitive assays with other proteins of similar size demonstrated good selectivity for the CRP. Using the same method the group copied the experiment with human serum albumin (HSA). This polymer demonstrated a relatively good recognition for its own template of 2.66 µg/cm2 compared with 0.27 µg/cm2 for CRP (78).
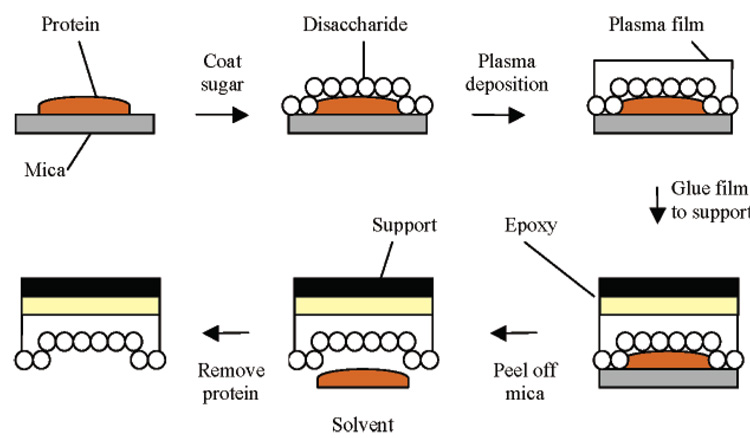
Another method of “protein stamping” was developed previous to this by Ratner, who used radio frequency glow-discharge (RFGD) plasma deposition to form polymeric thin films around proteins coated with disaccharide molecules (65, 66, 98, 99). A protein is fixed onto a mica surface and exposed to a disaccharide solution. During sample dehydration multiple hydrogen bonds are formed between the hydroxyl groups of the disaccharide and polar residues on the surface of the protein. When exposed to the plasma-induced polymer film, these disaccharides form covalent bonds, creating upon protein removal polysaccharide-like cavities that exhibit highly selective recognition for the template protein (Figure 6). This is then fixed to a support. The mica is then removed, and the protein is decomposed using a pronase and eluted with a basic solution (NaOH/NaClO), leaving a thin film with protein-imprinted cavities fixed onto a support. By using this sugar coat the protein can be incorporated into the plasma film with minimal damage. The RFGD plasma deposition of fluoropolymers (e.g., hexafluoropropylene) yields a smooth film that is mechanically and chemically stable, and since mica is atomically flat, the imprint surface reflects only the topographic features arising from the template protein. Using this technique, several proteins including BSA, immunoglobulin G (IgG), Lzy, RNase, and streptavidin were imprinted (66). Template recognition was assessed by radiolabelled protein adsorption from single solutions and binary mixtures, detergent elution of surface-bound proteins, and displacement of surface-adsorbed proteins by solution-phase proteins (65). Specificity in competitive assays was shown for BSA imprints (MW 66k) (5–10 times greater uptake of BSA over IgG) and IgG imprints (150k) (4–7 times greater uptake of IgG over BSA); and more impressively between Lzy (MW 14.5k) and RNase (13.7k), two proteins that are comparable in molecular weight and dimension (20 and 26 times greater uptake in favor of template respectively).
Figure 6.
RFGD plasma deposition used for surface imprinting of proteins adapted from Shi et al. (66).
The mechanism of recognition was mainly attributed to hydrogen bonding between the precisely positioned hydroxyl groups from the immobilized sugars and the surface polar residues of the protein. Hydrophobic interactions and van der Waals forces were also thought to be involved. The extent to which elution/decomposition of the protein may alter the imprint site or leave fragments of protein behind is unknown, but loss of protein structure during dehydration also appears to be minimal as inferred by the selectivity upon rebinding.
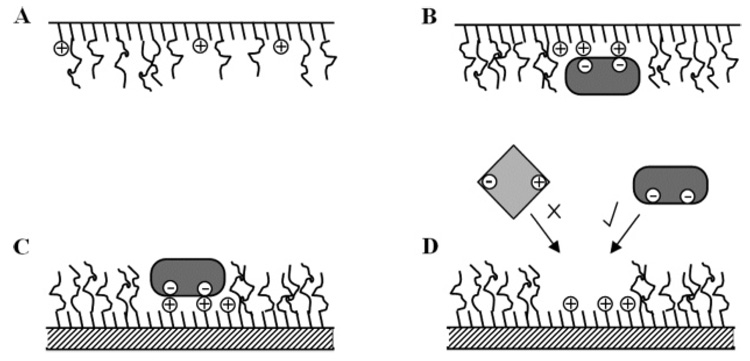
It is noted that many of these techniques are variants of bulk imprinting methods that have been restricted to an interface. A more biomimetic approach has been followed by allowing proteins to adsorb to a fluid interface containing the recognition elements, such as multicomponent lipid monolayers, at the air/water interface (81, 100, 101) or supported bilayers (102). For this class of 2D imprinting the protein must “select” the recognition elements (e.g., through electrostatic interactions between protein residues and charged lipid head groups) and induce a local demixing of the lipids as predicted through a mean-field theory (103).
The binding of proteins to monolayers and at interfaces has been well documented (80, 104); however, recent attention has been drawn toward the possibility of using these interactions to produce imprints at the air/water interface. The potential for using lipid monolayer films as protein imprint matrices is evident in template-mediated crystal nucleation and biomineralization, where monolayers serve as fluid lattices that laterally restructure to favor epitaxial crystal nucleation and growth (105, 106). This concept can be extended to impart recognition sites through adsorption-induced restructuring (patterning or templating) of fluid monolayers, as has been investigated using metal chelating lipid monolayers to target His-tagged proteins (101). A multivalent interaction of protein with functionalized lipids such as lectin-mannose or avidin-biotin is desirable for enhancing the affinity and specificity of the patterned lipid film for the target protein. However, patterning of mixed lipid monolayers not containing a population of lipids presenting known ligands for the target protein offers a more generic imprinting platform, in which the pattern of residues on the protein surface must induce a complimentary pattern in the lipid head groups.
Applying this concept, Britt adsorbed the acidic protein ferritin to mixed cationic/non-ionic lipid monolayers in an effort to induce charge patterns (i.e., local demixing) in the fluid monolayers (100). It was noted that as the monolayer charge density was decreased through “diluting” the cationic lipid with the neutral lipid, ferritin binding increased, reaching a maximum value for a neutral/cationic lipid ratio of 6:1. However, if the monolayers were first immobilized on a solid support, protein adsorption was maximal for the pure cationic lipid film. These opposing protein adsorption trends on fluid versus immobilized monolayers demonstrated the strong influence of lipid lateral mobility in protein adsorption to a heterogeneous film, supporting the applicability toward protein-induced monolayer patterning.
Building upon this work, Du investigated whether the initial binding event induced a protein-specific imprint through rebinding kinetics (81). Successive adsorption/desorption cycles revealed that fluid monolayers, capable of laterally restructuring during the initial protein adsorption event, bound up to 60% more ferritin (dependent on the cationic/nonionic lipid ratios) as compared to monolayers that were immobilized on a hydrophobic support during this first adsorption step. These findings suggest lipid restructuring producing local charge patterns complementary to the fixed distribution of acidic amino acids on the ferritin coat. The success of this method was limited by a large fraction of irreversibly bound protein on the patterned monolayers, attributed in part to strong protein-protein interactions (aggregation) during the patterning process. The latest advancement in this area has demonstrated the prevention of this aggregation by the addition of poly(ethylene glycol) (PEG) bearing phospholipids to the monolayers. These inhibit protein aggregation by forming an inert brush layer separating the specific binding pockets (79). The binding/rebinding of ferritin to these layers has been studied using QCM. Mixed layers were prepared at the air/water interface, with and without ferritin, and transferred to a hydrophobically modified QCM crystal (82). The results demonstrated increased binding to the imprinted films over the non-imprinted. AFM of the resultant surfaces showed pockets formed in the templated monolayer, formations not seen in the blank. Cross-reactivity and stability is yet to be studied.
3. Discussion
The evidence presented above demonstrates that imprinting proteins is a reality. However, protein imprinting accounts for <2% of the total published material in the field, the focus of most molecular imprinting still being small bioactives. As discussed, the inherent difficulties associated with protein imprinting account for this imbalance, especially when competing technologies such as antibody recognition, immunoaffinity columns, ELISA, and separation gels are commercially available and well-developed.
The size of the template and its steric influences are major obstacles. Working with a macromolecular template requires, in bulk, high porosity for mass transfer. In turn, this porosity weakens the structure of any polymer, leading to stability and practical issues. In general, bulk acrylate materials imprinted for proteins tend to rely on loose-linked polymers with specific sites embedded in them (49, 51–53, 67). This low cross-linking affords reasonable mass transfer and, as demonstrated with hydrogels, can offer swelling as an additional factor for template removal and rebinding. The ability to control pore size by environmental conditions is of great benefit, as demonstrated clearly in the acrylate beads developed by Pang, which showed clear macroporous structure (67) at the working range of his protocol. The advent of hydrogel technology has brought the possibility of combining protein encapsulation and specific imprinting. For example, use of these materials as drug delivery agents means that degradation or porosity change could be tailored to release template at a known rate by altering the environmental conditions. However, for other uses such as separation, this instability and loss of function would be detrimental. By forming dense matrices to obtain stability and maintain function, the chances of template entrapment are greatly increased. This in turn, lowers any practical use (possible template leaching and decreased total capacity). More traditional highly cross-linked polymers as described by Venton (61), albeit in this case a sol-gel, have shown that they can be used for protein recognition; however, it was clear from this work that a high percentage of template remained in the polymer confirming the importance of porosity.
Working with a thin film or imprinted surface greatly reduces the steric influence of the template but can lead to potential problems, with partial imprinting causing site heterogeneity. The stability issues seen with 3D gels are negated by the presence of a support, be it beads (71), glass (78), or polystyrene (72), and the demonstrated integration with sensor platforms is also advantageous (73, 76). However, 2D imprinting also reduces the total capacity of a material, which limits practical uses in some circumstances, such as high volume separations.
The reliance on aqueous solvent for the imprinting step limits the choice of monomer, cross-linker, and polymerization method. Water is far from the ideal solvent for conventional molecular imprinting, as it competes for and disrupts potential hydrogen-bonding sites on the template and monomers (13). Any produced polymer will be required to work in conditions that are considered, in theory, unavorable to binding, such as the competition of water in binding pockets. The additional and often cumulative effects caused by changes in environment (pH, ionic strength, temperature) will also affect polymer performance (7, 72).
The approaches described in this paper aim to overcome these problems, either by utilizing protocols known to be aqueous friendly (53, 61, 72), or by using traditional organic techniques to target a peptide chain so it may be used as an anchor point for total protein recognition (59). Disruption of the binding pocket by the presence of water does not seem to greatly affect the protein/polymer interactions. This could be attributed to the size of the protein and the high number of interactions present, which may aid in forcing water out.
It is accepted that MIPs exhibit greatest selectivity when the number of points of interaction is small but these interactions are strong. Multiple weak interactions are less likely to produce a highly specific MIP and tend to favor nonspecific binding (14). It is also theorized that the thermodynamic considerations of imprinting indicate that a less rigid template such as a protein will yield less defined recognition sites in its corresponding matrix (107). The increased surface area of a protein will in theory lead to a higher number of interactions, which will generally be weaker and less specific. This leads to site heterogeneity, as a high number of potential interactions between a protein and corresponding polymer will lead to an increase in nonspecific interactions.
It is clear from the literature that there are two schools of thought regarding the best methods to form imprints. Hjerten proposed that the weak but many approach produces selectivity as steric factors found with a large template aids the recognition and alignment process. This has some experimental support from his 3D gel work (53) and others relying on massed electrostatic interactions (69). The method shown by Dhruv (79, 82) relies on hydrophobic interactions with steric factors playing a huge part in obtaining enhanced binding of their chosen template. These are in contrast to other protocols that rely on focused interactions to obtain specificity. For example, the epitope approach relies on a target region to provide recognition (59). The APBA (72) and metal chelation (77) polymers each have their own specific targets on a polymer surface (hydroxyl groups and His residues, respectively), leading to a few strong but specific interactions. Neither method has demonstrated its superiority.
The majority of work within the field of molecular imprinting has concentrated on small bioactive molecules. The imprinting of these materials has resulted in the development of materials with exceptionally high affinity for their target templates. For example, the dissociation constant (KD) for biotin using computationally designed MIPs ranged from 1.4 to 16.8 nM (108); for microcystin-LR, 0.3 nM (32); and for estradiol, 0.44–6.6 nM (109). In comparison Bossi demonstrated KD’s of 1.5 µM for microperoxidase, 0.54 µM for lactoperoxidase, and 0.056 µM for hemoglobin, using an APBA matrix (72). The obtained values for protein detection are an order of magnitude lower than those gained for small molecules. This is not surprising as the lack of study in this field has not lead to optimization of the protein systems, in comparison to those found with the technically easier small molecule systems. Likewise, specific recognition of template protein compared to other macromolecules has been demonstrated in several systems (66, 74), but so far limited success has been attained in separations of template protein from a mixture of competing macromolecules (63), unlike in the case of small molecules where enantiomers have been separated by MIPs (110, 111). All of the systems presented in this paper show enhanced rebinding of original templates, but this competitive rebinding to demonstrate true imprints for the target template is the more definitive metric. There are several cases where enhanced selectivity for the template is hidden by high cross-reactivity (60, 71). This reduces the practical value of the corresponding materials. It has also been shown that cooperative binding (73) and aggregation of template (81) are also factors that can affect the formation of any imprint. A specific recognition site for a monomeric protein will be greatly different from that of an aggregated particle. Forming an imprint against an aggregate will lead to site heterogeneity and loss of function. Again, environmental factors during the imprinting phase have to be taken into consideration, as even minor changes in phase, pH, or salt concentration can cause proteins to aggregate.
In the examples presented here, the removal of proteins tends to rely on fairly harsh methods (acid washes, detergent, pronases), which can affect polymer structure, properties, and ability (53, 66). Indeed, incubating a polymer with pronase for 160 h does not lead to a practical protocol for commercialization (60). The steps required to return a polymer back to a working level are also of importance (e.g.,, detergents such as SDS must be removed completely to ensure no bearing on rebinding). Milder approaches tend to leave a high percentage of template behind (52).
4. Conclusions and Future Outlook
The 3D and 2D protein imprints presented in this review are elegant in design (51, 66, 77) and have demonstrated fair affinity and specificity. However, several of these protocols are complex and labor-intensive (66), lack generic application (50), and have yet to demonstrate long-term stability. At present no universal method exists for generic protein imprinting. This is due simply to the wide and varying properties that proteins exhibit (the very properties that make them so important in biology). Certain methods have demonstrated versatility in imprinting several targets, but each has its drawbacks. Further examples of each technique are required, using a wider range of proteins. The potential demonstrated with sol-gels is high (93), but as yet only three protocols have been reported (60, 62, 75).
As yet, only MIPs for smaller biomolecules have yielded commercial success (112, 113). Areas such as medical devices, biomarker sensing, drug delivery, and peptide separations would benefit greatly from materials with the recognition properties of antibodies but superior robustness and reusability. Therefore the continued study of methods to imprint recognition sites in artificial matrices, especially for important biomolecules such as proteins, must continue as the practical benefits of such systems will be great indeed.
Figure 7.
Schematic of Langmuir monolayer imprinting. The unordered surfactant layer at the air/water interface (A) interacts with the protein, inducing demixing, allowing specific binding pockets to form (B). The layer is transferred to a hydrophobic support, which hinders further lateral movement of the monolayer components hence holding the defined pockets in place (C). Specific rebinding of protein is demonstrated (D), upon removal of the template.
Acknowledgment
N.W.T. and D.W.B. gratefully acknowledge support from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Servicegrant no. 2005-35603-15829 and NSF-NER Division of Bioengineering & Environmental Systems grant no. 0404262. N.W.T. would also like thank Michael Whitcombe, Miruna Petcu, and Sergey Piletsky for discussions.
Notation
- AA
acrylic acid
- AAm
acrylamide
- APBA
3-aminophenylboronic acid
- BisA
N,N′-ethylenebis(acrylamide)
- BSA
bovine serum albumin
- CRP
C-reactive protein
- CTrp
chymotrypsin
- CyC
cytochrome c
- DMPA
N-[3-(dimethylamino)propyl]methacrylamide
- EGDMA
ethylene glycol dimethacrylate
- ELISA
enzyme-linked immunosorbent assay
- GO
glucose oxidase
- GPX
glutathione peroxidase
- Hb
hemoglobin
- His
histidine
- HgH
human growth hormone
- HOAC
acetic acid
- HPLC
high performance liquid chromatography
- HSA
human serum albumin
- IgG
immunoglobulin G
- Lzy
lysozyme
- MAA
methacrylic acid
- Mb
myoglobin
- MBisA
N,N′-methylenebisacrylamide
- MIP
molecularly imprinted polymer
- OA
ovalbumin
- PEG
poly(ethylene glycol)
- QCM
quartz crystal microbalance
- RFGD
radio frequency glow-discharge plasma deposition
- SDS
sodium dodecyl sulfate
- SPE
solid-phase extraction
- Tf
transferrin
- TLC
thin layer chromatography
- TRP
trypsin
- VBIDA
N-(4-vinyl)-benzyl iminodiacetic acid
- 2D
two-dimensional
- 3D
three-dimensional
- 4-VP
4-vinylpyridine
References and Notes
- 1.Cram DJ. The design of molecular hosts, guests and their complexes. Angew. Chem., Int. Ed. Engl. 1988;27:1009–1020. [PubMed] [Google Scholar]
- 2.Schneider HJ. Mechanisms of molecular recognition–investigatons of organic host guest complexes. Angew. Chem., Int. Ed. Engl. 1991;30:1417–1446. [Google Scholar]
- 3.Clackson T, Lowman HB, editors. Phage Display: A Practical Approach. 1st ed. Oxford: Oxford University Press; 2004. p. 240. [Google Scholar]
- 4.Dyck ASM, Kisiel U, Bohne C. Dynamics for the assembly of pyrene-γ-cyclodextrin host-guest complexes. J. Phys. Chem. B. 2003;107:11652–11659. [Google Scholar]
- 5.Kohn F, Hofkens J, Wiesler UM, Cotlet M, van der Auweraer M, Mullen K, de Schryver F. Single-molecule spectroscopy of a dendrimer-based host-guest system. Chemistry. 2001;7:4126–4133. doi: 10.1002/1521-3765(20011001)7:19<4126::aid-chem4126>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong DW, Ward TJ, D AR, Beesley TE. Separation of drug stereoisomers by the formation of beta-cyclodextrin inclusion complexes. Science. 1986;232:1132–1135. doi: 10.1126/science.3704640. [DOI] [PubMed] [Google Scholar]
- 7.Wulff G. Molecular imprinting in cross-linked materials with the aid of molecular templates–a way towards artificial antibodies. Angew. Chem., Int. Ed. Engl. 1995;34:1812–1832. [Google Scholar]
- 8.Nicholls IA, Rosengren JP. Molecular imprinting of surfaces. Bioseperations. 2002;10:301–305. doi: 10.1023/a:1021541631063. [DOI] [PubMed] [Google Scholar]
- 9.Byrne ME, Oral E, Hilt JZ, Peppas NA. Networks for recognition of biomolecules: Molecular imprinting and micropatterning poly(ethylene glycol)-containing films. Polym. Adv. Technol. 2002;13:798–816. [Google Scholar]
- 10.Shea KJ. Molecular imprinting of synthetic network polymers: The de novo synthesis of macrolmolecular binding and catalytic sites. Trends Polym. Sci. 1994;2:166–173. [Google Scholar]
- 11.Mosbach K. Toward the next generation of molecular imprinting with emphasis on the formation, by direct molding, of compounds with biological activity (biomimetics) Anal. Chim. Acta. 2001;435:3–8. [Google Scholar]
- 12.Ansell RJ, Ramström O, Mosbach K. Towards artificial antibodies prepared by molecular imprinting. Clin. Chem. 1996;42:1506–1512. [PubMed] [Google Scholar]
- 13.Ramström O, Ansell RJ. Molecular imprinting technology: Challenges and prospects for the future. Chirality. 1998;10:195–209. [Google Scholar]
- 14.Sellergren B. The non-covalent approach to molecular imprinting. In: Sellergren B, editor. Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and Their Applications in Analytical Chemistry. Amsterdam: Elsevier; 2001. pp. 113–183. [Google Scholar]
- 15.Komiyama M, Takeuchi T, Asanuma H. Molecular Imprinting: From Fundamentals to Applications. Weinheim: Wiley-VCH; 2002. [Google Scholar]
- 16.Yan M, Ramström O, editors. Molecularly Imprinted materials: Science and Technology. 1st ed. New York: Dekker; 2005. [Google Scholar]
- 17.Piletsky S, Turner A, editors. Molecular Imprinting of Polymers. Austin, TX: Landes Bioscience; 2004. [Google Scholar]
- 18.Piletsky SA, Alcock S, Turner AP. Molecular imprinting: At the edge of the third millennium. Trends Biotechnol. 2001;19:9–12. doi: 10.1016/s0167-7799(00)01523-7. [DOI] [PubMed] [Google Scholar]
- 19.Bowman MAE, Allender CJ, Brain KR, Heard CM. A high-throughput screening technique employing molecularly imprinted polymers as biomimetic selectors. Methodol. Surv. Bioanal. Drugs. 1998;25:37–43. [Google Scholar]
- 20.Takeuchi T, Matsui J. Molecular imprinting: An approach to “tailor-made” synthetic polymers with biomimetic functions. Acta Polym. 1996;47:471–480. [Google Scholar]
- 21.Kempe M. Antibody-mimicking polymers as chiral stationary phases in HPLC. Anal. Chem. 1996;68:1948–1953. doi: 10.1021/ac9512160. [DOI] [PubMed] [Google Scholar]
- 22.Vlatakis G, Andersson LI, Muller R, Mosbach K. Drug assay using antibody mimics made by molecular imprinting. Nature. 1993;361:645–647. doi: 10.1038/361645a0. [DOI] [PubMed] [Google Scholar]
- 23.Spivak DA, Shea KJ. Investigation into the scope and limitations of molecular imprinting with DNA molecules. Anal. Chim. Acta. 2001;435:65–74. [Google Scholar]
- 24.Siemann M, Andersson LI, Mosbach K. Selective recognition of the herbicide atrazine by noncovalent imprinted polymers. J. Agric. Food Chem. 1996;44:141–145. [Google Scholar]
- 25.Wulff G. The covalent and other stoichiometric approaches. In: Yan M, Ramström O, editors. Molecularly Imprinted Materials: Science and Technology. New York: Dekker; 2005. pp. 59–92. [Google Scholar]
- 26.Wulff G, Schauhoff S. Enzyme-analog built polymers. 27. Racemic resolution of free sugars with macroporous polymers prepared by molecular imprinting–selectivity dependence on the arrangement of functional groups versus requirements. J. Org. Chem. 1991;56:395–400. [Google Scholar]
- 27.Alexander C, Smith CR, Whitcombe M, Vulfson EN. Imprinted polmers as protecting groups for regioselective modification or polyfunctional substrates. J. Am. Chem. Soc. 1999;121:6640–6651. [Google Scholar]
- 28.Cheong SH, McNiven S, Rachkov A, Levi R, Yano K, Karube I. Testosterone receptor binding mimic constructed using molecular imprinting. Macromolecules. 1997;30:1317–1322. [Google Scholar]
- 29.Dhal PK, Kulkarni MG, Mashelkar RA. Bio-imprinting: polymeric receptors with and for biological macromolecules. In: Sellergren B, editor. Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and Their Applications in Analytical Chemistry. New York: Elsevier; 2001. p. 23. [Google Scholar]
- 30.Mallik S, Plunkett SD, Dhal PK, Johnson RD, Pack D, Shnek D, Arnold FH. Towards materials for the specific recognition and separation of proteins. New J. Chem. 1994;18:299–304. [Google Scholar]
- 31.Ramström O, Ye L, Mosbach K. Artificial antibodies to corticosteroids prepared by molecular imprinting. Chem. Biol. 1996;3:471–477. doi: 10.1016/s1074-5521(96)90095-2. [DOI] [PubMed] [Google Scholar]
- 32.Chianella I, Lotierzo M, Piletsky SA, Tothill IE, Chen B, Karim K, Turner APF. Rational design of a polymer specific for microcystin-LR using a computational approach. Anal. Chem. 2002;74:1288–1293. doi: 10.1021/ac010840b. [DOI] [PubMed] [Google Scholar]
- 33.Kugimiya A, Matsui J, Takeuchi T. Sialic acid-imprinted polymers using noncovalent interactions. Mater. Sci. Eng., C. 1997;4:263–266. [Google Scholar]
- 34.Haupt K, Dzgoev A, Mosbach K. Assay system for the herbicide 2,4-dichlorophenoxyacetic acid using a molecularly imprinted polymer as an artificial recognition element. Anal. Chem. 1998;70:628–631. doi: 10.1021/ac9711549. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Moral N, Whitcombe MJ, Vulfson EN. Molecularly imprinted nanoparticles prepared by core-shell emulsion polymerization. J. Appl. Polym. Sci. 2000;77:1851–1859. [Google Scholar]
- 36.Perez-Moral N, Whitcombe MJ, Vulfson EN. Surface imprinting of cholesterol on submicrometer core shell particles. Macromolecules. 2001;34:830–836. [Google Scholar]
- 37.Carter SR, Rimmer S. Surface molecular imprinting in aqueous medium on polymer core-shell particles. Abstr. Pap. Am. Chem. Soc. 2002;224 363-COLL. [Google Scholar]
- 38.Carter SR, Rimmer S. Molecular recognition of caffeine by shell molecular imprinted core-shell polymer particles in aqueous media. Adv. Mater. 2002;14:667–670. [Google Scholar]
- 39.Han M, Kane R, Goto M, Belfort G. Discriminate surface molecular recognition sities on a microporous substrate: A new approach. Macromolecules. 2003;36:4472–4477. [Google Scholar]
- 40.Dirion B, Cobb Z, Schillinger E, Andersson LI, Sellergren B. Water-compatible molecularly imprinted polymers obtained via high-throughput synthesis and experimental design. J. Am. Chem. Soc. 2003;125:15101–15109. doi: 10.1021/ja0355473. [DOI] [PubMed] [Google Scholar]
- 41.Mathew J, Buchardt O. Molecular imprinting approach for the recognition of adenine in aqueous medium and hydrolysis of adenosine 5′-triphosphate. Bioconjugate Chem. 1995;6:524–528. doi: 10.1021/bc00035a004. [DOI] [PubMed] [Google Scholar]
- 42.Batra D, Shea KJ. Molecular imprinting of oligonucleotides. Abstr. Pap. Am. Chem. Soc. 2001;221 158-ORGN. [Google Scholar]
- 43.Hart BR, Shea KJ. Molecular imprinting for the recognition of N-terminal histidine peptides in aqueous solution. Macromolecules. 2002;35:6192–6201. [Google Scholar]
- 44.Diaz-Garcia ME, Laino RB. Molecular imprinting in sol-gel materials: Recent developments and applications. Microchim. Acta. 2005;149:19–36. [Google Scholar]
- 45.Zhang Z, Liao HP, Li H, Nie L, Yao S. Stereoselective histidine sensor based on molecularly imprinted sol-gels films. Anal. Biochem. 2005;336:108–116. doi: 10.1016/j.ab.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 46.Makote R, Collinson M. Template recognition in inorganicorganic hybrid films prepared by the sol-gel process. Chem. Mater. 1998;10:2440–2445. [Google Scholar]
- 47.Baggiani C, Giovannoli C. Bioimprinting. In: Piletsky S, Turner A, editors. Molecular Imprinting of Polymers. Austin, TX: Landes Bioscience; 2004. [Google Scholar]
- 48.Huang C-S. Aspira Biosystems, Burlington, CA; Compositions and methods for capturing, isolating, detecting, analyzing and quantifying macromolecules. 6,979,573. U.S. Patent. 2005
- 49.Huang J, Zhang J, Zhang J, Zheng S. Template imprinting amphoteric polymer for the recognition of proteins. J. Appl. Polym. Sci. 2005;95:358–361. [Google Scholar]
- 50.Umeno D, Kawasaki M, Maeda M. Imprinting of proteins on polymer-coated DNA for affinity seperation with enhanced selectivity. In: Bartsch RA, Maeda M, editors. Molecular and Ionic Recognition with Imprinted Polymers. Washington DC: American Chemical Society; 1998. pp. 202–216. ACS Symposium Series 703. [Google Scholar]
- 51.Vaidya AA, Lele BS, Kulkarni MG, Mashelkar RA. Creating a macromolecular receptor by affinity imprinting. J. Appl. Polym. Sci. 2001;81:1075–1083. [Google Scholar]
- 52.Ou SH, Wu MC, Chou TC, Liu CC. Polyacrylamide gels with electrostatic functional groups for the molecular imprinting of lysozyme. Anal. Chim. Acta. 2004;504:163–166. [Google Scholar]
- 53.Hjerten S, Liao JL, Nakazato K, Wang Y, Zamaratskaia G, Zhang HX. Gels mimicking antibodies in their selective recognition of proteins. Chromatographia. 1997;44:227–234. [Google Scholar]
- 54.Liao JL, Wang Y, Hjerten S. Novel support with artificially created recognition for the selective removal of proteins and for affinity chromatography. Chromatographia. 1996;42:259–262. [Google Scholar]
- 55.Tong D, Hetenyi C, Bikadi Z, Gao JP, Hjerten S. Some studies of the chromatographic properties of gels (‘artificial antibodies/ receptors’) for selective adsorption of proteins. Chromatographia. 2001;54:7–14. [Google Scholar]
- 56.Ou SH, Chou TC, Liu CC. Molecularly Imprinted Polymer Science and Technology. La Grande Motte, France: 2nd International Workshop on Molecular Imprinting; 2002. Sep 15–19, Polyacrylamide gels with electrostatic functional groups for the molecular imprinting of lysozyme; p. 13. [Google Scholar]
- 57.Rachkov A, Hu MJ, Bulgarevich E, Matsumoto T, Minoura N. Molecularly imprinted polymers prepared in aqueous solution selective for Sar(1), Ala(8) angiotensin II. Anal. Chim. Acta. 2004;504:191–197. [Google Scholar]
- 58.Rachkov A, Minoura N. Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J. Chromatogr. A. 2000;889:111–118. doi: 10.1016/s0021-9673(00)00568-9. [DOI] [PubMed] [Google Scholar]
- 59.Rachkov A, Minoura N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim. Biophys. Acta. 2001;1544:255–266. doi: 10.1016/s0167-4838(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 60.Venton DL, Gudipati E. Influence of protein on polysiloxane polymer formation–Evidence for induction of complementary protein-polymer interactions. Biochim. Biophys. Acta. 1995;1250:126–136. doi: 10.1016/0167-4838(95)00080-e. [DOI] [PubMed] [Google Scholar]
- 61.Venton DL, Gudipati E. Entrapment of enzymes using organofunctionalized polysiloxane copolymers. Biochim. Biophys. Acta. 1995;1250:117–125. doi: 10.1016/0167-4838(95)00079-a. [DOI] [PubMed] [Google Scholar]
- 62.Glad M, Norrlow O, Sellergren B, Siegbahn N, Mosbach K. Use of silane monomers for molecular imprinting and enzyme entrapment in polysiloxane-coated porous silica. J. Chromatogr. A. 1985;347:11–23. [Google Scholar]
- 63.Guo TY, Xia YQ, Wang J, Song MD, Zhang BH. Chitosan beads as molecularly imprinted polymer matrix for selective seperation of proteins. Biomaterials. 2005;26:5737–5745. doi: 10.1016/j.biomaterials.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 64.Guo TY, Xia YQ, Hao GJ, Song MD, Zhang BH. Adsorptive seperation of hemoglobin by molecularly imprinted polymers. Biomaterials. 2004;25:5905–5912. doi: 10.1016/j.biomaterials.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 65.Shi HQ, Ratner BD. Template recognition of protein-imprinted polymer surfaces. J. Biomed. Mater. Res. 2000;49:1–11. doi: 10.1002/(sici)1097-4636(200001)49:1<1::aid-jbm1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 66.Shi HQ, Tsai WB, Garrison MD, Ferrari S, Ratner BD. Template-imprinted nanostructured surfaces for protein recognition. Nature. 1999;398:593–597. doi: 10.1038/19267. [DOI] [PubMed] [Google Scholar]
- 67.Pang X, Cheng G, Lu S, Tang E. Synthesis of polyacrylamide gel beads with electrostatic functional groups for the molecular imprinting of bovine serum albumin. Anal. Bioanal. Chem. 2006;384:225–230. doi: 10.1007/s00216-005-0147-x. [DOI] [PubMed] [Google Scholar]
- 68.Burow M, Minoura N. Molecular imprinting: Synthesis of polymer particles with antibody-like binding characteristics for glucose oxidase. Biochem. Biophys. Res. Commun. 1996;227:419–422. doi: 10.1006/bbrc.1996.1522. [DOI] [PubMed] [Google Scholar]
- 69.Hirayama K, Burow M, Morikawa Y, Minoura N. Synthesis of polymer-coated silica particles with specific recognition sites for glucose oxidase by the molecular imprinting technique. Chem. Lett. 1998:731–732. [Google Scholar]
- 70.Hirayama K, Kameoka K. Synthesis of polymer particles with specific binding sites for lysozyme by a molecular imprinting technique and its application to a quartz crystal microbalance sensor. Bunseki Kagaku. 2000;49:29–33. [Google Scholar]
- 71.Hirayama K, Sakai Y, Kameoka K. Synthesis of polymer particles with specific lysozyme recognition sites by a molecular imprinting technique. J. Appl. Polym. Sci. 2001;81:3378–3387. [Google Scholar]
- 72.Bossi A, Piletsky SA, Piletska EV, Righetti PG, Turner APF. Surface-grafted molecularly imprinted polymers for protein recognition. Anal. Chem. 2001;73:5281–5286. doi: 10.1021/ac0006526. [DOI] [PubMed] [Google Scholar]
- 73.Rick J, Chou TC. Imprinting unique motifs formed from protein–protein associations. Anal. Chim. Acta. 2005;542:26–31. [Google Scholar]
- 74.Rick J, Chou TC. Enthalpy changes associated with protein binding to thin films. Biosens. Bioelectron. 2005;20:1878–1883. doi: 10.1016/j.bios.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 75.Shiomi T, Matsui M, Mizukami F, Sakaguchi K. A method for the molecular imprinting of hemoglobin on silica surfaces using silanes. Biomaterials. 2005;26:5564–5571. doi: 10.1016/j.biomaterials.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 76.Ramanaviciene A, Ramanavicius A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004;20:1076–1082. doi: 10.1016/j.bios.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 77.Kempe M, Glad M, Mosbach K. An approach towards surface imprinting using the enzyme ribonuclease A. J. Mol. Recognit. 1995;8:35–39. doi: 10.1002/jmr.300080106. [DOI] [PubMed] [Google Scholar]
- 78.Chou J, Rick J, Chou TC. C-reactive protein thin-film molecularly imprinted polymers formed using a micro-contact approach. Anal. Chim. Acta. 2005;542:20–25. [Google Scholar]
- 79.Dhruv H, Pepalla R, Taveras M, Britt DW. Protein insertion and patterning of PEG bearing Langmuir monolayers. Biotechnol. Prog. 2006;22:150–155. doi: 10.1021/bp050173y. [DOI] [PubMed] [Google Scholar]
- 80.Britt DW, Jogikalmath G, Hlady V. Protein Interactions with Monolayers at the Air/Water Interface. In: Malmsten M, editor. Biopolymer at Interfaces II. New York: Dekker; 2002. [Google Scholar]
- 81.Du X, Hlady V, Britt DW. Langmuir monolayer approaches to molecular recognition through molecular imprinting. Biosens. Bioelectron. 2005;20:2053–2060. doi: 10.1016/j.bios.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 82.Turner NW, Wright BE, Dhruv H, Hlady V, Britt DW. Proceedings of Biosensors 2006. Canada: Toronto; 2006. May 10–12, Protein imprinting in langmuir monolayers; p. 068. [Google Scholar]
- 83.Ahmed H. Principles and Reactions of Protein Extraction, Purification and Characterization. 1st ed. London: CRC Press; 2004. p. 387. [Google Scholar]
- 84.Conrad PG, II, Shea KJ. Use of metal co-ordination for controlling imprinted polymers. In: Yan M, Ramström O, editors. Molecularly Imprinted Materials: Science and Technology. New York: Dekker; 2005. pp. 123–180. [Google Scholar]
- 85.Karmalkar RN, Kulkarni MG, Mashelkar RA. Molecularly imprinted hydrogels exhibit chymotrypsin-like activity. Macromolecules. 1996;29:1366–1368. [Google Scholar]
- 86.Parmpi P, Kofinas P. Biomimetic glucose recognition using molecularly imprinted polymer hydrogels. Biomaterials. 2004;25:1969–1973. doi: 10.1016/j.biomaterials.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 87.Wizeman WJ, Kofinas P. Molecularly imprinted polymer hydrogels displaying isomerically resolved glucose binding. Biomaterials. 2001;22:1485–1491. doi: 10.1016/s0142-9612(00)00303-3. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe M, Akahoshi T, Tabata Y, Nakayama D. Molecular specific swelling change of hydrogels in accordance with the concentration of guest molecules. J. Am. Chem. Soc. 1998;120:5577–5578. [Google Scholar]
- 89.Reddy S. [accessed July 9, 2006];Hydrogels for Molecular Imprinting of Proteins. http://www.survey.ac.uk/SBMS/ACADEMICS_homepage/reddy_sub/mips.html.
- 90.Peppas NA, Huang Y, Torres-Lugo M, Ward JH, Zhang J. Physiocochemical foundations and structural design of hydrogels in medicine and biology. Annu. ReV. Biomed. Eng. 2000;2:9–29. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- 91.Peppas NA. Intelligent biomaterials in protein delivery, molecular imprinting and micropatterning. Proc. Controlled Release Soc. 2002;29 [Google Scholar]
- 92.Gill I, Ballesteros A. Bioencapsulation within synthetic polymers (Part 2): Non sol-gel protein-polymer biocomposites. Trends Biotechnol. 2000;18:469–479. doi: 10.1016/s0167-7799(00)01493-1. [DOI] [PubMed] [Google Scholar]
- 93.Gill I, Ballesteros A. Bioencapsulation within synthetic polymers (Part 1): sol-gel encapsulation biologicals. Trends Biotechnol. 2000;18:282–296. doi: 10.1016/s0167-7799(00)01457-8. [DOI] [PubMed] [Google Scholar]
- 94.Gill I. Bio-doped nanocomposite polymers: Sol-gel bioencapsulates. Chem. Mater. 2001;13:3403–3421. [Google Scholar]
- 95.Gill I, Ballesteros A. Encapsulation of biologicals within silicate, siloxane and hybrid sol-gel polymers: an efficient and generic approach. J. Am. Chem. Soc. 1998;120:8587–8598. [Google Scholar]
- 96.Walcarius A, Mandler D, Cox J, Collinson M, Lev O. Exciting new directions in the intersection of functionalized sol-gel materials with electrochemistry. J. Mater. Chem. 2005;15:3663–3689. [Google Scholar]
- 97.AspiraBiosystems. [accessed July 9, 2006];ProteinPrint Technology. http://www.aspirabio.com/2.3_protein_print.html.
- 98.Ratner BD. The engineering of biomaterials exhibiting recognition and specificity. J. Mol. Recognit. 1996;9:617–625. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<617::aid-jmr310>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 99.Ratner BD, Shi HQ. Recognition templates for biomaterials with engineered bioreactivity. Curr. Opin. Solid State Mater. Sci. 1999;4:395–402. [Google Scholar]
- 100.Britt DW, Möbius D, Hlady V. Ferritin adsorption to multicomponent monolayers: Influence of lipid charge density, miscibility and fluidity. Phys. Chem. Chem. Phys. 2000;20:4594–4599. [Google Scholar]
- 101.Pack D, Arnold FH. Langmuir monolayer characterization of metal-chelating lipids for protein targeting to membranes. Chem. Phys. Lipids. 1997;86:135–152. doi: 10.1016/s0009-3084(97)02662-5. [DOI] [PubMed] [Google Scholar]
- 102.Bondurant B, Last JA, Waggoner TA, Slade A, Sasaki DY. Optical and scanning probe analysis of glycolipid reorganization upon concanavalin A binding to mannose-coated lipid bilayers. Langmuir. 2003;19:1829–1837. [Google Scholar]
- 103.May S, Harries D, Ben-Shaul A. Lipid demixing and protein-protein interactions in the adsorption of charged proteins on mixed membranes. Biophys. J. 2000;79:1747–1760. doi: 10.1016/S0006-3495(00)76427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simon S. Current Topics in Membranes. Amsterdam: Elsevier; 2002. Peptide-lipid interactions; p. 52. [Google Scholar]
- 105.Litvin AL, Samuelson LA, Charych DH, Spevak W, Kaplan DL. Influence of supramolecular template organisation on mineralisation. J. Phys. Chem. 1995;100:17708–17708. [Google Scholar]
- 106.Gidalevitz D, Weissbuch I, Kjaer K, Alsneilsen J, Leiserowitz L. Design of two dimensional crystals as models for probing the structure of solid-liquid interface. J. Am. Chem. Soc. 1994;116:3271–3278. [Google Scholar]
- 107.Nicholls IA. Thermodynamic considerations for the design of and ligand recognition by molecularly imprinted polymers. Chem. Lett. 1995:1035–1036. [Google Scholar]
- 108.Piletska E, Piletsky S, Karim K, Terpetschnig E, Turner A. Biotin-specific synthetic receptors prepared using molecular imprinting. Anal. Chim. Acta. 2004;504:179–183. [Google Scholar]
- 109.Giraudi G, Giovannoli C, Tozzi C, Baggiani C, Anfossi L. Molecular recognition properties of peptide mixtures obtained by polymerisation of amino acids in the presence of estradiol. Anal. Chim. Acta. 2003;481:41–53. [Google Scholar]
- 110.Sellergren B. Molecular Imprinting by noncovalent interactions–Enantioselectivity and binding capacity of polymers prepared under conditions favouring the formation of template complexes. Makromol. Chem.-Macromol. Chem. Phys. 1989;190:2703–2711. [Google Scholar]
- 111.Yu C, Ramström O, Mosbach K. Enantiomeric recognition by molecularly imprinted polymers using hydrophobic interactions. Anal. Lett. 1997;30:2123–2140. [Google Scholar]
- 112.MIP-Technologies. [accessed July 9, 2006];MIP Technologies. http://www.miptechnologies.se/
- 113.MRM. [accessed July 9, 2006];Molecular Recognition Materials. http://www.recognitionmaterials.com.