Abstract
In Ciona intestinalis, leprecan was identified as a target of the notochord-specific transcription factor Ciona Brachyury (Ci-Bra) (Takahashi et al., 1999). By screening ~14 kb of the Ci-leprecan locus for cis-regulatory activity, we have identified a 581-bp minimal notochord-specific cis-regulatory module (CRM) whose activity depends upon T-box binding sites located at the 3’-end of its sequence. These sites are specifically bound in vitro by a GST-Ci-Bra fusion protein, and mutations that abolish binding in vitro result in loss or decrease of regulatory activity in vivo. Serial deletions of the 581-bp notochord CRM revealed that this sequence is also able to direct expression in muscle cells through the same T-box sites that are utilized by Ci-Bra in the notochord, which are also bound in vitro by the muscle-specific T-box activators Ci-Tbx6b and Ci-Tbx6c. Additionally, we created plasmids aimed to interfere with the function of Ci-leprecan and categorized the resulting phenotypes, which consist of variable dislocations of notochord cells along the anterior-posterior axis. Together, these observations provide mechanistic insights generally applicable to T-box transcription factors and their target sequences, as well as a first set of clues on the function of Leprecan in early chordate development.
Keywords: Ciona, Brachyury, notochord, muscle, ascidian, leprecan, T-box, Tbx6, prolyl 3-hydroxylase, cis-regulation
INTRODUCTION
The tunicate ascidian Ciona intestinalis (hereinafter Ciona) provides an exquisitely simplified model for studies of notochord development and differentiation in chordate embryos, as well as for the identification of cis-regulatory sequences (Jiang and Smith, 2007; Kusakabe, 2005; Passamaneck and Di Gregorio, 2005; Satoh et al., 2003). The Ciona notochord is invariantly composed of 40 cells, is easily distinguishable and its development requires the single-copy Brachyury (Ci-Bra) gene, which encodes a DNA-binding transcription factor expressed exclusively in this tissue (Corbo et al., 1997). This fortunate combination of experimental advantages has been successfully exploited to identify at least 50 notochord genes whose expression is influenced by Ci-Bra (Takahashi et al., 1999; Di Gregorio and Levine, 1999; Hotta et al., 2000; Oda-Ishii and Di Gregorio, 2007; Hotta et al., 2008; Kugler et al., 2008). Ci-Bra is able to bind sequences with palindromic symmetry, likely as a dimer, as well as non-palindromic sites, which are referred to as half-sites or monomeric sites (Di Gregorio and Levine, 1999; Tada and Smith, 2001).
Here we analyze in detail the mechanism by which Ci-Bra controls transcription of one its early target genes, Ciona leprecan (Ci-leprecan; Hotta et al., 2000), and report the function of Ci-Leprecan in notochord formation.
Leprecan (leucine proline-enriched proteoglycan) was first described in rat yolk sac carcinoma L2 cells as a basement membrane-associated chondroitin sulfate proteoglycan-encoding gene (Wassenhove-McCarthy and McCarthy, 1999) and one of its human orthologs, located on chromosome 1, was shortly thereafter reported to act as a potential tumor suppressor (and was also named Gros1, from growth suppressor 1; Kaul et al., 2000). Subsequent sequence profile searches predicted Leprecan to be a novel member of the iron-binding 2-oxoglutarate family of dioxygenases (Aravind and Koonin, 2001) and the characterization of the enzymatic activity of its chick ortholog validated these predictions (Vranka et al., 2004). In humans and mice, in addition to the LEPRE1 gene, two additional Leprecan orthologs, Leprecan-like1 (LEPREL1) and Leprecan-like 2 (LEPREL2) were subsequently identified (Järnum et al., 2004). The prolyl 3-hydroxylase (P3H) encoded by the LEPRE1 locus (called P3H1) and its interacting factors, Cartilage-Associated Protein (CRTAP) and Cyclophilin B (CYPB), have been shown to form a complex that hydroxylates unfolded procollagen type I and type II, in particular at proline 986 of their α1 chains, likely ensuring its proper folding and secretion (Marini et al., 2007). Mutations in either LEPRE1 or CRTAP have been shown to be responsible for human recessive osteogenesis imperfecta, also known as brittle bone disease (Baldridge et al., 2008; Cabral et al., 2007). Leprecan proteins are highly conserved across vertebrates, sharing common features such as an N-terminal signal cleavage peptide and a C-terminal KDEL endoplasmic reticulum retrieval signal (Järnum et al., 2004). Their C-terminal region contains a conserved prolyl 3-hydroxylase domain, which was identified by its high sequence similarity to the prolyl 4-hydroxylase alpha (P4Hα) domain (Aravind and Koonin, 2001). The prolyl 3-hydroxylase domain contains evolutionarily conserved amino acid residues which are able to coordinately bind iron and are essential for the enzymatic activity (Clifton et al., 2001).
Our group has recently analyzed the expression patterns of the three Leprecan orthologs found in the mouse genome. These studies have shown that all three genes are expressed in the developing notochord of mid-gestation mouse embryos, in addition to being expressed in various additional tissues (Capellini et al., 2008). The evolutionary conservation of the notochord expression of the Leprecan genes prompted us to investigate both the transcriptional regulation and the developmental role of the single-copy, notochord-specific Ci-leprecan gene. By scanning ~14 kb of the Ci-leprecan genomic locus, we identified a cis-regulatory module (CRM) able to recapitulate Ci-leprecan expression in the notochord. We show that this CRM is controlled by overlapping minimal sequences specifically bound in vitro by the notochord-specific Ci-Bra as well as by two additional, highly related muscle-specific T-box transcription factors, Ci-Tbx6b and Ci-Tbx6c. Consistent with the in vitro analysis, these sequences are not only required for notochord activity, but are also able to direct a genuine reporter gene expression in muscle cells in vivo. However, the activation by Ci-Tbx6b and Ci-Tbx6c is counterbalanced by tissue-specific repressive signals, so that the transcriptional output in muscle cells is null.
We also employed Ciona to gain a first insight on the function of Leprecan in notochord formation, which was yet to be investigated in any chordate. We created constructs aimed to study the effects of Ci-Leprecan over-expression and knock-down on notochord development, and to analyze the phenotypes associated with the expression of a dominant-negative form of Ci-Leprecan. We report that the over-expression of Ci-Leprecan in developing notochord cells produces little or no phenotypic effect. The expression in notochord cells of the dominant-negative Ci-Leprecan, however, reproducibly results in the disruption of their linear, single-file arrangement with respect to the AP axis. Similar defects are observed in Ci-leprecan knock-down embryos. We discuss these results in light of the current knowledge of Leprecan function and propose a model of its potential role in notochord formation.
MATERIALS AND METHODS
Embryo rearing and electroporations
Adult Ciona intestinalis were purchased from Marine Research and Educational Products (M-REP; Carlsbad, CA) and kept at 18°C in recirculating artificial sea water. Embryos were cultured at temperatures ranging from 15°C to 21°C; no phenotypic effect due to the culturing temperature was observed.
Plasmid construction
Unless otherwise specified, all constructs used for enhancer analysis were cloned into the XhoI and XbaI sites of the pFBΔSP6 vector (Oda-Ishii and Di Gregorio, 2007). All fragments shown in Fig. 1 were obtained from Ciona intestinalis genomic DNA via PCR amplification. After the 581-bp Ci-leprecan notochord enhancer was identified, all the further truncation and/or mutation plasmids were generated by PCR amplification, using the 581-bp Ci-leprecan plasmid as the template DNA. The sequences of the primers used for amplification of genomic DNA fragments, as well as the sequences of the primers employed to create constructs containing mutations and truncations are listed in Supplemental Table 1. Subsequent to its amplification via RT-PCR, the full-length Ci-leprecan 2.7-kb cDNA was cloned into the pCR-2.1-TOPO vector (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, and the resulting plasmid was named LepTOPO. To generate the Ci-leprecan over-expression or dominant-negative constructs, a modified version of pFBΔSP6 lacking the Ci-fkh basal promoter and the LacZ open reading frame was constructed by excising an XbaI/EcoRI fragment and by re-ligating the resulting 2.6-kb vector backbone in the presence of the following annealed oligos:
Linker.F: 5’-CTAGAGCTCTGTACATCACGGTACCG-3’ and
Linker.R: 5’-AATTCGGTACCGTGATGTACAGAGCT-3’
which contain recognition sequences for SacI, BsrGI and KpnI. The resulting plasmid was named pΔSP6-Linker. As a next step the full-length Ci-Bra enhancer/promoter region, a 3.35-kb XhoI/SacI fragment, was excised from the 3.5-kb Ci-Bra->LacZ construct (Corbo et al., 1997) and ligated into pΔSP6-Linker. This resulted in a vector containing the full-length notochord-specific Ci-Bra enhancer/promoter region followed by a minimal multicloning site, which was named pCi-Bra-Linker.
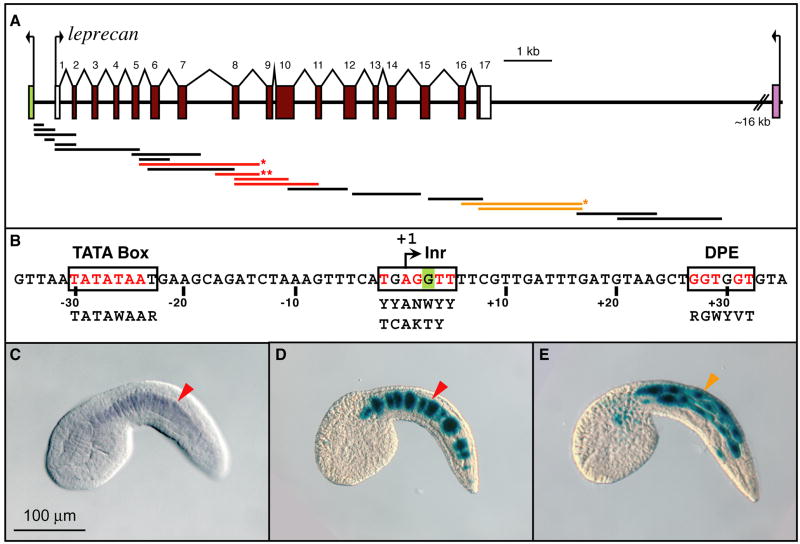
Figure 1. Identification of distinct cis-regulatory modules within the Ciona leprecan genomic locus.
(A) Schematic representation of the Ci-leprecan genomic locus and its surroundings. Exons are indicated by colored rectangles, introns by lines, untranslated regions by white rectangles. The fragments that were cloned and tested in vivo for cis-regulatory activity are shown as thick lines. Black, red and orange lines indicate fragments that were found to be either inactive, or active in notochord or muscle, respectively. The red line with one asterisk represents the plasmid employed in (D); the red line with two asterisks represents the 581-bp CRM (see Fig. 2). (B) Sequences of the core promoter elements identified within the Ci-leprecan locus. Inr: initiator; DPE: downstream promoter element. The “G” highlighted in green is the first nucleotide of the Ci-leprecan cDNA (see Methods). Consensus sequences for human and Drosophila elements (Butler and Kadonaga, 2002), respectively, are shown in black below the Ci-leprecan promoter sequence. Exact matches between Ciona and the published consensus sequences are shown in red.
(C) Microphotograph of a Ciona intestinalis whole-mount mid-tailbud embryo hybridized in situ with a Ci-leprecan antisense RNA probe. Signal is detected exclusively in notochord cells (red arrowhead). (D) A Ciona mid-tailbud embryo electroporated at the 1-cell stage with a plasmid containing the genomic fragment indicated by a red asterisk in (A), then fixed and stained to detect β-galactosidase activity. Staining is detected in most of the notochord cells (red arrowhead); some of the notochord cells are blank due to mosaic incorporation of the transgene. (E) A Ciona mid-tailbud embryo electroporated at the 1-cell stage with a plasmid containing the genomic fragment indicated by an orange asterisk in (A), then fixed and stained as described. Staining is detected in muscle cells (orange arrowhead). Embryos are oriented with anterior to the left and dorsal on top.
The Ci-leprecan over-expression construct was generated by amplifying the entire 2.7-kb Ci-leprecan cDNA from LepTOPO using the following primers:
Lep.Full.Sac.F: 5’-ACTGGAGCTCGTTTTCGTTGATTTGATGTAAGCTGG-3’ and
Lep.KpnI.R: 5’-TTCAGGTACCATGCATGCTCGAGCGGCCGCCAGTGTG-3’
and subcloning it into the appropriate restriction sites of the pCi-Bra-Linker vector. The Ci-leprecan dominant-negative RFP fusion was generated as follows: the first 2.055 kb of the Ci-leprecan cDNA were PCR-amplified using the primers Lep.Full.Sac.F and Lep.RTTV.RFP.R:
5’-GGAGGAGGCCATAACAGTTGTCCTGCACACAAGG-3’.
This latter primer appends the initial 12 bp of the monomeric RFP (mRFP) cDNA (Rhee et al., 2005 and references therein) onto the 3’-end of the amplified product. In parallel, mRFP was amplified using the primers:
Lep.RTTV.RFP.F: 5’-AGGACAACTGTTATGGCCTCCTCCGAGGACGTCA-3’ and
mRFP.Kpn.R: 5’-AACTGGGTACCTTAGGCGCCGGTGGAGTGGC-3’.
In this case, bp 2044-2055 of the Ci-leprecan cDNA were appended to the 5’-end of the amplified product. The resulting PCR products, now with 24 bp of identity at their 3’ and 5’ termini respectively, were purified and used to seed an additional PCR using the Lep.Full.Sac.F and mRFP.Kpn.R primers. The product of this new PCR round was then cloned into the appropriate sites in the pCi-Bra-Linker vector.
The Ci-leprecan shRNA plasmids (shLEP-767 and shLEP-1426) were constructed according to Nishiyama and Fujiwara (2008). For the construction of shLEP-767, which targets 21 nucleotides of the Ci-leprecan coding sequence starting at nucleotide 767, the following oligonucleotides were used:
shLEP767.A1: 5’-CAGGGCATGCCATGTAGCATTTTT-3’
shLEP767.A2: 5’-AATTAAAAATGCTACATGGCATGCCCTGGGAC-3’
shLEP767.S1: 5’-AATGCTCCATGTCATGTCCTGGTGTGCTGTCC-3’ and
shLEP767.S2: 5’-AGCACACCAGGACATGACATGGAGCATT-3’.
To construct shLEP-1426, which targets 21 nucleotides of the Ci-leprecan coding sequence starting at nucleotide 1426, we utilized the following oligonucleotides:
shLEP1426.A1: 5’-CCTAGCATTGTAAGCATCCTTTTT-3’
shLEP1426.A2: 5’-AATTAAAAAGGATGCTTACAATGCTAGGGGAC-3’
shLEP1426.S1: 5’-AAGGGTGCTTGCAATGTTAGGGTGTGCTGTCC-3’ and
shLEP1426.S2: 5’-AGCACACCCTAACATTGCAAGCACCCTT-3’.
Plasmid purifications, electroporations, fixation and staining were carried out as described previously (Oda-Ishii and Di Gregorio, 2007 and references therein). After each electroporation, embryos were stained and preserved and reporter gene expression in notochord and muscle was quantified under a dissecting microscope. Each plasmid was tested at least five times, and for inclusion in the quantitative analysis a minimum of three independent experiments demonstrating representative staining and containing ≥100 scored embryos were averaged.
Whole-mount in situ hybridization
Whole-mount in situ hybridization experiments were performed as previously described (Oda-Ishii and Di Gregorio, 2007). A digoxigenin-labeled antisense RNA was synthesized in vitro from the linearized full-length Ci-leprecan cDNA (Capellini et al., 2008) according to the manufacturers’ instructions (Roche, Indianapolis, IN), followed by alkaline degradation for 30 min. at 65°C (Corbo et al., 1997). Probes for Ciona intestinalis orthologs of CRTAP (ci0100133696 http://genome.jgi-psf.org/Cioin2/Cioin2.home.html Dehal et al., 2002) and CYPB (ci0100136100) were obtained from EST clones GC17h21 and GC19n23 (Satou et al., 2002), respectively, and labeled as described above, without alkaline degradation.
Electrophoretic mobility shift assays (EMSA)
For the Ci-Bra in vitro DNA-binding assays we used a previously described GST-Ci-Bra fusion protein (Di Gregorio and Levine, 1999). For the Ci-Tbx6b and Ci-Tbx6c EMSA we used GST-Ci-Tbx6b and GST-Ci-Tbx6c fusion proteins that were cloned and prepared accordingly to the previously described procedure (Yagi et al., 2005). For the Ci-FoxA-a EMSA we employed the GST-Ci-FoxA-a fusion protein previously described as GST-Ci-Fkh (Di Gregorio et al., 2001) (Ci-Fkh was recently renamed Ci-FoxA-a; Imai et al., 2004). The following double-stranded oligonucleotides were used (only the 5’-3’ strand is reported):
5’-GTATCTAAGTGTGATACTTTGTGT-3’ (466-489),
5’-GTATCTAATGGTGCTACTTTGTGT-3’ (466-489 m1),
5’-GTATCTAATGGTGATACTTTGTGT-3’ (466-489 m2),
5’-GTATCTAATGAGACGACTTTGTGT-3’ (466-489 m3),
5’-TGATACTTTGTGTAAACTGTAAAA-3’ (477-500),
5’-TGATACTTTTCTTAGACTGTAAAA-3’ (477-500 T-box4 mut),
5’-CCTATTTACTTGGTATTTGTTAACTCTTA-3’ (532-560),
5’-CTACACCCATATTTGAAGACCAAA-3’ (27-52 Fox),
5’-CTACACCCAGCGAGGAAGACCAAA-3’ (27-52 Fox mut) and
5’-CCTAGCGACTTGGTAGCGGTTAACTCTTA-3’ (532-560 Fox mut)
Complementary oligonucleotides were annealed, then radioactively labeled as previously described (Corbo et al., 1998) and purified using G-25 Sephadex columns (Roche, Indianapolis, IN).
Protein-DNA complexes were formed on ice for 30 min., essentially as previously described (Corbo et al., 1998), in the presence of 3 × 104 cpm of each probe and 80 ng of the respective GST fusion proteins, unless otherwise specified. The complexes were fractionated on 5% polyacrylamide/0.5x TBE gels and visualized by autoradiography.
Confocal imaging
Embryos electroporated with fluorescent constructs were fixed and mounted as previously described (Oda-Ishii and Di Gregorio, 2007). Laser scanning confocal images were obtained with a Zeiss LSM 510 on a Zeiss Axiovert 200 wide-field microscope, employing a 25X oil immersion lens, at the Weill Cornell Medical College Optical Microscopy Core Facility.
RESULTS
Identification of cis-regulatory modules in the Ci-leprecan genomic locus
The Ci-leprecan coding region (gene model: ci0100131532; Hotta et al., 2000) spans ~11.3 kb and is flanked by the ci0100149251 (pontin) gene and by the distant neighbor ci0100149186 (Ephrin type-A receptor 7 precursor, or Eph4; Imai et al., 2004) (Fig. 1A). By combining RT-PCR experiments (Capellini et al., 2008) with the analysis of the available EST clones (Satou et al., 2001), we were able to extend the previously published 1236-bp cDNA sequence by over 1 kb, resulting in a cDNA sequence 2684 bp long. The analysis of the Ci-leprecan genomic locus directly upstream of this longer cDNA sequence elucidated the presence of landmark sequences usually associated with RNA polymerase II transcription start sites (Fig. 1B), including a canonical TATA box, a conserved initiator (Inr) and downstream promoter element (DPE) sequences (boxed in Fig. 1B; Butler and Kadonaga, 2002). Furthermore, the first nucleotide of our additional cDNA sequence maps within the initiator sequence (green “G” in Fig. 1B).
The extended Ci-leprecan sequence encodes CxxxC motifs, which have been hypothesized to serve as sources of intramolecular disulfide bridges (Hassell et al., 1980), alpha-helical stretches which presumably correspond to the tetratricopeptide repeat protein (TPR) domains (Wassenhove-McCarthy and McCarthy, 1999) and a putative N-terminal signal cleavage peptide, indicating that the newly identified full-length Ci-Leprecan protein is more closely related to its vertebrate orthologs than was originally thought (Fig. S1).
To identify a CRM(s) able to recapitulate Ci-leprecan expression in the notochord (Fig. 1C), 20 different genomic fragments from the Ci-leprecan locus, ranging in size from 222 bp to 2.3 kb and spanning from position -433 to +13,772, were cloned in the pFBΔSP6 vector (Oda-Ishii and Di Gregorio, 2007) and independently tested in vivo by electroporation in Ciona intestinalis zygotes. Four genomic fragments spanning exons 5-11 were found to direct strong staining in notochord cells (red bars in Fig. 1A); a representative mid-tailbud embryo electroporated with a 2.3-kb fragment (indicated by a red asterisk in Fig. 1A) is shown in Fig. 1D. Two fragments covering the 3’-end of the coding region (orange bars in Fig. 1A) were found to direct only muscle expression; one, spanning 2.5 kb, yielded the staining seen in the embryo shown in Fig. 1E (indicated by an orange asterisk in Fig. 1A). Due to their lack of notochord activity, these latter fragments were not analyzed further.
A cluster of T-box half-sites is required for the activity of the main Ci-leprecan CRM in both notochord and muscle
By comparing the activity of the four constructs active in notochord cells (red bars in Fig. 1A), we were able to select the shortest fragment that retained the ability to recapitulate the notochord expression of Ci-leprecan: a 581-bp sequence located at position +3246 from the transcription start site, which we named the 581-bp CRM (indicated by two red asterisks in Fig. 1A). Initial sequence searches of the 581-bp CRM (represented by a beige horizontal bar in Fig. 2) uncovered putative binding sites for known notochord transcriptional activators, namely T-box (minimal core sequence: TNNCAC) and Fox (minimal core sequence: TRTTKR) binding sites (Di Gregorio and Levine, 1999; Di Gregorio et al., 2001). We therefore sought to examine whether activity of the 1-581 CRM is controlled by these transcription factors via the creation of 29 additional constructs, bearing truncations and/or point mutations (Figs. 2 and S2). The binding sites identified in the regions that were found to be relevant for activity are depicted as colored vertical boxes in Fig. 2.
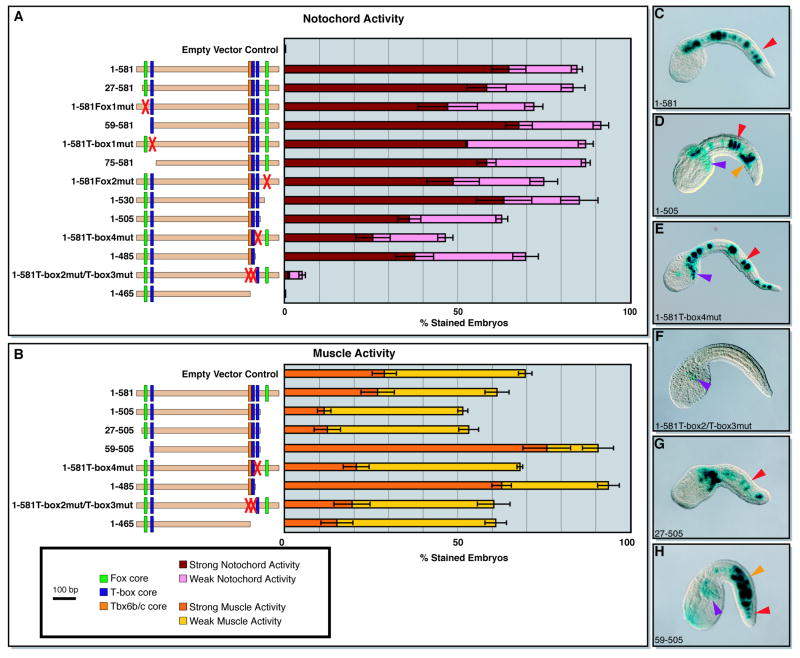
Figure 2. Semi-quantitative analysis of the cis-regulatory activity of different Ci-leprecan genomic fragments in notochord and muscle cells.
Fifteen different plasmids containing various truncations and mutations of the minimal 581-bp notochord CRM were tested in vivo for cis-regulatory activity in notochord and muscle cells and then subjected to a quantitative analysis. “Weak” staining is defined here as staining seen in ≤10% of the cells of the tissue analyzed, while “strong” notochord staining is defined as activity in >10% of the cells, regardless of the qualitative intensity of the coloration.
(A) Left side: schematic representation of the Ci-leprecan genomic fragments tested (beige horizontal bars). Transcription factors binding sites are shown as color-coded vertical boxes. Sites that were ablated by mutations are replaced by a red “X”. The empty vector control is the pFBΔSP6 plasmid (see Methods). Right side: graph showing the notochord activity of each of the plasmids listed on the left side as the percent of embryos showing staining. (B) Left side: schematics of the plasmids scored for muscle staining, as described above. Right side: graph showing the muscle activity of the plasmids listed on the left side. (C-H) Microphotographs of representative embryos electroporated at the 1-cell stage with some of the most informative constructs analyzed. Arrowheads are color-coded as follows: red: notochord cells; purple: mesenchyme cells; orange: muscle cells. In the graphs, error bars correspond to the Standard Error of the Mean; in all cases n≥3 independent experiments containing >100 well-developed stained embryos.
The analysis of an initial series of 5’ truncation constructs generated to examine the contributions of the 5’ T-box and Fox sites (blue and green vertical boxes) showed that their removal did not significantly reduce notochord activity (Fig. 2A, 27-581 through 75-581). The analysis of 3’ truncations (Fig. 2A) demonstrated that while removal of the 3’-most T-box half-site (T-box4) and of the 3’ Fox site attenuated the activity of the 1-581 CRM in the notochord, both sites are overall dispensable (Fig. 2A, compare 1-530, 1-505, and 1-485 to 1-581). However, the deletion of a further 20 bp from the 3’-end, containing the overlapping T-box2 and T-box3 half-sites, resulted in a complete loss of notochord activity (Fig. 2A, 1-465). Comparable results were obtained when these T-box sites were ablated via sequence-specific point mutations (Fig. 2A, 1-581T-box2mut/T-box3mut), which were sufficient to eliminate almost all the notochord activity of the 1-581 CRM. Additionally, no notochord staining was detected when the T-box2 and T-box3 sites were mutated along with the T-box4 half-site (Fig. S2), suggesting that these three T-box half-sites are the main source of the notochord activity of the 1-581 CRM.
In addition to identifying the sequences responsible for notochord activity, the analysis of the 3’ truncations also uncovered ectopic activity of this CRM in muscle cells, which was significantly above the levels attributable to the pFBΔSP6 vector alone (Fig. 2B, empty vector control). The muscle activity remained at background levels with the truncation of up to 76 bp of the 1-581 CRM 3’ sequence (Fig. 2B, 1-505). However, the truncation of either 20 bp from the 3’-end of the 1-505 construct (Fig. 2B, 1-485), or of 59 bp from its 5’-end (Fig. 2B, 59-505) abruptly increased muscle activity. Truncation of the first 26 bp from the 5’-end of the 1-505 construct did not have any effect on muscle activity (Fig. 2B, compare 27-505 with 59-505). These results could be explained by the presence of binding sites for a transcriptional repressor(s) within these regions (e.g., Mannervik et al., 1999). Neither sequence contained evident binding sites for the well-characterized muscle repressor Ci-Snail (Fujiwara et al., 1998) and our studies on other candidate repressors were not conclusive (data not shown); however, each of the two regions required for repression contains a 7-bp sequence, TAAACTG, which might help future searches. As in the case of the notochord, we found that either deletion or mutation of the overlapping T-box2 and T-box3 half-sites was sufficient to eliminate the ectopic muscle activity. In an effort to identify any additional sequences which might be contributing to the muscle activity, we also mutated the only E-box (consensus: CANNTG; e.g., Sartorelli et al., 1990) found within the 581-bp sequence, since bHLH transcription factors such as MyoD/MRF and Tbx15/18/22 have been shown to activate gene expression in muscle cells in ascidian embryos (e.g., Erives and Levine, 2000; Meedel et al., 2007), but no visible effect was obtained on the activity of the CRM (Fig. S2).
The T-box half-sites in the 3’ region of the main Ci-leprecan CRM are bound in vitro by Ci-Bra, Ci-Tbx6b and Ci-Tbx6c
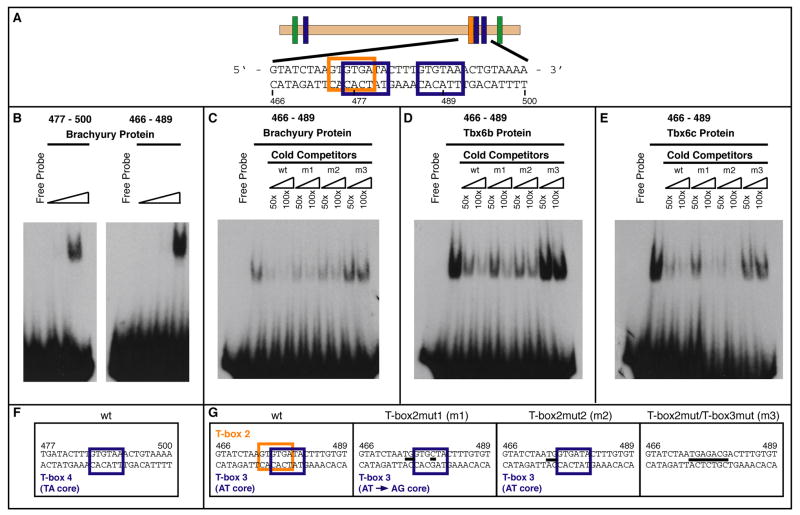
The main sequence required for activity of the Ci-leprecan 581-bp CRM spans 34 bp (Fig. 3A) and contains three T-box half-sites (generic sequence: TNNCAC), two of which are overlapping (boxed in orange and blue in Fig. 3A). One of the two, indicated as T-box2 (boxed in orange in Fig. 3A,G), shares a 8/10 match with the consensus binding site previously identified in vitro for two related muscle activators, Ci-Tbx6b and Ci-Tbx6c (GWTCACACCT; Yagi et al., 2005), while the other, indicated as T-box3 (boxed in blue in Fig. 3A,G), shares a 6/6 match with Ci-trop-prox, one of the Ci-Bra sites found in the 114-bp Ci-tropomyosin-like (Ci-trop) notochord CRM (Di Gregorio and Levine, 1999). Another site, T-box4, located 8 bp downstream (also boxed in blue in Fig. 3A,G) of T-box2 and T-box3, displays a 5/6 match with Ci-trop-dist, another Ci-Bra site found in the Ci-trop notochord CRM (Di Gregorio and Levine, 1999). For these reasons, we tested the binding to these sites of all three proteins, Ci-Bra, Ci-Tbx6b and Ci-Tbx6c by EMSA.
Figure 3. In vitro binding of Ci-Bra, Ci-Tbx6b and Ci-Tbx6c to the T-box sites found in the 3’ region of the 581-bp Ci-leprecan CRM.
(A) Schematic representation of the 581-bp Ci-leprecan CRM, with the generic T-box binding sites indicated by blue vertical boxes; an orange vertical box represents the region matching the Tbx6b/c core consensus sequence; green vertical boxes denote Fox binding sites (see Fig. S3). (B) EMSA performed employing two different 24-bp radioactive oligonucleotides encompassing either the two distal-most T-box binding sites (466-489; T-box2 and T-box3) or the 3’-most T-box half-site (477-500; T-box4) and increasing amounts of a GST-Ci-Bra fusion protein (0.27 ng, 2.67 ng, 80 ng, respectively). (C) Competition assay performed using the GST-Ci-Bra fusion protein, the radiolabeled 24-bp oligonucleotide 466-489 and unlabeled 24-bp competitor oligonucleotides containing different mutations (T-box2mut1, T-box2mut2 and T-box2mut/T-box3mut) in the respective T-box binding sites. (D) Competition assay performed using a GST-Ci-Tbx6b fusion protein and the probe and competitors described in (C). (E) Competition assay involving a GST-Ci-Tbx6c fusion protein and the probe and competitors described in (C). (F) Sequences of the oligonucleotides which were employed in the assay. Mutations are indicated by thick black dashes.
We found that a GST-Ci-Bra fusion protein efficiently binds in vitro the 477-500 sequence (Fig. 3A,F), which contains the “TA core” (i.e., TTACAC) 3’-most T-box half-site, T-box4 (Fig. 3B, left panel). Oligonucleotides spanning the 466-489 sequence, which contains two partially overlapping T-box half-sites in tandem orientation, T-box2 and T-box3 (Fig. 3A,G), are bound more avidly than the T-box4 sequence when the same amount of protein is employed (Fig. 3B, right panel, and data not shown).
To assess the relative affinity of each protein for the different binding sites, we carried out competition assays (Fig. 3C-E). In the case of Ci-Bra, as expected, the competition exerted by the unlabeled (cold) wt 466-489 double-stranded oligonucleotide was highly efficient. When the competition was performed using two unlabeled double-stranded oligonucleotides carrying mutations that either obliterate the T-box2 site changing the T-box3 ‘NN’ core from AT to AG (T-box2mut1; Fig. 3C,G) or that obliterate the T-box2 site leaving the T-box3 site unchanged (T-box2mut2; Fig. 3C,G) the radiolabeled DNA-protein complex was drastically reduced, although not completely eliminated (Fig. 3C). Finally, when the competition reaction was performed in the presence of an unlabeled double-stranded oligonucleotide containing a mutation that ablates both sites, T-box2 and T-box3 (T-box2mut/T-box3mut in Fig. 3G), the radiolabeled complex remained unaffected, suggesting that the T-box sites are specifically required for the binding of this sequence by Ci-Bra.
In a different experiment, the same amount of the GST-Ci-Tbx6b fusion protein (Fig. 3D) was utilized for EMSA involving the 466-489 sequence and its mutant versions. In this case, a competition slightly lower but comparable to that exerted by the unlabeled wt sequence was observed when both the T-box2mut1 and the T-box2mut2 unlabeled double-stranded oligonucleotides were used. Also in this case, the unlabeled T-box2mut/T-box3mut double-stranded oligonucleotide did not affect the complex formed by Ci-Tbx6b and the wt 466-489 probe, again suggesting that the T-box sites are required for binding of this sequence by Ci-Tbx6b.
Finally, we found that the 466-489 sequence was bound also by Ci-Tbx6c (Fig. 3E). In this case, the radioactive DNA-protein complex was efficiently competed by both the unlabeled wt and T-box2mut2 double-stranded oligonucleotides and it was weakened by the T-box3mut double-stranded oligonucleotide, suggesting the possibility that the binding by Ci-Tbx6c to the 466-489 sequence was not as specific as it was in the case of Ci-Bra and Ci-Tbx6b.
To better assess the effects of point mutations on the interaction between the 466-489 sequence and the three T-box transcription factors, we carried out the reciprocal in vitro binding assays by mixing the radioactively labeled double-stranded mutant oligonucleotides T-box2mut, T-box2mut2 and T-box3mut with the GST fusion proteins described above. As shown in Figure S2A, Ci-Bra does not bind the T-box2mut1 mutant probe while both Ci-Tbx6b and Ci-Tbx6c are able to bind this sequence, suggesting that both Tbx6 transcription factors might be able to bind the residual “AG core” T-box (Fig. S2A, m1 panel). Similar results were obtained when the radiolabeled mutant probe T-box2mut2 was tested (Fig. S2A, m2 panel). Finally, the T-box2mut/T-box3mut sequence was not bound by any of the T-box proteins tested (Fig. S2A, m3 panel).
The intensity of notochord staining directed by the 1-581Fox1mut and 1-581Fox2mut constructs, which bear mutations in the 5’ and 3’ Fox sites, respectively, appeared slightly decreased when compared to the wt 581-bp CRM (Fig. 2A). This observation prompted us to verify the interaction between a transcription factor of the Fox family to the putative Fox sites found in the 5’-and 3’-end of the 581-bp CRM. Both putative Fox binding sites were tested in vitro using a fusion protein containing the helix-winged-helix DNA-binding domain of Ci-FoxA-a (formerly Ci-Fkh/HNF-3beta) (Di Gregorio et al., 2001). Interestingly, both sequences are bound by Ci-FoxA-a, although with very different affinities (Fig. S2B). The distal site (27-52 probe in Fig. S2B) is bound weakly by Ci-FoxA-a; the radiolabeled complex is disrupted by the cold wt competitor, but is not affected by a cold competitor oligonucleotide containing the same mutations shown in Fig. 2A. The proximal site (532-566 probe in Fig. S2B) is bound intensely by the same amount of Ci-FoxA-a, and also in this case the DNA-protein complex is competed efficiently only by the cold wt probe, but not by the cold mutant competitor containing the mutations indicated in Fig. 2A, suggesting that the binding is specific. These results suggest that a transcription factor of the Fox family might play a minor role in activating the 581-bp CRM in notochord cells.
Point mutation analysis highlights individual nucleotides required for the tissue-specific activity of the T-box binding sites in vivo
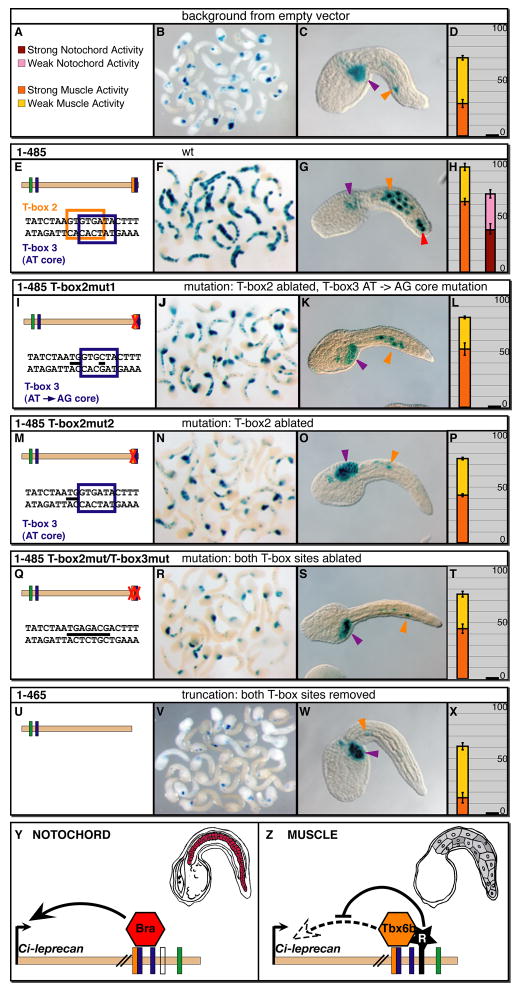
After assessing in vitro the binding of different T-box transcription factors to the previously identified T-box binding sites, we introduced the mutations previously tested by EMSA into the Ci-leprecan CRM sequence, in order to test in vivo their respective effects on the cis-regulatory activity (Fig. 4). We used the pFBΔSP6 vector to derive a baseline, since this vector is active in mesenchyme cells and, weakly, in muscle cells (Fig. 4A-D). For each of the constructs analyzed, at least 100 well-developed embryos were pooled (examples are shown in Fig. 4B,F,J,N,R,V) and scored (Fig. 4D,H,L,P,T,X), with at least 3 such experiments averaged. To analyze these mutations, we used as a starting point the 1-485 construct (Fig. 4E), a shorter version of the 581-bp CRM, since in addition to being consistently active in notochord cells, it is active well above background in muscle cells (Fig. 4F-H). When the wt sequence of the two T-box sites was changed to the T-box2mut1 sequence (Fig. 4I), the notochord staining was completely abolished, and the muscle staining was reduced to slightly above background levels (Fig. 4K,L). The T-box2mut2 mutation (Fig. 4M) had a similar effect on both notochord and muscle activity (Fig. 4N-P), and the more aggressive T-box2mut/T-box3mut mutation (Fig. 4Q) reduced the intensity of the residual muscle staining even further (Fig. 4S,T). Finally, a truncation which removed the 20-bp sequence harboring the two T-box sites (construct 1-465, Fig. 4U) led to the loss of activity above baseline levels in both tissues (Figs. 2A,B and 4V-X).
Figure 4. In vivo effects of point mutations in the T-box sites found in the main Ci-leprecan CRM.
Ciona zygotes were electroporated with the various plasmids indicated on top of each row, then fixed at the mid-tailbud stage and stained to detect reporter gene expression. (A) the color scheme employed in the graphs. (B,F,J,N,R,V) Low-magnification microphotographs of stained mid-tailbud embryos. (C,G,K,O,S,W) High-magnification views of representative embryos; arrowheads are color-coded as follows: purple: mesenchyme; orange: muscle; red: notochord. Dorsal is up, anterior to the left. (D,H,L,P,T,X) Graphs reporting the activity of each plasmid as the percentage of stained embryos. Error bars were determined as in Fig. 2. (E,I,M,Q,U) Sequences of the regions that were mutagenized in order to assess the relative contributions of different T-box sites to muscle and notochord activity. Mutations are indicated by thick black dashes. The empty vector pFBΔSP6 (see Methods), which directs a consistent background staining in mesenchyme and a sporadic staining in muscle cells, was used as a control in all the experiments described. (Y,Z) A model for the regulation of the Ci-leprecan CRM by related T-box proteins in different tissues. In notochord cells (Y) the cluster of T-box half-sites found in the 581-bp CRM is employed by Ci-Bra to activate transcription of Ci-leprecan (red notochord cells in the mid-tailbud embryo schematic). In muscle cells (Z) the same cluster of sites is able to recruit the Ci-Tbx6b/c activators; however, due to the presence of a repressor bound nearby (black star), the resulting transcriptional output is zero and Ci-leprecan expression is not observed (grey muscle cells in the mid-tailbud embryo schematic). For simplicity, only one of the two regions responsible for repression is depicted.
These results are consistent with those obtained in both the competition assays (Fig. 3C-E) and the binding assays (Fig. S2). In fact, the binding assays shown in Figure S2 indicate that all three mutations abolish binding of Ci-Bra to the CRM, consistent with the loss of notochord activity observed in all three cases; conversely, the residual binding of both Ci-Tbx6b and Ci-Tbx6c to the mutant sites likely explains the residual muscle staining detected above background levels in mid-tailbud embryos electroporated with the 1-485 construct carrying either the T-box2mut1 (Fig. 4I-L) or the T-box2mut2 mutation (Fig. 4M-P). Our observations are summarized in the model shown in Fig. 4Y,Z; in notochord cells, the cluster of T-box sites found in the 581-bp Ci-leprecan CRM is bound by Ci-Bra to activate transcription, while in muscle cells, where Ci-Bra is not present, the sites can be bound by Ci-Tbx6b and/or Ci-Tbx6c. In muscle cells, however, the presence of a transcriptional repressor(s) zeroes the contribution of these activators, and as a result no expression of Ci-leprecan is detected.
A dominant-negative form of Ci-Leprecan affects the structural integrity of the notochord
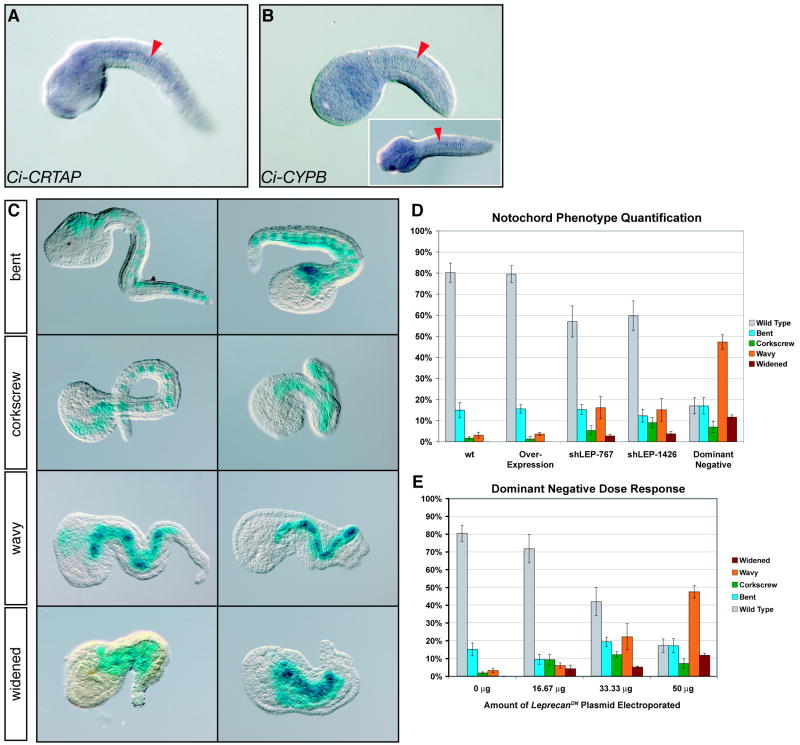
Given that human P3H1, the product of LEPRE1, forms a collagen-modifying complex together with CRTAP and CYPB (Vranka et al., 2004), we sought to determine whether the putative Ci-Leprecan interacting partners were present in the Ciona genome, and whether their expression overlapped with that of Ci-leprecan. By whole-mount in situ hybridization, we observed that Ci-CRTAP is expressed in the notochord of mid-tailbud embryos (Fig. 5A); the antisense Ci-CYPB RNA probe produced a more widely diffused signal (Fig. 5B) while no signal was observed when a sense probe was tested (data not shown), suggesting a nearly ubiquitous expression of this gene, although some embryos showed a slightly stronger expression in notochord cells (inset in Fig. 5B). The nearly ubiquitous expression of Ci-CYPB is in agreement with what had been previously reported in Ciona (http://ghost.zool.kyoto-u.ac.jp/cgi-bin3/txtgetr2.cgi?CLSTR00900; Satou et al., 2001) and with the expression observed so far throughout eukaryotes (Stamnes et al., 1990). These results suggest that Ci-Leprecan, Ci-CRTAP and Ci-CYPB might form a collagen-modifying complex similar to those seen in vertebrates.
Figure 5. Effects of Ci-leprecan gain-of-function and loss-of-function on notochord development.
(A, B) Whole-mount in situ hybridizations performed on mid-tailbud embryos with antisense RNA probes specific for Ci-CRTAP (A) and Ci-CYPB (B). The notochord is indicated by red arrowheads. The inset in (B) shows a dorsal view. (C) Classification of the notochord phenotypes based on tail morphology. Different phenotypes are arranged in order of increasing severity from top to bottom. All panels show mid/late-tailbud embryos co-electroporated with the marker construct 434-bp Ci-Bra->LacZ and the Ci-Bra->Ci-leprecanDN∷RFP construct. (D) Quantification of the occurrence of notochord phenotypes in embryos electroporated with the 434-bp Ci-Bra->LacZ construct alone (wt) or in combination with Ci-Bra->Ci-leprecan (over-expression), shLEP-767 or shLEP-1426 (both shRNA), or Ci-Bra->Ci-leprecanDN∷RFP (dominant-negative). The different phenotypes are color-coded as shown by the key on the right side of each graph. (E) Graph plotting the variation in the relative fractions of the different phenotypes seen in response to increasing concentrations of the Ci-Bra->Ci-leprecanDN∷RFP transgene. For experimental consistency, the “0 μg” point represents embryos electroporated with 50 μg of the 434-bp Ci-Bra->LacZ construct.
These observations prompted us to create a truncated form of Ci-Leprecan lacking the last 129 amino acid residues of its C-terminus (Fig. S1), which is expected to be still capable of interacting with Ci-CRTAP and Ci-CYPB and to bind its collagen substrate but to not be able to modify it due to the truncation of its P4Hα catalytic domain (Myllyharju and Kivirikko, 1997). The truncated Ci-Leprecan was expressed in notochord cells by means of the Ci-Bra promoter region (Corbo et al., 1997). To provide an internal control for the electroporation efficiency and to unequivocally label the notochord cells expressing the dominant-negative Ci-Leprecan, we tagged it with RFP, and named the resulting plasmid Ci-Bra->Ci-leprecanDN∷RFP. In addition, to rule out the phenotypic contribution of the possible folding constraints imposed by the RFP tag, we tested in parallel an identical, untagged version named Ci-Bra->Ci-leprecanDN.
We also constructed the Ci-Bra->Ci-leprecan plasmid, aimed to over-express Ci-leprecan in the notochord, and two shLEP plasmids, targeting two different regions of the Ci-leprecan coding sequence, to induce short hairpin RNA (shRNA)-mediated Ci-leprecan knock-down (Nishiyama and Fujiwara, 2008). These plasmids were electroporated in parallel in Ciona zygotes along with the well-characterized, developmentally neutral 434-bp Ci-Bra->LacZ construct (Corbo et al., 1997), which was used individually as a control or in combination with the over-expression and knockdown constructs. After the staining reaction was complete, only the stained (i.e., reporter-expressing) embryos were scored under a dissecting microscope, and the different notochord phenotypes were grouped into 4 categories based on their increasing degree of severity, as shown in Fig. 5C. In addition, the relative incidence of each phenotypical category was quantified in the Ci-Bra->LacZ wt control embryos as well as in embryos transfected with either Ci-Bra->Ci-leprecan, shLEP-767, shLEP-1426, or Ci-Bra->Ci-leprecanDN∷RFP (Fig. 5D). The mildest phenotype, designated “bent” in Fig. 5C, is attributable to one or more misshapen notochord cells; the “corkscrew” phenotype indicates that the notochord is bent in more than two places, causing the tail to twirl around its AP axis; the “wavy” phenotype is observed in embryos where in addition to bends, the notochord contains various cells that end up aligned side-by-side instead of adopting the normal “stack of coins” configuration seen in mid-tailbud embryos at the end of intercalation. Finally, we also observed an extreme phenotype, which we tentatively indicated as “widened” (Fig. 5C, bottom panels), whereby the majority of the notochord cells were both misshapen and misaligned.
The quantitative analysis of these phenotypical classes revealed that all phenotypes occur in identical or comparable percentages in wt and in embryos transfected with the Ci-Bra->Ci-leprecan plasmid (first two bars in the graph in Fig. 5D). Noticeably, the “widened” phenotype is extremely rare or virtually absent in both wt and over-expression embryos in electroporations performed under optimal developmental conditions, e.g. unbiased by seasonal and/or batch-to-batch fluctuations in fertility of the animals employed. Conversely, in embryos transfected with either the shRNA plasmids or the dominant-negative plasmid, all phenotypes are detected with a considerably higher incidence (last three bars in the graph in Fig. 5D). In particular, the more intense phenotypes, “wavy” and “widened”, are quite frequently seen in embryos transfected with the Ci-Bra->Ci-leprecanDN∷RFP plasmid.
A close observation of embryos showing a notochord phenotype suggested a direct correlation between the levels of the transgenes incorporation and the severity of the phenotypes, as can be seen in Fig. 5C, where the embryos with a lower number of stained notochord cells usually display milder phenotypes. To better quantify the dose-dependency of the phenotypes observed, we employed different concentrations of the Ci-Bra->Ci-leprecanDN∷RFP plasmid on a single batch of embryos and quantified the results as described above (Fig. 5E). To normalize for phenotypes that might have been induced by the electroporation procedure, the “0 μg” dose-point was analyzed in embryos electroporated with the developmentally neutral 434-bp Ci-Bra->LacZ plasmid (Corbo et al., 1997). This analysis shows a direct proportionality between the dose of plasmid employed and the occurrence of the severe phenotypes.
As internal controls for the shRNA experiments we employed either an shRNA plasmid aimed to knock-down GFP expression in Ci-Bra->GFP transfected embryos or double-stranded RNA (dsRNA) electroporation (data not shown). The clearest results were obtained when we used as a control the previously published Ci-tyrosinase shRNA plasmid, shTYR (Nishiyama and Fujiwara, 2008; kindly provided by Dr. S. Fujiwara). Ci-tyrosinase is expressed in the precursors of the melanized sensory organs, otolith and ocellus, but not in notochord cells (Caracciolo et al., 1997; Sato et al., 1997); therefore, we did not expect any reproducible notochord phenotype to derive from the electroporation of shTYR. We also performed the triple electroporation of shTYR with shLEP-767 in the presence of the 434-bp Ci-Bra->LacZ marker, and we did detect a fraction of embryos showing both lack of pigmentation and notochord phenotype (Fig. S4); however, in this case, the overall development was suboptimal, possibly due to the larger amount of DNA transfected and the multiple shRNA species that were originated.
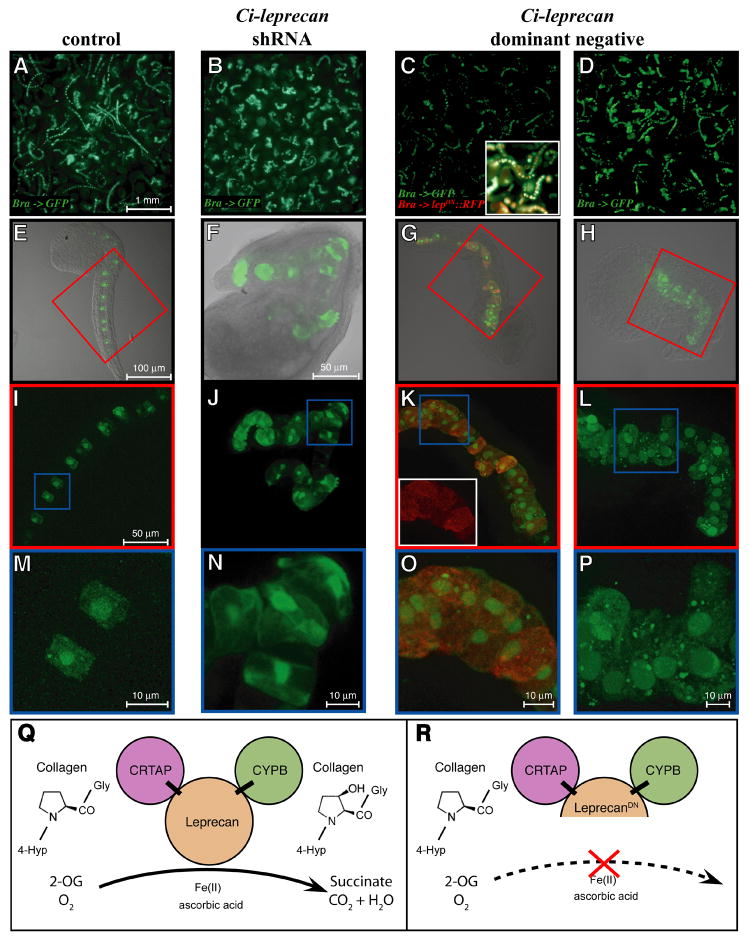
Confocal analysis of the Ci-Leprecan notochord phenotypes
We employed laser-scanning confocal microscopy to image notochord cells in mid/late tailbud control Ciona embryos transfected with the Ci-Bra->GFP plasmid (Fig. 6A,E,I,M), as well as in embryos containing, in addition to this marker, shLEP-767 (Fig. 6B,F,J,N), Ci-Bra->Ci-leprecanDN∷RFP (Fig. 6C,G,K,O), or Ci-Bra->Ci-leprecanDN (Fig. 6D,H,L,P), respectively.
Figure 6. Analysis of the notochord phenotypes induced by the over-expression, dominant-negative and knock-down of Ci-leprecan.
Comparative microscopic study of Ciona embryos electroporated with the Ci-Bra->GFP reporter alone (A,E,I,M) or in combination with either of the following constructs: shLEP-767 (B,F,J,N), Ci-Bra->Ci-leprecanDN∷RFP (C,G,K,O) and Ci-Bra->Ci-leprecanDN (D,H,L,P). (A-D) Embryos photographed at low magnification using an upright microscope. (E-P) Collapsed image stacks obtained with a confocal microscope. The inset in (C) shows a small area of the same image with the red and green channels merged. (E-H) Merge of the bright-field and dark-field images of representative individual embryos. (I,K,L) Dark-field high-magnification images of the areas boxed in red in panels E,G,H and (J) dark-field high-magnification view of the whole embryo shown in (F). The inset in (K) shows the red channel image of the area boxed in blue. (M-P) Dark-field high-magnification images of the areas boxed in blue in panels I-L. (Q,R) A simplified model for the mechanism of action of the native Ci-Leprecan protein (Q) and of the dominant-negative protein employed in this study (R). The dominant-negative Ci-Leprecan lacks the iron-binding region of the P4Hα domain, hence it is not expected to be able to modify collagen, but rather to interfere with the native Ci-Leprecan by sequestering its collagen substrate and its binding partners, CRTAP and CYPB. Abbreviations: 4-Hyp = 4-hydroxyproline; 2-OG = 2-oxoglutarate.
Control embryos transfected only with Ci-Bra->GFP predominantly showed long, well-extended tails (Fig. 6A,E), with columnar-shaped cells aligned in a single row (Fig. 6I, M). In the loss-of-function experiments, roughly half of the successfully transfected embryos displayed a notochord phenotype (Fig. 6B; see also Fig. 5D) consistent with the results obtained when the shRNA technique was employed to knockdown expression of other genes (Nishiyama and Fujiwara, 2008). Fig. 6F,J,N shows progressively higher magnifications of a “corkscrew” embryo, with recurring bends in the notochord (Fig. 6J) and irregularly shaped cells (Fig. 6N). The analysis of embryos electroporated with either Ci-Bra->Ci-leprecanDN∷RFP (Fig. 6C,G,K,O) or the untagged Ci-Bra->Ci-leprecanDN (Fig. 6D,H,L,P) provided two comparable examples of the “widened” phenotype (Fig. 6K,L) induced by the presence of various segments of the notochord containing two cells flanking each other (Fig. 6O,P). High-magnification confocal images of notochord cells transfected with Ci-Bra->Ci-leprecanDN∷RFP show that Ci-leprecanDN∷RFP is localized to the cytoplasm and is excluded from the nuclei (white inset in Fig. 6K). Similar results were obtained when we used a Ci-Leprecan∷GFP-KDEL fusion containing GFP fused in-frame to the full-length Ci-Leprecan, immediately upstream of its C-terminal KDEL ER-retention signal (data not shown). In sum, the analysis of the phenotypes presented in Figs. 5 and 6 suggests that the notochord defects displayed by the shRNA knock-down and the dominant-negative embryos might be dependent on the impairment caused by the loss of function of Leprecan on collagen hydroxylation (model in Fig. 6Q,R).
DISCUSSION
Since their identification, Leprecan genes and the prolyl 3-hydroxylases they encode have been the focus of increasing attention, due to their multi-faceted nature and function and to their involvement in recessive osteogenesis imperfecta (Wassenhove-McCarthy and McCarthy, 1999; Cabral et al., 2007). Previous work by our group has highlighted the evolutionary conservation of the notochord expression in the three Leprecan mouse orthologs (Capellini et al., 2008). However, both the transcriptional regulation and the function of Leprecan proteins in notochord formation were yet to be studied in any chordate. Here we have exploited the experimental advantages offered by the Ciona model system to address these points. We have shown that expression of the single-copy, notochord-specific Ci-leprecan is directly controlled by Ci-Bra through a compact CRM, which is also responsive to the muscle-specific T-box transcription factors Ci-Tbx6b and Ci-Tbx6c. We have described the phenotypes caused by over-expression and shRNA-mediated knock-down of Ci-leprecan, as well as the effects caused by the expression of a presumptive dominant-negative version of this protein on notochord formation.
The Ci-leprecan genomic locus harbors two distinct cis-regulatory modules
Ci-leprecan is expressed in notochord cells and their precursors starting from the neural plate stage (Hotta et al., 2000). An extensive analysis of ~14 kb from the Ci-leprecan genomic locus led us to the identification of two distinct regions with cis-regulatory activity, of which only one was able to direct gene expression in notochord cells and was therefore subjected to further analysis. This systematic survey of the Ci-leprecan genomic locus suggests that there is roughly 1 CRM/7 kb; these results provide a reasonable prediction for similar loci, and are in agreement with the findings obtained from a previous survey of two genomic domains covering the loci of 5 members of the Ciona Hox cluster, whereby 14 distinct CRMs were identified within a region spanning ~100 kb (Keys et al., 2005).
Interestingly, while expression of the Ci-leprecan close genomic neighbor, pontin, has only been reported in pharyngeal gills and stomach of juveniles but not in notochord (http://ghost.zool.kyoto-u.ac.jp/cgi-bin3/txtgetr2.cgi?CLSTR10840; Ogasawara et al., 2002; our data not shown), the distant neighbor Eph4 has been previously reported to be expressed in notochord cells at early, middle and late tailbud stages (citb036k05; Imai et al., 2004) suggesting the possibility that the Ci-leprecan notochord CRM might also participate, as a long-acting enhancer, in the regulation of Eph4 expression in notochord cells. Surprisingly, the 422-bp intergenic region located immediately 5’ of Ci-leprecan and Ci-pontin did not reveal any cis-regulatory activity in our assays, even when we subdivided it into shorter fragments to rule out the possible contribution from insulator sequences. However, it is possible that this region might be active only after metamorphosis, considering that so far pontin expression has only been detected in juveniles (Ogasawara et al., 2002).
Related T-box transcription factors employ the same cluster of T-box half-sites to direct gene expression in notochord and muscle
Ci-leprecan was originally identified in a subtractive screen between embryos that were both mis-expressing Ci-Bra in neural and endodermal cells and over-expressing it in notochord cells, and therefore it was indicated as a bona fide Ci-Bra transcriptional target (Takahashi et al., 1999). Ci-Bra has been previously shown to directly control a notochord-specific CRM associated with another of its target genes, Ci-trop, which is also expressed in notochord cells from the neural plate stage (Di Gregorio and Levine, 1999). The semi-quantitative analysis of serial truncations narrowed the Ci-leprecan notochord CRM to a 581-bp fragment; additional truncations and mutations unveiled its potential to activate transcription also in muscle cells.
Interestingly, activity in both notochord and muscle turned out to be dependent upon a small cluster of three T-box half-sites, located at the 3’-end of the 581-bp CRM. One of these sites matches the consensus sequence previously identified for two muscle activators, Ci-Tbx6b and Ci-Tbx6c (Yagi et al., 2005), while another half-site matches one of the Ci-Bra binding sites found in the minimal Ci-trop notochord CRM (Di Gregorio et al., 1999). Electrophoretic mobility shift assays showed that all T-box half-sites are bound in vitro by Ci-Bra, Ci-Tbx6b and Ci-Tbx6c. The study of the half-sites mutations in vivo indicates that notochord activity is lost when the T-box site matching the Tbx6b/c consensus is knocked out, suggesting that this site is not only bound by Ci-Bra in vitro but it is also occupied by this transcriptional activator in vivo. This hypothesis is reinforced by the previous observation that a site with an identical core, located 172 bp downstream of the 114-bp minimal Ci-trop notochord CRM, is specifically bound by Ci-Bra (Di Gregorio and Levine, 1999) and by the sequence comparisons with the mouse and frog Bra core consensus binding sites (Casey et al., 1998; Kispert et al., 1995; Tada and Smith, 2001). These considerations are of interest since Ci-Bra is only expressed in notochord and Ci-Tbx6b and Ci-Tbx6c are exclusively expressed in muscle cells (Corbo et al., 1997; Takatori et al., 2004). We conclude that the rigorously compartmentalized expression of related T-box factors in the Ciona embryo potentially allows for the same cis-regulatory sequences to activate gene expression simultaneously in different tissues. Furthermore, the action of tissue-specific repressors restricts and refines gene expression to distinct cell populations, thus creating an additional layer of transcriptional sophistication (model in Fig. 4).
A role for Ci-Leprecan in notochord development
In ascidians, as well as in vertebrates, the notochord is surrounded by a basement membrane composed of extracellular matrix proteins, such as Laminins (Scott and Stemple, 2005; Veeman et al., 2008) and by a notochordal sheath, which envelops the notochord and counteracts the increasing turgor caused by its expanding vacuoles (Miyamoto and Crowther, 1985; Stemple, 2005). Fibrillar collagen is a main component of the vertebrate notochordal sheath (e.g., Grotmol et al., 2006; Platz, 2006) and mutations in genes encoding collagen-modifying enzymes, such as the lysyl oxidases, have been shown to compromise the integrity of this structure causing notochord distortions (Anderson et al., 2007; Gansner et al., 2007). Similarly, our group has shown that two lysyl oxidases are expressed in the Ciona notochord and/or its precursors (Kugler et al., 2008). In addition, ultrastructural studies have described in the notochordal sheath of various ascidian species the presence of a conspicuous fibrillar component (Cloney, 1964, 1969), which presumably includes collagen fibrils, and through transmission electron microscopy we have observed fibrillar structures resembling collagen fibrils in the notochordal sheath of Ciona intestinalis late tailbuds and larvae (unpublished observations).
The co-expression of Ci-Leprecan and its putative interacting factors, Ci-CRTAP and Ci-CYPB, described here, suggests the possibility that these proteins might interact in notochord cells and participate to the maturation and secretion of collagen molecules destined for the notochordal sheath. To investigate this possibility, we expressed in the developing notochord a truncated version of Ci-Leprecan, as well as shRNAs against Ci-leprecan. The truncated Ci-Leprecan presumably is still able to bind its partners and substrate, but it is expected to be unable to modify its collagen substrate(s) since it lacks the C-terminal half of the prolyl 4-hydroxylase alpha (P4Hα) domain, which in turn contains the iron-binding residues required for enzymatic activity. Therefore, it is conceivable that the truncated Ci-Leprecan might act as a “dominant-negative” by sequestering the interacting partners and substrate from the full-length wt Ci-Leprecan, ultimately preventing collagen hydroxylation (model in Fig. 6). The notochord phenotypes produced by the Ci-Bra->Ci-leprecanDN∷RFP, and by the version lacking the RFP tag are comparable to those produced by the shLEP transgenes and are, as expected, dose-dependent; higher amounts of each construct cause a higher incidence of the more severe notochord-specific phenotypes. The phenotypes observed are temperature-independent and resemble those seen in preliminary morpholino oligonucleotide experiments (data not shown). The phenotypes presented in Figs. 5 and 6, together with the observation that in the majority of cases the notochord appears mostly as a single rod, suggest that the notochord deformities are not induced by a block in intercalation, but rather by the loss of structural integrity of the notochord basement membrane and of the nascent notochordal sheath. This hypothesis is reinforced by the results of time-course experiments, which show that intercalation is occurs normally in transgenic embryos (data not shown).
Human mutations affecting either CRTAP or LEPRE1 result in recessive osteogenesis imperfecta (Cabral et al., 2006; Baldridge et al., 2008). Interestingly, at least 5 of the human LEPRE1 mutations identified so far introduce premature stop codons, causing the synthesis of a shorter protein (Cabral et al., 2007; Baldridge et al., 2008). In particular, two of these mutations truncate the C-terminal region of LEPRE1, leaving 552 and 681 amino acid residues out of 708 total (Baldridge et al., 2008). Skin fibroblasts from an individual carrying the 1-681 LEPRE1 truncation were found to contain highly reduced levels of 3-hydroxylation of proline 986 in the α chains of collagen type I (Baldridge et al., 2008). The 1-552 truncation removes part of the P4Hα domain of the LEPRE1 protein, and one of its three iron-binding residues; similarly, the truncation that we created in the Ci-LeprecanDN protein removes roughly half of this domain and all three iron-binding residues, suggesting that Ci-LeprecanDN might be unable to modify fibrillar collagen.
Finally, the over-expression and temporal mis-expression of Ci-leprecan, caused by the Ci-Bra promoter region, did not induce considerable effects on notochord development, similar to the results observed in the case of other Ci-Bra target genes (Hotta et al., 2007). In the case of Ci-leprecan, the lack of an overt over-expression phenotype might be ascribed to the stoichiometric requirements for its interacting partners, CRTAP and CYPB. This is conceivable since although P3H1 has been shown to be capable of hydroxylating collagen in the absence of its partners in vitro (Vranka et al., 2004), CRTAP is needed for its function in vivo (Tiainen et al., 2008).
Leprecan proteins in chordate evolution
Although Leprecan genes have only recently been identified (Wassenhove-McCarthy and McCarthy, 1999; Järnum et al., 2004), it has been long known that vertebrate prolyl 3-hydroxylases utilize the peptide sequence Gly-Pro-4-Hydroxyproline found in collagen α-chains as their specific substrate (Risteli et al., 1977; Tryggvason et al., 1976, 1979). However, 3-hydroxyproline residues have been identified in non-collagenous proteins extracted from the fluke worm Fasciola hepatica (Wijffels et al., 1994) and a proline 3-hydroxylase able to modify free L-proline has been purified from Streptomyces (Mori et al., 1997). We have recently reported the phylogenetic relationships of the Leprecan family of P3H proteins extending from the sea anemone Nematostella throughout the metazoans (Capellini et al., 2008); however, the relationship between the Leprecan P3H proteins and these “non-collagen” prolyl 3-hydroxylases remains unclear.
In addition, the presence of a single Leprecan prolyl 3-hydroxylase in Ciona raises interesting questions, such as those concerning its substrate specificity. Recent studies on human Leprecan (P3H1) and Leprecan-like1 (P3H2) hydroxylases have shown that P3H1 is responsible for the hydroxylation of collagen I, while P3H2 preferentially hydroxylates collagen IV (Tiainen et al., 2008). Ciona collagen IV and collagen I are both expressed in notochord and numerous other tissues of tailbud embryos (http://ghost.zool.kyoto-u.ac.jp/cgi-bin3/txtgetr2.cgi?CLSTR00837; Satou et al., 2001; Wada et al., 2006; our unpublished results), hence no prediction can be made at this point with regard to the substrate specificity of Ci-Leprecan. One possibility is that the substrate specificity arose as genome duplications occurred during chordate evolution and novel leprecan genes appeared and started diverging from each other, as did fibrillar collagen genes. This might be the case in the mouse, where the three leprecan genes found in the genome are all at some point expressed in notochord cells but are also widely expressed in additional tissues and embryonic structures (Capellini et al., 2008).
Another interesting open question concerns the dichotomy between the ER-sequestered P3H1 and the secreted Leprecan. In humans, at least two transcripts derive from the LEPRE1 gene, one encoding a C-terminal KDEL ER-retention signal peptide, which is likely to be sequestered in the ER and form a complex with CYPB and CRTAP, the other, lacking the KDEL peptide, which is secreted and becomes a component of the basement membrane (Baldridge et al., 2008). We obtained only a single evident band in RT-PCR experiments performed using oligo-dT and Ci-leprecan specific 5’-end primers (data not shown), however this result does not completely rule out the possibility that alternatively spliced forms of Ci-leprecan might exist also in Ciona. This hypothesis seems supported by the presence of multiple ESTs of various lengths covering this region; future analyses will further explore this possibility.
In conclusion, this study has provided new insights on the molecular mechanisms employed by related T-box transcription factors to control gene activity during mesoderm formation in Ciona and a working hypothesis for the role of Leprecan proteins in notochord development.
Supplementary Material
Acknowledgments
We thank Dr. Yutaka Nibu, Dr. Dianella G. Howarth and the members of the Di Gregorio and Nibu labs for helpful discussion and critical comments on the manuscript. We are indebted to Stefan Gazdoiu, Jamie Kugler and Eamon Monaghan for their valuable technical help and suggestions and to Leona Cohen-Gould for her precious assistance with the confocal microscopy. We thank Dr. Shigeki Fujiwara for the shTYR and the U6 polymerase constructs and Dr. Nori Satoh for the Ciona intestinalis cDNA collection. This work was supported by NIH/NICHD grant R01HD050704 and by grant no. 1-FY08-430 from the March of Dimes Birth Defects Foundation to A.D.G.
A.D.G. is an Irma T. Hirschl Scholar.
Abbreviations
- AP
anterior-posterior
- bp
base pair(s)
- CNS
central nervous system
- ER
endoplasmic reticulum
- EST
expressed sequence tag
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- kb
kilobase(s), or 1000 base pairs
- PCR
polymerase chain reaction
- RFP
red fluorescent protein
- wt
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson C, Bartlett SJ, Gansner JM, Wilson D, He L, Gitlin JD, Kelsh RN, Dowden J. Chemical genetics suggests a critical role for lysyl oxidase in zebrafish notochord morphogenesis. Mol Biosyst. 2007;3:51–59. doi: 10.1039/b613673g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate-and iron-dependent dioxygenases. Genome Biol. 2001;2(3) doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. 0001-0007.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, Lambert D, Green A, Robinson H, Michelson M, Houge G, Lindman C, Martin J, Ward J, Lemyre E, Mitchell JJ, Krakow D, Rimoin DL, Cohn DH, Byers PH, Lee B. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008;29:1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39:359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini TD, Dunn MP, Passamaneck YJ, Selleri L, Di Gregorio A. Conservation of notochord gene expression across chordates: insights from the Leprecan gene family. Genesis. 2008;46:683–696. doi: 10.1002/dvg.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracciolo A, Gesualdo I, Branno M, Aniello F, Di Lauro R, Palumbo A. Specific cellular localization of tyrosinase mRNA during Ciona intestinalis larval development. Dev Growth Differ. 1997;39:437–444. doi: 10.1046/j.1440-169x.1997.t01-3-00004.x. [DOI] [PubMed] [Google Scholar]
- Casey ES, O’Reilly MA, Conlon FL, Smith JC. The T-box transcription factor Brachyury regulates expression of eFGF through binding to a non-palindromic response element. Development. 1998;125:3887–3894. doi: 10.1242/dev.125.19.3887. [DOI] [PubMed] [Google Scholar]
- Clifton IJ, Hsueh LC, Baldwin JE, Harlos K, Schofield CJ. Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases. Eur J Biochem. 2001;268:6625–6636. doi: 10.1046/j.0014-2956.2001.02617.x. [DOI] [PubMed] [Google Scholar]
- Cloney RA. Development of the ascidian notochord. Acta Embryol Morphol Exp. 1964;7:111–130. [Google Scholar]
- Cloney RA. Cytoplasmic filaments and morphogenesis: the role of the notochord in ascidian metamorphosis. Z Zellforsch Mikrosk Anat. 1969;100:31–53. doi: 10.1007/BF00343819. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Fujiwara S, Levine M, Di Gregorio A. Suppressor of Hairless activates Brachyury expression in the Ciona embryo. Dev Biol. 1998;203:358–368. doi: 10.1006/dbio.1998.9067. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian, Ciona intestinalis. Development. 1999;126:5599–5609. doi: 10.1242/dev.126.24.5599. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Corbo JC, Levine M. The regulation of forkhead/HNF-3beta expression in the Ciona embryo. Dev Biol. 2001;229:31–43. doi: 10.1006/dbio.2000.9964. [DOI] [PubMed] [Google Scholar]
- Erives A, Levine M. Characterization of a maternal T-Box gene in Ciona intestinalis. Dev Biol. 2000;225:169–178. doi: 10.1006/dbio.2000.9815. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Corbo JC, Levine M. The Snail repressor establishes a muscle/notochord boundary in the Ciona embryo. Development. 1998;125:2511–2520. doi: 10.1242/dev.125.13.2511. [DOI] [PubMed] [Google Scholar]
- Gansner JM, Mendelsohn BA, Hultman KA, Johnson SL, Gitlin JD. Essential role of lysyl oxidases in notochord development. Dev Biol. 2007;307:202–213. doi: 10.1016/j.ydbio.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotmol S, Kryvi H, Keynes R, Krossøy C, Nordvik K, Totland GK. Stepwise enforcement of the notochord and its intersection with the myoseptum: an evolutionary path leading to development of the vertebra? J Anat. 2006;209:339–357. doi: 10.1111/j.1469-7580.2006.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell JR, Robey PG, Barrach HJ, Wilczek J, Rennard SI, Martin GR. Isolation of a heparan sulfate-containing proteoglycan from basement membrane. Proc Natl Acad Sci USA. 1980;77:4494–4498. doi: 10.1073/pnas.77.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Asakura T, Saitoh B, Takatori N, Satou Y, Satoh N. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev Biol. 2000;224:69–80. doi: 10.1006/dbio.2000.9765. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Satoh N, Gojobori T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol Dev. 2008;10:37–51. doi: 10.1111/j.1525-142X.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Hotta K, Yamada S, Ueno N, Satoh N, Takahashi H. Brachyury-downstream notochord genes and convergent extension in Ciona intestinalis embryos. Dev Growth Differ. 2007;49:373–382. doi: 10.1111/j.1440-169X.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Jiang D, Smith WC. Ascidian notochord morphogenesis. Dev Dyn. 2007;236:1748–1757. doi: 10.1002/dvdy.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul SC, Sugihara T, Yoshida A, Nomura H, Wadhwa R. Gros1, a potential growth suppressor on chromosome 1: its identity to basement membrane-associated proteoglycan, leprecan. Oncogene. 2000;19:3576–3583. doi: 10.1038/sj.onc.1203696. [DOI] [PubMed] [Google Scholar]
- Keys DN, Lee BI, Di Gregorio A, Harafuji N, Detter JC, Wang M, Kahsai O, Ahn S, Zhang C, Doyle SA, et al. A saturation screen for cis-acting regulatory DNA in the Hox genes of Ciona intestinalis. Proc Natl Acad Sci USA. 2005;102:679–683. doi: 10.1073/pnas.0408952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JE, Passamaneck YJ, Feldman TG, Regnier TW, Di Gregorio A. Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis. 2008;46:697–710. doi: 10.1002/dvg.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T. Decoding cis-regulatory systems in ascidians. Zoolog Sci. 2005;22:129–146. doi: 10.2108/zsj.22.129. [DOI] [PubMed] [Google Scholar]
- Mannervik M, Nibu Y, Zhang H, Levine M. Transcriptional coregulators in development. Science. 1999;284:606–609. doi: 10.1126/science.284.5414.606. [DOI] [PubMed] [Google Scholar]
- Marini JC, Cabral WA, Barnes AM, Chang W. Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development. Cell Cycle. 2007;6:1675–1681. doi: 10.4161/cc.6.14.4474. [DOI] [PubMed] [Google Scholar]
- Medzihradszky KF. Characterization of site-specific N-glycosylation. Methods Mol Biol. 2008;446:293–316. doi: 10.1007/978-1-60327-084-7_21. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Chang P, Yasuo H. Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev Biol. 2007;302:333–344. doi: 10.1016/j.ydbio.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DM, Crowther RJ. Formation of the notochord in living ascidian embryos. J Embryol Exp Morphol. 1985;86:1–17. [PubMed] [Google Scholar]
- Mori H, Shibasaki T, Yano K, Ozaki A. Purification and cloning of a proline 3-hydroxylase, a novel enzyme which hydroxylates free L-proline to cis-3-hydroxy-L-proline. J Bacteriol. 1997;179:5677–5683. doi: 10.1128/jb.179.18.5677-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Characterization of the iron-and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Fujiwara S. RNA interference by expressing short hairpin RNA in the Ciona intestinalis embryo. Dev Growth Differ. 2008;50:521–529. doi: 10.1111/j.1440-169X.2008.01039.x. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I, Di Gregorio A. Lineage-independent mosaic expression and regulation of the Ciona multidom gene in the ancestral notochord. Dev Dyn. 2007;236:1806–1819. doi: 10.1002/dvdy.21213. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Sasaki A, Metoki H, Shin-i T, Kohara Y, Satoh N, Satou Y. Gene expression profiles in young adult Ciona intestinalis. Dev Genes Evol. 2002;212:173–85. doi: 10.1007/s00427-002-0230-7. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Di Gregorio A. Ciona intestinalis: chordate development made simple. Dev Dyn. 2005;233:1–19. doi: 10.1002/dvdy.20300. [DOI] [PubMed] [Google Scholar]
- Platz F. Structural and experimental investigations of the functional anatomy and the turgor of the notochord in the larval tail of anuran tadpoles. Ann Anat. 2006;188:289–302. doi: 10.1016/j.aanat.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Rentzsch A, Krummeck-Weiss G, Hofer A, Bartuschka A, Ostermann K, Rödel G. Mitochondrial copper metabolism in yeast: mutational analysis of Sco1p involved in the biogenesis of cytochrome c oxidase. Curr Genet. 1999;35:103–108. doi: 10.1007/s002940050438. [DOI] [PubMed] [Google Scholar]
- Rhee JM, Oda-Ishii I, Passamaneck YJ, Hadjantonakis AK, Di Gregorio A. Live imaging and morphometric analysis of embryonic development in the ascidian Ciona intestinalis. Genesis. 2005;43:136–147. doi: 10.1002/gene.20164. [DOI] [PubMed] [Google Scholar]
- Risteli J, Tryggvason K, Kivirikko KI. Prolyl 3-hydroxylase: partial characterization of the enzyme from rat kidney cortex. Eur J Biochem. 1977;73:485–492. doi: 10.1111/j.1432-1033.1977.tb11341.x. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Webster KA, Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- Sato S, Masuya H, Numakunai T, Satoh N, Ikeo K, Gojobori T, Tamura K, Ide H, Takeuchi T, Yamamoto H. Ascidian tyrosinase gene: its unique structure and expression in the developing brain. Dev Dyn. 1997;208:363–374. doi: 10.1002/(SICI)1097-0177(199703)208:3<363::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Satoh N, Satou Y, Davidson B, Levine M. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 2003;19:376–381. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Yamada L, Mochizuki Y, Hamaguchi M, Ishikawa H, Chiba S, Imai K, Kano S, Murakami SD, et al. Gene expression profiles in Ciona intestinalis tailbud embryos. Development. 2001;128:2893–2904. doi: 10.1242/dev.128.15.2893. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, et al. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Scott A, Stemple DL. Zebrafish notochordal basement membrane: signaling and structure. Curr Top Dev Biol. 2005;65:229–253. doi: 10.1016/S0070-2153(04)65009-5. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Zuker CS. Peptidyl-prolyl cis-trans isomerases, cyclophilin, FK506-binding protein, and ninaA: four of a kind. Curr Opin Cell Biol. 1990;2:1104–1107. doi: 10.1016/0955-0674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Tada M, Smith JC. T-targets: clues to understanding the functions of T-box proteins. Dev Growth Differ. 2001;43:1–11. doi: 10.1046/j.1440-169x.2001.00556.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller RW, Levine M, Satoh N. Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 1999;13:1519–1523. doi: 10.1101/gad.13.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori N, Hotta K, Mochizuki Y, Satoh G, Mitani Y, Satoh N, Satou Y, Takahashi H. T-box genes in the ascidian Ciona intestinalis: characterization of cDNAs and spatial expression. Dev Dyn. 2004;230:743–753. doi: 10.1002/dvdy.20082. [DOI] [PubMed] [Google Scholar]
- Tiainen P, Pasanen A, Sormunen R, Myllyharju J. Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem. 2008;283:19432–19439. doi: 10.1074/jbc.M802973200. [DOI] [PubMed] [Google Scholar]
- Tryggvason K, Majamaa K, Risteli J, Kivirikko KI. Partial purification and characterization of chick-embryo prolyl 3-hydroxylase. Biochem J. 1979;183:303–307. doi: 10.1042/bj1830303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryggvason K, Risteli J, Kivirikko KI. Separation of prolyl 3-hydroxylase and 4-hydroxylase activities and the 4-hydroxyproline requirement for synthesis of 3-hydroxyproline. Biochem Biophys Res Commun. 1976;76:275–281. doi: 10.1016/0006-291x(77)90722-7. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Nakatani Y, Hendrickson C, Ericson V, Lin C, Smith WC. chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development. 2008;135:33–41. doi: 10.1242/dev.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranka JA, Sakai LY, Bächinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279:23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- Wada H, Okuyama M, Satoh N, Zhang S. Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol Dev. 2006;8:370–377. doi: 10.1111/j.1525-142X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- Wassenhove-McCarthy DJ, McCarthy KJ. Molecular Characterization of a Novel Basement Membrane-associated Proteoglycan, Leprecan. J Biol Chem. 1999;274:25004–25017. doi: 10.1074/jbc.274.35.25004. [DOI] [PubMed] [Google Scholar]
- Wu TD, Brutlag DL. Discovering empirically conserved amino acid substitution groups in databases of protein families. Proc Int Conf Intell Syst Mol Biol. 1996;4:230–240. [PubMed] [Google Scholar]
- Yagi K, Takatori N, Satou Y, Satoh N. Ci-Tbx6b and Ci-Tbx6c are key mediators of the maternal effect gene Ci-macho1 in muscle cell differentiation in Ciona intestinalis embryos. Dev Biol. 2005;282:535–549. doi: 10.1016/j.ydbio.2005.03.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.