Abstract
T-cell interaction with a target cell is a key event in the adaptive immune response and primarily driven by T-cell receptor (TCR) recognition of peptide-MHC (pMHC) complexes. TCR avidity for a given pMHC is determined by number of MHC molecules, availability of coreceptors, and TCR affinity for MHC or peptide, respectively, with peptide recognition being the most important factor to confer target specificity. Here we present high-resolution crystal structures of 2 Fab antibodies in complex with the immunodominant NY-ESO-1157–165 peptide analogue (SLLMWITQV) presented by HLA-A*0201 and compare them with a TCR recognizing the same pMHC. Binding to the central methionine-tryptophan peptide motif and orientation of binding were almost identical for Fabs and TCR. As the MW “peg” dominates the contacts between Fab and peptide, we estimated the contributions of individual amino acids between the Fab and peptide to provide the rational basis for a peptide-focused second-generation, high-affinity antibody library. The final Fab candidate achieved better peptide binding by 2 light-chain mutations, giving a 20-fold affinity improvement to 2–4 nM, exceeding the affinity of the TCR by 1,000-fold. The high-affinity Fab when grafted as recombinant TCR on T cells conferred specific killing of HLA-A*0201/NY-ESO-1157–165 target cells. In summary, we prove that affinity maturation of antibodies mimicking a TCR is possible and provide a strategy for engineering high-affinity antibodies that can be used in targeting specific pMHC complexes for diagnostic and therapeutic purposes.
Keywords: antibody engineering, crystallography, tumor immunology
The complex formed by the major histocompatibility complex (MHC) class I molecule and a peptide primarily derived from a cytoplasmic protein (pMHC) represents a unique, cell surface-exposed marker (1). T-cell receptors (TCRs) as natural ligands of pMHC complexes can target the pMHC efficiently (2) but generally have low binding affinities, preventing their use as therapeutic reagents (3). Therefore, high-affinity recombinant TCRs would be ideal candidates for diagnostic and even therapeutic purposes (4). Indeed, affinity maturation strategies have been successfully developed and have shown dramatic increases of TCR affinity (5, 6). However, random mutagenesis was sometimes hampered by loss of peptide specificity, resulting in cross-reactivity with control pMHC complexes. Gain of affinity was the direct consequence of tighter binding to the MHC backbone (7), indicating that the knowledge of the precise TCR-pMHC interaction could be critical for the development of specific, high-affinity TCRs.
We previously determined the structure of the NY-ESO-1157–165/HLA-A*0201 complex recognized by a specific TCR (1G4) with moderate affinity of 3–5 μM (8). Though the 1G4 TCR binds in a classical diagonal orientation with respect to the peptide axis, it notably centers on the peptide's hydrophobic prominently exposed methionine (M) and tryptophan (W) side chains at residues 4 and 5 of the NY-ESO-1157–165 peptide. This MW motif is enveloped by the TCR's CDR loops and contributes ≈50% of the buried surface area contacted between TCR and peptide. Both M and W are encoded by single codons each, making the combination of these amino acids rare in the human proteome and an uncommon motif found in peptides presented by MHC molecules. To develop a reagent with diagnostic or even therapeutic potential, random mutagenesis of the 1G4 TCR was performed and resulted in an impressive 220,000-fold increase of binding affinity (9). However, tighter binding was again associated with loss of peptide specificity when tested in a cellular assay (4). Therefore, new approaches replacing the TCR as a binding tool have to be established. One option could be the use of antibodies, as they share structural homology with TCRs regarding target recognition and usually confer higher affinity with excellent specificity (10).
Indeed, phage display libraries have recently enabled the rapid isolation of human Fab fragments highly specific to pMHC molecules (11). The structure of one Fab fragment (Hyb3) bound to the HLA-A*1-MAGE-A1 peptide complex has been solved demonstrating considerable deviation form a normal TCR-like binding footprint (12), though detailed comparison of Hyb3 and HLA-A*1-MAGE-A1-specific TCR binding modes was precluded because of the lack of a representative TCR-pMHC structure. Hyb3 was generated by random affinity maturation, raising the question that the recognition focus on the MHC alpha-1 (α1) helix, rather than peptide, might be the result of this process. T cells grafted with Hyb3 as recombinant TCR had lost peptide specificity and killed HLA-A*1-positive cells irrespective of peptide presentation (13). Therefore, shifting the binding affinity from the MHC helices to the peptide and keeping peptide specificity is the critical issue, even in antibody-based approaches when aiming for high-affinity pMHC binders. Here we describe a 2-step procedure where we first compared the high-resolution structure of 2 HLA-A*0201/NY-ESO-1157–165-specific Fab antibodies (3M4E5 and 3M4F4 Fab) bound to the complex with the structure of the corresponding 1G4 TCR. In the second step, these structural data were used to generate a new antibody library on the basis of Fab 3M4E5. Key residues making contact to the MW motif were conserved and others were randomized where individual side chains could be optimized to interact with the peptide but not with the MHC backbone. To our knowledge, this is the first time that a rational strategy like this has been successfully followed for the affinity maturation of a T-cell receptor-like antibody.
Results
TCR-Like Binding Properties of Phage-Derived Fabs to HLA-A*0201-NY-ESO-1 Complexes.
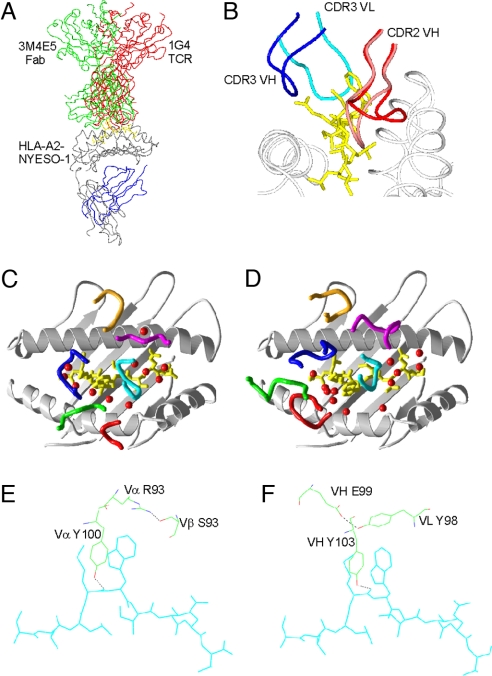
The distinctive, exposed MW motif of NY-ESO-1157–165 antigen (Fig. 1 A and B) may facilitate the generation of suitably peptide-focused Fabs. We had selected from a naïve antibody phage library (14) NY-ESO-1157–165/HLA-A*0201-specific Fabs (3M4E5 and 3M4F4) with 100-fold-higher affinities than the soluble 1G4 TCR, principally from much slower Koff rates (Table 1). The sequences of both Fabs were very similar with identical CDR3 VH and CDR3 VL loops and only a small number of relatively conservative amino acid differences in the CDR1 VH, CDR2 VH, and CDR2 VL loop regions. Crystal structures of the Fab-pMHC complexes and Fab 3M4E5 in isolation at 1.9 Å, 2.9 Å, and 2.3 Å resolution (supporting information (SI) Table S1) revealed 1G4 TCR-like binding modes. The VH and VL domains of the bound Fabs broadly superposed with the 1G4 TCR Vα and Vβ domains (Fig. 1 A, C, and D), respectively, with diagonal binding angles of 69° for the 1G4 TCR (8) and 40° for the Fab 3M4E5. Like the TCR, the Fabs focused the central hotspot of their binding footprint onto the MW peg with the CDR2 VH, CDR3 VH, and CDR3 VL loops playing similar roles to the equivalent TCR Vα/Vβ loops (Fig. 1 A and D and Table S2). In particular, TCR CDR3α residue Y100 and Fab VH CDR3 residue Y103 made hydrogen bonds to the peptide main chain carbonyl oxygen of M4 and their aromatic tyrosine rings interacted closely with the hydrophobic MW motif (Fig. S1). Additionally, in both TCR and Fabs, the hydrophobic portion of a large residue (TCR R93α and Fab Y98VL) arched over the MW motif and was stabilized through hydrogen bonds to the counterpart CDR3 loop (TCR S93β and Fab E99VH; Fig. 1 E and F).
Fig. 1.
Structural comparison of TCR and Fab. (A) Comparison of positions of 1G4 TCR (red) and 3M4E5 Fab in complex with HLA-A*0201/NY-ESO-1157–167. (B) Positions of the CDR3 loops (blue and magenta) bound to the peptide (cyan) and MHC (gray), and the 2 positions of the CDR2 VH loop from the 3M4F4 complex in the up (red) and down conformations. (C) Schematic of CDR loops of 1G4 TCR in complex with HLA-A*0201/NY-ESO-1157–167 with loops colored as follows: Vα CDR1, green; Vα CDR2, red; Vα CDR3, blue; Vβ CDR1, magenta; Vβ CDR2, orange; and Vβ CDR3, cyan. Water molecules within 4 Å of both TCR and peptide are indicated as red spheres. (D) Diagram of CDR loops of 3M4E5 Fab in complex with HLA- A*0201/NYESO-1157–167 with loops colored as follows: VH CDR1, green; VH CDR2, red; VH CDR3, blue; VL CDR1, magenta; VL CDR2, orange; and VL CDR3, cyan. Water molecules within 4 Å of both TCR and peptide are indicated as red spheres. (E) Structure of the 1G4 TCR residues forming the “roof” residues in the cavity that binds the peptide (cyan) MW side chains. (F) The analogous 3M4E5 Fab side chains that form the peptide MW binding cavity. Hydrogen bonds are shown in black dashed lines.
Table 1.
SPR affinity measurements for TCR 1G4 and Fabs 3M4E5, 3M4F4, and Fab T1
| Analyte | A2-NY-ESO-1 peptide | Ka, mol-1 s-1 | Kd, s-1 | KD, nM |
|---|---|---|---|---|
| 1G4 TCR | 9C | 87,500 | 0.357 | 4,080 |
| 9V | 53,425 | 0.078 | 1,460 | |
| 3M4E5 | 9C | 93,700 | 0.004225 | 47 |
| 9V | 80,850 | 0.00362 | 46 | |
| 3M4F4 | 9C | 68,800 | 0.00651 | 95 |
| 9V | 73,700 | 0.00467 | 63 | |
| Fab T1 | 9C | 138,375 | 0.000674 | 2 |
| 9V | 249,000 | 0.000835 | 4 |
NY-ESO-1157–165 peptides carried a cysteine (9C) or valine(9V) at position 9.
The Fabs and 1G4 TCR shared many contact residues with HLA-A*0201 (R65, K66, A69, Q72, T73, A150, and Q155), including hydrogen bonds to the same atoms (R65 NE, T73 OG, A150 O, and Q155 E1). Fab 3M4E5 had significantly lower protein-protein shape complementarity to the peptide (Sc = 0.85 with waters, 0.74 without waters) compared with that of the 1G4 TCR interface (Sc = 0.87 with waters, 0.81 without waters), and Fab 3M4E5 trapped more water molecules at the interface, notably 4 located above the hydrophobic MW motif (Fig. 1 C and D). Multiple copies of the Fab-pMHC complexes in the crystallographic asymmetric unit, when superposed, revealed some “rocking” in relative orientation of Fab and pMHC is permitted (Fig. S2A). We and others have observed similar evidence of flexion in TCR-pMHC complexes (15, 16). Comparison of the crystal structures of liganded and unliganded 3M4E5 Fab revealed only very small changes in the antigen-combining site (Fig. S3), with the largest change being where the Q32HC side chain flips 180° to make space for the MW motif. However, comparison of the 4 3M4F4 Fab-HLA-A*0201/NY-ESO-1157–165 complexes revealed 2 VH CDR2 conformations: an upward and a downward position, differing by a maximum of 6.7 Å in the main chain (Fig. 1B and Fig. S2 B and C).
Selecting Amino Acid Positions for Affinity Maturation.
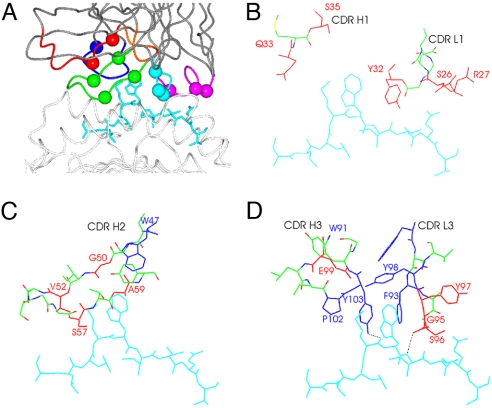
Because the NY-ESO-1157–165/HLA-A*0201-specific Fabs 3M4E5 and 3M4F4 were of moderate affinity (Table 1) compared with therapeutic antibodies used in the clinic, we aimed to affinity mature the Fab 3M4E5 based on our structural data. Previous studies had indicated that the hotspot of the interaction to a protein antigen remained conserved during affinity maturation (17). Gains in binding energy could be achieved by (i) stabilizing the bound conformation of any flexible CDR loops in the unliganded Fab and (ii) optimizing interactions in the periphery of the binding interface (i.e., around the hotspot). The first option looked a less promising route to affinity maturation because the structural data indicated a rather limited induced fit on binding of Fab 3M4E5 to the NY-ESO-1157–165/HLA-A*0201 complex. Therefore, we focused on the interacting amino acids around the hotspot MW motif, and residues directly interacting with the MW motif were held invariant. We randomized amino acids at positions where side chains could be optimized for direct peptide interaction (Fig. 2 A–D and Table S3) to enhance Fab affinity while retaining peptide specificity. Residues orientated away from the peptide or toward the MHC helices were not included, to avoid ineffective changes or unwanted increase of affinity to the MHC helices. For the light chain, the structural data suggested residues 26S, 27R, 32Y, 95G, 96S, and 97Y as candidates for variation.
Fig. 2.
Structural basis for a second-generation library. (A) Side view of 3M4E5 Fab in complex with HLA-A*0201/NY-ESO-1157–167, with residues randomized in the second-generation phage display library highlighted as spheres. Colors are as in Fig. 1D. (B–D) Illustrations of 3M4E5 Fab CDR residues that were randomized (red) in the vicinity of the peptide, which displayed nonoptimal contacts with the peptide. Residues making key contacts with the peptide MW side chains and lining the cavity in the CDR loops that interacts with the MW peg are colored in blue. The NYESO-1157–167 peptide is colored cyan, and hydrogen bonds are represented as dashed black lines.
Residue identification for the heavy chain was more complicated because a less clear-cut discrimination between residues interacting with peptide and with HLA-A*0201 could be established (Fig. 2 A–D and Table S3). In the Fab 3M4E5 complex structure, the large water-filled cavity within the MW binding pocket, occupied by 4 water molecules, was surrounded by residues Q33VH, S35VH, G50VH, and E99VH (Figs. 1D and 2 B–D). The higher B factors for these waters (average 37 Å2, n = 4), compared with other water molecules trapped at the interface (average 30.5 Å2, n = 8), suggested that these waters were not stably positioned within the interface, and thus side chain alterations in this pocket could stabilize the association of the Fab to the peptide. In addition, positions V52VH, S57VH, and A59VH were identified as candidate positions to enhance affinity by changing the conformational stability of the CDR2 loop inferred from the 3M4F4 Fab-HLA-A*0201/NY-ESO-1157–165 complex.
Identifying Higher-Affinity Fabs from the Second-Generation Library.
The final antibody library contained 108 independent clones, and after the third round of selection, 480 candidate clones were identified, of which 172 revealed, as soluble phage particles, specific binding to the HLA-A*0201/NY-ESO-1157–165 complex. Sequence data of strong binders were grouped by cluster analysis and 3 types (Fab T1–3) of repeatedly selected mutants could be identified by phylogenetic tree analysis (Fig. S4). The most dominant mutations present in all 3 types of mutants included S26E (LC CDR1) and S96G (LC CDR3), respectively. Fab T1 contained only these amino acid changes, whereas Fab T2 and T3 had additional heavy-chain mutations (Table S4).
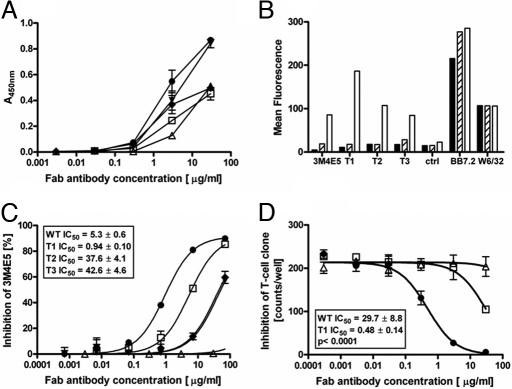
All 3 Fab candidates exhibited an equivalent or stronger binding signal by ELISA on NY-ESO-1157–165/HLA-A*0201 complexes at different concentrations when compared with Fab 3M4E5 (Fig. 3A). The strongest signal was detected for Fab T1. All 3 Fab candidates (T1–T3) conserved the precise binding specificity of Fab 3M4E5 and did not cross-react to other HLA-A*0201 complexes displaying an irrelevant peptide (data not shown). Moreover, Fabs T1–T3 reacted only with HLA-A*0201+ T2 cells presenting the NY-ESO-1157–165 peptide but not any partially overlapping NY-ESO-1 peptide sequences (Fig. 3B). Inhibition experiments on NY-ESO-1157–165 peptide pulsed T2 cells (Fig. 3C) using nonsaturating amounts of biotinylated 3M4E5 tetramer indicated differences in binding affinities for T1–T3. Fab T1 exhibited the highest binding affinity by achieving a significantly stronger inhibitory signal when compared with Fab 3M4E5 or Fab T2 and T3, respectively. The same pattern of inhibition was observed in a cellular assay with the strongest T-cell inhibition capacity, for Fab T1, significantly exceeding the parental Fab 3M4E5 (Fig. 3D). These functional data were in line with affinity and kinetic measurements showing a 20-fold affinity improvement for Fab T1, achieving 2- to 4-nM affinities as anticipated (Table 1).
Fig. 3.
Binding characteristics of 3M4E5-evolved Fab antibodies T1–3. (A) Comparison of binding characteristics as detected by ELISA on HLA-A*0201/NY-ESO-1157–165 complexes. Fab type 1 T1 (filled circles), type 2 T2 (diamonds), type 3 T3 (filled triangles), control Fab (open triangles) with weak binding characteristics, and Fab WT 3M4E5 (open squares) were titrated over the indicated concentration range. (B) Confirmation of Fab binding specificity was achieved by incubation with minigene transfected T2 cells. Reactivity of 3M4E5 and Fabs T1–T3 molecules with target HLA-A*0201/NY-ESO-1157–165 complex (white column) and control complexes (black HLA-A*0201/NY-ESO-1157–167 and gray HLA-A*0201/NY-ESO-1155–163) is depicted. HLA-A*0201 expression was confirmed for all cell lines by BB7.2 and w6/32 antibody staining, respectively. (C) Inhibition of 3M4E5-b binding by competition FACS analysis. Coincubation of PE-labeled biotinylated 3M4E5-b tetramers and different concentrations (50, 5, 0.5, 0.05, 0.005 0.0005, 0 μg/mL) of nonlabeled 3M4E5 (open squares), T1 (filled circles), T2 (filled triangles), T3 (filled triangles), or control Fab (open triangles) with peptide-pulsed T2 cells. (D) Inhibition of NY-ESO-1157–165-specific CTL responses by indicated Fab concentrations of T1 (filled circles), Fab 3M4E5 (open squares), or irrelevant Fab (open triangles). IFN-γ response of NY-ESO-1157–165-specific CTL to T2 cells pulsed with SLLMWITQC peptide (10−6 M) was detected by ELISPOT. All assays were done in triplicate.
Translating Affinity into Increased T-Cell Activity.
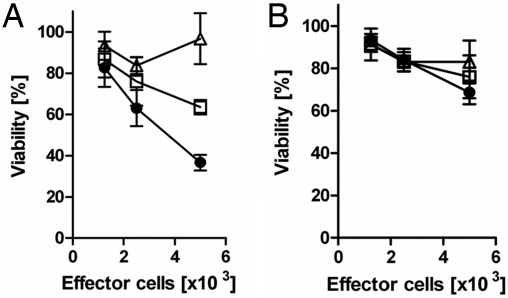
Recombinant single-chain TCRs were generated to demonstrate the biological relevance of Fab 3M4E5 and T1 in T-cell-based assays. For that purpose, CD3+ T cells were isolated from the peripheral blood of healthy donors and retrovirally transduced in vitro with single-chain (sc)-3M4E5 TCR, sc-T1 TCR, or sc-anti-CEA TCR as control (18). Transduction efficacy varied between 40% and 50%, and correct functional assembly of immune receptors was confirmed by HLA-A*0201/NY-ESO-1157–165 tetramer staining, exhibiting adequate expression levels for sc-3M4E5 TCR and sc-T1 TCR, respectively (Fig. S5). Sc-3M4E5 TCR and sc-T1 TCR grafted T cells specifically recognized and killed HLA-A*0201+ T2 cells (Fig. 4) presenting the NY-ESO-1157–165 peptide but not any HLA-A*0201+ T2 cells loaded with Flu peptide (14). Sc-T1 TCR revealed a significantly higher level of IFN-γ release (data not shown) and cytotoxicity (Fig. 4 A and B) when compared with the sc-3M4E5 TCR. In addition, both receptors did not lose peptide specificity and killed in a pMHC-restricted fashion.
Fig. 4.
Specific lysis of HLA-A*0201-positive T2 cells by recombinant immunoreceptors. CD3+ T cells were grafted by retroviral gene transfer with different recombinant immunoreceptors [T1 (filled circles), 3M4E5 (open squares), anti-CEA scTCR (open triangles)] and cocultivated (1.25–10 × 103/well) with HLA-A*0201-positive T2 cells (5 × 104/well) expressing either the NY-ESO-1157–165 (A) or IMP58–66 Flu peptide (B). Viability of target cells was recorded by a XTT-based viability assay as described in Materials and Methods. All assays were performed at least in triplicate with T1 always achieving a significantly (P < 0.05) higher cytotoxic activity.
Discussion
We describe the structures of 2 Fabs recognizing the HLA-A*0201-NY-ESO-1 complex to determine the structural basis of their specificity to the NY-ESO-1 peptide, and examine how the structures can assist in generating high-affinity Fab variants keeping their specificity.
Both Fab complex structures bind in a remarkably similar way compared with the 1G4 TCR. By centering on the peptide MW motif, the Fab is positioned similarly to the TCR and adopts a diagonal binding orientation that generates very similar MHC contact points on the helices. The structures contrast with the HLA-A*01-MAGE-A1-Hyb3 complex, which binds more to the MHC α1 helix and samples a smaller portion of the peptide (12). The constraints on MHC class I binding for Fabs derived from phage libraries seem therefore less stringent than for TCRs, although the 3M4E5 and 3M4F4 Fabs adopt similar docking positions as the HLA-A*0201-NY-ESO-1-specific 1G4 TCR. The diagonal orientation of all TCRs and the 3M4E5 and 3M4F4 Fabs appear to sample the peptide at an orientation that positions the receptors into a “saddle” in the MHC helices. At this diagonal orientation, much of the peptide can be sampled, particularly the central region, whereas the MHC helices stabilize the interaction through hydrogen bonds, salt bridges, and van der Waal's bonds (Table S3). The rocking motion on pMHC observed with these Fabs, and indeed for some TCRs (JM22 and SB27) (16, 19, 20), is generally in line with the axis of the saddle, highlighting the axis of flexibility on the pMHC surface that demarcates the 2 lower regions of the α1 and α2 helices (Fig. S2A). These data suggest that there are structural constraints that preferentially define a diagonal binding orientation found in most pMHC-receptor complexes, but does not exclude the possibility that CD8/CD4/CD3 coreceptors may define the conserved Vα/Vβ position and the general TCR binding position.
The NY-ESO-1 peptide in HLA-A*0201 is a relatively unique epitope in that it presents the M and W side chains for recognition by incoming receptors (8). These side chains form a much larger available binding surface area than conventional HLA-A*0201 peptides, where often one bulky residue forms a key central motif exploited for functional recognition. The large MW motif, therefore, assists in generating peptide-specific interactions and is an excellent focal feature against which to develop therapeutic Fabs with high specificity and affinity. Because the antibodies selected from the primary phage selection process resulted in good specificity with moderate affinity (14), the footprint of TCR and Fab binding to the MHC backbone and the NY-ESO-1157–165 peptide directed the design of the second-generation library as described previously. Our strategy of keeping key MW motif contact residues conserved while randomizing only individual side chains interacting with the peptide but not with the MHC backbone was proven experimentally to be correct. The final affinity-matured antibody achieved its 20-fold improved affinity by just 2 light-chain mutations. The double-mutation S26E and S96G showed a clear route forward for light-chain optimization because it allowed the residues of the VL CDR1 and CDR3 loops to optimize interactions with the I6, T7, and Q8 peptide residues in accordance with the principle of enhancing affinity through improved binding peripheral to the hotspot (17). Interestingly, the usage of residues in the second-generation Fab for this region of the interface appeared to have moved toward that of the TCR, which has greater interaction with this C-terminal region of the peptide than that of the original Fab 3M4E5. The most frequently identified heavy-chain sequence resulting in highest binding affinity was identical to the original Fab 3M4E5 in amino acid sequence. This might be a result of the underlying nature of the antibody library used for the initial selection of Fab 3M4E5. The library is based on the human germline VH3–23 fragment with synthetic CDR1 and CDR2 domains exceeding the natural diversity of germline VH and of Ig sequences found in mature B cells (21). Therefore, VH positions identified in our model to be of relevance for antigen binding had already been under high selection pressure in the selection process of Fab 3M4E5 and could not been further optimized.
Our data are in line with previous attempts on affinity maturation of TCRs, especially those focusing on the HLA-A*0201/NY-ESO-1157–165 complex (22). Random affinity maturation of multiple amino acid positions within the 1G4 TCR resulted in dramatic increase of affinity but loss of specificity (4, 9). The affinity increase was primarily due to tighter binding of the MHC backbone and not peptide focused. As a consequence, the same group generated 1G4 TCR variants with single or dual amino acid substitutions to selectively improve peptide binding (23). A relatively modest increase in TCR affinity to 280 nM was achieved while maintaining antigen-specific reactivity of CD8+ T cells. We confirm these data because affinity maturation by replacement of 2 amino acids in the light did not change peptide specificity when analyzed in T-cell assays. Both Fab 3M4E5 and T1 grafted as recombinant scTCR on T cells kept their pMHC specificity, and the higher affinity of Fab T1 translated into a significantly enhanced cytotoxic activity.
In conclusion, we show that pMHC-specific Fabs can mimic the binding mode of a TCR, and that these structural data can be used to generate by rational design antibody variants with single-digit nanomolar binding affinity, keeping the specificity unchanged. NY-ESO-1 in particular is an attractive target antigen because its expression pattern is restricted to germ cells (lacking MHC class I molecules) and a wide variety of hematological and solid organ tumors (24). However, it is expressed in the nucleus and cytoplasm, and therefore not accessible for most targeted approaches (25). In contrast, NY-ESO-1-derived peptides such as the NY-ESO-1157–165 peptide described here are unique cell surface markers when expressed in the appropriate MHC context (14). The rational, peptide-focused approach as described resulted in Fab fragments with low nanomolar affinities and will hopefully be a model for the generation of pMHC-specific diagnostic and therapeutic reagents.
Materials and Methods
Antibody Crystallization and Data Collection.
Nanoliter-scale, sitting drop (100 nL protein solution and 100 nL reservoir solution), vapor-diffusion crystallization trials were set up in 96-well plates using Cartesian Technologies pipetting robots (26). Single 3M4E5-A2-NYESO-1 and 3M4F4-A2-NYESO-1 complex crystals grew from stoichiometric mixtures (final concentration 10 mg/mL) of purified Fab and HLA-A*0201 proteins at 21 °C. The detailed analysis procedure is described in SI Text.
Structure Determination and Refinement.
All structures were determined by molecular replacement in PHASER (27) using the peptide-MHC class I molecule from the HLA-A*0201-NYESO-1–1G4 TCR complex (PDB ID code 2BNQ) and the Fab fragment from PDB entry 1RZF as search coordinates. Initial rigid-body refinement of individual domains (α1α2, α3, β2M, peptide, VΗ, VL, CH, and CL) followed by restrained TLS refinement was performed in REFMAC5 (28). Manual rebuilding was carried out in COOT (29), and water picking in the final stages of refinement was performed with ARPw/ARP (30). All regions of the Fabs and HLA-A*0201-NY-ESO-1 were included in the final models, except for residues 154–159 from the light chain (chain L) of the 3M4E5-HLA-A*0201-NYESO-1 complex where no electron density was detectable. Crystallographic statistics for the final models are given in Tables S2 and S3. Figures were generated in CCP4MG (31).
Phage Display Selection.
For the library construction, an antibody phagemid library was generated by gene synthesis (GENART), including random codon mutations (to NNB) at indicated positions and subcloned into the pCES vector. Selection of phage Fab particles was done as previously described (14), and detailed information is provided in SI Text.
Antibody Characterization.
The specificity of individual phage clones and soluble Fab antibodies was assessed by ELISA at RT with indirectly coated MHC-class I antigen peptide complexes (14). The binding affinity was measured by surface plasmon resonance as described (SI Text). Flow cytometry was performed to determine recognition of pMHC complexes on living cells (SI Text). Finally, the capacity of antibodies to block recognition of peptide-pulsed target cells by HLA-A2/NY-ESO-1157–165-specific T cells was analyzed using an ELISPOT assay (SI Text).
Generation and Functional Analysis of Recombinant T-Cell Receptors.
Candidate Fab antibodies were converted into scFv fragments and grafted onto human CD3 zeta and CD28 signaling domains to study functional activity as recombinant T-cell receptors (18). Receptor design and analysis of T-cell activity is described in detail (SI Text).
Supplementary Material
Acknowledgments.
We thank staff at the European Synchrotron Radiation Facility and the European Molecular Biology Laboratory outstation (Grenoble, France) for assistance with data collection. We thank Immanuel Luescher and Philippe Guillaume (Ludwig Institute for Cancer Research, Lausanne, Switzerland) for providing pMHC complexes. We thank Natko Nuber, Petra Schuberth, and Manuela Bienemann for construct generation and help with the T-cell assays. We acknowledge the Medical Research Council funded Oxford Protein Production Facility and the European Commission Integrated Program (SPINE2; LSHG-CT-2006–031220) for crystallization facilities. This work was supported in part by grants from the Wilhelm-Sander Stiftung, the Cancer Research Institute, Cancer Research U.K. (C399/A2291), and the Medical Research Council. This work was conducted as part of the Atlantic Philanthropies/Ludwig Institute for Cancer Research Clinical Discovery Program. E.Y.J. is a Cancer Research U.K. Principal Research Fellow. E.S. was supported by a Rhodes scholarship and by the National Institute of Allergy and Infectious Diseases, Division of Intramural Research, National Institutes of Health, Department of Health and Human Services.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes HLA-A2-NYESO-1–3M4E5, 3GJE; HLA-A2-NYESO-1–3M4F4, 3GJF; and 3M4E5 Fab, 3GJG).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901425106/DCSupplemental.
References
- 1.Mak TW. The T cell antigen receptor: “The Hunting of the Snark.”. Eur J Immunol. 2007;37(Suppl 1):S83–S93. doi: 10.1002/eji.200737443. [DOI] [PubMed] [Google Scholar]
- 2.Chames P, Hufton SE, Coulie PG, Uchanska-Ziegler B, Hoogenboom HR. Direct selection of a human antibody fragment directed against the tumor T-cell epitope HLA-A1-MAGE-A1 from a nonimmunized phage-Fab library. Proc Natl Acad Sci USA. 2000;97(14):7969–7974. doi: 10.1073/pnas.97.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konig R. Interactions between MHC molecules and co-receptors of the TCR. Curr Opin Immunol. 2002;14(1):75–83. doi: 10.1016/s0952-7915(01)00300-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol. 2007;179(9):5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4(1):55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 6.Weber KS, Donermeyer DL, Allen PM, Kranz DM. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc Natl Acad Sci USA. 2005;102(52):19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129(1):135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 8.Chen JL, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201(8):1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23(3):349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 10.Greenspan NS. Affinity, complementarity, cooperativity, and specificity in antibody recognition. Curr Top Microbiol Immunol. 2001;260:65–85. doi: 10.1007/978-3-662-05783-4_5. [DOI] [PubMed] [Google Scholar]
- 11.Denkberg G, Reiter Y. Recombinant antibodies with T-cell receptor-like specificity: Novel tools to study MHC class I presentation. Autoimmun Rev. 2006;5(4):252–257. doi: 10.1016/j.autrev.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Hulsmeyer M, et al. A major histocompatibility complex-peptide-restricted antibody and T cell receptor molecules recognize their target by distinct binding modes: Crystal structure of human leukocyte antigen (HLA)-A1-MAGE-A1 in complex with FAB-HYB3. J Biol Chem. 2005;280(4):2972–2980. doi: 10.1074/jbc.M411323200. [DOI] [PubMed] [Google Scholar]
- 13.Willemsen RA, Ronteltap C, Chames P, Debets R, Bolhuis RL. T cell retargeting with MHC class I-restricted antibodies: The CD28 costimulatory domain enhances antigen-specific cytotoxicity and cytokine production. J Immunol. 2005;174(12):7853–7858. doi: 10.4049/jimmunol.174.12.7853. [DOI] [PubMed] [Google Scholar]
- 14.Held G, et al. Dissecting cytotoxic T cell responses towards the NY-ESO-1 protein by peptide/MHC-specific antibody fragments. Eur J Immunol. 2004;34(10):2919–2929. doi: 10.1002/eji.200425297. [DOI] [PubMed] [Google Scholar]
- 15.Lee JK, et al. T cell cross-reactivity and conformational changes during TCR engagement. J Exp Med. 2004;200(11):1455–1466. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6(11):1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat Struct Biol. 2003;10(6):482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- 18.Hombach A, et al. Tumor-specific T cell activation by recombinant immunoreceptors: CD3 zeta signaling and CD28 costimulation are simultaneously required for efficient IL-2 secretion and can be integrated into one combined CD28/CD3 zeta signaling receptor molecule. J Immunol. 2001;167(11):6123–6131. doi: 10.4049/jimmunol.167.11.6123. [DOI] [PubMed] [Google Scholar]
- 19.Ishizuka J, et al. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28(2):171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 20.Tynan FE, et al. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. 2005;280(25):23900–23909. doi: 10.1074/jbc.M503060200. [DOI] [PubMed] [Google Scholar]
- 21.Hoet RM, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol. 2005;23(3):344–348. doi: 10.1038/nbt1067. [DOI] [PubMed] [Google Scholar]
- 22.Derre L, et al. Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157–165 by interacting with its central Met/Trp residues. Proc Natl Acad Sci USA. 2008;105(39):15010–15015. doi: 10.1073/pnas.0807954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins PF, et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol. 2008;180(9):6116–6131. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YT, Old LJ. Cancer-testis antigens: Targets for cancer immunotherapy. Cancer J Sci Am. 1999;5(1):16–17. [PubMed] [Google Scholar]
- 25.Chen YT, et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94(5):1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 6):651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 31.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.