Abstract
Cytochrome P450s form a large and diverse family of heme-containing proteins capable of carrying out many different enzymatic reactions. In both mammals and plants, some P450s are known to carry out reactions essential for processes such as hormone synthesis, while other P450s are involved in the detoxification of environmental compounds. In general, functions of insect P450s are less well understood. We characterized Drosophila melanogaster P450 expression patterns in embryos and 2 stages of third instar larvae. We identified numerous P450s expressed in the fat body, Malpighian (renal) tubules, and in distinct regions of the midgut, consistent with hypothesized roles in detoxification processes, and other P450s expressed in organs such as the gonads, corpora allata, oenocytes, hindgut, and brain. Combining expression pattern data with an RNA interference lethality screen of individual P450s, we identify candidate P450s essential for developmental processes and distinguish them from P450s with potential functions in detoxification.
Keywords: detoxification genes, in situ hybridization, insecticide resistance, multigene family, RNAi
Cytochrome P450s form a diverse and important gene superfamily present in virtually all organisms. Genome sequencing projects have identified a large number of P450 sequences in eukaryotes (1–5). Originally identified as monooxygenases, P450 enzymes are now known to catalyze an extremely diverse range of chemical reactions important both in developmental processes and in the detoxification of foreign compounds (6–8).
P450s that perform essential functions in mammals and plants have been identified. For example, in humans, CYP17A1 and CYP19A1 catalyze steps in the production of androgens and estrogens (9). CYP26B1 metabolizes retinoic acid and is involved in the fate of germ cells in the testes of mice (10). CYP4F2 is involved in postabsorptive elimination of gamma-tocopherol forms of vitamin E (11). Mutations in human P450s also lead to various diseases (12, 13). Plant P450s are essential for catalyzing steps in the synthesis of many compounds, including phenylpropanoids, lipids, phytohormones, and carotenoids (14, 15). Roles for P450s in the detoxification of xenobiotics, for example P450-mediated drug metabolism in mammals (16), are also well characterized.
Compared to plants and mammals, much less is known about functions of the different insect P450 enzymes. One exception is the involvement of P450s in the biosynthesis of the major insect hormone 20-hydroxyecdysone (20H) from plant sterols, where in Drosophila melanogaster at least six P450s are involved (17). Cyp302a1, Cyp306a1, Cyp315a1, and Cyp307a2 are all expressed in the prothoracic glands of late embryos and larvae and catalyze steps in the ecdysone biosynthetic pathway (18–21), while Cyp314a1 (shade), responsible for catalyzing the conversion from ecdysone to 20H, is expressed in the midgut, Malpighian tubules, and the fat body (22). Cyp307a1 (spook) has also been suggested to play a role in the synthesis of ecdysone, but it is not expressed in larvae (21). P450s involved in other important endogenous processes in D. melanogaster have also been identified, including Cyp303a1, required for the development and structure of sensory bristles (23), and Cyp4g1, an oenocyte-specific P450 required for correct triacylglycerol composition (24). P450s are also involved in behavioral phenotypes in D. melanogaster, with Cyp6a20 associated with aggressive behavior in males (25, 26) and Cyp4d21 necessary for efficient male mating (27). In the cockroach Diploptera punctata, Cyp15a1 is involved in the biosynthesis of juvenile hormone III (28).
It has been suggested that the large complement of P450s in insect genomes (most insects possess around 100 different P450s) is necessary to protect the insect from the diverse array of harmful compounds in its environment (29). Insect P450s capable of metabolizing allelochemicals have been identified (30, 31). For example, CYP6B1v1 and CYP6B3v1 from the black swallowtail butterfly, Papilio polyxenes, can metabolize the furanocoumarins it is exposed to in its diet (32, 33). Numerous individual P450s have also been implicated in insecticide resistance in different insect populations (34–38).
Approaches, such as sex-specific expression of P450s, induction capacity of P450s by various exogenous compounds, and transcriptional profiling have been useful in characterizing P450 functions (39–42). In an effort to distinguish P450s with essential endogenous functions from those involved in detoxification, we characterized expression patterns of D. melanogaster P450s by in situ hybridization and performed a P450 RNA interference (RNAi) knockdown screen.
Results
The sequenced, isogenic D. melanogaster strain y; cn bw sp (43) was used in this study, as the complement of P450s in this strain has been annotated (4). In y; cn bw sp, most of the 86 P450 genes were experimentally distinguishable from paralogs, except for the recently described duplication of Cyp12d1 (Cyp12d1-d and Cyp12d1-p), which only differ by 3 nucleotides in the coding region (www.flybase.org). We classified both Cyp12d1-d and Cyp12d1-p as Cyp12d1, bringing the number of P450s analyzed to 85. Using RT-PCR with P450 specific primers, 70 P450s were amplified from cDNA isolated from embryos and 81 P450s were amplified from cDNA isolated from third instar larvae, with 69 P450s amplified from both embryos and larvae. Cyp6t3, Cyp308a1, and Cyp313a2 were the only P450s that could not be amplified from either embryos or larvae. Cyp308a1 and Cyp313a2 could not be amplified from cDNA isolated from embryos, larvae, or adults, while Cyp6t3 could only be amplified from cDNA isolated from adults. Using DIG-labeled RNA probes for the 82 P450s amplified, in situ hybridization was performed on the different stages of embryogenesis, feeding third instar larval stage and wandering third instar larval stage. Expression patterns were obtained for 50 P450s during embryogenesis and 58 P450s during the third instar larval stages (Table S1).
Many P450s Are Expressed in the Larval Midgut, Malpighian Tubules, and Fat Body.
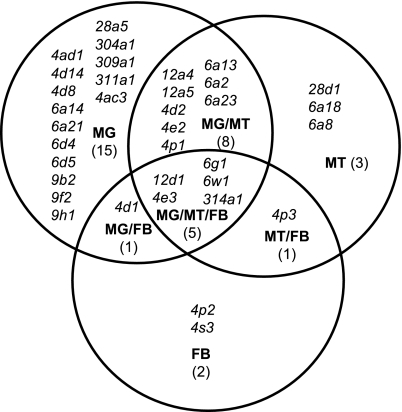
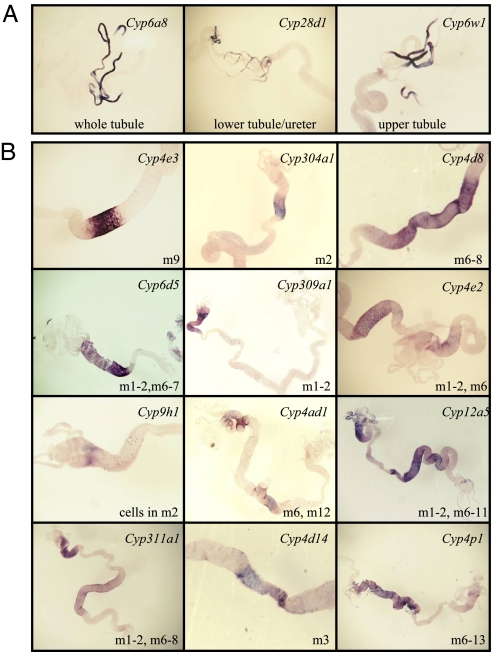
Hybridization signals for 35 P450s were detected in combinations of the midgut, Malpighian tubules, and fat body during third instar larval stages (Fig. 1; see Table S1). This is consistent with a presumed function of P450s in detoxification processes, as the metabolism of both exogenous and endogenous compounds is likely to occur in these tissues (44, 45). Seventeen P450s were detected in the larval Malpighian tubules (see Fig. 1 and Table S1), which are suggested to be the primary organs of excretion in insects (46). It has recently been demonstrated that the Malpighian tubules are important for the metabolism and detoxification of xenobiotics, and might also be involved in immunity (44, 47). There are at least 5 defined domains in the Malpighian tubules (48). Of the 17 P450s detected in the Malpighian tubules, 15 are expressed in all whole tubules, except Cyp28d1, which is expressed only in the lower tubule and ureter, and Cyp6w1 which is expressed only in the main segment and the transitional segment (Fig. 2A). Because of the initial segments of the tubules often being lost during dissection and in situ hybridization, expression in the initial segment was not characterized. In contrast to the relatively high number of P450s expressed in the larval tubules, only Cyp6a8 and Cyp6a18 are detected in Malpighian tubules of embryos (Fig. S1). Nine P450s are detected in the fat body, including the previously characterized Cyp6g1, Cyp12d1, and Cyp314a1 (22, 40, 49). Cyp4p2 and Cyp4s3 are the only P450s detected exclusively in the fat body. Cyp4d1 is alternatively spliced, giving rise to 2 different isoforms, Cyp4d1-PA and Cyp4d1-PB. The DIG-labeled RNA probe used in this study hybridizes to both isoforms and detected expression in the midgut and fat body. RT-PCR of individual tissues using exon-specific primers showed that Cyp4d1-PA is preferentially expressed in the midgut while Cyp4d1-PB is preferentially expressed in the fat body (Fig. S2).
Fig. 1.
Venn diagram representing unique and overlapping tissue expression of P450 genes expressed in the 3 key metabolic tissues of third instar larvae. FB, fat body; MG,midgut; MT, Malpighian tubules.
Fig. 2.
P450s expressed in the midgut and Malpighian tubules are detected in specific compartments by in situ hybridization. (A) Spatial patterns of P450s expressed in Malpighian tubules. (B) Spatial patterns of P450s in the midgut.
The Larval Midgut Contains Different Regions of P450 Expression.
The role of the D. melanogaster midgut in food digestion is well accepted (50) and contains at least 13 different distinct regions of gene expression, likely to be composed of multiple cell types (51). The anterior portion of the midgut (m1–4) is suggested to play some role in immunity (52, 53), although the functions of many midgut regions are not well established (54, 55). Using in situ hybridization, we detected expression of different P450s in a number of specific but different midgut regions in third instar larvae (Fig. 2B), with a predominance of P450s in the anterior (m1–m4) (55% of all midgut expressed P450s) and middle (m5–m10) midgut regions (49% of all midgut expressed P450s expressed).
While expression patterns for the majority of P450s is consistent between individual larvae, expression patterns for 2 P450s, Cyp6d4 and Cyp28a5, are not. In different larvae reared under the same conditions, expression in different compartments of the midgut for these 2 genes was consistently observed. When the upstream regulatory region of Cyp28a5 was cloned upstream of a GFP reporter [p-Stinger (56)] and transformed into flies (SI Methods), GFP expression was observed in different midgut regions in different larvae from an individual transgenic line, even when larvae of the same life stage grown under the same conditions were compared (Fig. S3). Specificity of P450 expression within in the embryonic midgut was also observed, with 15 P450s expressed in different regions of the midgut at different stages throughout embryogenesis (see Fig. S1).
Some P450s Have Specific Expression Patterns.
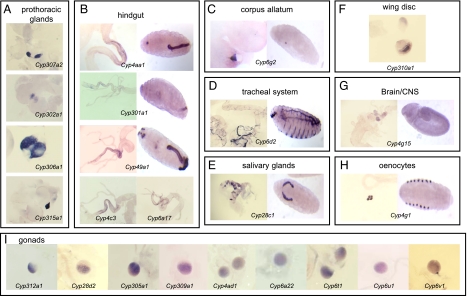
A subset of P450s was found to have specific tissue-expression patterns. Cyp302a1, Cyp306a1, Cyp307a2, and Cyp315a1 were detected in the prothoracic cells of the ring gland in embryos and larvae (Fig. 3A), as per previous studies (18, 19, 21, 57). Five P450s were detected in the hindgut (Cyp6a17, Cyp301a1, Cyp4c3, Cyp49a1, and Cyp4aa1) (Fig. 3B). Cyp301a1, Cyp49a1, Cyp4aa1 are also expressed in the embryonic hindgut (see Fig. 3B). Cyp6g2 is the only P450 detected in the corpora allata (Fig. 3C). Cyp6d2 (Fig. 3D), and Cyp4e3 are expressed in the tracheal system, although Cyp4e3 is also expressed in other tissues. Cyp28c1 is detected in the salivary glands (Fig. 3E). Cyp310a1 is expressed in the larval wing disc (Fig. 3F), Cyp4g15 expressed in the embryonic and larval brain and central nervous system (Fig. 3G), and Cyp4g1 is expressed in the embryonic and larval oenocytes (Fig. 3H), confirming previous studies (24, 58, 59), Nine P450s were detected in the larval gonads (Cyp305a1, Cyp28d2, Cyp309a2, Cyp312a1, Cyp4ad1, Cyp6a22, Cyp6t1, Cyp6u1, and Cyp6v1) (Fig. 3I). These P450s were not detected in the embryonic gonads (data not shown) (60). Refer to Fig. S4 for complete P450 in situ hybridization patterns in embryos and larvae.
Fig. 3.
(A–I) P450s with specific expression patterns in third instar larvae as determined by in situ hybridization.
Some P450 Show Differential Expression Between Feeding and Wandering Stages of Third Instar Larvae.
As a minor 20H peak thought to induce wandering behavior in third instar larvae had been shown to alter the expression of certain metabolic genes (61), in situ hybridization experiments were performed on both feeding and wandering larvae. A number of P450s were found to have expression changes between feeding and wandering larval stages. Four P450s expressed in the hindgut, Cyp301a1, Cyp49a1, Cyp4aa1, and Cyp4c3 were only detected in feeding larvae. Cyp6a2, implicated in insecticide resistance (35), is expressed in the midgut and Malpighian tubules in feeding larvae and not detected in wandering larvae. Similarly, Cyp4e3, expressed in specific regions of the midgut, and Cyp28a5, expressed in the midgut in a dynamic fashion (see Fig. S3), are expressed only in feeding but not wandering larvae. Other P450s were more readily detected in wandering larvae. These include the P450s involved in the synthesis of ecdysone, Cyp302a1, Cyp306a1, Cyp307a2, Cyp314a1, and Cyp315a1. Cyp6g1 is detected in the midgut and Malpighian tubules in both feeding and wandering third instar larvae, with additional expression detected in the fat body of wandering larvae (49). Expression for a subset of these P450s was confirmed by semiquantitative PCR (SI Methods; Fig. S5).
RNAi Knockdown Screen Indicates that Some P450s Are Essential for Viability.
A conditional knockdown screen of P450s using RNAi was performed to identify P450s essential for viability. Using available strains from the Vienna Drosophila RNAi Centre (62), we assayed for viability after RNAi knockdown of individual P450s in all tissues using the binary GAL4/UAS system and a tubulin-GAL4 strain (63). Most RNAi strains selected do not have off-target matches [defined as one or more 19-mer match (62)]. We were conservative in our approach, and only selected strains with off-targets that were other P450s. Out of the 59 different P450s targeted, 9 were found to be lethal by RNAi. As expected, RNAi of P450s in the ecdysone biosynthetic pathway (Cyp306a1 and Cyp314a1) resulted in lethality. RNAi of Cyp4g1 also resulted in lethality, giving a similar phenotype to Cyp4g1 knockout mutants (24). Six P450s (Cyp4c3, Cyp4d2, Cyp6g2, Cyp28c1, Cyp309a1, Cyp311a1) not as yet functionally characterized, were found to result in lethality by RNAi (Table S2). Phenotypic characterization of lethality revealed these P450s to be lethal at larval or pupal stages. Additional RNAi constructs targeting Cyp4g1 and Cyp4c3, constructed using the pWIZ vector (SI Methods), were also lethal. Fifty-one P450s are viable when targeted by RNAi. Of these, 3 P450s (Cyp318a1, Cyp4s3, Cyp4ac2) had lower numbers of progeny emerge than expected, suggesting a potential fitness deficit may exist when these individual P450s are targeted (see Table S2). As RNAi silencing can be variable, we cannot conclude that all of these P450s are involved in nonessential processes (62). Along with inefficient silencing, some P450s may be involved in survival on alternate food sources to the standard laboratory media used.
Discussion
Insect P450s are a large gene family, with many P450s evolving rapidly. Few P450 orthologs are identifiable in insect genomes of species 250 million years diverged (5, 29, 64–66). We undertook a systematic approach to studying this gene family in the model organism D. melanogaster, characterizing expression patterns in embryos and larvae, and investigating function with an RNAi screen. Expression patterns for 68 out of 85 P450s were obtained from either embryos, larvae, or both life stages. We found that most P450s are expressed in the larval midgut, Malpighian tubules, and fat body, suggesting potential roles in detoxification processes and protection from harmful exogenous compounds. Other P450s were identified to have specific expression patterns (e.g., corpora allata, brain, salivary glands, hindgut, and oenocytes). These expression patterns are conserved between life stage, and these P450s potentially have specific biological functions within these tissues.
The number of P450s successfully detected by in situ hybridization is less than the number of P450 that was amplified by PCR from each life stage. This is likely because of limitations in the in situ hybridization technique. Tissues, such as the cuticle, are waterproof and would not allow the entry of reagents. P450s expressed in late embryos also escape detection because of deposition of the cuticle (67). Additionally, P450s expressed at very low levels, which are detected by PCR, may not be detected by the conventional in situ hybridization protocol, but development of fluorescence in situ hybridization techniques might improve detection sensitivity (68).
Identification of a P450 Expressed in the Corpora Allata.
Cyp6g2 is the only P450 detected in the corpora allata of embryos and larvae, the site of juvenile hormone (JH) biosynthesis in insects (69). Given this expression pattern, Cyp6g2 might possibly be involved in the production of JH. There is precedent for the involvement of P450s in JH biosynthesis. Cyp15a1 from the cockroach D. punctata is expressed selectively in the corpora allata, and encodes a P450 enzyme that catalyses the conversion of methyl farnesoate to JH III (28). No clear orthologue to Cyp15a1 is identifiable in D. melanogaster (28). However, unlike in D. punctata, JH III bisepoxide (JHB3) is thought to be the major JH produced in D. melanogaster (70). JHB3 is synthesized by an alternate pathway possibly involving 2 epoxidation reactions catalyzed by a P450, converting farnesoic acid to JHB3 (71). It is possible that this P450 is CYP6G2.
Temporal Expression Differences of P450 Between Embryos and Larvae.
Out of the 68 P450s that we were able to detect by in situ hybridization, 9 are detected only in embryos, 15 are detected only in larvae, and the expression of another 32 P450s are not completely conserved between embryos and larvae (see Table S1). The P450s detected only in embryos are mostly expressed in structures such as the yolk cells, or are maternal transcripts, whereas the P450s detected in larvae only are mostly expressed in tissues involved in detoxification, such as the midgut, Malpighian tubules, and the fat body. Cyp6g1 and Cyp12a4, which confer insecticide resistance when over-expressed (36, 38, 49), are expressed in the midgut and Malpighian tubules in feeding third instar larvae, but show no detectable expression in embryos. RNAi of these 2 genes showed no mortality under laboratory conditions. In contrast, Cyp309a1 is expressed in the midgut in both embryos and larvae. RNAi of Cyp309a1 results in a lethal phenotype, suggesting that it might have an essential endogenous function. Genes with known developmental functions, such as the ecdysone biosynthesis genes, Cyp302a1, Cyp306a1, Cyp307a2, and Cyp315a1 are expressed in the prothoracic glands in both embryos and larvae. Cyp4g1 is also expressed in the oenocytes in both embryos and larvae. These genes are essential for the viability of the fly (24, 72). It is possible that P450s expressed in the same tissue throughout development are more likely to be involved in essential endogenous processes, while P450s expressed at only larval or adult stages are more likely to be involved in detoxification processes, although an overlap of these processes cannot be ruled out.
Comparison with Microarray Results.
Microarrays on dissected tissues detect a high number of P450s expressed in the larval midgut (54, 73), the larval Malpighian tubules, and the larval fat body (74). Comparing microarrays to our in situ hybridization results, we find a high number of P450s are detected using both methods. A microarray of mosquito (Anopheles gambiae) midgut regions show P450s to be highly enriched in the anterior midgut (75), consistent with our finding of a higher proportion of P450s in the anterior and middle midgut regions in D. melanogaster. A genome-wide study of embryonic gene expression patterns also characterized 30 P450s (67). Compared to the current study, expression pattern differences were found for 7 P450s. Any differences between datasets, microarray, and in situ hybridization, may be because of strain differences, detection methods, or rearing conditions.
Other microarray experiments performed by challenging D. melanogaster to a wide variety of conditions, such as exposure phenobarbital (40–42, 76), ethanol (77, 78), starvation (79), and looking at transcriptome changes in response to aging (80) identified a number of P450s that alter expression levels in response to variations in these parameters. We highlight 2 examples in which tissue-expression data can be used to extend microarray results. First, it has been shown that the transcription factor dGATAe is expressed in the Malpighian tubules and midgut, where it has roles in the development of the endoderm (81) and tissue-specific immune response (52). When dGATAe was misexpressed in the larval fat body, whole-genome microarray analysis revealed 291 genes to be up-regulated when compared to fat body of control larvae. This set of genes includes 21 P450s (52), 16 of which we have identified as being expressed in the larval midgut (Table S3). It is possible that dGATAe has a role in the regulation of these P450s in the larval midgut. Secondly, exposure of y; cn bw sp larvae to phenobarbital had shown that 21 P450s are induced by this regime (40). This list of 21 P450s includes 16 P450s, which are either expressed in the midgut or Malpighian tubules, suggesting that the induction response mainly occurs in these tissues (Table S4).
Evolutionary Dynamics of Drosophila P450s.
Many insect P450s are rapidly evolving, with orthologs to D. melanogaster P450s being difficult to identify outside of Drosophila (29, 64). Phylogenetic analysis of P450s in vertebrates has separated P450s into stable and unstable groups. Stable P450s have endogenous substrates, implying these P450 have essential functions. Most of the unstable P450s encode enzymes that function as xenobiotic detoxifiers (82). Evolutionary analysis of P450s found in 12 sequenced Drosophila genomes has also identified stable and unstable P450s across the species (83) (Lydia Gramzow, Robert Good and Charles Robin, unpublished data). For the 58 P450s with characterized expression patterns in third instar larvae, 80% of the P450s expressed in the midgut, Malpighian tubules, and fat body are classified as unstable, while only 27% of P450s expressed in other tissues are unstable, suggesting that the large complement of rapidly evolving P450s could be an evolutionary consequence of the wide variety of xenobiotics that D. melanogaster are exposed to. Our RNAi screen also shows that out of the 9 P450s with essential endogenous functions identified, 8 are categorized as evolutionarily stable (Table 1), suggesting that conserved P450s are likely to be have essential functions.
Table 1.
A higher Proportion of P450s lethal by RNAi are evolutionarily stable
| Stable P450s | Unstable P450s | Total | |
|---|---|---|---|
| RNAi lethal P450s | 8 (36.4%) | 1 (2.6%) | 9 |
| RNAi sub-viable P450s | 1 (4.5%) | 2 (5.3%) | 3 |
| RNAi viable P450s | 13 (59.1%) | 34 (92.1%) | 47 |
| Total | 22 | 38 | 59 |
Percentages refer to P450s in each class (stable or dynamic) for the 60 individual P450s assayed by RNAi.
In this study, the combination of tissue expression analysis and RNA interference allowed us to infer the general functions of P450s in D. melanogaster. Further evolutionary and functional studies of this rapidly evolving gene family may provide insights into various developmental and habitat defining processes in insects.
Materials and Methods
Drosophila Strains.
Strains were maintained on a glucose, semolina, and yeast medium at 25 °C with constant light. All strains were obtained from the Bloomington Drosophila Stock Center, Indiana, except strains used in RNAi experiments, which were obtained from the Vienna Drosophila RNAi Center. y; cn bw sp was used in all in situ hybridization experiments.
Synthesis of cDNA and in Situ Hybridization.
Synthesis of cDNA was performed using standard techniques as reported previously (38), using total RNA extracted using TRIzol reagent (Invitrogen) from either mixed stage embryos or a pool of ≈30 feeding and 30 wandering third instar larvae. PCR were performed using primers spanning the complete ORF of each P450.
In Situ Hybridization.
DIG-labeled RNA probes were synthesized as per standard protocol using the PCR products amplified from cDNA. In situ hybridization in embryos and larvae was performed as described previously (49, 84).
RNAi of P450s.
UAS-RNAi strains were individually crossed to tubulin-GAL4/TM3 (63), resulting in RNAi knockdown in a ubiquitous pattern. Reciprocal crosses were performed at 29 °C. The sex and phenotype of emerging adults was scored. Stubble bristles were used to indicate the presence of the TM3, Sb chromosome in progeny, and therefore the absence of the tubulin-GAL4 chromosome. CHI square analysis of sex and genotype of emerging progeny was performed. Strains where the RNAi knockdown resulted in lethality or reduced numbers of flies in both reciprocal crosses were characterized further by defining life-stage of lethality.
Supplementary Material
Acknowledgments.
The Australian Drosophila Biomedical Research Support Facility is acknowledged for providing Drosophila services. The Vienna Drosophila RNAi Center and Bloomington Drosophila Stock Center are acknowledged for providing Drosophila stocks. We thank Charles Robin, Robert Good, and Lydia Gramzow (University of Melbourne) for valuable discussion and sharing unpublished data, Alex Andrianopoulos (University of Melbourne) for valuable discussion, and Gary Hime (University of Melbourne) for help with microscopy. This work was supported by the Australian Research Council through its funding of the Centre of Environmental Stress and Adaptation Research and an Australian Research Fellowship (to P.J.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812141106/DCSupplemental.
References
- 1.Le Goff G, et al. Microarray analysis of cytochrome P450 mediated insecticide resistance in Drosophila. Insect Biochem Mol Biol. 2003;33:701–708. doi: 10.1016/s0965-1748(03)00064-x. [DOI] [PubMed] [Google Scholar]
- 2.Lewis DF. 57 varieties: the human cytochromes P450. Pharmacogenomics. 2004;5:305–318. doi: 10.1517/phgs.5.3.305.29827. [DOI] [PubMed] [Google Scholar]
- 3.Paquette SM, Bak S, Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;19:307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- 4.Tijet N, Helvig C, Feyereisen R. The cytochrome P450 gene superfamily in Drosophila melanogaster: annotation, intron-exon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/s0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- 5.Strode C, et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2008;38:113–123. doi: 10.1016/j.ibmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems–biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Mansuy D. The great diversity of reactions catalyzed by cytochromes P450. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:5–14. doi: 10.1016/s0742-8413(98)10026-9. [DOI] [PubMed] [Google Scholar]
- 9.Hakki T, Bernhardt R. CYP17- and CYP11B-dependent steroid hydroxylases as drug development targets. Pharmacol Ther. 2006;111:27–52. doi: 10.1016/j.pharmthera.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Bowles J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 11.Sontag TJ, Parker RS. Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. Novel mechanism of regulation of vitamin E status. J Biol Chem. 2002;277:25290–25296. doi: 10.1074/jbc.M201466200. [DOI] [PubMed] [Google Scholar]
- 12.Li A, et al. Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am J Hum Genet. 2004;74:817–826. doi: 10.1086/383228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K. Carotenoid hydroxylation–P450 finally! Trends Plants Sci. 2004;9:515–517. doi: 10.1016/j.tplants.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Annu Rev Plant Biol. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- 16.Guengerich FP. Cytochromes P450, drugs, and diseases. Mol Interv. 2003;3:194–204. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert LI. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol. 2004;215:1–10. doi: 10.1016/j.mce.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Chavez VM, et al. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development. 2000;127:4115–4126. doi: 10.1242/dev.127.19.4115. [DOI] [PubMed] [Google Scholar]
- 19.Warren JT, et al. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:11043–11048. doi: 10.1073/pnas.162375799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren JT, et al. phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem Mol Biol. 2004;34:991–1010. doi: 10.1016/j.ibmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Ono H, et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol. 2006;298:555–570. doi: 10.1016/j.ydbio.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Petryk A, et al. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci USA. 2003;100:13773–13778. doi: 10.1073/pnas.2336088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willingham AT, Keil T. A tissue specific cytochrome P450 required for the structure and function of Drosophila sensory organs. Mech Dev. 2004;121:1289–1297. doi: 10.1016/j.mod.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2006;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 25.Dierick HA, Greenspan RJ. Molecular analysis of flies selected for aggressive behavior. Nat Genet. 2006;38:1023–1031. doi: 10.1038/ng1864. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci USA. 2008;105:5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujii S, Toyama A, Amrein H. A male-specific fatty acid {omega}-hydroxylase, SXE1, is necessary for efficient male mating in Drosophila melanogaster. Genetics. 2008;180:179–190. doi: 10.1534/genetics.108.089177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helvig C, Koener JF, Unnithan GC, Feyereisen R. CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata. Proc Natl Acad Sci USA. 2004;101:4024–4029. doi: 10.1073/pnas.0306980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claudianos C, et al. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 31.Danielson PB, MacIntyre RJ, Fogleman JC. Molecular cloning of a family of xenobiotic-inducible drosophilid cytochrome p450s: evidence for involvement in host-plant allelochemical resistance. Proc Natl Acad Sci USA. 1997;94:10797–10802. doi: 10.1073/pnas.94.20.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JS, Berenbaum MR, Schuler MA. Amino acids in SRS1 and SRS6 are critical for furanocoumarin metabolism by CYP6B1v1, a cytochrome P450 monooxygenase. Insect Mol Biol. 2002;11:175–186. doi: 10.1046/j.1365-2583.2002.00323.x. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc Natl Acad Sci USA. 2004;101:2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzov VM, Unnithan GC, Chernogolov AA, Feyereisen R. CYP12A1, a mitochondrial cytochrome P450 from the house fly. Arch Biochem Biophys. 1998;359:231–240. doi: 10.1006/abbi.1998.0901. [DOI] [PubMed] [Google Scholar]
- 35.Bergé JB, Feyereisen R, Amichot M. Cytochrome P450 monooxygenases and insecticide resistance in insects. Philos Trans R Soc Lond B Biol Sci. 1998;353:1701–1705. doi: 10.1098/rstb.1998.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daborn PJ, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Chen S, Wu S, Yue L, Wu Y. Constitutive overexpression of multiple cytochrome P450 genes associated with pyrethroid resistance in Helicoverpa armigera. J Econ Entomol. 2006;99:1784–1789. doi: 10.1603/0022-0493-99.5.1784. [DOI] [PubMed] [Google Scholar]
- 38.Bogwitz MR, et al. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasai S, Tomita T. Male specific expression of a cytochrome P450 (Cyp312a1) in Drosophila melanogaster. Biochem Biophys Res Commun. 2003;300:894–900. doi: 10.1016/s0006-291x(02)02950-9. [DOI] [PubMed] [Google Scholar]
- 40.Willoughby L, et al. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem Mol Biol. 2006;36:934–942. doi: 10.1016/j.ibmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Le Goff G, et al. Xenobiotic response in Drosophila melanogaster: Sex dependence of P450 and GST gene induction. Insect Biochem Mol Biol. 2006;36:674–682. doi: 10.1016/j.ibmb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 42.King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 44.Dow JA, Davies SA. The Malpighian tubule: rapid insights from post-genomic biology. J Insect Physiol. 2006;52:365–378. doi: 10.1016/j.jinsphys.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Hoshizaki DK. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 2. Amsterdam: Elsevier; 2005. pp. 315–345. [Google Scholar]
- 46.Denholm B, Skaer H. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 2. Amsterdam: Elsevier; 2005. pp. 291–314. [Google Scholar]
- 47.Yang J, et al. A Drosophila systems approach to xenobiotic metabolism. Physiol Genomics. 2007;30:223–231. doi: 10.1152/physiolgenomics.00018.2007. [DOI] [PubMed] [Google Scholar]
- 48.Sozen MA, Armstrong JD, Yang M, Kaiser K, Dow JA. Functional domains are specified to single-cell resolution in a Drosophila epithelium. Proc Natl Acad Sci USA. 1997;94:5207–5212. doi: 10.1073/pnas.94.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terra WR, Ferreira C. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 4. Amsterdam: Elsevier; 2005. pp. 171–244. [Google Scholar]
- 51.Murakami R, Takashima S, Hamaguchi T. Developmental genetics of the Drosophila gut: specification of primordia, subdivision and overt-differentation. Cell Mol Biol. 1999;45:661–676. [PubMed] [Google Scholar]
- 52.Senger K, Harris K, Levine M. GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc Natl Acad Sci USA. 2006;103:15957–15962. doi: 10.1073/pnas.0607608103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tzou P, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 54.Li H-M, et al. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol Biol. 2008;17:325–339. doi: 10.1111/j.1365-2583.2008.00808.x. [DOI] [PubMed] [Google Scholar]
- 55.Nakagoshi H. Functional specification in the Drosophila endoderm. Dev Growth Differ. 2005;47:383–392. doi: 10.1111/j.1440-169X.2005.00811.x. [DOI] [PubMed] [Google Scholar]
- 56.Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–728. 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 57.Niwa R, et al. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279:35942–35949. doi: 10.1074/jbc.M404514200. [DOI] [PubMed] [Google Scholar]
- 58.Maibeche-Coisne M, Monti-Dedieu L, Aragon S, Dauphin-Villemant C. A new cytochrome P450 from Drosophila melanogaster, CYP4G15, expressed in the nervous system. Biochem Biophys Res Commun. 2000;273:1132–1137. doi: 10.1006/bbrc.2000.3058. [DOI] [PubMed] [Google Scholar]
- 59.Jacobsen TL, et al. Functional analysis of genes differentially expressed in the Drosophila wing disc: role of transcripts enriched in the wing region. Genetics. 2006;174:1973–1982. doi: 10.1534/genetics.106.056788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shigenobu S, Kitadate Y, Noda C, Kobayashi S. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:13728–13733. doi: 10.1073/pnas.0603767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riddiford LM. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Vol. 2. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 899–940. [Google Scholar]
- 62.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 63.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 64.Feyereisen R. Evolution of insect P450. Biochem Soc Trans. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- 65.Richards S, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 66.Ranson H, et al. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- 67.Tomancak P, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Wigglesworth VB. Insect Hormones. San Francisco: W.H. Freeman; 1970. [Google Scholar]
- 70.Richard DS, et al. Juvenile hormone bisepoxide biosynthesis in vitro by the ring gland of Drosophila melanogaster: a putative juvenile hormone in the higher Diptera. Proc Natl Acad Sci USA. 1989;86:1421–1425. doi: 10.1073/pnas.86.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moshitzky P, Applebaum SW. Pathway and regulation of JHIII-Bisepoxide biosynthesis in adult Drosophila melanogaster corpus allatum. Arch Insect Biochem Physiol. 1995;30:225–237. doi: 10.1002/arch.940300211. [DOI] [PubMed] [Google Scholar]
- 72.Huang X, Warren JT, Gilbert LI. New players in the regulation of ecdysone biosynthesis. J Genet Genomics. 2008;35:1–10. doi: 10.1016/S1673-8527(08)60001-6. [DOI] [PubMed] [Google Scholar]
- 73.Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 74.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 75.Neira Oviedo M, Van Ekeris L, Cornea-Mcleod MDP, Linser PJ. A microarray-based analysis of transcriptional compartmentalization in the alimentary canal of Anopheles gambiae (Diptera: Culicidae) larvae. Insect Mol Biol. 2008;17:61–72. doi: 10.1111/j.1365-2583.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 76.Sun W, et al. Genome-wide analysis of phenobarbital-inducible genes in Drosophila melanogaster. Insect Mol Biol. 2006;15:455–464. doi: 10.1111/j.1365-2583.2006.00662.x. [DOI] [PubMed] [Google Scholar]
- 77.Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morozova TV, Anholt RR, Mackay TF. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai CQ, Parnell LD, Lyman RF, Ordovas JM, Mackay TF. Candidate genes affecting Drosophila life span identified by integrating microarray gene expression analysis and QTL mapping. Mech Ageing Dev. 2007;128:237–249. doi: 10.1016/j.mad.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 81.Murakami R, Okumura T, Uchiyama H. GATA factors as key regulatory molecules in the development of Drosophila endoderm. Dev Growth Differ. 2005;47:581–589. doi: 10.1111/j.1440-169X.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- 82.Thomas JH. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 2007;3:e67. doi: 10.1371/journal.pgen.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 84.Sztal T, et al. Two independent duplications forming the Cyp307a genes in Drosophila. Insect Biochem Mol Biol. 2007;37:1044–1053. doi: 10.1016/j.ibmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.