Abstract
Since publication of the first Thermotogales genome, Thermotoga maritima strain MSB8, single- and multi-gene analyses have disagreed on the phylogenetic position of this order of Bacteria. Here we present the genome sequences of 4 additional members of the Thermotogales (Tt. petrophila, Tt. lettingae, Thermosipho melanesiensis, and Fervidobacterium nodosum) and a comprehensive comparative analysis including the original T. maritima genome. While ribosomal protein genes strongly place Thermotogales as a sister group to Aquificales, the majority of genes with sufficient phylogenetic signal show affinities to Archaea and Firmicutes, especially Clostridia. Indeed, on the basis of the majority of genes in their genomes (including genes that are also found in Aquificales), Thermotogales should be considered members of the Firmicutes. This result highlights the conflict between the taxonomic goal of assigning every species to a unique position in an inclusive Linnaean hierarchy and the evolutionary goal of understanding phylogenesis in the presence of pervasive horizontal gene transfer (HGT) within prokaryotes. Amino acid compositions of reconstructed ancestral sequences from 423 gene families suggest an origin of this gene pool even more thermophilic than extant members of this order, followed by adaptation to lower growth temperatures within the Thermotogales.
Keywords: classification, horizontal (lateral) gene transfer, thermoadaptation, taxonomy, phylogenomic
The 1999 publication of the genome sequence of Thermotoga maritima strain MSB8 brought horizontal (or lateral) gene transfer (HGT or LGT) to the attention of genome biologists (1) and at the same time marked the beginning of a long quest for this hyperthermophilic organism's true phylogenetic home or taxonomic position. That report suggested that up to 24% of Tt. maritima genes, many clustered in its chromosome, were acquired by HGT from archaea: almost as many (21%) showed Firmicute affinities. Although rRNA phylogenies most often placed Tt. maritima (and Aquifex aeolicus, another hyperthermophilic bacterium) at the base of the bacterial tree, there was little consistent support for this from protein-coding genes. Indeed, Nelson et al. (1) concluded that “the phylogenetic position of Aquifex and Thermotoga, and the nature of the deepest branching eubacterial species, should be considered ambiguous.” This situation has not changed much in the ensuing 10 years: many single- or multigene analyses put Tt. maritima (sometimes with A. aeolicus as its sister) deepest in the bacterial tree, but several convincing reports (again including several multigene studies) place Tt. maritima with bacterial taxa not generally thought of as deep (most often Firmicutes, frequently among Clostridia). These results have been used to support claims for (i) a hyperthermophilic ancestry for Bacteria (or indeed for all life), (ii) the retention by Thermotogales and Aquificales, as basal lineages, of ancestral genes kept otherwise only in Archaea, or (iii) their adaptation to high-temperature life by import of genes from hyperthermophilic archaea. Two recent developments are relevant here: first that mesophilic Thermotogales have been discovered (2), raising the possibility that hyperthermophily is not ancestral to the group, and second that a thorough analysis of A. aeolicus shows that, although many of its informational genes support sisterhood with Tt. maritima, substantial exchange of some of these genes has occurred with ε-proteobacteria (3).
Although genome data derived from Tt. maritima have found wide use, this sequence provides only a single data set to represent all Thermotogales, of which at least 25 named species have been isolated from diverse geothermal features worldwide. Consequently, there is a potentially large genomic resource available to more thoroughly examine the extent to which HGT from archaea or other groups has contributed to the evolution of these organisms. We present here an analysis of genomes from this pivotal lineage, expanded by the addition of 4 sequences completed at the Joint Genome Institute. These are from Tt. petrophila RKU-1, Tt. lettingae TMO, Thermosipho melanesiensis BI429, and Fervidobacterium nodosum Rt17-B1, species spanning much of the known Thermotogales phylogenetic spectrum and isolated from sites around the world. All are extreme thermophiles or hyperthermophiles and grow primarily on sugars. We use these new sequences to revisit the role of HGT from Archaea and Firmicutes in the origin of Thermotogales, consider the meaning of such chimerism for positioning the group, and examine properties of the ancestral Thermotogales proteome by investigating the amino acid composition of ancestral protein sequences.
Results
Genome Characteristics.
The Thermotogales considered here include 2 close relatives (Tt. maritima and Tt. petrophila) and 3 with genus-level divergence. All have genome sizes similar to Tt. maritima [1.86 Mbp; supporting information (SI) Table S1] (1) with Tt. lettingae having the largest genome in this group (2.14 Mbp). Of the 5 genomes, only those from Tt. maritima strain MSB8 and Tt. petrophila showed strong synteny over their entire lengths, with 3 inversions (Fig. S1). PSI-TBLASTN analysis revealed many putative or fragmentary insertion sequence (IS) elements in the 5 sequenced Thermotogales genomes and a tendency of certain genomes to accumulate specific families of IS elements over others (Table S1). Tt. lettingae has the fewest IS elements, with only 3 identifiable IS605 elements. F. nodosum has 49 IS elements, the largest number and the greatest diversity (4 represented IS families) of the 5 genomes. Twelve IS elements of F. nodosum were detectable only using a PSI-TBLASTN analysis, an approach that does not rely on ORF prediction and also recognizes pseudogenes. All 5 genomes have elements from the IS605 family, whereas IS6 family transposons are unique to F. nodosum.
No prophages were found in these genomes. The only evidence of a prophage remnant is 2 adjacent Ts. melanesiensis genes, Tmel_1472 and Tmel_1473, annotated as encoding an endonuclease-like protein and a RecT-like protein, respectively, and reminiscent of the prophage rac recET genes in Escherichia coli (4). The presence of clustered regularly interspaced palindromic repeat (CRISPR) elements (Table S2) nevertheless suggests that these organisms may be subject to phage infection in their environments. Several of these elements are unique to these genomes (Table S2), perhaps an indication of phage host range specificity.
Ribosomal RNA (rRNA) genes all occur in operons in the order 5S-23S-tRNA-tRNA-16S. Tt. maritima, Tt. petrophila, and Tt. lettingae all have 1 rRNA operon on their minus strands. Tt. lettingae has an intron in its 23S rRNA gene that is 99% identical to the one in Tt. subterranea (5). In F. nodosum 2 operons are on the minus strand and the 5S and 16S rRNA genes in these are identical. An IS element is present in the 23S rRNA gene of the distal operon that may disrupt its function, especially given that the sequences of the 23S rRNA genes additionally differ by 2 nucleotide transitions that map to stem regions. Ts. melanesiensis has 4 rRNA operons on its minus strand that are identical in sequence. Two adjacent operons have the typical 5S-23S-tRNA-tRNA-16S organization while the others lack the tRNA genes.
Intralineage HGT by Orthologous Replacement Between the 5 Genomes.
Analysis of 1,115 gene families shared by at least 4 of 5 Thermotogales genomes identified a strong tree-like signal supporting the relationship among these genomes previously indicated by a 16S rRNA gene tree (Fig. S2). This was not surprising, given that 2 of the 5 genomes are very closely related compared to the other 3. The recovered plurality topology grouped the genomes according to their GC content, which might raise the suspicion that the observed relationships could be an artifact due to skewed amino acid composition (6). A compositional homogeneity test showed that this is unlikely, because all but 73 gene families passed the test (data not shown). Additionally, the same tree topology was recovered with 2 independent reconstruction methods (gene presence/absence and rearrangement distance, see Methods, data not shown). Fifty-eight gene families strongly disagree with the plurality topology (not shown), supporting the alternative relationships shown as red lines in Fig. S2. The majority (40 gene families) are from “metabolism” and “poorly characterized” functional supercategories. Better taxonomic sampling will be required to identify cases of intralineage transfer within the Thermotogales, suggested in refs. 7–9. Additionally, our methodology detects only cases of HGT that resulted in orthologous replacement, while ignoring cases of transfer that resulted in gene gain.
Genome Contents and Unusual Physiologies.
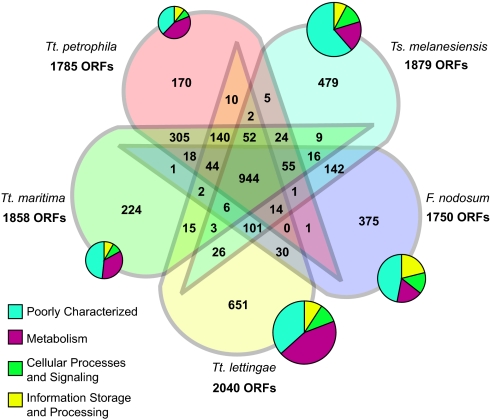
The 5 genomes share 944 orthologous gene families (the core), comprising 46–54% of protein-coding genes per genome (Fig. 1). The remaining genes are shared among only some of these 5 genomes, are unique for a specific genome, or form additional (paralogous or xenologous) copies of genes. The unique genes comprise 10–32% of these genomes, a disproportionately large fraction in functional categories termed metabolism and poorly characterized (Fig. 1). In many cases such patchily distributed genes might be held responsible for physiological differences between the species [as has been shown, for instance, for different ecotypes of Prochlorococcus spp. (ref. 10)].
Fig. 1.
Pan-genome of 5 members of Thermotogales. The Venn diagram shows the number of genes shared by different subsets of genomes, as determined by BLAST+BRANCHCLUST (see SI Methods for details). Pie charts show the distribution of genes unique for each genome across functional supercategories. The size of a pie chart is proportional to the number of genes.
For example, although the fermentative catabolism of the Thermotogales species appears uniform, their genome sequences reveal interesting differences in carbon and electron transfer pathways. All 5 catabolize glucose via the Embden–Meyerhof–Parnas pathway with likely contributions by the Entner–Doudoroff pathway as has been demonstrated for Tt. maritima (11). Pyruvate is converted to an intermediate of a partial tricarboxylic acid pathway, using either the malic enzyme (Tt. maritima, Tt. petrophila, Tt. lettingae, F. nodosum) or pyruvate carboxylase (Tt. lettingae, Ts. melanesiensis, F. nodosum). Tt. maritima and Tt. petrophila appear to catabolize malate in the oxidative direction to produce succinyl-CoA while the remaining species catabolize malate or oxalacetate in the reductive direction to 2-oxoglutarate. Tt. maritima and Tt. petrophila can also produce succinate from malate in the reductive direction. These differences suggest different needs to expel excess reducing power, particularly in Tt. maritima and Tt. petrophila when using the oxidative arm of the tricarboxylic acid pathway.
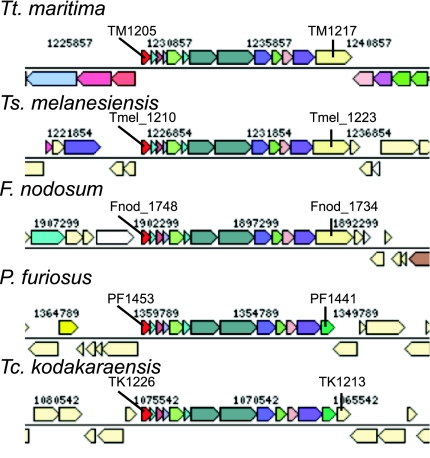
An operon annotated as a putative membrane-associated proton-translocating ferredoxin:NAD(P)H oxidoreductase is shared between archaea and the Thermotogales (Fig. 2). Horizontal acquisition of this operon from the Thermococcales has been previously noted (1, 12). An analysis of phylogenies of several of the encoded proteins indicates that these genes likely evolved as an operon (data not shown). Because the operon is found in genomes from across the clade, it appears to have been lost by Tt. petrophila and Tt. lettingae or their direct ancestors. The orthologous mbx operon in Pyrococcus furiosus encodes an oxidoreductase thought to couple the oxidation of ferredoxin (reduced by electrons from sugar catabolism) with the reduction of NADP+. NADPH then transfers electrons to a sulfur reductase when elemental sulfur is available (13, 14). A similar role is not indicated for the Thermotogales because all of these species, even those lacking the operon, can reduce elemental sulfur. This operon may encode a membrane-bound NAD:methyl viologen oxidoreductase like that observed in Tt. neapolitana (15). That enzyme did not use ferredoxin as a substrate, so the cofactor naturally paired with NAD under physiological conditions is unknown.
Fig. 2.
The ferredoxin:NAD(P)H oxidioreductase operon shared by Thermotogales and Thermococcales species. The homologous genes in P. furiosus compose the mbx operon. The promoter-proximal and -distal locus names are indicated.
Tt. lettingae is the only species of the 5 that contains a gene annotated as encoding a homolog of the large subunit of ribulose bisphosphate carboxylase (RuBisCO), Tlet_1684. In Bacillus subtilis, a homologous gene has been shown to encode 2,3 diketo-5-methylthiopentyl-1-phosphate enolase, an enzyme of a methionine salvage pathway (16). These RuBisCO-like proteins (RLPs) are related to true RuBisCO proteins, and an evolutionary scheme has been proposed that suggests that the RuBisCO large subunit and RLP arose in the archaea and were subsequently transferred to an ancestral bacterial lineage via HGT (17). Phylogenetic analysis shows that Tlet_1684 belongs to the group that Tabita et al. called “IV-Deep Ykr,” containing proteins from an eclectic mixture of organisms including alpha proteobacteria, Archaeoglobus, some clostridia, and a green alga (Fig. S3A and ref. 17). The group also contains sequences derived from the Global Ocean Sampling expedition (18). We conducted a phylogenetic analysis using only group IV-Deep Ykr sequences and others with similarity to Tlet_1684 (Fig. S3A). Poor resolution did not allow us to reliably place the Tt. lettingae sequence relative to other IV-Deep Ykr sequences. However, additional evidence for the inclusion of Tlet_1684 as a member of the group IV-Deep Ykr clade is provided by gene synteny around Tlet_1684. Four genes encoding enzymes that are likely part of a methionine salvage pathway are syntenic in the genomes of members of the IV-Deep Ykr clade, Tt. lettingae, Oceanicola granulosus, and Ochrobactrum anthropi (Fig. S3B). These encode the RLP, 2 separately encoded transketolase domains, and a methylthioribose-1-phosphate isomerase. The first 2 genes are also adjacent in the genomic fragment sequence from Beggiatoa sp. PS. A fifth gene, 5-methylthioribose kinase, is also syntenic in Tt. lettingae and O. anthropi (Fig. S3B). No other Thermotogales genome examined to date contains this RLP gene, suggesting that Tt. lettingae acquired this gene via HGT.
Multiple Gene Histories in Thermotogales.
Since the genomewide analysis of Tt. maritima in 1999 (1), GenBank has grown substantially, offering much better taxonomic sampling for such BLAST-based analyses. We performed similar BLAST-based analyses for the five Thermotogales genomes (which included the Tt. maritima genome analyzed in ref. 1), using the nonredundant (nr) database as a reference and recording highest-ranking matches (other than Thermotogales) with E-values <10−4. Only 7.7–11.0% of genes retain closest affiliation with Archaea (Table 1), in agreement with more sophisticated DarkHorse-based results (19). That this number is substantially lower than reported by Nelson et al. (1) likely reflects differential growth of the bacterial (vs. archaeal) database. Notably, the largest proportion of genes affiliate Thermotogales with Firmicutes (42.3–48.2%), especially Clostridiales. Furthermore, thermophilic members of Firmicutes were among the top-scoring BLAST hits and the proportion of thermophilic Firmicutes top-scoring hits was higher for the genomes of Thermotogales with higher optimal growth temperatures (Table 1). More striking was the much lower number of top-scoring BLAST hits to Aquificales, because this group appears as sister to Thermotogales in many analyses. This may reflect the fact that Firmicutes and Thermotogales are heterotrophs and Aquificales are autotrophs.
Table 1.
Taxonomic affiliations of top-scoring BLAST hits of Thermotogales ORFs found in the GenBank nonredundant (nr) database
| Tt. maritima | Tt. petrophila | Tt. lettingae | Ts. melanesiensis | F. nodosum | |
|---|---|---|---|---|---|
| Bacteria | 1,379 (74%) | 1,355 (76%) | 1,633 (80%) | 1,345 (71%) | 1,369 (78%) |
| Firmicutes | 821 (44%) [66] | 816 (46%) [65] | 985 (48%) [59] | 794 (42%) [60] | 844 (48%) [58] |
| Class Clostridia | 680 | 670 | 785 | 644 | 710 |
| Order Thermoanaerobacterales | 273 | 269 | 279 | 217 | 259 |
| Class Bacilli (Order Bacillales only) | 117 | 119 | 174 | 126 | 111 |
| Proteobacteria | 211 | 207 | 276 | 247 | 215 |
| Aquificae | 46 | 43 | 36 | 38 | 45 |
| Chloroflexi | 61 | 52 | 52 | 23 | 31 |
| Deinococcus-Thermus | 37 | 35 | 29 | 23 | 32 |
| Bacteroidetes | 30 | 32 | 40 | 38 | 29 |
| Cyanobacteria | 43 | 40 | 44 | 42 | 36 |
| Actinobacteria | 26 | 25 | 46 | 22 | 18 |
| Planctomycetes | 22 | 16 | 28 | 21 | 15 |
| Acidobacteria | 10 | 7 | 16 | 10 | 9 |
| Spirochaetes | 10 | 12 | 14 | 12 | 15 |
| Archaea | 204 (11%) | 197 (11%) | 187 (9%) | 168 (9%) | 135 (8%) |
| Euryarchaeota | 171 | 155 | 148 | 138 | 111 |
| Thermococcales | 95 | 80 | 66 | 51 | 58 |
| Archaeoglobales | 18 | 18 | 13 | 13 | 7 |
| Methanococcales | 18 | 17 | 13 | 20 | 12 |
| Methanosarcinales | 20 | 21 | 30 | 29 | 16 |
| Crenarchaeota | 27 | 35 | 31 | 26 | 22 |
| Thermoproteales | 12 | 15 | 13 | 9 | 12 |
| Desulfurococcales | 8 | 12 | 10 | 11 | 3 |
| Sulfolobales | 5 | 6 | 6 | 4 | 6 |
| Unclassified Archaea | 6 | 7 | 8 | 4 | 2 |
| Eukaryotes | 16 | 17 | 13 | 12 | 11 |
| Viruses | 1 | 1 | 0 | 5 | 2 |
| Others | 6 | 5 | 6 | 6 | 1 |
| Thermotogales specific* | 252 | 210 | 201 | 343 | 232 |
| ORFans† | 52 | 22 | 81 | 170 | 71 |
All percentage values are fractions of the total number of ORFs for that genome. Numbers refer to the number of ORFs in each taxonomic category (only selected major contributors are shown). Within each major taxonomic group only the groups with largest number of genes are shown. Numbers in brackets indicate the percentage of thermophilic Firmicutes among the reported number of top-scoring BLAST hits to Firmicutes.
*, Homologs found in Thermotogales genomes, but not in the nr database.
†, Homologs found neither in the other Thermotogales genomes nor in the nr database.
The number of top-scoring BLAST hits per taxonomic group can be affected by the number of genomes sequenced from it. At the time of our analyses (August 2008) Aquificales were represented only by 2 sequenced genomes (comprising 3,281 genes), while 231,386 genes in the nr database represented Clostridiales. To test if the low number of top-scoring BLAST hits between the Thermotogales and Aquificales is an artifact of underrepresentation of Aquificales genes in GenBank, we created a reduced nr database by removal of all Clostridiales sequences and reintroduction of 2 randomly chosen Clostridiales genomes (10 replicates were generated, see Table S3; note that Thermoanaerobacteriales is an order within the class Clostridiales). The assignment of taxonomic affiliations of top-scoring hits to ORFs in the Tt. maritima genome was repeated for each database replicate as described above. The number of top-scoring BLAST hits to the Aquificales did not increase above 60 (even in the case where the database did not contain any Clostridiales), but an increase was noted for the number of hits to other Firmicutes, Proteobacteria, and Archaea (Table S3). There were always more hits to clostridial sequences than to Aquificales, indeed 3–4 times as many when a Thermoanaerobacter was included as one of the clostridial contributors. Thus the low number of top-scoring BLAST hits to Aquificales is not a consequence of database sampling biases.
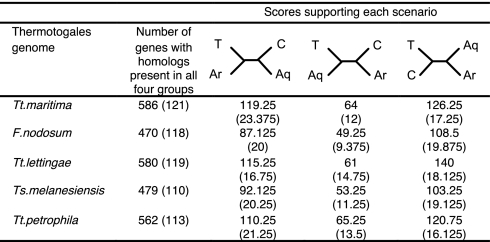
Because the top-scoring BLAST hit does not always correspond to the closest phylogenetic neighbor (20), we performed phylogenetic analyses as well. To avoid sampling bias because of using only 2 completely sequenced genomes from the Aquificales, we additionally selected only 2 genomes from both Archaea and Clostridiales and asked the following question: In individual gene trees, does the Thermotogales sequence group closer to that from the Archaea, Aquificales, or Clostridiales? We evaluated embedded quartets (up to 8 quartets if all 6 genomes had homologs to a query gene from the Thermotogales). While ≈50% of data sets did not produce sufficiently resolved signals, those that did agreed with top-scoring BLAST hit results: the majority of the genes prefer to group with those from the Clostridiales, while those from the Aquificales tend to group with the Thermotogales sequences in the least number of data sets (Fig. 3). Although these analyses do not preclude the possibility that Thermotogales and Aquificales might be considered independently deep branches in the Bacterial tree, they are not, for the clear majority of genes we could look at, sister taxa.
Fig. 3.
Examination of evolutionary relationships between genes in the Thermotogales genomes and their homologs in the Archaea, Aquificales, and Clostridia. For each scenario, all embedded quartets in a tree made from Thermotogales, 2 Archaea, 2 Aquificales, and 2 Clostridia homologs were evaluated. A score per scenario was calculated as the ratio of the number of embedded quartets significantly supporting the scenario to the total number of evaluated quartets (for more details see SI Methods). Numbers in parentheses refer to subsets of data from the “Information Storage and Processing” functional supercategory. A quartet topology is unrooted and does not allow distinguishing which 2 taxa are responsible for the observed relationship (i.e., the inferred topology could be because of gene flow either to/from Thermotogales or to/from Aquificales); analyses in Table 1 provide some evidence for directionality. Abbreviations: T, Thermotogales genome; C, Clostridiales; Aq, Aquificales; Ar, Archaea.
The Affiliation of the Thermotogales with Aquificales Based on Ribosomal Protein Data.
Ribosomal proteins are often used to derive the phylogenetic position of a group of organisms, because they are thought to be infrequently transferred (however, see refs. 21 and 22) and are highly conserved in sequence. Phylogenetic analysis of 29 concatenated bacterial ribosomal proteins provides a high level of support for the monophyly of the Thermotogales and 100% support for Aquificales as a sister group (Fig. S4A).
To determine if the phylogeny of individual ribosomal proteins supports this sister relationship, we compared the significantly supported bipartitions of individual ribosomal trees and the concatenated tree. Individual ribosomal protein trees often disagreed with the concatenated tree or did not resolve the relationships (Fig. S4B), most likely because of the relatively short length of most ribosomal proteins. Only 2 individual ribosomal proteins significantly support the branch grouping Thermotogales with Aquificales, but none show significant conflict (Fig. S4B). However, even if these 2 ribosomal proteins are deleted, the grouping of Thermotogales with Aquificales remains robustly supported, indicating a strong but distributed phylogenetic signal for this grouping in the ribosomal proteins.
It has been suggested that the deep position of Aquifex and Thermotoga results from long branch attraction (LBA) to Archaea due to saturation in rRNA caused by multiple substitutions (23), and the same might be the case for the protein data in their support of the sisterhood of these 2 taxa. We therefore used a nonhomogeneous model that is known to deal better with LBA (24) and still obtained strong support for the grouping of Aquificales and Thermotogales (Fig. S4A). We further investigated the possibility of the LBA artifact using the slow–fast method (25), separately analyzing subsets of the concatenated ribosomal protein alignment that contained faster evolving sites only. When only sites that vary by at least 50% in amino acid composition (i.e., representing more saturated sites, probably with multiple substitutions per site) were used, the recovered tree supports the grouping of Thermotogales with members of Firmicutes and Proteobacteria with 83% bootstrap support (Fig. S4C). This suggests that while the more reliable conserved sites unambiguously group Thermotogales with Aquificales, sequence saturation might artificially bring Thermotogales closer to Firmicutes. However, this does not explain the strong affinity for Clostridia for the majority of genes other than those encoding ribosomal proteins (Fig. 3). Separate analysis of the slow sites of the 100–150 genes supporting the sisterhood of Thermotogales and Clostridia retained that relationship and did not associate these genes with those of Aquificales. Thus if phylogenetic classification were based on the majority phylogenetic signal within the proteome, each of these members of the Thermotogales and the order as a whole should be considered members of class Clostridia within the Firmicutes—from both BLAST and phylogenetic analyses (Table 1 and Fig. 3).
The Thermophilic Ancestral Proteome of the Thermotogales.
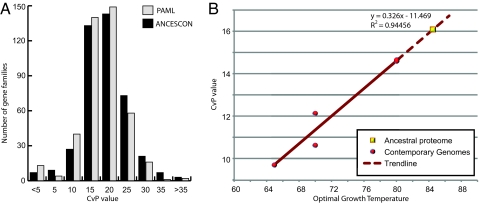
Recently, 2 compositional features of protein sequences have been suggested to be indicators of optimal growth temperature of an organism: overrepresentation of charged amino acid residues over polar ones (CvP bias) (26) and overrepresentation of IVYWREL amino acids (27). Application of both methods of analysis revealed linear correlations between optimal growth temperatures and compositional features of the proteins (Fig. S5A). Distribution of CvP values is unimodal for proteins within each genome (Fig. S5B), providing evidence against the hypothesis that thermophily was brought very recently to Thermotogales through HGT from Archaea (23). Because the above-described compositional features correlate so well with optimal growth temperatures, we used them to examine the nature of the ancestral proteome of these 5 Thermotogales species. We identified ancestral sequences of the most recent common node of the 5 Thermotogales in each gene family (see Methods) and inferred CvP values for all of the gene families for which we could reconstruct their ancestral sequence. The distribution, with peak CvP values of 15–20, suggests that the ancestral proteome of Thermotogales contained mostly thermophilic proteins (Fig. 4A). The thermophilic ancestral proteome inferred here was not necessarily the product of any single ancestral genome or perhaps even of a contemporary population of genomes, because HGT can affect coalescence times of individual gene histories (28). Furthermore, even if the gene families were from a single ancestral genome (as the plurality phylogenetic tree in Fig. S2 may suggest), the inferred ancestor does not necessarily represent the lineage at the root of all Thermotogales. Nevertheless, by establishing a correlation between optimal growth temperature and the median CvP values obtained with the extant Thermotogales species (Fig. 4B), we can extrapolate the ancestral proteome belonging to organisms with an optimal growth temperature of ≈84.5 °C, higher than that of any known modern member of the Thermotogales.
Fig. 4.
CvP values indicate the ancestor of the Thermotogales was an extreme thermophile. (A) Distribution of CvP values for predicted proteins of Thermotogales' ancestral proteome. The results of reconstruction with 2 different programs are shown (see Methods). (B) Extrapolation of optimal growth temperature for the ancestral Thermotogales proteome. Red points represent median CvP values and optimal growth temperature of 5 contemporary Thermotogales genomes. A strong linear correlation is observed between CvP values and optimal growth temperature. The median CvP value of the ancestral proteome is based on 423 inferred ancestral protein sequences.
The GC content of ribosomal rRNA also correlates with optimal growth temperature (29). Using a more extended 16S rRNA gene data set of both mesophilic and thermophilic members of Thermotogales, we reconstructed ancestral sequences and GC content at the ancestral nodes of the 16S rRNA gene phylogenetic tree (Fig. S5C). This analysis suggests that the ancestral 16S rRNA also belonged to a thermophile, favoring a scenario of secondary loss of thermophily within Thermotogales, a process carried farthest in the recently recognized “mesotoga” lineage (2). However, the position of the root of Bacteria is not certain and the phylogenetic position of Thermotogales within Bacteria cannot be easily pinpointed (see also discussion below). Thus the inferred thermophilic character of the group of organisms that gave rise to the 5 Thermotogales does not provide evidence for the commonly accepted thermophily of early Bacteria.
Discussion
Thermotogales genomes have complex and incongruent evolutionary histories, with compositions appearing to be more the product of HGT than of vertical descent, as these are traditionally defined. Particularly prominent “highways of gene sharing” (30) link Thermotogales to thermophilic Firmicutes and, to a lesser but significant extent, Archaea. Indeed, the majority of the genes in each of the 5 genomes examined here appear to be derived from these sources (Table 1 and Fig. 3). A high level of between-phylum HGT between Thermotogales and Firmicutes is in fact to be expected, since members of the Firmicutes frequently cohabit with Thermotogales in natural environments (31–33). Indeed, Thermotogales and the Firmicute genera Thermoanaerobacter and Desulfotomaculum are the only bacteria thought to be indigenous to high-temperature oil reservoirs (32, 33). This situation might thus be contrasted to that of the more physiologically restricted cyanobacteria, which tend to exchange more genes within their phylum (34).
A likely very much smaller portion (informational genes, here represented by ribosomal proteins) strongly supports a position of Thermotogales as sister group to Aquificales. A similar and reciprocal analysis of the genomes of Aquificales, while showing complex evolutionary histories for some informational genes (3), also supported this sisterhood.
What then is the true “phylogenetic position” of this group? That term is usually interpreted from a taxonomic perspective to refer to an unambiguous location within an inclusive Linnaean hierarchy. In the case of the order Thermotogales, which may so far be unique for a group of this rank, a relatively few genes commonly taken to be “phylogenetic markers” favor one answer, while a clear majority of the proteome gives another. Any decision to see Thermotogales' “true position” as sisters to Aquifex based only on the former genes thus cannot escape a certain arbitrary flavor.
In their discussion of the placement of Aquificales, Boussau et al. conclude cautiously that “if a tree of vertical descent can be reconstructed for Bacteria, our results suggest Aquificales should be placed close to Thermotogales” (3). We take these authors' “should” as normative. It is only because of a convention backed by an hypothesis—the complexity hypothesis (35)—that informational genes (and indeed only a flexibly defined subset of these) are privileged in determining a species' “true phylogenetic position”. Some convention like this may be necessary if our goal is an inclusively hierarchical (tree-like) classification of all living things, but as the Thermotogales make especially clear, such a classification should not be taken as a recapitulation of evolutionary history. In the real world of prokaryote evolution (36) phylogenetic positions are multidimensional relationships nuanced by a relative number of genes contributed by multiple lineages. As was first suggested by Nelson et al. in 1999 (1), Thermotogales (not just the species but as we conclude here, the whole “order”) pose an especially strong challenge to the Linnaean paradigm, as they can be made to conform to an inclusive hierarchy only by ignoring the majority of the evolutionary information their proteomes contain.
Methods
Genomes were sequenced by the Joint Genome Institute and annotated at Oak Ridge National Laboratory. The genomes were examined for synteny and were searched for various mobile and repetitive elements using established procedures as indicated in Results. To infer the phylogenetic position of each gene, they were subjected to BLAST-based and embedded quartet analyses (34). Ribosomal protein phylogeny was reconstructed using maximum-likelihood and Bayesian methods. Thermophilic signatures were assessed using CvP bias (26), IVYWREL bias (27) for proteins, and GC content for rRNA. Details of all analyses are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank the University of Connecticut Biotech Center for computational support. The genome sequencing and annotation work was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory (contract DE-AC02–05CH11231), Lawrence Livermore National Laboratory (DE-AC52–07NA27344), and Los Alamos National Laboratory (DE-AC02–06NA25396). Genomic DNA was prepared by J. L. DiPippo and I. U. Nieves. O.Z. was supported by a Canadian Institutes of Health Research postdoctoral fellowship and Canadian Institutes of Health Research Grant MOP-4467 (to W.F.D.), and C.L.N. was supported by the Norwegian Research Council (Grant 180444/V40). This work was also funded by the National Aeronautics and Space Administration Exobiology program Grants NNX08AQ10G and NNG05GN41G (jointly to K.M.N. and J.P.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank (accession nos. CP000702, CP000812, CP000716, and CP000771).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901260106/DCSupplemental.
References
- 1.Nelson KE, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 2.Nesbø CL, Dlutek M, Zhaxybayeva O, Doolittle WF. Evidence for existence of “mesotogas,” members of the order Thermotogales adapted to low-temperature environments. Appl Environ Microbiol. 2006;72:5061–5068. doi: 10.1128/AEM.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boussau B, Gueguen L, Gouy M. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evol Biol. 2008;8:272. doi: 10.1186/1471-2148-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark AJ, et al. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesbø CL, Doolittle WF. Active self-splicing group I introns in 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles. Proc Natl Acad Sci USA. 2003;100:10806–10811. doi: 10.1073/pnas.1434268100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer GAC, Hickey DA. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol Biol Evol. 2000;17:1581–1588. doi: 10.1093/oxfordjournals.molbev.a026257. [DOI] [PubMed] [Google Scholar]
- 7.Nesbø CL, Nelson KE, Doolittle WF. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J Bacteriol. 2002;184:4475–4488. doi: 10.1128/JB.184.16.4475-4488.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nesbø CL, Dlutek M, Doolittle WF. Recombination in Thermotoga: Implications for species concepts and biogeography. Genetics. 2006;172:759–769. doi: 10.1534/genetics.105.049312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mongodin EF, et al. Gene transfer and genome plasticity in Thermotoga maritima, a model hyperthermophilic species. J Bacteriol. 2005;187:4935–4944. doi: 10.1128/JB.187.14.4935-4944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman ML, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 11.Selig M, Xavier KB, Santos H, Schonheit P. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium. Thermotoga Arch Microbiol. 1997;167:217–232. doi: 10.1007/BF03356097. [DOI] [PubMed] [Google Scholar]
- 12.Calteau A, Gouy M, Perriere G. Horizontal transfer of two operons coding for hydrogenases between bacteria and archaea. J Mol Evol. 2005;60:557–565. doi: 10.1007/s00239-004-0094-8. [DOI] [PubMed] [Google Scholar]
- 13.Sapra R, Bagramyan K, Adams MW. A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapra R, Verhagen M, Adams MWW. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2000;182:3423–3428. doi: 10.1128/jb.182.12.3423-3428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Käslin S, Childers SE, Noll KM. Membrane-associated redox enzymes in Thermotoga neapolitana. Arch Microbiol. 1998;170:297–303. doi: 10.1007/s002030050645. [DOI] [PubMed] [Google Scholar]
- 16.Sekowska A, Danchin A. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2002;2:8. doi: 10.1186/1471-2180-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabita FR, et al. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podell S, Gaasterland T. DarkHorse: A method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 21.Brochier C, Philippe H, Moreira D. The evolutionary history of ribosomal protein RpS14: Horizontal gene transfer at the heart of the ribosome. Trends Genet. 2000;16:529–533. doi: 10.1016/s0168-9525(00)02142-9. [DOI] [PubMed] [Google Scholar]
- 22.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: Recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. RESEARCH 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brochier C, Philippe H. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature. 2002;417:244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- 24.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- 26.Suhre K, Claverie JM. Genomic correlates of hyperthermostability, an update. J Biol Chem. 2003;278:17198–17202. doi: 10.1074/jbc.M301327200. [DOI] [PubMed] [Google Scholar]
- 27.Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhaxybayeva O, Gogarten J Peter. Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 2004;20:182–187. doi: 10.1016/j.tig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Galtier N, Tourasse N, Gouy M. A nonhyperthermophilic common ancestor to extant life forms. Science. 1999;283:220–221. doi: 10.1126/science.283.5399.220. [DOI] [PubMed] [Google Scholar]
- 30.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonch-Osmolovskaya EA, et al. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl Environ Microbiol. 2003;69:6143–6151. doi: 10.1128/AEM.69.10.6143-6151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahle H, Garshol F, Madsen M, Birkeland NK. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie Leeuwenhoek. 2008;93:37–49. doi: 10.1007/s10482-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 33.Magot M. In: Petroleum Microbiology. Ollivier B, Magot M, editors. Washington, DC: ASM Press; 2005. pp. 21–33. [Google Scholar]
- 34.Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: Quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.