Abstract
The recognition by a viral envelope of its cognate host-cell receptor is the initial critical step in defining the viral host-range and tissue specificity. This study combines a single-round of selection of a random envelope library with a parallel cDNA screen for receptor function to identify a distinct retroviral envelope/receptor pair. The 11-aa targeting domain of the modified feline leukemia virus envelope consists of a constrained peptide. Critical to the binding of the constrained peptide envelope to its cellular receptor are a pair of internal cysteines and an essential Trp required for maintenance of titers >105 lacZ staining units per milliliter. The receptor used for viral entry is the human GPR172A protein, a G-protein-coupled receptor isolated from osteosarcoma cells. The ability to generate unique envelopes capable of using tissue- or disease-specific receptors marks an advance in the development of efficient gene-therapy vectors.
Keywords: library screening, retroviral entry, viral envelope, viral receptor
The binding of the retroviral envelope protein (Env) to its cognate host receptor is the critical first step in infection. The expression profile of a given receptor therefore regulates the viral host-range and tissue specificity. Viral receptors that are broadly and abundantly expressed may be a desirable trait in nature and for general gene delivery. This distribution, however, represents a significant obstacle in the development of retroviral-based gene-therapy vectors, where a targeted vector, which utilizes a receptor that is specific to the target tissue or pathologic state (cancerous vs. healthy tissue), is desired for safety concerns. Recent gene-therapy protocols have successfully circumvented this problem through ex vivo infection and reimplantation of the treated cells (1). This method, however, is applicable only to a small fraction of the disorders that could potentially benefit from gene delivery. A major research goal is to be able to develop alternative Env proteins, which provide the necessary specificity while still maintaining highly efficient gene delivery compatible with the viral entry pathway.
In gamma-retroviruses, including murine leukemia virus (MuLV) and feline leukemia virus (FeLV), receptor binding is mediated through a variable region (VRA) within the N-terminal half of the surface (SU) Env protein. In FeLV, replacing the VRA region with that of another switches receptor usage accordingly (2). For targeted entry, studies have looked into inserting binding domains (3, 4) and single-chain antibody regions (5) into Env backbones to alter receptor usage to specific proteins. Two common roadblocks to these methods are that (i) these Env may fail to be incorporated into virions (4, 5), and (ii) adding bulky constituents like antibodies can interfere with postbinding events (3, 6). Rather than direct viral entry, the approach used in our studies randomizes a 10- to 11-aa region within the VRA of FeLV Env (7–9). This creates an Env library of high complexity that can be screened for isolates capable of productive infection. In this method, the unique binding region is presented in the context of the Env backbone and in its natural conformation during the initial screening process. Using this method, Env proteins have been identified which use unique host-cell receptors outside the known viral interference groups (10).
Key to the development of new retroviral Envs is an understanding of the potential host receptors, as not all surface proteins can provide the necessary structural and functional requirements to permit the latter steps in viral entry. Each FeLV subgroup (11–14), like all other known gamma-retroviruses (15–21), utilizes multipass transmembrane receptors for entry. Furthermore, in some cases, multiple gamma-retroviruses use the same receptor. MuLV amphotrophic Env, Gibbon-ape leukemia virus, and FeLV-B for instance, all use pit-1 as their host receptor (14, 15, 19). This suggests that while Envs can be targeted to bind to virtually any protein on the cell surface, not all provide the proper features to mediate fusion and subsequent viral entry.
This study demonstrates the development and screening of a unique Env library generated through the randomization of 11 aa within the VRA of a FeLV-A and -C chimera. The 11-aa receptor targeting constrained peptide (CP) encodes unique structural features required for the presentation of an essential Trp residue. The receptor for this Env isolate was identified as the G-protein-coupled receptor, GPR172A, which serves as the human gamma-hydroxybutyrate (GHB) receptor (22), as well as the human receptor for porcine endogenous retrovirus A (PERV-A) (16). Furthermore, GPR172A has been shown to be commonly up-regulated in human malignancies (23). Thus, a single-round of selection and infection has yielded high titer virus (1.4 × 105 LSU/ml on the human osteosarcoma cell line 143B), which has independently selected, as a cell-surface receptor, a protein that has coevolved as the receptor for an endogenous virus from a distant species. The implications of this finding with respect to receptor usage and gene therapy will be discussed.
Results
Developing an 11-aa Random Env Library Within a FeLV A/C Backbone.
Retroviral entry involves a complex series of interactions between the viral Env protein and the host-cell receptor. Receptor binding is the initial step of entry, triggering a series of conformational changes that ultimately yield membrane fusion. For multiple retroviral Env proteins, various domains of SU have been implicated in receptor binding (24–28). Within the FeLV-A backbone, however, the primary epitope lies within the VRA region alone (2). Through randomizing a limited primary sequence within this VRA region, a library of Env proteins have been generated and screened for productive infection into target cells. Through this process, it has been established that retroviral entry can be targeted to novel host-cell receptors (7, 8, 10). To understand the scope of the potential receptor usage and the correlation with the evolution of known endogenous and exogenous retroviruses, a new FeLV library was generated randomizing 11 amino acids within the VRA region.
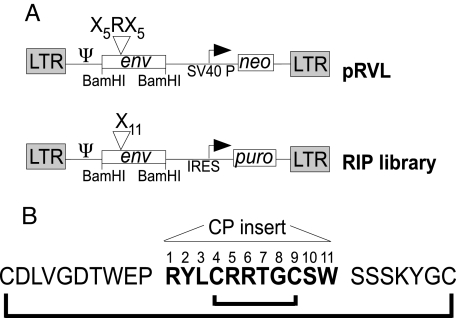
Fig. 1 outlines the modified vector system. Two highlights of the modified vector (RIP) are that (i) the drug selection cassette has been modified to encode an IRES-puromycin cassette rather than an internal SV40 promoter expressing neo, and (ii) the bias for an arginine was removed from the library at position 6 (7). A constitutive library expressing the random Env cassettes was generated through a 2-step process. The Env library was transiently expressed in 293TCeB cells (expressing Gag/Pol) in the presence of vesicular stomatitis virus G glycoprotein (VSV-G) and the viral particles released were used to infect TECeBF2 cells (29) at a low multiplicity of infection to ensure no more than 1 viral particle per infected cell. The complexity of the library was 2 × 105. Viral supernatants from the constitutive library were then used to infect the Caki-1 renal carcinoma cell line, resulting in ≈100 puror Caki-1 colonies, which were maintained as a population. PCR analysis of the bulk population yielded 1 predominant Env isolate with the 11-aa insert shown (see Fig. 1B). The general features of the insert are outlined in Fig. 1B. The randomized insert (indicated in bold) contains 3 arginine residues as well as the potential for an alternative cysteine bond.
Fig. 1.
Schematic of the Env library vector and CP-targeting peptide. (A) Schematic of the randomized Env-IRES-puro (RIP) library vector. The SV40 promoter and the neomycin selection marker from pRVL (7) were replaced by an expression cassette in which an internal ribosomal entry site (IRES) drives the expression of puro. Position of the LTRs and viral genes are marked; Ψ, packaging signal. (B) Sequence of the Env isolated from Caki-1. The 11-aa encoding the CP new binding insert are numbered in bold and flanked by the conserved regions of the FeLV envelope. The putative cystine bonds are represented and alternative configurations of bonds between cysteines are possible. The cysteine loop in the parental FeLV Env consists of the outer residues.
Host Range and Tissue Specificity.
To determine the host and tissue specificity of the CP Env, cell lines derived from a range of species and tissue types were challenged with viral particles bearing CP and packaging lacZ as a marker of gene transfer (Tables 1 and 2). Titers were measured as lacZ-staining units per milliliter of viral supernatant (LSU/ml).
Table 1.
Animal cell-line host range and tissue specificity
| Cell lLine | Host | Tissue (pathology) | Titer (LSU/ml) |
|---|---|---|---|
| D17 | Dog | Bone (osteosarcoma) | 1.2 ± 1.6 × 105 |
| MDCK | Dog | Kidney (normal) | 2.2 ± 3.4 × 101 |
| AH927 | Cat | Fibroblast (normal) | <10 |
| UMR-106 | Rat | Bone (osteosarcoma) | <10 |
| B16F1 | Mouse | Skin (melanoma) | <10 |
| MC3T3 | Mouse | Bone (pre-osteoblast) | <10 |
| NIH3T3 | Mouse | Fibroblast (normal) | <10 |
| Primary Calvaria OB | Mouse | Bone (osteoblast) | <10 |
Table 2.
Human cell-line and tissue specificity
| Cell lLine | Human tissue (pathology) | Titer (LSU/ml) |
|---|---|---|
| Caki-1 | Kidney (carcinoma) | 2.1 ± 1.6 × 103 |
| Caki-2 | Kidney (carcinoma) | <10 |
| A498 | Kidney (carcinoma) | 7.7 ± 1.5 × 103 |
| ACHN | Kidney (carcinoma) | 3.9 ± 3.8 × 102 |
| 293T | Kidney (normal) | 4.0 ± 2.0 × 105 |
| TE671 | Muscle (rhabdomyosarcoma) | 2.5 ± 1.3 × 105 |
| 143B | Bone (osteosarcoma) | 1.4 ± 0.5 × 105 |
| 1.3 ± 0.3 × 108a | ||
| HOS (CRL1543) | Bone (osteosarcoma) | 8.9 ± 0.9 × 104 |
| MG63 (CRL1427) | Bone (osteosarcoma) | 2.2 ± 0.2 × 105 |
| HeLa | Cervix (carcinoma) | 1.6 ± 1.3 × 105 |
| HepG2 | Liver (carcinoma) | 3.5 ± 2.9 × 105 |
| Huh-7 | Liver (carcinoma) | 2.0 ± 0.9 × 104 |
| DLD-1 | Colon (carcinoma) | 8.3 ± 6.9 × 103 |
| PC3 | Prostate (metastatic carcinoma) | 2.7 ± 1.5 × 104 |
| DU-145 | Prostate (carcinoma) | 7.0 ± 4.7 × 101 |
| HFF | Foreskin fibroblasts (normal) | 5.1 ± 1.8 × 103 |
aVirus stock concentrated 56-fold by centrifugation.
The infectivity of virus bearing the CP Env was tested on murine, rat, feline, canine, and human cell lines. Significantly, no infection was detected on the feline fibroblast cell line AH927, the permissive cell line for both FeLV-A and -C viruses. This indicates that the CP Env is using a receptor outside of the known interference groups of FeLV. Similarly, no infection was observed on murine tail fibroblasts (NIH/3T3), eliminating receptor usage of the known ecotropic, polytropic, and amphotropic MuLV viruses (18, 20, 21). High titer was observed on the canine D17 osteosarcoma cells (1.2 × 105), but not on the canine kidney cell line MDCK (2.2 × 101), indicating tissue specificity within the canine family. The rat osteosarcoma UMR-106 cells were nonpermissive to infection with CP-pseudotyped virus (<10 LSU/ml), although infection with VSV-G pseudotyped virus was efficient, eliminating the possibility of a postentry block to infection (data not shown).
Productive infection into a wide-range of human cell lines was observed. Interestingly, although VSV-G was used to establish the constitutive Env producer cells, titers on the 2 cell lines that were used to create the Env library, 293T and TE671, were high (4 × 105 and 2.5 × 105 LSU/ml, respectively). Similar titers, ranging from 8.9 × 104 to 2.2 × 105 LSU/ml, were found on all 3 of the human osteosarcoma cell lines that were tested: 143B, MG63, and HOS. Furthermore, titers of 1.2 × 108 LSU/ml on 143B cells were readily obtained with CP viral preparations concentrated by ultracentrifugation. High titers (>104 LSU/ml) were also seen on other cancer cell lines, including the cervical carcinoma line HeLa, and the liver carcinoma cell lines Huh-7 and HepG2. Despite poor titers (<102 LSU/ml) on the prostate cancer cell line DU-145, much higher titers (3 × 104 LSU/ml) were seen on the PC-3 cell line, which is a prostate cancer line isolated as a bone metastasis. Interestingly, titers in the range of 103 or lower were found on all 4 of the renal carcinoma cell lines that were tested, Caki-1, Caki-2, ACHN, and A498, despite the fact that the library was initially screened on the Caki-1 cell line.
Mutation Analysis of the CP Env.
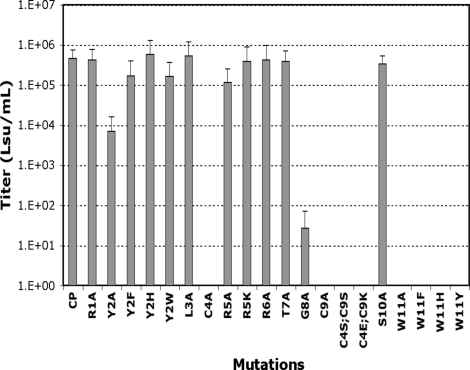
The 11-aa insert in the CP Env presented several interesting characteristics (see Fig. 1), including a putative cysteine bond and a clustering of basic residues. To assess the role that each of the 11 residues plays as it relates to viral entry, alanine scanning was performed followed by site-directed mutagenesis of the residues that were found to be necessary for infection. The relative activity of each Env mutant was measured by its ability to mediate gene transfer of the lacZ marker as compared to wild-type CP (Fig. 2). All of the Envs that had lost infectivity were still incorporated into viral particles at levels comparable to that of wild-type CP, as demonstrated by Western blot (data not shown).
Fig. 2.
Titer of the CP Env mutants. The assay is quantifying the transfer of the lacZ marker via wild-type CP or mutant CP Envs. The figure represents the average of the 3 values, with error bars representing 1 SD. Mutations are numbered according to their position in the randomized region of the wild-type CP insert.
Replacement of the cysteines in position 4 or 9 with alanine (C4A or C9A) resulted in a complete loss of titer (see Fig. 2). Furthermore, the more conservative substitution of both cysteines with serines in these positions (C4S;C9S) was unable to rescue infectivity (see Fig. 2). To assess whether a disulfide bond existed between these 2 positions, the residues in positions 4 and 9 were replaced with glutamate and lysine, respectively (C4E;C9K) to determine whether a salt bridge between these positions could rescue Env function. This mutation resulted in no measurable titer (see Fig. 2), suggesting that these residues are forming disulfide bonds, but with residues outside of the randomized region. The glycine in position 8 also proved to be necessary for proper infectivity, as its replacement with alanine (G8A) resulted in a greater than 4 log decrease in titer (see Fig. 2). The positioning of this flexible residue immediately next to a cysteine (position 9) may provide the necessary rotation to permit cystine bond formation, which could be sterically constrained by alanine in the G8A mutant.
Beyond the amino acids implicated in the structural backbone, the tryptophan in position 11 proved to be essential for Env function. Replacement with alanine (W11A) resulted in a complete loss of function (see Fig. 2). Significantly, conservative replacement with other aromatic residues (W11F, W11H, and W11Y) yielded similar results. Substitution of the second aromatic residue, tyrosine in position 2 with alanine (Y2A) resulted in a 1 log decrease in titer (see Fig. 2), which could be restored by replacement with other aromatic residues (Y2F, Y2H, Y2W) (see Fig. 2).
Although sequence scanning highlighted the presence of a cluster of basic residues in the amino terminus of the insert, replacement of any of the individual basic residues with alanine (R1A, R5A, R6A) did not result in any appreciable decrease in titer (see Fig. 2), suggesting that these individual charges are not necessary for binding. Additional mutations having no significant effect on binding or entry include L3A, R5K, T7A, and S10A (see Fig. 2.).
Binding Assay.
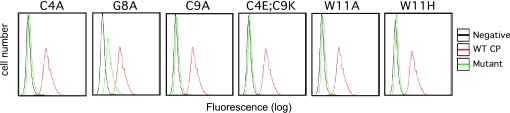
To confirm that loss of infectivity associated with mutants C4A, C9A, C4E;C9K, G8A, and W11A were the result of a loss of receptor binding and not a result of a defect in any later step in viral entry, the ability of each of these viral Envs to bind to the cell surface was measured (Fig. 3). The virus bound to the cell surface was detected with FACS analysis using antibodies recognizing a conserved epitope of the FeLV Env, outside of the receptor-binding domain conjugated to an FITC secondary Ab (30). Using this assay, viral-binding correlated with viral titer. No significant viral-binding was observed for C4A, C9A, C4E;C9K, W11H, and W11A. The G8A mutant, which had a measurable but significantly decreased level of infectivity (see Fig. 2), similarly had a detectable but significantly lower binding than that observed by wild-type CP. These data demonstrate that the loss of infectivity noted with these Env mutants was directly related to a decrease in binding to the host receptor.
Fig. 3.
Binding studies of virus bearing mutant CP on human 143B osteosarcoma cells. Virus-binding to 143B cells was detected by flow-cytometric analysis using the C11D8 mAb recognizing the FeLV Env backbone and goat anti-mouse IgG conjugated with FITC. For each panel, the black line represents the control assay performed in the absence of virus and the red line represents the binding of the wild-type CP virus to the 143B cells. The green line represents the virus bearing the mutated CP Env as indicated on each panel.
Isolation of the CP Receptor.
The host-range of virus bearing the CP Env indicated that it utilizes a receptor outside the known receptors of the FeLV and MuLV. To isolate the host receptor for the CP Env, a cDNA library was derived from the permissive cell line 143B. The library was inserted into the retroviral backbone pMX1 and transduced into the nonpermissive cell line UMR-106 (see Table 1). Two independent screens for the CP receptor were performed, differing in the quantity of 143B cDNA introduced into the UMR-106 cells. UMR-106 cells expressing the 143B cDNA library were challenged with virus bearing the CP Env and packaging the GFP-IRES-puro vector. UMR-106 cells expressing the CP receptor will be permissive to infection and be identified as puror, GFP+ colonies. After selection with puromycin, 8 GFP+ colonies from the first screen and 84 from the second were isolated and subjected to a secondary challenge with virus bearing the CP Env and packaging lacZ to confirm that these cells were now permissive to infection. From the first screen, 2 of the 8 colonies resulted in titers of 1.5 × 103 and 3.3 × 103 LSU/ml, respectively, upon secondary challenge with CP/lacZ virus. Using pMX-specific primers, a common 2-kb band was amplified from genomic DNA from both colonies (clones 4 and 5). Sequence analysis of the PCR products revealed identical cDNA inserts encoding the G-protein coupled receptor 172A (GPR172A) (Accession # NM024531.3). This protein has been reported to function as the GHB receptor (22) as well as one of the receptors for PERV-A (16). An additional 4 clones, which were puror, GFP+, and lacZ+, were identified from the second cDNA library screening when challenged with virus bearing CP Env. PCR analysis of these 4 colonies amplified a similar 2-kb product using the pMX primers. Sequence analysis indicated that 2 of these clones encoded the identical 1941 base GPR172A cDNA (Fig. 4, isolate 1). In addition, 2 clones encoded a GPR172A cDNA of 1961 bases (Fig. 4, isolate 2). The 2 cDNAs differ by 20 bases in their 5′ UTR.
Fig. 4.
Schematic diagram of the GPR172A mRNA from 143B cells. The GPR172A gene is encoded on chromosome 8 at position q24.3 (top line). Shown is the 5′ terminus of the gene including the 5′ UTR, with the Met start encoded at position 731 within the gene. Multiple mRNA start sites and splice junctions have been reported within the 5′ UTR for the GPR172A mRNA and are individually listed. Key positions with respect to the transcription start site in the chromosome, as well as the sequence of the 5′ terminus of the mRNAs, are indicated. The 2 cDNAs isolated from 143B cells are shown in comparison with the reported GPR172A mRNAs.
Confirmation that the GPR172A cDNA functions as the receptor for virus bearing the CP Env was obtained through reconstruction and reintroduction of the 1941 base cDNA into nonpermissive cells. The cDNA was reinserted into the pMX1 backbone, and viral particles packaging the GPR172A cDNA were introduced into the nonpermissive cell lines UMR-106 and AH927 [CP titer of <10 and 1.5 × 101 LSU/ml, respectively (see Table 1)]. For UMR-106 cells, a GPR172A+ population was selected, whereas total GPR172A-transduced AH927 cells were used for CP titer analysis. In the presence of GPR172A cDNA, the CP titers were 1.3 × 104 and 2.2 × 104 LSU/ml for AH927 and UMR-106 cells, respectively (Fig. 5A).
Fig. 5.
GPR172A functions as CP Env viral receptor. (A) Introduction of GPR172A cDNA renders cells permissive to CP infection. Human GPR172A cDNA was introduced into the nonpermissive AH927 cells and UMR-106 cells by retroviral infection (see Materials and Methods). Cells were challenged with virus bearing the CP Env and packaging lacZ. Titers of CP/lacZ virus (LSU/ml) are indicated: nonpermissive cells (light gray); GPR172A expressed in nonpermissive cells (Gray); control 143B permissive cells (black). (B) Introduction of shRNA against GPR172A in permissive 143B cells. VSV-G pseudotyped lentiviral vectors packaging shRNA were used to infected 143B cells, selected for puror and challenged with virus bearing the CP Env and packaging lacZ. Viral titer on 143B was 3.0 × 105 LSU/ml. Relative rates of infectivity are shown for shRNAs 11421 and 8272, as well as for the empty vector control (pLKO.1) compared to that of wild-type 143B cells. The figures represent the average of the 3 values with error bars representing 1 SD.
Further confirmation that GPR172A served as the CP receptor was obtained by shRNA knockdown in 143B cells. Knockdown of GPR172A expression by 2 specific shRNAs (11421 and 8272) resulted in a 98.4% and 99.5% reduction in viral titer in 143B cells, respectively. RT-PCR analysis showed a marked decrease in GPR172A expression although detectable levels still remained after shRNA treatment (data not shown). The empty vector control (pLKO.1) had minimal effect on CP viral titer (Fig. 5B).
Discussion
The results of these studies highlight the remarkable flexibility as well as the limitations of retroviral receptor usage. By providing the virus with an appropriate backbone in which a short primary sequence can determine receptor binding, a single-round of infection can identify unique functional Env isolates of high titer. Through this Env library system, viral receptor usage has been expanded to use cell-surface proteins previously not reported as viral receptors (10). In the current study, the receptor identified is one used by a porcine endogenous retrovirus.
In this Env library screen as well as in the screening of a previously derived library (10), the receptors isolated were multipass transmembrane receptors, consistent with that reported for all known gamma-retroviruses. This demonstrates an advantage for viral entry to proceed through this class of proteins. However, the independent selection of an identical receptor from a single-round of infection implies an additional benefit for this particular protein. This could include readily facilitating membrane fusion or perhaps a preference for the intracellular pathway used for recycling and trafficking of the receptor. The use of identical receptors by different viral species is not unusual; the Pit receptors are used by murine, feline, and gibbon ape leukemia viruses (14, 15, 19).
Of particular interest is the fact that the VRA regions between the PERV-A and CP Envs are unrelated, although alternative domains within PERV SU could contribute to receptor recognition (25, 26). The GPR172A protein, beyond its function as a PERV-A receptor, has also been discovered to serve as the human GHB receptor. From an evolutionary point of view, the endogenous retroviruses might have evolved to bind this receptor without altering the function of the host protein, whereas the CP Env does not have this constraint and could function similarly to known substrates or inhibitors. It is of interest to examine if the CP acts as an antagonist or agonist to the known physiological responses characteristic of GHB receptors.
All 6 of the cDNAs isolated from receptor-positive cells encoded the GPR172A gene, although they differed in their 5′ termini. It is not known whether these reflect previously unidentified start sites or incomplete cDNAs. The splicing pattern corresponds with a cDNA that was previously isolated from Ramos cells, a human Burkitt's lymphoma cell line (CR606512). As studies have indicated that GPR172A is commonly up-regulated in human cancers (23), it would be of interest to determine whether this 5′UTR results in an altered level of expression or mRNA stability in the osteosarcoma cell line or other transformed cells. The absence of the cDNA for the GPR172B gene, which also functions as the PERV-A receptor (16), is intriguing. It is possible that the remaining titers seen in the shRNA-treated cells are the result of additional receptors including GPR172B. However, the isolation of multiple cDNAs encoding GPR172A from 2 independent library screenings in concert with the >98% decrease in titer observed upon GPR172A-targeted shRNA treatment, confirms that in the 143B cell line GPR172A is the predominant receptor responsible for entry of virus bearing CP Env.
Published data using PERV-A Env pseudotyped onto MuLV-based particles have generally yielded titers in the range of 104 on HeLa and TE671 cells (31), while the CP Env produced titers greater than 105 LSU/ml. This single round of molecular evolution and selection has yielded an Env that is capable of using a known retroviral receptor with higher efficiency than its naturally evolved counterpart, and which can be easily concentrated to produce extremely high titers on human cancer cell lines. This method of retargeting has now been shown to be able to create unique and functionally efficient retroviral Envs with therapeutic potential.
Materials and Methods
Library Plasmids.
The vector pRVL-SV40-neo (7) used for the initial library construction was modified by replacing the SV40-neo expression cassette with IRES-puro (see SI Text and Table S1 for a list of primers and sequences). The new vector, named RIP, was used for the creation of the randomized envelope library. The plasmid pHIT-G (32) expresses VSV-G.
Cell Lines.
All of the cell lines used for the assays were maintained in media containing 10% FBS and 10 μl/ml of antibiotic-antimycotic mixture (Gibco). The producer cell lines 293TCeB (7), TECeBF2, and TELCeB (29) were maintained in DMEM containing 9 μg/ml of Blasticidin S (Invivogen). The cell line 293TCeB/GIP was created as previously described (10) and maintained in DMEM containing 2.5 μg/ml puromycin, 400 μg/ml G418, and 9 μg/ml Blasticidin S. The cell lines used for screening and host- and tissue-specificity assays were maintained as follows: Caki-1 and Caki-2 in McCoy's 5A medium; CRL1543, CRL1427, Huh7, UMR106, NIH 3T3, D17, and MDCK in DMEM; 143B and AH927 in MEM with 2 mM L-glutamine; PC3 and DU145 in RPMI with nonessential amino acids; ACHN, A498, Hep-G2, HeLa, and DLD-1 in MEM with 2 mM L-glutamine, nonessential amino acids, 1 mM sodium pyruvate, and 1.5 g/L sodium bicarbonate; MC3T3 and primary calvaria preosteoblasts in αMEM medium and human foreskin fibroblasts (HFF) in DMEM/F12 medium. For all infections, the media was filtered (0.45 μm filters) and performed in the presence of 8 μg/ml polybrene. Viral concentration was performed by overnight utracentrifugation as previously described (8).
RIP Library Construction.
The 11 random amino acid library with the RIP vector was generated through insertion within 2 back-to-back BbsI sites using 3 oligonucleotides, as previously described (7). The oligonucleotides encoding the random library contained the sequence 5′-GTGGGAGACACCTGGGAACCT(NNB)11TCCTCCTCAAAATAT-GGA-3′, where B stands for T, C, or G. The elimination of A in the third position of the codon biases the library against stop codons. The library plasmid DNA was introduced into Electromax DH10B competent cells (Invitrogen) by electroporation. Ampr colonies were harvested and their plasmids were purified by CsCl gradient. DNA analysis of 10 individual colonies indicated 6 contained random inserts, with the remaining 4 representing the parental pRIP vector encoding a stop codon between the BbsI sites, and thus would not yield functional Env protein.
A TECeB-based cell line constitutively expressing the randomized Env library was obtained as previously described (8), with the modification that transfections were performed with lipofectamine 2000 and cells were selected with 2.5 μg/ml puromycin. After drug selection, the number of colonies was counted and the complexity of the library was determined to be 2 × 105.
Screening of the RIP Library and Identification of a Unique Env Peptide.
The RIP library was used to screen for envelopes capable of productively infecting the human renal cell carcinoma cell line Caki-1. Viral supernatants from TECeBF2 cells expressing the RIP library were collected in the absence of puromycin and applied to the target cells at 15 ml per 15-cm plate. A total of 100 puror colonies grew and were maintained as a population.
Genomic DNA was extracted from puromycin-resistant Caki-1 cells using the DNeasy kit (Qiagen). The Env was PCR amplified with the FeLV-specific primers FeLV1 and FeLV2, and sequenced using FeLV1 primer. The Env isolate (named CP) was subsequently reconstructed through PCR amplification, with the primers RANBBS5 and RANBBS3 flanking the randomized 11 aa, as previously described (7), and reinserted into the pRVL-SV40-neo plasmid.
Mutations of CP Env Isolate.
Mutagenesis of the 11 randomized amino acids in the CP Env isolate was performed by overlapping PCR for mutations T7A and G8A and PCR for the remaining mutations (see SI Text and Table S2). Reinsertion into the Env backbone was facilitated by the presence of an AgeI site within the 33-bp portion encoding the randomized region and by HpaI and ApaI sites flanking this region. The mutations were verified by sequencing with the primer FeLV1.
Viral Titers.
A stable producer cell line capable of producing virus particles pseudotyped with CP-Env and transferring the lacZ marker has been obtained via transfection of the TELCeB cell-line with pRVL-SV40-neo-CP and subsequent G418 selection. Viral titers were determined as previously described (7).
Titration with the mutant CP envelopes was performed by transfecting a subconfluent 10-cm plate of TELCeB cells with the 10 μg of pRVL-SV40-neo-CP(mut) or pRVL-SV40-neo-CP with lipofectamine 2000 (Invitrogen). The next day, the cells were induced with 10-mM sodium butyrate. Viral supernatant was collected 24 h after induction and used to infect 143B cells at 0 and 1/10 dilutions.
Virus Binding Assay.
The binding assay was performed as previously described on 143B cells (30), with the modification that the cells were not fixed with paraformaldehyde. The binding analysis was performed on a Beckman Coulter Cytomics FC500 at the Cytometry Core Facility at the Environmental and Occupational Health Sciences Institute of the University of Medicine and Dentistry of New Jersey.
Construction of 143B cDNA Library.
Poly(A)+ mRNA was isolated and purified from 5 × 107 143B cells using an mRNA isolation kit (Stratagene) and 5 μg was used as template for cDNA synthesis using a cDNA synthesis kit (Stratagene) in the absence of radioactive dNTPs.
The pMX vector was modified as follows to allow for insertion of the library. The vector was digested with BstXI and a linker region encoding a cloning site containing EcoRI and XhoI restriction sites was inserted. Next, pMX+EcoRI/XhoI was digested with EcoRI and XhoI and a 2-kB stuffer region was inserted to reduce background of singly cut vector to produce pMX-EcoRIstufferXhoI. The cDNA library was inserted into this vector after digestion with EcoRI and XhoI, electroporated into DH10B electromax ultracompetent cells according to the manufacturers protocol, and plated on four 245 × 245-mm plates containing LB-carbenecillin (100 ng/ml). After overnight incubation, the plates yielded ≈2 × 106 colonies. The resulting colonies were harvested and plasmids were purified using the Endofree Maxiprep kit.
To determine that the cDNA library contained full-length cDNAs, regions corresponding to the 5′ and 3′ ends of the clathrin mRNA-coding region as well as to the 5′ end of the GAPDH mRNA-coding region were amplified after first-strand cDNA synthesis, second-strand synthesis, as well as from the completed library after endotoxin-free plasmid preparation, as previously described (33). Amplification of the cDNA regions that correspond to the 5′ and 3′ portions of the clathrin mRNA-coding region assures that at each step the library contained full length cDNAs up to at least 6 kb.
cDNA Library Screening for the CP Receptor.
Viral particles were assembled as described above by transfecting 30 to 60 μg of the cDNA library and 30 μg of pHIT-G in each of 3 15-cm plates of 293TCeB. This viral supernatant was added to each of 3 15-cm plates of UMR-106 cells overnight.
Two days after infection, the cells were challenged with virus bearing the CP Env and packaging GIP (25 ml viral supernatant/15 cm plate) overnight. This virus was generated by transfecting 293TCeB/GIP cells with 30 μg of pRVL-SV40-neo-CP, as described above. The viral supernatant was harvested and used to infect the library containing UMR-106 cells. Beginning 2 days after infection, the UMR-106 cells were submitted to selection with puromycin (2.5 μg/ml). Surviving colonies were then isolated and challenged with virus packaging the lacZ gene and bearing the CP Env (see Viral Titers, above).
PCR and Sequence Analysis of cDNA Inserts.
Genomic DNA was harvested from ≈5 × 106 cells using the DNeasy kit (Qiagen). The viral insert was then PCR amplified with virus-specific primers MX11 and MXcDNA R. The PCR products were isolated and sequenced using the primers MXcDNA F, GPR172A603 F, and GPR172A1066 F.
Receptor cDNA Cloning.
The receptor cDNA was PCR amplified from genomic DNA isolated from the 2 clones (numbers 4 and 5) derived from our first library screening with primers MX11 and MXcDNA R using Fidelitaq Polymerase (USB) and subcloned into the Topo2.1 vector (Invitrogen). The insert was removed using EcoRI and XhoI yielding a 0.8 kB 5′ EcoRI/XhoI cDNA fragment and a 1.1 kB 3′ XhoI/XhoI cDNA fragment. The 2 fragments were inserted sequentially into pMX1+EcoRIstufferXhoI. The resulting pMX-GPR172A was sequenced as described above. The pMX+GPR172A vector was packaged into viral particles by transfecting 293TCeB cells with 10 μg each of pMX-GPR172A and pHIT-G, as described above. The viral supernatant was used to infect UMR-106 and AH927 cells. Two days later, lacZ titers were measured on the AH927 cells while the UMR-106 cells were infected with CP-GIP and placed under puromycin selection. LacZ titers were then measured on the puror GFP+ UMR-106 population.
shRNA Knockdown of GPR172A.
Receptor expression was knocked down using the lentiviral based plasmid pLKO.1 expressing the GPR172A specific shRNA's TRCN0000008272 (8272) and TRCN0000011421 (11421) (Open Biosystems). Lentiviral particles were produced by transfecting 293T cells with 5 μg each of pHIT-G, pCMVΔR8.2Δvpr (34), and pLKO.1 either containing the shRNA or as an empty vector control. The viral supernatant was used to infect 143B cells. After puromycin selection, titers based on lacZ transfer were measured on the shRNA+ population as described above.
Sequence Analysis and Alignment.
DNA and protein sequences were analyzed using the Basic Local Alignment and Search Tool provided by the National Institutes of Health (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Protein sequence alignment was performed using the MultAlin software (35).
Supplementary Material
Acknowledgments.
This work was supported by Grant 1RO1CA49932 (to M.J.R.) and Fellowship 707060 from the New Jersey Commission on Cancer Research (to P.M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809741106/DCSupplemental.
References
- 1.Hacein-Bey-Abina S, et al. Sustained correction of x-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 2.Rigby MA, et al. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol. 1992;73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 3.Cosset F-L, et al. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnierle BS, Moritz D, Jeschke M, Groner B. Expression of chimeric envelope proteins in helper cell lines and integration into Moloney murine leukemia virus particles. Gene Ther. 1996;3:334–342. [PubMed] [Google Scholar]
- 5.Benedict CA, et al. Targeting retroviral vectors to CD34-expressing cells: Binding to CD34 does not catalyze virus-cell fusion. Hum Gene Ther. 1999;10:545–557. doi: 10.1089/10430349950018625. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Identification of the block in targeted retroviral-mediated gene transfer. Proc Natl Acad Sci USA. 1999;96:4005–4010. doi: 10.1073/pnas.96.7.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bupp K, Roth M. Altering retroviral tropism using a random display envelope library. Mol Ther. 2002;5:329–335. doi: 10.1006/mthe.2002.0546. [DOI] [PubMed] [Google Scholar]
- 8.Bupp K, Roth MJ. Targeting a retroviral vector in the absence of a known cell-targeting ligand. Hum Gene Ther. 2003;14:1557–1564. doi: 10.1089/104303403322495061. [DOI] [PubMed] [Google Scholar]
- 9.Bupp K, Sarangi A, Roth MJ. Selection of feline leukemia virus envelope proteins from a library by functional association with a murine leukemia virus envelope. Virology. 2006;351:340–348. doi: 10.1016/j.virol.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 10.Sarangi A, Bupp K, Roth MJ. Identification of a retroviral receptor used by an envelope protein derived by peptide library screening. Proc Natl Acad Sci USA. 2007;104:11032–11037. doi: 10.1073/pnas.0704182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza R, Anderson MM, Overbaugh J. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol. 2006;80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quigley JG, et al. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95:1093–1099. [PubMed] [Google Scholar]
- 13.Tailor CS, Willett BJ, Kabat D. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol. 1999;73:6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi Y, et al. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albritton LM, Tweng L, Scadden D, Cunningham JM. A putative murine retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 16.Ericsson TA, et al. Identification of receptors for pig endogenous retrovirus. Proc Natl Acad Sci USA. 2003;100:6759–6764. doi: 10.1073/pnas.1138025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavanaugh MP, et al. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller DG, Edwards RH, Miller AD. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Hara B, et al. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 20.Tailor CS, et al. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.vanZeijl M, et al. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andriamampandry C, et al. Cloning and functional characterization of a gamma-hydroxybutyrate receptor identified in the human brain. FASEB J. 2007;21:885–895. doi: 10.1096/fj.06-6509com. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, et al. Common human cancer genes discovered by integrated gene expression analysis. PLoS ONE. 2007;2:e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tailor XS, Nouri A, Kabat D. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J Virol. 2000;74:237–244. doi: 10.1128/jvi.74.1.237-244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe R, Miyazawa T, Matsuura Y. Cell-binding properties of the envelope proteins of porcine endogenous retroviruses. Microbes Infect. 2005;7:658–665. doi: 10.1016/j.micinf.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Argaw T, Figueroa M, Salomon DR, Wilson CA. Identification of residues outside of the receptor binding domain that influence infectivity and tropism of porcine endogenous retrovirus. J Virol. 2008;82:7483–7491. doi: 10.1128/JVI.00295-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rey MA, Prasad R, Tailor CS. The C domain in the surface envelope glycoprotein of subgroup C feline leukemia virus is a second receptor-binding domain. Virology. 2008;370:273–284. doi: 10.1016/j.virol.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Boomer S, Eiden M, Burns CC, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosset F, et al. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadan MJ, Sturm S, Anderson WF, Eglitis MA. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi Y, et al. Host range and interference studies of the three classes of pig endogenous retroviruses. J Virol. 1998;72:9986–9991. doi: 10.1128/jvi.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouchier RAM, et al. HIV-1 infection of non-dividing cells: Evidence that the amino-terminal basic region of the viral matrix protein is important for gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai Y, Sugawara R, Ohinata H, Takai A. Two types of non-selective cation channel opened by muscarinic stimulation with carbachol in bovine ciliary muscle cells. J Physiol. 2004;559(Pt 3):899–922. doi: 10.1113/jphysiol.2004.065607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart SA, Poon B, Jowett JBM, Xie Y, Chen ISY. Lentiviral delivery of HIV-1 Vpr protein induces apoptosis in transformed cells. Proc Natl Acad Sci USA. 1999;96:12039–12043. doi: 10.1073/pnas.96.21.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corpet F. Multiple sequence alignment with heirarchical clustering. Nucleic Acids Res. 1988;18:1206–1207. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.