Abstract
The walls of grasses and related members of the Poales are characterized by the presence of the polysaccharide (1,3, 1,4)-β-D-glucan (β-glucan). To date, only members of the grass-specific cellulose synthase-like F (CSLF) gene family have been implicated in its synthesis. Assuming that other grass-specific CSL genes also might encode synthases for this polysaccharide, we cloned HvCSLH1, a CSLH gene from barley (Hordeum vulgare L.), and expressed an epitope-tagged version of the cDNA in Arabidopsis, a species with no CSLH genes and no β-glucan in its walls. Transgenic Arabidopsis lines that had detectable amounts of the epitope-tagged HvCSLH1 protein accumulated β-glucan in their walls. The presence of β-glucan was confirmed by immunoelectron microscopy (immuno-EM) of sectioned tissues and chemical analysis of wall extracts. In the chemical analysis, characteristic tri- and tetra-saccharides were identified by high-performance anion-exchange chromatography and MALDI-TOF MS following their release from transgenic Arabidopsis walls by a specific β-glucan hydrolase. Immuno-EM also was used to show that the epitope-tagged HvCSLH1 protein was in the endoplasmic reticulum and Golgi-associated vesicles, but not in the plasma membrane. In barley, HvCSLH1 was expressed at very low levels in leaf, floral tissues, and the developing grain. In leaf, expression was highest in xylem and interfascicular fiber cells that have walls with secondary thickenings containing β-glucan. Thus both the CSLH and CSLF families contribute to β-glucan synthesis in grasses and probably do so independently of each other, because there is no significant transcriptional correlation between these genes in the barley tissues surveyed.
Keywords: (1,3;1,4)-β-glucan endo-hydrolase, (1,3;1,4)-β-d-glucan synthase, cell wall biosynthesis, grasses
The 1,3;1,4-β-glucans, homopolymers of (1,3)-β– and (1,4)-β–linked glucose (Glc) residues, are found characteristically in the walls of grasses (Poaceae) and in related families from the Poales (1, 2). β-Glucans transiently accumulate in the elongating walls of vegetative tissues, are also found in secondary walls of the vasculature (3, 4), and are the major components of the endosperm walls of cereal grains, where they are known to influence downstream processing and nutritional properties (5, 6). Although the tissue distribution, structure, and physicochemical properties of the β-glucans are well defined, the enzymes and encoding genes responsible for their synthesis remain poorly characterized.
Current evidence suggests that synthases encoded by members of the cellulose synthase (CESA)/cellulose synthase-like (CSL) superfamily and the glucan synthase-like (GSL) gene family are involved in the synthesis of most β-linked wall polysaccharides. The GSL genes encode (1,3)-β-d-glucan synthases (callose synthases) (7, 8), and the CESA genes encode cellulose synthases (9). The CSLs are believed to encode enzymes that synthesize the backbone of various non-cellulosic β-linked polysaccharides of the wall (10–13). They have been classified into 8 gene families, designated CSLA to CSLH; the CSLF and CSLH families are restricted to the grasses (12, 14). A third group of grass-specific CSL genes, designated CSLJ, has been identified recently (11). Because of their restricted taxonomic distribution, grass-specific CSLs are seen as candidate genes for enzymes that synthesize polysaccharides such as the β-glucans that are characteristic of the grasses. Consistent with this hypothesis, rice CSLF genes were shown to encode proteins capable of mediating β-glucan synthesis when expressed in transgenic Arabidopsis, a species that lacks this polysaccharide in its walls (15). However, given the low levels of β-glucan made by Arabidopsis lines that expressed rice CSLF genes and other evidence accumulated in cereals, it was concluded that other proteins probably are also associated with β-glucan synthesis (15).
Thus, we tested whether members of another grass-specific CSL gene family also were capable of mediating the synthesis of β-glucan by expressing a barley CSLH gene in Arabidopsis. Here we show that the HvCSLH1 protein is responsible for the β-glucan deposited in the walls of transgenic Arabidopsis plants. We conclude that the CSLH family encodes a second class of (1,3;1,4)-β-d-glucan synthases.
Results
Barley Has only a Single CSLH Gene.
Candidate CSLH genes in barley were identified initially by querying online public EST databases with rice CSLH sequences. All CSLH-related ESTs from barley could be aligned into a single contiguous sequence of ≈1,500 bp that included the entire 3′ untranslated region and a region encoding the COOH-terminal 488 (of an expected ≈750) amino acid residues of the protein (Table S1). We designated this gene HvCSLH1. Screening of a barley BAC library with HvCSLH1-derived probes identified several genomic clones, all containing HvCSLH1, from which the missing 5′ end was obtained (SI Text). A PCR-amplified 2,430-bp HvCSLH1 cDNA fragment contains a single 2,256-bp ORF and encodes a protein with a predicted molecular mass of 82.6 kDa and a pI of 7.0 (Fig. S1A). Analysis of the conceptual translation of this sequence found between 5 and 9 transmembrane domains, with the consensus being 2 NH2-terminal and 4 COOH-terminal transmembrane domains (Fig. S1B) and with both termini of the mature protein predicted to be cytoplasmic. This topology places the large, central domain containing the D,D,D,QFKRW motif within the cytoplasm (Fig. S1C). At the nucleotide level, HvCSLH1 shares 68% to 74% identity (62%–69% amino acid identity) with the 3 rice CSLH genes (14). Sequence alignments, the position and phase of exon-intron splice sites (Fig. S1A), and a phylogenetic reconstruction (Fig. S2) suggest HvCSLH1 is the likely barley orthologue of OsCSLH1. Genetic mapping of HvCSLH1 using a Sloop × Halcyon doubled haploid population (16) showed that HvCSLH1 is on the short arm of chromosome 2H, ≈1.5 centimorgans from a cluster of 4 HvCSLF genes (HvCSLF3, -4, -8, -10) shown to be within a major quantitative trait locus controlling β-glucan content in barley grain (17, 18) (Fig. S3).
Expression of HvCSLH1 in Arabidopsis Results in Deposition of (1,3;1,4)-β-d-Glucan.
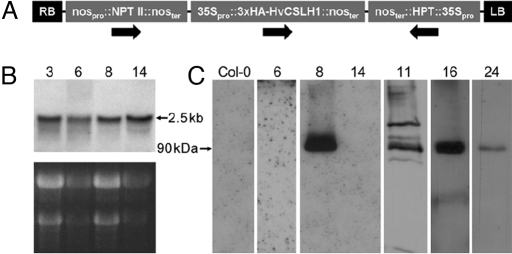
For heterologous expression in Arabidopsis, the HvCSLH1 ORF was cloned into the Gateway-enabled binary vector pGWB15 (19), which placed HvCSLH1 under the control of the CaMV 35S promoter and added a 3×HA epitope tag to the encoded protein's NH2-terminal end (Fig. 1A). Initial selection of transformed Arabidopsis seeds identified a number of putative transgenic seedlings which PCR confirmed contained HvCSLH1. RNA blot analysis of these T1 plants showed that ≈ 90% accumulated HvCSLH1 transcripts in rosette leaves (Fig. 1B). Western blotting using an anti-HA tag antibody was used to detect HvCSLH1 protein in these lines (Fig. 1C). A mixed microsomal membrane fraction (50,000–100,000 × g pellet) was prepared from pooled 3-week-old kanamycin-resistant T2 seedlings. Western blotting with the anti-HA antibody showed that only 4 of 28 lines containing HvCSLH1 transcripts accumulated a polypeptide of the expected size (≈90 kDa, Fig. 1C). Occasionally proteins of higher and lower molecular mass were detected also (e.g., lane 11). The 90-kDa protein was not observed in total protein extracts (data not shown) or in mixed-membrane fractions prepared from untransformed Arabidopsis plants (Fig. 1C, Lane Col-0). It is not known why HA-tagged HvCSLH1 accumulated in only some of the plant lines that expressed HvCSLH1 mRNA or why little correlation was apparent between HvCSLH1 protein levels and either HvCSLH1 transcript levels (compare Fig. 1 B and C) or the number of HvCSLH1 transgenes present in a plant (data not shown), although this lack of correlation has been observed previously (15). Lines 8, 11, 16, and 24, which expressed the HA-tagged HvCSLH1, and line 6, which did not express detectable levels of the protein (control), were selected for subsequent experiments.
Fig. 1.
(A) Schematic of the transfer DNA of the HvCSLH1::pGBW15 construct used in gain-of-function experiments in Arabidopsis. Arrows indicate the direction of gene transcription. (B) Northern blot showing transcript levels in mature leaves of HvCSLH1 transgenic T1 plants. (Upper) X-ray film exposure. (Lower) Corresponding ethidium bromide-stained gel. The observed 2.5-kb transcript corresponds to the expected size of the tagged HvCSLH1 mRNA. (C) Western blot showing 3×HA-tagged HvCSLH1 protein levels in 3-week-old pooled seedlings of HvCSLH1 transgenic T2 lines. Mixed microsomal membrane protein (30 μg/lane) was loaded, and blots were probed with the anti-HA antibody. (B and C) Numbers refer to transgenic lines. Col-0 is the wild-type untransformed line. Lines 8 and 14 are from the same blot; all other lines are from different blots.
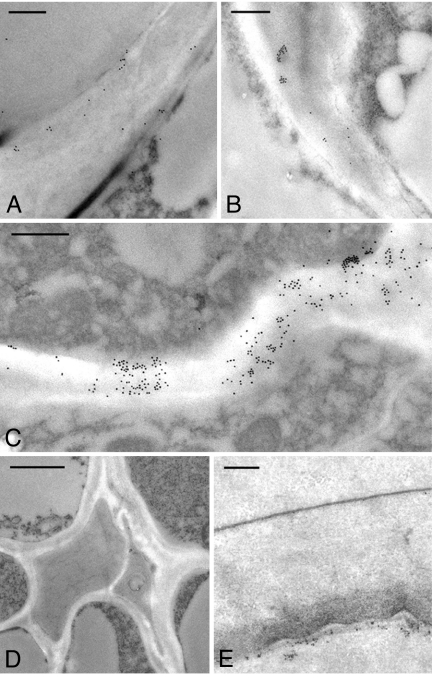
Immuno-electron microscopy (immuno-EM) was used to determine whether the walls of the transgenic Arabidopsis plants accumulated detectable levels of β-glucan. Mature leaf sections from self-pollinated progeny of lines 8, 11, 16, 24, and 6 (T2 generation) were probed with a monoclonal antibody specific for β-glucan (20), followed by detection using a secondary antibody conjugated to 18-nm gold. Labeling was evident in walls of the HA-tagged HvCSLH1-positive lines 8, 11, and 16 (Fig. 2A, C, and B, respectively) but not in the walls of either line 24 (data not shown), which also expressed HvCSLH1, or line 6 (Fig. 2E), which had no detectable HvCSLH1 protein (Fig. 1C). No labeling was seen in untransformed Arabidopsis leaves (Fig. 2D). As previously observed for the CSLF genes (15), each positive transgenic line showed a different pattern of tissue labeling. In line 8, patchy labeling was observed in the walls of epidermal cells and occasionally in xylem walls (Fig. 2A), whereas in line 11 epidermal and vascular tissue walls were labeled only lightly, but heavier (albeit more patchy) labeling was observed in mesophyll walls (Fig. 2C). Broadly distributed, light labeling was present in all walls of the mature leaf of line 16 (Fig. 2B). Overall, the level of β-glucan labeling in the walls of the CSLH lines appeared similar to that seen in the Arabidopsis CSLF transgenics (15), although the heavier labeling seen in outer periclinal walls of the epidermis of CSLF transgenic lines was not so apparent in the CSLH lines. Labeling was virtually abolished in leaf sections of transgenic plants that had been preincubated with a (1,3;1,4)-β-glucan-specific endo-hydrolase (β-glucan hydrolase) (Fig. S4) (15). None of the transgenic Arabidopsis lines exhibit major phenotypic differences compared with wild type.
Fig. 2.
Transmission electron micrographs showing detection of β-glucan in walls of HvCSLH1-expressing lines with a β-glucan antibody (20). (A–C) Lines 8, 16, and 11, respectively; (D) wild-type Col-0 control; (E) line 6. A and D show cells of the vascular bundle; B and C show mesophyll cells; E shows epidermal cells. Scale bar represents 0.5 μm in A–C and E and 1 μm in D.
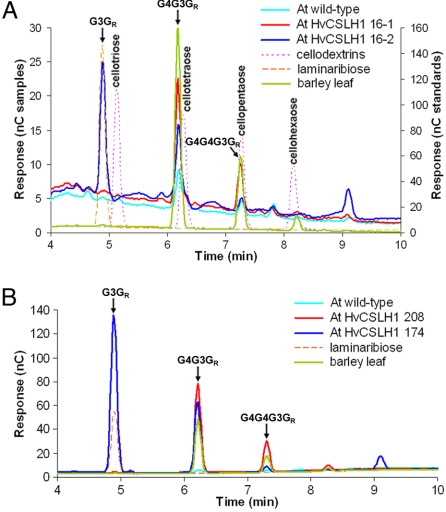
To provide biochemical confirmation of the presence of β-glucan in transgenic Arabidopsis walls and to examine its fine structure, pooled leaf and/or stem material was collected from the progeny of lines 8, 11, 174, 208, and 2 homozygous lines derived from plant 16 (lines 16–1 and 16–2). Lines 174 and 208 were identified by the high-performance anion-exchange chromatography (HPAEC) method (as discussed later) from among the kanamycin-resistant T2 lines that accumulated CSLH transcripts. Walls of alcohol-insoluble residue (AIR) were prepared and digested with a linkage-specific β-glucan hydrolase and the released oligosaccharides profiled by HPAEC and MALDI-TOF MS. β-Glucan hydrolase hydrolyses (1,4)-β-Glc linkages when these linkages are on the reducing-end side of a (1,3)-β-Glc residue, yielding a series of oligosaccharides. These oligosaccharides are diagnostically the tri-saccharide G4G3GR and the tetra-saccharide G4G4G3GR (in which G is β-d-Glcp, 3 and 4 indicate (1,3) and (1,4) linkages, respectively, and GR is the reducing terminal residue). Upon digestion of AIR samples, 2 types of HPAEC profiles were found among the CSLH transgenics. The first type, which released variable quantities of G4G3GR and G4G4G3GR, is represented by lines 16–1 (Fig. 3A) and 208 (Fig. 3B). The ratio of G4G3GR to G4G4G3GR (DP3:DP4) released from these lines was similar to the DP3:DP4 ratio of 3.6 released from barley leaf β-glucan. Oddly, peaks of low abundance with similar retention times to the β-glucan–derived oligosaccharides were detected in wild-type Arabidopsis when comparable AIR concentrations were used. These peaks were always much smaller than those present in the transgenic lines (Fig. 3 A and B). The oligosaccharides present in these wild-type peaks cannot be derived from an endogenous β-glucan, because no β-glucan labeling was detected in untransformed Arabidopsis by immuno-EM (Fig. 2D) (15). These oligosaccharides must be derived instead from another polysaccharide and co-elute with β-glucan–derived oligosaccharides. For example, cellopentaose co-elutes with G4G4G3GR under the conditions used (Fig. 3A). In the second type of profile, found in most CSLH lines, the major peak among the digestion products co-eluted with laminaribiose, G3GR (Figs. 3 A and B and S5A). The presence of laminaribiose in these samples was not brought about by a contaminant in the β-glucan hydrolase preparation, by an endogenous disaccharide, or by the activity of an uncharacterized Arabidopsis enzyme, because it either was absent or was present in minute quantities in the barley and Arabidopsis controls and in lines 16–1 and 208 (Figs. 3 A and B and S5A). The sizes of oligosaccharides in each peak in this profile were confirmed further by MALDI-TOF MS analysis, which showed the presence of hexose di-, tri- and tetra-saccharides in ratios similar to those observed in the HPAEC profile (Fig. S5B). The amounts of β-glucan in the transgenic lines, as estimated from the peak areas of each oligosaccharide relative to the barley control (0.4% [wt/wt] β-glucan), were between 0.00015% (lines 8 and 11) and 0.016% (line 174) of total wall (wt/wt).
Fig. 3.
HPAEC profiles of oligosaccharides released upon β-glucan hydrolase digestion of total walls (AIR) prepared from HvCSLH1 transgenic Arabidopsis lines 16–1 and 16–2 (A) and174 and 208 (B). Samples were taken from plants at 145 d (16–1, leaf; 16–2: leaf and stem), 21 d (174, entire seedlings) and 56 d (208, leaf), respectively. AIR positive control: barley mature leaf (entire sheath); AIR negative control: wild-type Arabidopsis Col-0 (mature leaves). AIR samples were loaded at similar concentrations. Laminaribiose (G3GR) and a cellodextrin series (degree of polymerization [DP] 3–6) were used as standards. Arabidopsis samples are plotted on the left y-axis; standards and positive control are plotted on the right y-axis. G3GR (DP2), G4G3GR (3-O-β-cellobiosyl-Glc, DP3), and G4G4G3GR (3-O-β-cellotriosyl-Glc, DP4) peaks are indicated, as are the cellodextrins.
HvCSLH1 Is Located in Endoplasmic Reticulum- and Golgi-Associated Vesicles but Not the Plasma Membrane of Transgenic Arabidopsis Plants Expressing HvCSLH1.
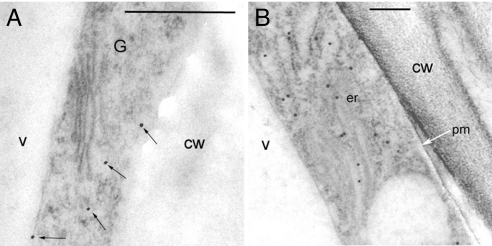
Sections of high-pressure–frozen leaves from line 11 were incubated with gold-labeled anti-HA antibody to determine the subcellular location of HvCSLH1. Labeling was seen in the endoplasmic reticulum (ER)- and Golgi-derived vesicles but not in the plasma membrane (Fig. 4 A and B). Similar results were observed in labeled sections of roots and seedlings (data not shown).
Fig. 4.
Transmission electron micrographs showing the detection of the 3×HA-tagged HvCSLH1 protein by a gold-labeled anti-HA antibody in sections of high-pressure-frozen leaves of Arabidopsis transgenic line 11. (A and B) Mesophyll cells. cw, cell wall; er, endoplasmic reticulum; G, Golgi body; pm, plasma membrane; v, vacuole. Scale bar represents 0.5 μm in A and 0.2 μm in B. Black arrows indicate Golgi-associated vesicle labeling; white arrow indicates plasma membrane.
HvCSLH1 Is Transcribed in Barley at Low Levels in Developing Grain, Floral Tissues and Cells of the Leaf Undergoing Secondary Wall Thickening.
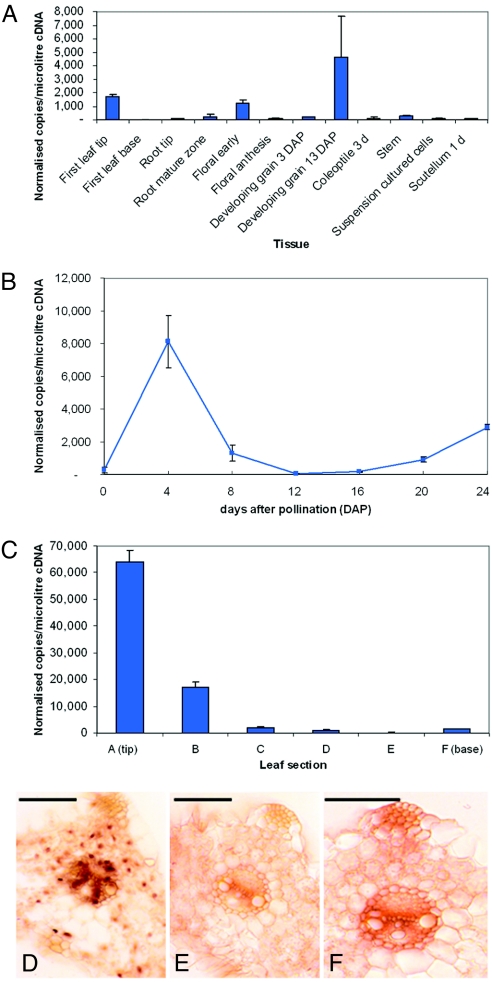
HvCSLH1 transcript levels in various barley tissues were determined by quantitative PCR (qPCR) using gene-specific primers (Table S2). Fig. 5 A–C shows that across a set of barley vegetative and floral tissues, HvCSLH1 transcripts accumulated to levels that routinely were less than 1,000 copies/μl cDNA. This level is lower than in some other barley CESAs and CSLs, in which values typically range from 10,000 to 100,000 copies/μl cDNA (18, 21). Levels of HvCSLH1 transcripts were relatively low in tissues composed of rapidly elongating cells, including coleoptile and leaf base, which are those cells actively synthesizing β-glucan (22). The highest levels of HvCSLH1 transcripts were in leaf tip, where cells no longer are actively growing and less β-glucan accumulates (2, 4). HvCSLH1 transcription in leaf was characterized further using RNAs isolated from 6 zones within the ≈13-cm-long leaves of 10-day-old seedlings, starting from the leaf tip composed of fully mature cells (zone A), to the leaf base composed of dividing cells (zone F) (21). In situ PCR (23) was used to identify cells in the leaf tip that contained the HvCSLH1 mRNA. Cells in which gene transcripts are detected stain purple to dark brown (Fig. 5D, 18S rRNA positive control), and cells where no transcription is detected stain light brown (Fig. 5E, negative control). HvCSLH1 was transcribed mostly in cells that are undergoing secondary wall thickening, such as interfascicular sclerenchymal fiber and xylem cells (Fig. 5F) known to contain β-glucan in their walls (4).
Fig. 5.
HvCSLH1 expression in barley as determined by qPCR and in situ PCR analyses. (A) Normalized levels of HvCSLH1 transcript in a range of tissues. Control genes for normalization were GAPDH, cyclophilin, and α-tubulin. (B) Normalized levels of HvCSLH1 transcript in developing endosperm 0–24 days after pollination. Control genes were GAPDH, α-tubulin, and EF1a. (C) Normalized levels of HvCSLH1 transcript in 10-day-old first leaf. Control genes were GAPDH, cyclophilin, and HSP70. Error bars on qPCR plots indicate SD. (D–F) In situ PCR images of the maturing zone of a 7-day-old first leaf using probes for 18S RNA (positive control, D), HvCSLH1 (F), and a negative control (E). Scale bar represents 100 μm.
HvCSLH1 transcript levels also were investigated in a 24-day developing endosperm series (Fig. 5B). HvCSLH1 expression was low throughout the starchy endosperm during development. Maximum transcript levels were reached at 4 days after pollination, ≈ 1 day before β-glucan was first detected in endosperm walls (24). This transcription pattern is similar to that of several barley CSLF genes (HvCSLF3, -4, -7, -8, and -10), although HvCSLF9 and -6 show much higher transcript levels (18).
Discussion
The data presented here indicate that the product of HvCSLH1, a member of the grass-specific CSLH gene family, mediates β-glucan biosynthesis in Arabidopsis and does so independently of the CSLF proteins previously shown to have this ability (15). Barley has only a single CSLH gene based on EST database analyses, genomic DNA blot analysis, and BAC library screening (SI Text). Within the Poaceae, barley belongs to the subfamily Pooideae, and EST surveys of other members of this subfamily such as bread wheat, Lolium, Festuca, and Brachypodium also have identified only a single CSLH gene in each species. Other subfamilies, such as the Panicoideae (maize, sorghum, and sugar cane) and Ehrhartoideae (rice), have more than a single CSLH gene (M.S.D., unpublished data).
HvCSLH1 was transcribed most highly in leaf tips, a tissue composed of fully mature cells, and was not high in elongating cells, such as the coleoptile or developing endosperm, which in barley are the tissues where β-glucan accumulates (22, 24). Although found in primary walls of vegetative tissues where it is implicated in the control of cell expansion (5) and possibly acts as a temporary energy source (33), β-glucan is also found in the lignified walls of xylem tracheary elements and sclerenchyma fibers in both the middle lamella region (primary wall) and the secondary wall (2, 4). Because in situ PCR showed HvCSLH1 transcripts in the leaf were restricted to cells such as interfascicular sclerenchymal fiber and xylem cells, we suggest that a major role of this gene is in β-glucan synthesis during secondary wall development, although we cannot exclude a role in primary wall β-glucan synthesis elsewhere in the plant.
When an epitope-tagged version of the HvCSLH1 cDNA was heterologously expressed in Arabidopsis, plant lines in which HvCSLH1 protein was detected accumulated a polysaccharide in their walls that was recognized by a β-glucan–specific monoclonal antibody. When walls of these and other HvCSLH1 transgenic lines were digested with a specific β-glucan hydrolase, the characteristic oligosaccharides, G4G3GR and G4G4G3GR, were detected, demonstrating that the transgenic plant lines contained β-glucan. Furthermore, epitope-tagged HvCSLH1 was found in the ER- and Golgi-derived vesicles in cells of transgenic plants, a location consistent with biochemical studies that showed this polysaccharide was synthesized in vitro by Golgi-enriched membrane fractions from Lolium suspension-cultured endosperm cells (25) and maize coleoptiles (26). These data, when considered in combination with previous observations that no β-glucan is present intracellularly (24), suggest a mechanism of polysaccharide assembly different from that of other matrix polysaccharides (27).
Although some transgenic plant lines seemed to produce a β-glucan similar to that in barley, the β-glucan hydrolase digestion of the walls of other lines produced laminaribiose (G3GR) in addition to the diagnostic G4G3GR and G4G4G3R products (Fig. 3 and Fig. S5). The appearance of G3GR at variable levels in these walls was associated with increased levels of G4G3GR relative to the G4G4G3GR and thus with increased DP3:DP4 ratios. The detection of G3GR in wall digests indicates the presence of a polysaccharide containing sections of alternating (1,3)-β- and (1,4)-β-linked Glc (-G3G4G3G4-). It is not known if these alternating β-linked Glc residues reside in a separate polysaccharide or constitute a portion of a β-glucan chain that also has the usual fine structural features. Alternating (1,3)-β-Glc and (1,4)-β-Glc residues are not common in cereal β-glucans (28) but are a significant component of the β-glucan from the non-flowering plant Equisetum (29, 30) and also are detected in β-glucans from a number of fungi, including basidiomycetes (31) and ascomycetes (32). It is possible that G3GR arises through misregulation of the β-glucan synthase in transgenic Arabidopsis, possibly because its membrane microenvironment is different or because an unknown factor that in barley suppresses (1,3)-β Glc linkage formation (or, alternatively, promotes (1,4)-β Glc linkage formation) is present at suboptimal levels in Arabidopsis. Minor variations in the level of this factor among the transgenic lines would account for the different β-glucan structures. Another possible explanation for the structural variability in the β-glucan may relate to subtle differences between grasses and Arabidopsis in postassembly processing, e.g., involving cellulases and/or xyloglucan endo-transglycosylases/hydrolases.
Regardless of how the fine structures of β-glucans are generated, it is clear that the CSLHs can mediate their synthesis in Arabidopsis, a finding that has implications for our understanding of how this polysaccharide is made. Any assembly mechanisms being considered must account for the synthesis of the predominant cellotriosyl and cellotetraosyl units, the random linking of these (1,4)-β units together by single (1,3)-β linkages, and the means by which the ratio of tri- to tetra-saccharide units is regulated (33, 34). At least 2 glycosyltransferase activities might act in concert: an activity that processively adds (1,4)-β-Glc residues to assemble the tri- and tetra-saccharides and a second activity that adds single (1,3)-β-Glc residues. The simplest explanation is that a single polypeptide is responsible for the synthesis of both glucosidic linkages. Our transgenic experiments indicate that both the CSLH (this study) and CSLF (15) proteins are able independently to make a β-glucan and therefore conceivably could make both types of β linkages. Both the CSLH and CSLF families are classified by the Carbohydrate-Active Enzymes (CAZy) database (http://www.cazy.org) as members of Glycosyltransferase Family 2 (GT2) (10), a family that includes enzymes capable of independently catalyzing the synthesis of either (1,3)-β or (1,4)-β linkages and also bifunctional enzymes, i.e., enzymes that can synthesize 2 types of glycosidic linkages. For example, hyaluronan synthases (HAS) synthesize a repeating disaccharide of (1,4)-β-glucuronic acid-(1,3)-β-N-acetylglucosamine units with both transferase activities residing in the single polypeptide. In mouse HAS1, the region that includes the D,D,D,QXXRW motif is responsible for both these activities (35). The active site of the CSLHs and CSLFs, which also contains this motif, might be similarly bifunctional. Hyaluronan is, however, a true disaccharide-repeating polysaccharide, whereas β-glucan comprises irregular repeats of the tri- and tetra-saccharide. Therefore some form of regulatory mechanism must be invoked that allows switching to occur between synthesis of the 2 glucosidic linkages to generate the tri- to tetra-saccharide units (and units with a higher degree of polymerization). Another possibility is that the CSLHs and CSLFs synthesize only a single type of glucosidic linkage and that another glucosyltransferase (GlcT), common to monocots and dicots, is responsible for synthesis of a second linkage. To date, other CSLs have been shown to be involved only in generating (1,4)-β linkages, for example, in the synthesis of the backbones of mannan (36) and xyloglucan (37). On the basis of these data, if the CSLHs and/or CSLFs contribute only a single activity, a (1,4)-β-activity would be expected. However, there are GT2 family enzymes that make (1,3)-β linkages, such as the Agrobacterium CrdS (38) that synthesizes curdlan. Therefore, it is possible that CSLH and/or CSLF proteins have (1,3)-β activity alone and that another GlcT synthesizes the cellodextrins. If the CSLH/F proteins could synthesize only (1,4)-β linkages, another GlcT would be involved in making the (1,3)-β linkage between cellodextrin units. Such a mechanism might involve the synthesis of cellodextrins in the ER/Golgi, where the CSLH protein has been located (Fig. 4), and their assembly into a polysaccharide either in the same location or at the plasma membrane. The observation from immuno-EM studies that β-glucan has not been detected in the endomembrane system (15, 24) could be explained by a post-Golgi assembly of the polysaccharide from oligosaccharides (27). Although such a mechanism of assembly is possible, it does invoke the need for the participation of multiple enzymes and an elaborate regulatory mechanism. Another model in which CSLH and CSLF proteins act coordinately, each catalyzing a single linkage type, is possible but is not favored by the experimental evidence, because in Arabidopsis independent expression of each protein is able to produce β-glucan, and we see no significant co-ordinate transcription of HvCSLH1 with any HvCSLF gene or among the HvCSLF genes themselves that would indicate that they are part of the same β-glucan synthase enzyme complex (18). Further experiments are required to determine whether both catalytic activities reside within CSLH and CSLF proteins or whether other enzymes must be recruited to provide either of these required catalytic activities.
In summary, we have identified 2 families of CSL genes involved in the assembly of β-glucan in grasses and have studied their temporal and spatial expression patterns. Although many questions about the mechanisms of assembly and the regulation of the synthesis of β-glucans and their deposition into the wall remain, we now can begin unraveling these processes at the molecular/biochemical level.
Materials and Methods
Binary Vector Construction and Plant Transformation.
The HvCSLH1 ORF was amplified from barley cv Schooner mature leaf tip cDNA with Herculase (Stratagene) using primers HvH1TOPOf and HvH1TOPOr (Table S3), and the PCR product was cloned into pENTR/D-TOPO (Invitrogen). Using the manufacturer's protocol (Invitrogen), the cDNA was cloned into the destination vector pGWB15 containing an NH2-terminal 3×HA tag (19), and the predicted sequence was confirmed by DNA sequencing. The construct was transferred from Escherichia coli into Agrobacterium tumefaciens strain AGL1 via triparental mating using the helper plasmid pRK2013. Arabidopsis Col-0 plants were transformed using the floral dip method (39).
RNA Blot Analysis.
Samples of total RNA (≈10 μg) extracted from mature rosette leaves of T1 plants using TRIzol (Invitrogen) were prepared and separated on a 1% wt/vol agarose-formaldehyde gel (40). RNA was transferred to Hybond N+ membranes, prehybridized and hybridized as described in the Gene Images CDP-Star detection module (Amersham-Biosciences). A HvCSLH1 fragment amplified with primers H1F2 and HvH1TOPOr (Table S3) was labeled using the Gene Images Random Prime labeling module (Amersham-Biosciences) and was used as the probe.
Quantitative PCR Analysis.
RNA extractions, cDNA syntheses, and qPCR were carried out as previously described (15, 21). The primer sequences of the barley control genes are listed in Table S2.
In Situ PCR.
In situ PCRs were conducted according to the method of Koltai and Bird (23) with modifications as outlined in SI Text.
Protein Analysis.
Preparation of mixed microsomal membranes and detection of HvCSLH1 protein by Western blotting is described in SI Text.
Immuno-Electron Microscopy.
Arabidopsis tissues were fixed and labeled with anti-(1,3, 1,4)-β-d-glucan antibody (20) according to Burton et al. (15). For labeling with anti-HA antibody, plant tissue was high-pressure-frozen and freeze-substituted as described by Brownfield et al. (41). Thin sections were incubated in a 1:200 dilution of rat anti-HA polyclonal antibody (Roche) in PBS, pH 7.4, containing 1% wt/vol BSA for 1 h at room temperature and then overnight at 4 °C. Grids were washed in PBS and then incubated in a 1:20 dilution of anti-rat secondary antibody conjugated to 18 nm gold (Jackson ImmunoResearch). Sections then were washed, poststained, and viewed by transmission electron microscopy (15).
(1,3;1,4)-β-D-Glucan Endo-Hydrolase Digestion.
AIR (10–100 mg prepared as described in SI Text) was extracted 3 times with 5 ml 20 mM NaPO4 buffer, pH 6.5, for 2 h at 50 °C in a shaking incubator (200 rpm), and walls were collected by centrifugation (3,400 × g, 5 min). The pelleted AIR then was resuspended in 5 ml NaPO4 buffer to which 100 μl β-glucan hydrolase (42) was added. The mixture was incubated for 2 h at 50 °C with continuous mixing, after which the supernatant was collected (as the β-glucan hydrolase-released oligosaccharides), desalted on a graphitized carbon cartridge (43), and dried.
HPAEC Analysis.
The released oligosaccharides were separated by HPAEC on a CarboPac PA1 column (Dionex) equilibrated with 50 mM NaOAc in 0.2 M NaOH using a Dionex BioLC ICS 300 system (Dionex) equipped with a pulsed amperometric detector (PAD). Oligosaccharides were eluted at 1 ml/min with a linear gradient of NaOAc from 50 mM in 0.2M NaOH to 350 mM in 0.2 M NaOH over 15 min. Laminaribiose (Seikagaku) and cellodextrins (Sigma) were run as standards.
Supplementary Material
Acknowledgments.
We thank Margie Pallotta and Neil Shirley for their assistance with gene mapping and qPCR, respectively, Dr. Anne Medhurst for her assistance during the early stages of this work, and Drs. Tsuyoshi Nakagawa (Shimane University, Japan) and Maria Hrmova for providing the pGWB vectors and β-glucan hydrolase for immuno-EM work, respectively. This work was supported by funding from the Grains Research and Development Corporation, the Australian Research Council, and the Commonwealth Scientific and Research Organization Flagship Collaborative Research Program, provided to the High Fibre Grains Cluster via the Food Futures Flagship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FJ459581).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902019106/DCSupplemental.
References
- 1.Harris PJ. Diversity in plant cell walls. In: Henry RJ, editor. Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Wallingford, Oxon, UK: CAB International Publishing; 2005. pp. 201–227. [Google Scholar]
- 2.Trethewey JAK, Campbell LM, Harris PJ. (1→3),(1→4)-β-D-Glucans in the cell walls of the Poales (sensu lato): An immunogold labeling study using a monoclonal antibody. Am J Bot. 2005;92:1660–1674. doi: 10.3732/ajb.92.10.1660. [DOI] [PubMed] [Google Scholar]
- 3.Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- 4.Trethewey JAK, Harris PJ. Location of (1→3),(1→4)-β-D-glucans in vegetative cell walls of barley (Hordeum vulgare) using immunogold labelling. New Phytol. 2002;154:347–358. doi: 10.1046/j.1469-8137.2002.00383.x. [DOI] [PubMed] [Google Scholar]
- 5.Fincher GB, Stone BA. Chemistry of nonstarch polysaccharides. In: Wrigley C, Corke H, Walker CE, editors. Encyclopedia of Grain Science. Vol 1. London: Academic Press; 2004. pp. 206–222. [Google Scholar]
- 6.Stone BA, Clarke AE. Chemistry and Biology of (1, 3)-β-Glucans. Melbourne: La Trobe Univ Press; 1992. [Google Scholar]
- 7.Brownfield L, et al. Proteomic and biochemical evidence links the callose synthase in Nicotiana alata pollen tubes to the product of the NaGSL1 gene. Plant J. 2007;52:147–156. doi: 10.1111/j.1365-313X.2007.03219.x. [DOI] [PubMed] [Google Scholar]
- 8.Li J, et al. Biochemical evidence linking a putative callose synthase gene with (1→3)-β-D-glucan biosynthesis in barley. Plant Mol Biol. 2003;53:213–225. doi: 10.1023/B:PLAN.0000009289.50285.52. [DOI] [PubMed] [Google Scholar]
- 9.Delmer DP. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 10.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 11.Farrokhi N, et al. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotech J. 2006;4:145–168. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- 12.Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richmond TA, Somerville CR. Integrative approaches to determining Csl function. Plant Mol Biol. 2001;47:131–143. [PubMed] [Google Scholar]
- 14.Hazen SP, Scott-Craig JS, Walton JD. Cellulose synthase-like genes of rice. Plant Physiol. 2002;128:336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton RA, et al. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 16.Read BJ, et al. Mapping and QTL analysis of the barley population Sloop x Halcyon. Aust J Agric Res. 2003;54:1145–1153. [Google Scholar]
- 17.Han F, et al. Mapping of β-glucan content and β-glucanase activity loci in barley-grain and malt. Theor Appl Genet. 1995;91:921–927. doi: 10.1007/BF00223901. [DOI] [PubMed] [Google Scholar]
- 18.Burton RA, et al. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008;146:1821–1833. doi: 10.1104/pp.107.114694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 20.Meikle PJ, et al. A (1→3)(1→4)-β-D-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3)(1→4)-β-D-glucans. Plant J. 1994;5:1–9. doi: 10.1046/j.1365-313x.1994.5010001.x. [DOI] [PubMed] [Google Scholar]
- 21.Burton RA, et al. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004;134:224–236. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibeaut DM, Pauly M, Bacic A, Fincher GB. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta. 2005;221:729–738. doi: 10.1007/s00425-005-1481-0. [DOI] [PubMed] [Google Scholar]
- 23.Koltai H, Bird DM. High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol. 2000;123:1203–1212. doi: 10.1104/pp.123.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson S, et al. Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta. 2006;224:655–667. doi: 10.1007/s00425-006-0244-x. [DOI] [PubMed] [Google Scholar]
- 25.Henry RJ, Schibeci A, Stone BA. Localization of β-glucan synthases on the membranes of cultured Lolium multiflorum (ryegrass) endosperm cells. Biochem J. 1983;209:627–633. doi: 10.1042/bj2090627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibeaut DM, Carpita NC. Synthesis of (1→3),(1→4)-β-D-glucan in the Golgi apparatus of maize coleoptiles. Proc Natl Acad Sci USA. 1993;90:3850–3854. doi: 10.1073/pnas.90.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fincher GB. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 2009;149:27–37. doi: 10.1104/pp.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perlin A, Suzuki S. The structure of lichenin: Selective enzymolysis studies. Can J Chem. 1962;40:50–56. [Google Scholar]
- 29.Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR. Mixed-linkage (1→3),(1→4)-β-D-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 2008;179:104–115. doi: 10.1111/j.1469-8137.2008.02435.x. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen I, et al. Mixed-linkage (1→3),(1→4)-β-D-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J. 2008;54:510–521. doi: 10.1111/j.1365-313X.2008.03453.x. [DOI] [PubMed] [Google Scholar]
- 31.Pacheco-Sanchez M, et al. A bioactive (1→3),(1→4)-β-D-glucan from Collybia dryophila and other mushrooms. Mycologia. 2006;98:180–185. doi: 10.3852/mycologia.98.2.180. [DOI] [PubMed] [Google Scholar]
- 32.Fontaine T, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem. 2000;275:27594–27607. doi: 10.1074/jbc.M909975199. [DOI] [PubMed] [Google Scholar]
- 33.Staudte RG, Woodward JR, Fincher GB, Stone BA. Water-soluble (1→3),(1→4)-β-D-glucans from barley (Hordeum vulgare) endosperm. III. Distribution of cellotriosyl and cellotetraosyl residues. Carbohydrate Polymers. 1983;3:299–312. [Google Scholar]
- 34.Wood PJ, Weisz J, Blackwell BA. Molecular characterization of cereal β-D-glucans. Structural analysis of oat β-D-glucan and rapid structural evaluation of β-D-glucans from different sources by high-performance liquid chromatography of oligosaccharides released by lichenase. Cereal Chemistry. 1991;68:31–39. [Google Scholar]
- 35.Yoshida M, Itano N, Yamada Y, Kimata K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. J Biol Chem. 2000;275:497–506. doi: 10.1074/jbc.275.1.497. [DOI] [PubMed] [Google Scholar]
- 36.Dhugga KS, et al. Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- 37.Cocuron J-C, et al. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnezis T, Epa VC, Stone BA, Stanisich VA. Topological characterization of an inner membrane (1→3)-β-D-glucan (curdlan) synthase from Agrobacterium sp strain ATCC31749. Glycobiology. 2003;13:693–706. doi: 10.1093/glycob/cwg093. [DOI] [PubMed] [Google Scholar]
- 39.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 40.Farrell RE., Jr . RNA Nethodologies: A Laboratory Guide for Isolation and Characterization. Inc., San Diego: Academic Press; 1993. [Google Scholar]
- 41.Brownfield L, et al. Molecular control of the glucan synthase-like protein NaGSL1 and callose synthesis during growth of Nicotiana alata pollen tubes. Biochem J. 2008;414:43–52. doi: 10.1042/BJ20080693. [DOI] [PubMed] [Google Scholar]
- 42.McCleary BV, Glennie-Holmes M. Enzymatic quantification of (1→3)(1→4)-β-D-glucan in barley and malt. Journal of the Institute of Brewing. 1985;91:285–295. [Google Scholar]
- 43.Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconjugate Journal. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.