Abstract
Outer membrane-specific lipoproteins in Escherichia coli are released from the inner membrane by an ATP-binding cassette transporter, the LolCDE complex, which causes the formation of a soluble complex with a periplasmic molecular chaperone, LolA. LolA then transports lipoproteins to the outer membrane where an outer membrane receptor, LolB, incorporates lipoproteins into the outer membrane. The molecular mechanisms underlying the Lol-dependent lipoprotein sorting have been clarified in detail. However, it remained unclear how Lol factors interact with each other to conduct very efficient lipoprotein transfer in the periplasm where ATP is not available. To address this issue, a photo-reactive phenylalanine analogue, p-benzoyl-phenylalanine, was introduced at various positions of LolA and LolB, of which the overall structures are very similar and comprise an incomplete β-barrel with a hydrophobic cavity inside. Cells expressing LolA or LolB derivatives containing the above analogue were irradiated with UV for in vivo photo-cross-linking. These analyses revealed a hot area in the same region of LolA and LolB, through which LolA and LolB interact with each other. This area is located at the entrance of the hydrophobic cavity. Moreover, this area in LolA is involved in the interaction with a membrane subunit, LolC, whereas no cross-linking occurs between LolA and the other membrane subunit, LolE, or ATP-binding subunit LolD, despite the structural similarity between LolC and LolE. The hydrophobic cavities of LolA and LolB were both found to bind lipoproteins inside. These results indicate that the transfer of lipoproteins through Lol proteins occurs in a mouth-to-mouth manner.
Keywords: E. coli, Lol proteins, molecular chaperone, photo-cross-link
The chromosome of Escherichia coli encodes at least 90 species of lipoprotein precursors (1), which are translocated to the outer leaflet of the inner membrane by a Sec translocon and then processed to mature forms (2). Mature lipoproteins are next sorted to the outer membrane or retained in the inner membrane, depending on the lipoprotein-sorting signal located at position 2. Asp at this position is the inner membrane retention signal, whereas other residues function as outer membrane signals (3).
The Lol system, consisting of 5 Lol proteins, LolA through LolE, catalyzes the sorting of lipoproteins to the outer membrane (3). An ATP-binding cassette (ABC) transporter, the LolCDE complex, releases the outer membrane-specific lipoproteins, leading to the formation of a soluble complex with a periplasmic chaperone, LolA. Asp at position 2 functions as a LolCDE avoidance signal, which causes the retention of lipoproteins in the inner membrane. The LolA-lipoprotein complex crosses the periplasm to reach the outer membrane, to which a lipoprotein receptor, LolB, is anchored. LolB accepts a lipoprotein from LolA and incorporates it into the outer membrane. A single E. coli cell possesses ≈106 molecules of the major outer membrane lipoprotein Lpp (4), whereas only a few hundred LolA (5) and LolB (6) molecules exist in a single cell. Nevertheless, the Lol system very efficiently mediates the outer membrane sorting of lipoproteins, and the localization intermediate is not accumulated in the periplasm under normal conditions.
The overall structures of LolA and LolB are very similar, and comprise a novel fold comprising 11 antiparallel β-strands folded into an incomplete β-barrel with 2 loops covering the barrel (7). The barrel and the loops, containing 3 α-helices, form a hydrophobic cavity. The hydrophobic cavity of LolA, and probably that of LolB too, was recently found to undergo opening and closing upon the binding and release of lipoproteins, respectively (8). The strength of the hydrophobic interaction with lipoproteins was found to be critically important for efficient one-way transfer of lipoproteins from LolA to LolB (9). Moreover, the binding of ATP to LolD and its hydrolysis were found to cause a conformational change of the LolCDE complex, thereby causing the transfer of lipoproteins to LolA (10, 11). These observations revealed the detailed molecular mechanisms underlying the Lol protein–mediated lipoprotein transfer. On the other hand, it remains unknown how Lol proteins interact with each other and catalyze the lipoprotein transfer reaction.
To clarify the mode of interaction between Lol proteins, the in vivo photo-cross-linking technique developed by Schultz and his collaborators (12, 13) was used. This technique enables the introduction of p-benzoyl-phenylalanine (pBPA), a photo-reactive phenylalanine derivative, at desired positions. We expressed various amber mutants of LolA and LolB in E. coli together with an amber suppressor, tyrosyl tRNA, and tyrosyl tRNA synthetase, both of which were derived from Methanococcus jannaschii and engineered to incorporate pBPA into an amber (TAG) codon. The carbonyl oxygen at the benzophenone group of pBPA preferentially reacts with nearby carbon-hydrogen bonds when irradiated with UV light. We report here the details of the interactions between Lol proteins, and between Lol proteins and lipoproteins.
Results
Construction of Amber Mutants of LolA and LolB.
We constructed amber mutants of FLAG-tagged LolA and hexahistidine (His6)-tagged mLolB, the latter of which is a derivative lacking the N-terminal acyl chain anchor (6) but compensates for the LolB function. We used mLolB instead of LolB because the lack of the N-terminal acyl chain prevents the complex formation with LolA as a substrate lipoprotein. The mutation positions were selected so as to be distributed throughout the structure (Fig. 1). A plasmid encoding one of these amber mutants was introduced into E. coli BL21(DE3) cells harboring pSup-BpaRS-6TRN(D286R), which encodes the amber suppressor tyrosyl tRNA and tyrosyl-tRNA synthetase mutated to incorporate pBPA into an amber codon (14). All amber mutants of LolA, except I93 amber, complemented the growth of KTA2 cells (ΔlolA) in the presence of pBPA (Fig. S1A and SI Text). It has been shown that I93 of LolA is an important residue and its replacement by G yields a dominant negative mutant (15). The growth of KT60 (ΔlolB) was complemented by all amber mutants of mLolB in a pBPA-dependent manner (Fig. S1B and SI Text). These results indicate that LolA and LolB derivatives containing pBPA are functional except the LolA one containing pBPA in place of I93. The levels of pBPA-containing LolA and mLolB did not significantly differ with the mutation points (Fig. S2).
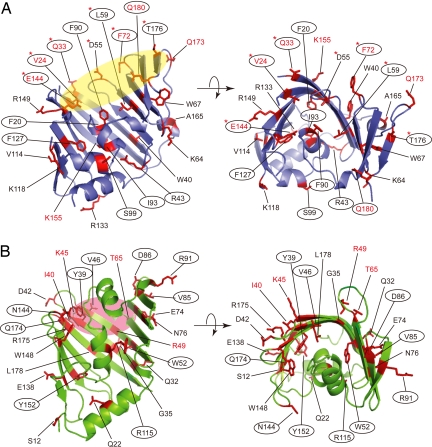
Fig. 1.
Residues of LolA and mLolB replaced by pBPA. The LolA (A) and mLolB (B) molecules each are shown as a ribbon model. The residues denoted by a 1-letter code with a number represent the positions of amber mutations introduced to incorporate pBPA and are shown as a red stick model. The left figures in A and B were rotated by ≈90° around the horizontal axis to give the right figures. Ovals in yellow (A) and pink (B) represent the entrance areas of the hydrophobic cavities of LolA and mLolB, respectively. (A) LolA derivatives having pBPA in place of the residues indicated in red letters cross-linked with mLolB, those circled cross-linked with Pal, and those indicated by asterisks cross-linked with LolC. (B) mLolB derivatives having pBPA in place of the residues indicated in red letters cross-linked with LolA and those circled cross-linked with Pal. The structural information on LolA (1IWL) and mLolB (1IWM) was obtained from the RCSB protein data bank (http://pdb.protein.osaka-u.ac.jp/pdb), and visualized with PyMOL ver. 0.98 (http://pymol.sourceforge.net/index.php).
Cross-linking Between LolA and mLolB.
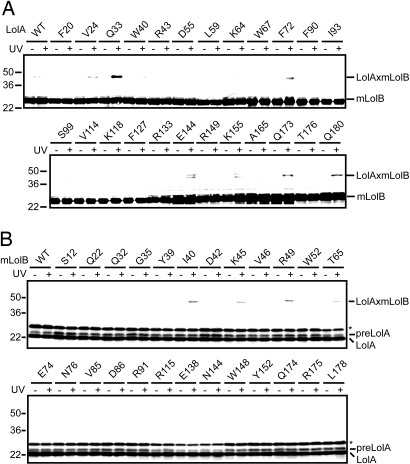
BL21(DE3) cells harboring pSup-BpaRS-6TRN(D286R) were further transformed with pSS1 encoding the wild-type or amber mutants of LolA and pSS24 encoding mLolB, and then grown in a medium supplemented with pBPA and isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were irradiated with UV light for 5 min, and then analyzed by SDS/PAGE and immunoblotting. Bands migrating to ≈45 kDa represented LolA-mLolB cross-linked products because they were only detected for UV-irradiated cells, and reacted with both anti-mLolB (Fig. 2A) and anti-LolA (see Fig. S3) antibodies. LolA derivatives with pBPA at positions 24, 33, 72, 144, 155, 173, and 180 (Fig. 1A, residues in red letters) generated cross-linked products with mLolB. All these positions except 155 were located around the entrance of the hydrophobic cavity of LolA (Fig. 1A, yellow oval). Crystallographic studies involving a LolA(R43L) derivative recently revealed the entrance of the hydrophobic cavity, which was open in this mutant (8). In contrast, the hydrophobic cavity of wild-type LolA was found to open upon lipoprotein binding (8). Importantly, the side chains of all these residues except that at position 155 were oriented toward the inside of the cavity. Among them, pBPA introduced at position 33 gave the most intense cross-linked band, suggesting that this residue most closely interacts with mLolB. It is postulated that the side chains of pBPA and the residue replaced by pBPA have the same orientation. Taken together, these results indicate that the internal surface of the hydrophobic cavity of LolA interacts with mLolB.
Fig. 2.
In vivo photo-cross-linking between LolA and mLolB. (A) Cells expressing mLolB-His6 and LolA-FLAG derivatives having pBPA in place of the indicated residues were UV-irradiated for 5 min or not irradiated, and then analyzed by SDS/PAGE and immunoblotting with anti-LolB antibodies, as described in Materials and Methods. As a control, cells expressing mLolB-His6 and wild-type (WT) LolA-FLAG were also examined. The cross-linked products generated from LolA and mLolB (LolAxmLolB) are indicated. LolA derivatives containing pBPA at positions 144 and 155 gave 2 cross-linked bands. The upper bands reacted with both anti-LolB and -LolA antibodies whereas the lower ones reacted only with anti-LolB antibodies. The properties of these materials are unknown at present. (B) Cells expressing LolA-FLAG and mLolB-His6 derivatives having pBPA in place of the indicated residues were irradiated as in (A), and then analyzed by SDS/PAGE and immunoblotting with anti-LolA antibodies. As a control, cells expressing LolA-FLAG and wild-type (WT) mLolB-His6 were also examined. The cross-linked products generated from LolA and mLolB (LolAxmLolB) are indicated. A precursor form of LolA (preLolA) was detected when wild-type LolA-FLAG was expressed from a very efficient plasmid (16). Bands migrating slightly slower than that of preLolA represent nonspecific proteins extensively expressed from the plasmid (indicated by asterisks).
To determine the region of mLolB involved in the interaction with LolA, amber mutants of mLolB and wild-type LolA were overexpressed in BL21(DE3)/pSup-BpaRS-6TRN(D286R) cells in the presence of pBPA, the cells were irradiated with UV light, and the gene products subjected to SDS/PAGE analysis (Fig. 2B). Among 25 positions mutated to yield amber codons, pBPA introduced at only 4 positions, 40, 45, 49, and 65 (Fig. 1B, residues in red letters), caused the generation of cross-linked products with LolA in an irradiation-dependent manner. These 4 residues were also located at the entrance of mLolB (Fig. 1B, pink oval), in which a mLolB crystal was previously found to contain polyethylene glycol monomethylether used in the crystallization process (7). Positions 39 and 46 are also located in the entrance area of mLolB (Fig. 1B), but pBPA at them did not generate cross-linked products (Fig. 2B). The side chains of the residues at 4 positions, 40, 45, 49, and 65, were oriented toward the outside whereas those at positions 39 and 46 were oriented toward the inside (Fig. 1B). It is therefore suggested that mLolB interacts with LolA on its external surface.
In vivo photo-cross-linking analyses thus revealed that the internal surface of the LolA entrance interacts with the external surface of the mLolB entrance (see Fig. 5A).
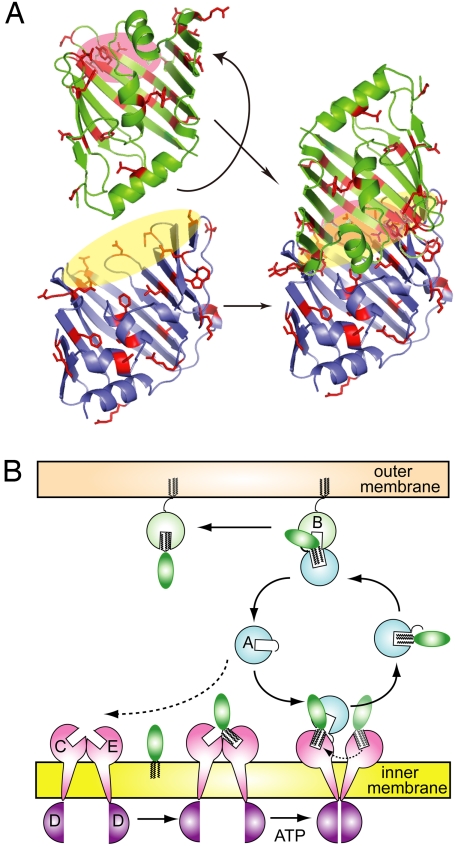
Fig. 5.
A mouth-to-mouth lipoprotein transfer model. (A) The LolA (lower) and mLolB (upper) molecules shown in Fig. 1A were rearranged to connect the respective entrances indicated by yellow and pink ovals. (B) As discussed in the text and Fig. S5, LolC and LolE might have hydrophobic cavities similar to those of LolA and LolB. It is speculated that LolE first binds a lipoprotein, which is then transferred to LolC. LolA preferentially interacts with liganded LolCDE and accepts a lipoprotein from LolC in an ATP-dependent manner. The LolA-lipoprotein complex crosses the periplasm and reaches LolB in the outer membrane. The lipoprotein transfer from LolA to LolB occurs in a mouth-to-mouth manner. A similar transfer mode may be applicable to the lipoprotein transfer from LolCDE to LolA.
Interaction with Pal.
It was recently shown that the hydrophobic cavity of LolA opens and closes upon the binding and release of lipoproteins, respectively (8). Moreover, LolA complexed with the outer membrane lipoprotein Pal was found to be accumulated in the periplasm when they were co-overexpressed from an efficient plasmid (16). To examine the interaction between LolA and Pal, we took advantage of this efficient expression system and introduced pBPA into the amber mutants of LolA, and then performed in vivo photo-cross-linking. The cross-linked products reacted with both anti-Pal (Fig. 3A) and anti-LolA antibodies and were detected at migration positions corresponding to ≈40 kDa. LolA derivatives possessing pBPA at positions 20, 24, 33, 43, 59, 72, 90, 93, 99, 127, 144, 176, and 180 (Fig. 1A, circled residues) were cross-linked with Pal. As expected, the side chains of these residues, except that at position 99, are located inside of the hydrophobic cavity and include 5 positions at which pBPA cross-linked with mLolB (Fig. 1A, residues in red letters).
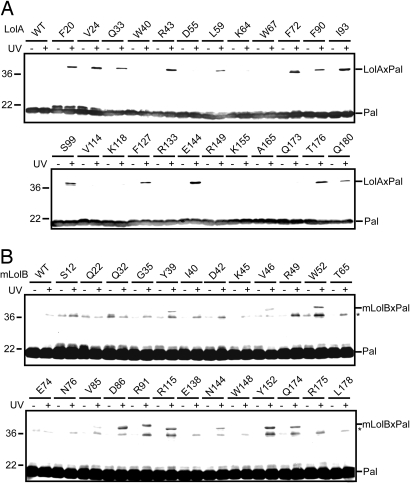
Fig. 3.
In vivo photo-cross-linking between Pal and LolA or mLolB. (A) Cells expressing Pal-strep and LolA-FLAG derivatives having pBPA in place of the indicated residues were UV-irradiated for 5 min as in Fig. 2A, and then subjected to SDS/PAGE and immunoblotting with anti-Pal antibodies. As a control, cells expressing Pal-strep and wild-type (WT) LolA-FLAG were also examined. (B) Cells expressing LolA-FLAG, Pal-strep, and mLolB-His6 derivatives having pBPA in place of the indicated residues were UV-irradiated for 5 min as in Fig. 2A, and then subjected to SDS/PAGE and immunoblotting with anti-Pal antibodies. As a control, cells expressing wild-type (WT) mLolB-His6 instead of pBPA-containing derivatives were also examined. Judging from the molecular masses of mLolB-His6 containing pBPA (22.1 kDa) and Pal-strep (17.6 kDa), the bands migrating as ≈40 kDa proteins represent the cross-linked products. In contrast, the bands migrating as ≈36 kDa proteins, indicated by asterisks, presumably represent the cross-linked Pal dimer, which was generated from a small amount of Pal containing mis-incorporated pBPA and detected upon extensive visualization of the blots.
To examine the interaction between mLolB and Pal, amber mutants of mLolB, Pal, and LolA were simultaneously overexpressed in the presence of pBPA. Co-expression of LolA was expected to facilitate the formation of the mLolB-Pal complex. Irradiated cells were then analyzed by SDS/PAGE and immunoblotting with anti-Pal (Fig. 3B) and anti-LolB antibodies. Cross-linked products, which migrated to positions corresponding to ≈40 kDa, were generated from mLolB derivatives with pBPA at positions 39, 46, 52, 85, 86, 91, 115, 144, 152, and 174 (Fig. 1B, circled residues). The residues at all these positions except 85 and 91 have side chains located inside of the hydrophobic cavity of mLolB, indicating that the hydrophobic cavity of mLolB is the binding site for the acyl chains of lipoproteins. Strikingly, however, 4 positions at which pBPA cross-linked with LolA (Fig. 1B, residues in red letters) were not included because the side chains of the residues at these positions were located outside of the mLolB molecule. The pBPA molecule introduced at position 85 or 91 might be cross-linked with the protein moiety of Pal or its acyl chains exposed to the outside of the molecule, because they are located near the entrance.
Interaction Between LolCDE and LolA.
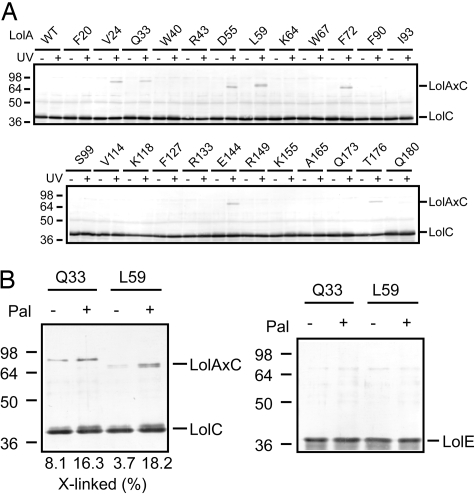
The LolCDE complex comprises 1 molecule each of membrane subunits LolC and LolE, and 2 copies of the ATP-binding subunit LolD (17). The predicted membrane topologies of LolC and LolE are very similar, i.e., both proteins span the membrane 4 times with a large region exposed to the periplasm. Although both LolC and LolE are required for growth (18), lipoprotein-releasing activity can be reconstituted into proteoliposomes with LolE and LolD without LolC (19), suggesting that the 2 membrane subunits play different roles in the lipoprotein sorting reaction. To determine whether LolA interacts with LolC or LolE or both, cells co-expressing LolCDE and amber mutants of LolA in the presence of pBPA were irradiated. Cross-linking was examined by SDS/PAGE and immunoblotting with anti-LolC (Fig. 4A), -LolA, -LolE, and -LolD antibodies. Despite the sequence similarity (26% identical) between LolC and LolE, only LolC generated cross-linked products with LolA. The migration positions of the cross-linked products slightly differed depending on the LolA derivative, as observed when the SecY-SecA interaction was examined by means of the same photo-cross-linking technique (20). LolA derivatives with pBPA at positions 24, 33, 55, 59, 72, 144, and 176 cross-linked with LolC (Fig. 1A, indicated by asterisks). These positions are located at the entrance of LolA, and 4 of them (Fig. 1A, residues in red letters) are involved in the interaction with mLolB, indicating that LolA utilizes an almost identical region for interaction with both LolC and LolB. LolD did not generate cross-linked products with LolA, as expected from its localization on the cytoplasmic side of the inner membrane. On the other hand, it is remarkable that LolE did not generate cross-linked products with LolA despite its structural similarity to LolC. These results indicate that the 2 membrane subunits indeed have distinct functions. This is discussed in more detail later.
Fig. 4.
In vivo and in vitro photo-cross-linking between LolA and LolCDE. (A) Cells expressing LolCDE and LolA-FLAG derivatives having pBPA in place of the indicated residues were UV-irradiated for 5 min as in Fig. 2A, and then subjected to SDS/PAGE and immunoblotting with anti-LolC antibodies. As a control, cells expressing LolCDE and wild-type (WT) LolA-FLAG were also examined. The cross-linked products generated from LolA and LolC (LolAxC) are indicated. (B) The purified LolA-FLAG derivative (0.32 μg) having pBPA in place of Q33 or L59 was UV-irradiated for 5 min with purified free-LolCDE (2.1 μg) or Pal-LolCDE (2.4 μg) in 0.01% DDM, as described in Materials and Methods. Samples were analyzed by SDS/PAGE and immunoblotting with anti-LolC (Left) or -LolE (Right) antibodies. The amounts of cross-linked LolC were densitometrically determined and expressed as percentages taking the total amount of LolC as 100.
LolA Preferentially Interacts with Liganded LolCDE.
Molecular mechanisms underlying the LolCDE-dependent lipoprotein release have been reported in detail (10, 11). Briefly, LolCDE interacts with outer membrane-directed lipoproteins in the inner membrane before ATP-binding, which causes an increase in the affinity of LolD for ATP. Subsequent ATP-binding to LolD causes a conformational change of LolC/LolE subunits and decreases the hydrophobic interaction between LolCDE and lipoproteins. ATP hydrolysis by LolD further causes a conformational change of LolC/LolE, enabling the transfer of lipoproteins to LolA, which accompanies the opening of the hydrophobic cavity. However, it was not known whether lipoprotein binding to LolCDE affects the interaction with LolA. To address this issue, the interaction between LolA and free or liganded LolCDE was examined in vitro. The LolCDE-Pal complex and free LolCDE were purified as reported (10) and then incubated with the purified LolA derivative with pBPA at position 33 or 59 in 0.01% n-dodecyl-β-d-maltopyranoside (DDM). Photo-cross-linking was analyzed by SDS/PAGE and immunoblotting after UV irradiation (Fig. 4B). The amount of the cross-linked product of LolA and LolC was higher with either LolA derivative when LolCDE contained Pal. It should be noted that no cross-linked product of LolA and LolE was generated. Moreover, the LolA-LolC cross-linking was completely dependent on UV irradiation. From these results, it is concluded that lipoprotein binding to LolCDE enhances the interaction between LolCDE and LolA.
Discussion
LolA and mLolB derivatives containing pBPA were functional and generated in vivo photo-cross-linked products with Lol proteins and lipoproteins. Based on the observations reported here, we propose that the transfer of lipoproteins from the inner to the outer membrane through Lol proteins takes place in a mouth-to-mouth manner (Fig. 5).
The crystal structures of free LolA, mLolB (7), and LolA(R43L) (8) together with the results of biochemical analyses (8, 9, 21) strongly suggest that LolA and LolB accommodate the acyl chains of lipoproteins in their hydrophobic cavities. Furthermore, the hydrophobic cavity of LolA was found to undergo opening and closing upon the binding and release of lipoproteins (8). It was then speculated that the hydrophobic cavity and its conformational change are critically important for the rapid and one-way transfer of lipoproteins from the inner to the outer membrane. The open conformation of a LolA(R43L) crystal (8) and an mLolB crystal containing polyethylene glycol monomethylether (7) revealed the entrance of the hydrophobic cavity through which lipoproteins are thought to enter the cavity. The study shown here revealed that this entrance plays a fundamental role in the interaction between LolA and LolB, and between LolA and LolC. Differences in the electrostatic properties of LolA (pI = ≈6) and mLolB (pI = ≈9) may be important for the interaction between these 2 proteins (7). Indeed, the entrance of LolA is enriched with negatively charged residues whereas that of mLolB has many positively charged residues, whose side chains are externally oriented (Fig. S4). When the hydrophobic cavities of LolA and LolB are aligned through connection of their entrances as proposed here, the distance between the acyl chain binding sites of LolA and LolB will be minimal, allowing very efficient transfer. The hydrophobic interaction between LolB and lipoproteins is stronger than that between lipoproteins and LolA, which drives the transfer of lipoproteins from LolA to LolB (9). Moreover, the hydrophobic cavity of LolA closes upon the transfer of lipoproteins, thereby preventing the backward transfer of lipoproteins.
According to the predicted membrane topology, each of LolC and LolE has a periplasmic region comprising ≈200 residues (17), which is slightly larger than LolA (182 residues) and LolB (186 residues) molecules. The periplasmic regions of both LolC (Fig. S5) and LolE exhibit ≈19% sequence identity with LolB, whereas the sequence identity between LolA and LolB is ≈14%, suggesting that the 2 membrane subunits have the LolA/LolB fold in their periplasmic regions. It seems therefore likely that the lipoprotein transfer from LolCDE to LolA also occurs in a mouth-to-mouth manner. It is noteworthy that LolA cross-linked with LolC but not LolE, although these 2 subunits are similar in structure. We previously reported that lipoprotein-releasing activity could be reconstituted into proteoliposomes from LolD and LolE without LolC (19). Taking these observations together, we speculate that lipoproteins are first accommodated in LolE and then transferred to LolC, which LolA targets to accept lipoproteins. This targeting step might be skipped in the reconstituted proteoliposomes because the reaction was examined in the presence of an excess amount of LolA.
The mode of interaction between LolA and mLolB was examined in the absence of overexpression of lipoproteins, which causes a conformational change of LolA (8, 21). The in vitro interaction between LolA and LolC was stimulated when the LolCDE complex bound Pal (Fig. 4). Because lipoproteins are rapidly transferred through Lol proteins, it is difficult to examine the effects of lipoprotein binding on the interaction between Lol proteins in vivo. However, the co-overproduction of Pal did not affect the mode of interaction between LolA and mLolB, indicating that the interaction site revealed by this study also functions when the Lol protein binds lipoproteins. Recent NMR analysis also revealed that the LolA-mLolB interaction occurs at a site which is essentially the same as that revealed here.
Materials and Methods
Materials.
A photo-reactive phenylalanine analogue, pBPA, was purchased from Bachem. TALON Co2+ affinity resin (Clontech) and FLAG M2 affinity gel-FLAG peptides (Sigma) were used to purify His6-tagged and FLAG-tagged proteins, respectively. DDM was purchased from Dojindo Laboratories. Antibodies against LolA (5), LolB (6), LolD (18), and Pal (22) were raised in rabbits as described. Anti-LolC and -LolE antibodies were raised in rabbits against in vitro synthesized peptides corresponding to the periplasmic regions of LolC and LolE (M48-G268 of LolC and M48-Y273 of LolE).
Bacterial Strains and Media.
E. coli strain BL21(DE3) [F−, dcm, ompT, hsdS(rB−mB−), gal, (λ DE3)] (23) was used for in vivo photo-cross-linking experiments and purification of proteins. DH5α (24) was used for gene manipulation. Cells were grown on LB broth. When required, 25 μg/mL chloramphenicol, 50 μg/mL ampicillin, or 50 μg/mL spectinomycin was added.
Construction of Plasmids.
To construct pOS141 carrying lolA-FLAG and pal-strep under PT7lac, the NcoΙ-EcoRΙ fragment carrying lolA-FLAG of pSS1 (PT7lac-lolA-FLAG) (16) was inserted into the same sites of pSS4–1 (PT7lac-pal-strep) (10). To construct pSS24 carrying mlolB-his under a T7 promoter and an lac operator, the lolB gene was amplified by PCR using the primers shown in Table S1 with the HMS174(DE3) [F−, recA, hsd(rk−mk+), rif, (λ DE3)] (25) chromosome as a template. The amplified fragment was digested with NcoI and XhoI and then inserted into the same sites of pET-22b(+) (Novagen). The plasmid thus constructed encodes mLolB-His6 fused to the PelB signal sequence. Mature mLolB expressed from this plasmid had M-A in place of C at the N terminus.
To construct pSS1-Xamber and pSS24-Xamber encoding amber mutants of FLAG-tagged LolA and His6-tagged mLolB, respectively, the specified codons at position X were replaced by the amber (TAG) codon, using a QuikChange site-directed mutagenesis kit (Stratagene) with pSS1 and pSS24 (PT7lac-mlolB-his) as templates. The pairs of oligonucleotides shown in Table S2 and Table S3 were used to construct pSS1-Xamber and pSS24-Xamber, respectively. pOS141-Xamber (PT7lac-lolA-FLAG-Xamber -pal-strep) was constructed as was pOS141 from pSS1-Xamber and pSS4–1. To construct pBADCDE carrying PBAD-lolCDE, the BamHI-HindIII fragment of pNASCDE (18) was inserted into the same sites of pBADHisA (Invitrogen). All of the mutations were confirmed by sequencing.
In Vivo Photo-Cross-Linking.
The reported method (20) was slightly modified. The BL21(DE3) cells harbored a plasmid, pSup-BpaRS-6TRN(D286R), encoding an amber suppressor tyrosyl tRNA and the engineered tyrosyl tRNA synthetase derived from M. jannaschii for the incorporation of pBPA into the amber codon. The cells harbored an additional 2 plasmids, one encoding an amber mutant of LolA-FLAG (pSS1-Xamber) or mLolB-His6 (pSS24-Xamber), and the other encoding the specified target proteins. When the interaction between LolA and Pal was examined, cells harbored a plasmid encoding both LolA-FLAG amber mutants, and Pal-strep (pOS141-LolA-FLAG-Xamber-Pal-strep) and pSup-BpaRS-6TRN(D286R). Cells were grown at 37 °C on LB broth supplemented with 1 mM pBPA to the midlog phase, and then the expression of amber-mutated proteins and target proteins were induced by the addition of 10 μM IPTG. LolCDE expression was induced at 30 °C with 10 μM IPTG and 0.2% arabinose. Aliquots (100–300 μL) of the cultures were transferred to micro titer plates, followed by irradiation with UV light at 365 nm for 5 min by using B-100AP (UV Products) at room temperature. The cells were harvested by centrifugation at 16,000 × g for 2 min, and then analyzed by SDS/PAGE and immunoblotting with antibodies against the specified proteins.
Purification of LolA Containing pBPA, Free LolCDE and Pal-LolCDE.
For in vitro photo-cross-linking studies, a FLAG-tagged LolA derivative containing pBPA at position 33 or 59 was overexpressed in BL21(DE3) cells harboring pSup-BpaRS-6TRN(D286R) and pSS1–33amber or pSS1–59amber, respectively. Cells were grown on LB-broth supplemented with 1 mM pBPA, 50 μg/mL spectinomycin, and 25 μg/mL chloramphenicol. When A660 of the culture reached 0.8, the cells were induced by the addition of 50 μM IPTG for 1.5 h at 37 °C, followed by conversion to spheroplasts. Periplasmic fractions obtained as spheroplast supernatants were adsorbed to an anti-FLAG M2 affinity column, and eluted with 50 mM Tris-HCl (pH 7.5) containing 100 μg/mL FLAG peptides. FLAG-tagged LolA proteins were purified to homogeneity as described previously (15) and kept at −80 °C in 50 mM Tris-HCl (pH 7.5) containing 10% glycerol after dialysis.
Free LolCDE and Pal-LolCDE were purified as reported (10) with a slight modification. BL21(DE3) cells harboring pKM402 carrying lolC and lolD-his under PBAD, pKM301 carrying lolE under tacPO, and pSS4–1 carrying pal-strep under a T7 promoter and an lac operator were grown on LB supplemented with 50 μg/mL ampicillin, 25 μg/mL chloramphenicol, and 50 μg/mL spectinomycin at 30 °C. When A660 reached 0.8, LolC and LolD were induced by the addition of 0.2% arabinose. After 2 h, LolE and Pal were induced with 1 mM IPTG for 1 h. A membrane fraction was prepared from the cells and then solubilized with 50 mM Tris-HCl (pH 7.5) containing 1% DDM, 5 mM MgSO4, and 10% glycerol with or without 2 mM ATP, for 30 min on ice. Pal-bound LolCDE was purified in the absence of ATP, whereas free LolCDE was obtained in the presence of ATP, as reported (10). Supernatants containing free LolCDE or Pal-LolCDE obtained by centrifugation at 100,000 × g for 40 min were applied on a TALON column equilibrated with buffer A (50 mM Tris-HCl, pH 7.5, containing 300 mM NaCl, 0.01% DDM, and 10% glycerol). After washing the column with buffer A supplemented with 10 mM imidazole, LolCDE and Pal-LolCDE were eluted with a linear gradient of imidazole (10–250 mM) in buffer A. The Pal-LolCDE complex was further purified through an anion exchange column of MonoQ (GE Healthcare) that had been equilibrated with 50 mM Tris-HCl (pH 7.5) containing 0.01% DDM and 10% glycerol. The complex was eluted with a linear gradient (0.1–0.5 M) of NaCl. The purified LolCDE and Pal-LolCDE were dialyzed against 50 mM Tris-HCl (pH 7.5) containing 0.01% DDM and 10% glycerol, and then kept at −80 °C.
In Vitro Photo-Cross-Linking.
Purified LolA-FLAG containing pBPA (0.5 μM, 0.32 μg) was incubated with free LolCDE (0.5 μM, 2.1 μg) or Pal-LolCDE (0.5 μM, 2.4 μg) in 30 μL of 50 mM Tris-HCl (pH 7.5) containing 0.01% DDM and 10% glycerol on ice. The mixtures were irradiated with UV light for 5 min as described for the in vivo photo-cross-linking. UV-irradiated samples were analyzed by SDS/PAGE and immunoblotting with antibodies against LolA, LolC, LolD, and LolE.
Other Methods.
SDS/PAGE was carried out according to Laemmli (26). Densitometric quantification was performed with an ATTO Densitograph.
Supplementary Material
Acknowledgments.
We thank Dr. Schultz (The Scripps Research Institute) for the pSup-BpaRS-6TRN(D286R), Dr. Shin-ichi Matsuyama (Rikkyo University) for the construction of E. coli KTA2, and Drs. Hiroyuki Mori (Virus Institute of Kyoto University) and Shoji Watanabe (RIKEN Institute) for valuable discussions. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan (to H.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900896106/DCSupplemental.
References
- 1.Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem. 2004;279:39807–39817. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 3.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1693:5–13. doi: 10.1016/j.bbamcr.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Braun V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta. 1975;415:335–377. doi: 10.1016/0304-4157(75)90013-1. [DOI] [PubMed] [Google Scholar]
- 5.Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda K, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 2003;22:3199–3209. doi: 10.1093/emboj/cdg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oguchi Y, et al. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J Biol Chem. 2008;283:25414–25420. doi: 10.1074/jbc.M804736200. [DOI] [PubMed] [Google Scholar]
- 9.Taniguchi N, Matsuyama S, Tokuda H. Mechanisms underlying energy-independent transfer of lipoproteins from LolA to LolB, which have similar unclosed β-barrel structures. J Biol Chem. 2005;280:34481–34488. doi: 10.1074/jbc.M507388200. [DOI] [PubMed] [Google Scholar]
- 10.Ito Y, Kanamaru K, Taniguchi N, Miyamoto S, Tokuda H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol Microbiol. 2006;62:1064–1075. doi: 10.1111/j.1365-2958.2006.05378.x. [DOI] [PubMed] [Google Scholar]
- 11.Taniguchi N, Tokuda H. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J Biol Chem. 2008;283:8538–8544. doi: 10.1074/jbc.M800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 14.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S, Matsuyama S, Tokuda H. Roles of the hydrophobic cavity and lid of LolA in the lipoprotein transfer reaction in Escherichia coli. J Biol Chem. 2006;281:3335–3342. doi: 10.1074/jbc.M509596200. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe S, Oguchi Y, Yokota N, Tokuda H. Large-scale preparation of the homogeneous LolA lipoprotein complex and efficient in vitro transfer of lipoproteins to the outer membrane in a LolB-dependent manner. Protein Sci. 2007;16:2741–2749. doi: 10.1110/ps.073101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 18.Narita S, Tanaka K, Matsuyama S, Tokuda H. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J Bacteriol. 2002;184:1417–1422. doi: 10.1128/JB.184.5.1417-1422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanamaru K, Taniguchi N, Miyamoto S, Narita S, Tokuda H. Complete reconstitution of an ATP-binding cassette transporter LolCDE complex from separately isolated subunits. FEBS J. 2007;274:3034–3043. doi: 10.1111/j.1742-4658.2007.05832.x. [DOI] [PubMed] [Google Scholar]
- 20.Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA. 2006;103:16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe S, Oguchi Y, Takeda K, Miki K, Tokuda H. Introduction of a lethal redox switch that controls the opening and closing of hydrophobic cavity in LolA. J Biol Chem. 2008;283:25421–25427. doi: 10.1074/jbc.M804737200. [DOI] [PubMed] [Google Scholar]
- 22.Tajima T, Yokota N, Matsuyama S, Tokuda H. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 1998;439:51–54. doi: 10.1016/s0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 23.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies of transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Campbell JL, Richardson CC, Studier FW. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci USA. 1978;75:2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.