Abstract
Noncoding RNAs (ncRNAs) are now recognized as transregulators of eukaryotic transcription, a role once attributed exclusively to protein factors. Two ncRNAs in mammalian cells have been shown to repress general mRNA transcription by RNA polymerase II (Pol II) in response to heat shock: mouse B2 RNA and human Alu RNA. B2 and Alu RNAs bind directly and tightly to Pol II and co-occupy the promoters of repressed genes along with the polymerase. Here, we identified the molecular mechanism by which mouse B2 RNA and human Alu RNA repress Pol II transcription. Biochemical assays to probe the network of protein–DNA interactions at the promoter revealed that B2 and Alu RNAs prevent Pol II from establishing contacts with the promoter both upstream and downstream of the TATA box during closed complex formation. Disruption of these contacts correlates with transcriptional repression. We conclude that B2 and Alu RNA prevent Pol II from properly engaging the DNA during closed complex formation, resulting in complexes with an altered conformation that are transcriptionally inert. In the absence of its normal contacts with the promoter, Pol II is likely held in these inactive complexes on DNA through interactions with promoter-bound TATA box-binding protein and transcription factor IIB.
Keywords: noncoding RNA, closed complex, open complex
Transcription is an intricate biological process in which DNA is copied into RNA; it is the critical first step in gene expression. In eukaryotes, the enzyme RNA polymerase II (Pol II) transcribes protein-encoding genes into mRNA with assistance of general transcription factors (GTFs; specifically TFIIA, TFIID, TFIIB, TFIIF, TFIIE, and TFIIH) that are thought to function at most promoters (1). Transcriptional regulation of specific genes occurs through the remarkably balanced interplay of auxiliary factors such as promoter-specific activators and repressors, coregulators, and chromatin-modifying complexes (1). Traditionally, it was thought that all of these factors were proteins, but now it is becoming clear that noncoding RNAs (ncRNAs) also play important roles in regulating transcription. Indeed, diverse ncRNAs have been identified as regulators of nearly every step in the process of mRNA transcription from controlling chromatin structure through regulating transcript elongation (2).
Our laboratory reported that 2 ncRNAs, mouse B2 RNA and human Alu RNA, repress mRNA transcription by binding to Pol II during the cellular heat shock response (3, 4). B2 and Alu RNAs are transcribed by RNA polymerase III from short interspersed elements (SINEs) (5). Upon heat shock, the levels of B2 RNA and Alu RNA increase (6, 7) and they function as general repressors of mRNA transcription (3, 4). Biochemical experiments showed that B2 and Alu RNAs bind directly to core Pol II with low nM affinity (4, 8). Other SINE RNAs have been identified in mammalian cells, including mouse B1 RNA and human scAlu RNA (5), the latter of which is likely derived from cleavage of full-length Alu RNA (9). The biological functions for B1 and scAlu RNAs are not known. In vitro both of these ncRNAs bind tightly to Pol II; however, they do not repress transcription (4).
Understanding the mechanisms by which B2 RNA and Alu RNA repress Pol II transcription would provide insight into this relatively new means of transcriptional control by 2 ncRNAs that are very different in sequence and overall secondary structure, yet share the same biological function. EMSAs showed that complexes containing Pol II, GTFs, and B2 RNA or Alu RNA can assemble on promoter DNA (4, 8). The polymerase in these complexes is transcriptionally inactive; all RNA synthesis is repressed. Consistent with this biochemical data, in mouse and human cells we found that B2 RNA and Alu RNA co-occupy with Pol II the promoters of repressed genes after heat shock (4). Therefore, B2 RNA and Alu RNA both repress a step in transcription that occurs after Pol II enters complexes at promoters, but before or at the point of initiation; however, the step repressed was not identified.
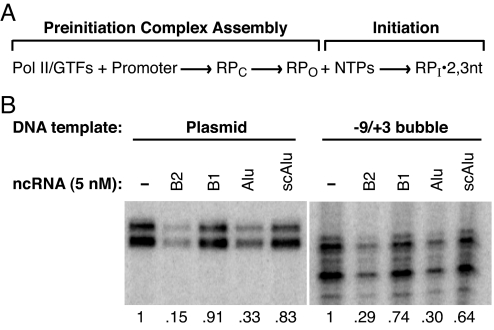
Fig. 1A shows a model for early steps in the transcription reaction, which provides a framework to determine the point at which B2 RNA and Alu RNA function. Pol II and the GTFs assemble on the promoter DNA to form an organized network of protein–protein and protein–DNA contacts that constitute a closed complex (10). Before complexes can initiate transcription, the promoter DNA melts around the start site to form the transcription bubble and the template strand enters the active site cleft on Pol II, resulting in open complexes (10–12). In vitro this can be facilitated by either the helicase TFIIH or negative supercoiling in the template DNA (13, 14). Next, transcription initiates and initiation complexes containing short (2–3 nt) RNAs form (13, 15). B2 RNA and Alu RNA, once incorporated into complexes at promoters, could potentially inhibit the formation of the proper protein–protein and/or protein-DNA contacts in a closed complex, the formation of open complexes, nucleotide binding, or synthesis of the first phosphodiester bond.
Fig. 1.
B2 RNA and Alu RNA repress transcription from a template containing a preformed transcription bubble. (A) A model depicting the early stages of the Pol II transcription reaction. RPC, closed complex; RPO, open complex; RPI2,3nt, initiation complex containing 2 or 3 nt RNA. (B) Transcriptional repression by B2 RNA and Alu RNA. (Left) Transcription from the AdMLP on a negatively supercoiled plasmid. (Right) Transcription from the AdMLP on a linear template containing a mismatched region from −9 to +3. Where indicated, ncRNAs (5 nM) were preincubated with Pol II, TFIIB, and TFIIF before the addition of promoter DNA with bound TBP. The number below each lane indicates the amount of transcript normalized to that produced in the absence of ncRNA.
The goal of this study was to identify the molecular mechanisms of transcriptional inhibition by B2 RNA and Alu RNA. We found that bypassing open complex formation by using promoters with a premelted transcription bubble did not alleviate transcriptional repression by B2 and Alu RNAs. To investigate how B2 and Alu RNAs affect interactions between proteins and the promoter DNA, we used UV cross-linking and DNase I footprinting. We found that B2 RNA and Alu RNA disrupted contacts between Pol II and the promoter DNA throughout the core promoter. Neither B1 RNA nor scAlu RNA had this effect. Moreover, the inhibitory effect of B2 RNA and Alu RNA was reversible; when RNase I removed the ncRNAs from complexes, contacts between Pol II and the promoter were restored. We conclude that B2 and Alu RNAs assemble into complexes at the promoter and prevent Pol II from establishing proper contacts with the DNA, thus rendering the complexes transcriptionally inert.
Results
B2 RNA and Alu RNA Repress Transcription from a Template Containing a Preformed Transcription Bubble.
Fig. 1B Left illustrates transcriptional repression by B2 and Alu RNAs in a highly purified human in vitro transcription system consisting of TATA box-binding protein (TBP), TFIIB, TFIIF, Pol II, and the adenovirus major late promoter (AdMLP) contained on a negatively-supercoiled plasmid. B1 and scAlu RNAs, included as negative controls, do not repress transcription. B2 and Alu RNAs repress transcription at a step occurring before or at the point of initiation (4, 8). Therefore, we considered the possibility that one or both of the ncRNAs inhibits promoter melting (formation of open complexes in Fig. 1A). We tested the ability of B2 RNA and Alu RNA to repress transcription from a heteroduplex promoter template, a linear template containing a mismatched region from −9 to +3 that simulates the bubble in an open complex (16). The presence of a preformed bubble alleviates the requirement for either TFIIH or negative superhelicity to observe promoter-specific transcription. We reasoned that if B2 RNA or Alu RNA represses transcription by blocking promoter melting, then the presence of the −9 to +3 bubble should abrogate transcriptional repression by the ncRNA. Fig. 1B Right shows that a premelted transcription bubble did not appreciably affect transcriptional repression by B2 and Alu RNAs. Therefore, bypassing the step of open complex formation does not alleviate transcriptional repression by either B2 RNA or Alu RNA.
Both B2 RNA and Alu RNA Block the Binding of Rpb1 and Rpb2 to Promoter DNA.
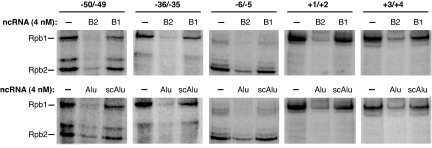
We next tested whether B2 and/or Alu RNA blocked transcription before open complex formation by preventing Pol II from establishing contacts with the promoter DNA within assembled complexes. To do so we monitored how the ncRNAs changed site-specific Pol II–DNA cross-linking. We constructed 5 32P-labeled AdMLP DNA fragments, each containing a single photoactivatable phenyl-azide located between positions −50 and −49, −36 and −35, −6 and −5, +1 and +2, or +3 and +4, all of which were found to cross-link to the Rpb1 and/or Rpb2 subunits of Pol II (17). On each of the DNA fragments we formed complexes containing TBP, TFIIB, TFIIF, and Pol II in the presence and absence of ncRNAs, UV-irradiated, and monitored cross-linking to the Rpb1 and/or Rpb2 subunits of Pol II. Because the complexes were assembled on linear DNA in the absence of TFIIE and TFIIH, promoter melting and open complex formation could not occur; hence these conditions allowed us to determine the effects of B2 and Alu RNAs on closed complex formation.
In the absence of B2 or Alu RNA, we observed cross-linking of the Rpb1 and/or Rpb2 subunits of Pol II to each template (Fig. 2), all of which we determined were TBP dependent. When B2 RNA or Alu RNA was present, the efficiency with which Rpb1 and Rpb2 cross-linked to all of the DNAs substantially decreased. Notably, B1 RNA and scAlu RNA did not appreciably affect the extent of cross-linking. These data show that Alu RNA and B2 RNA disrupt specific contacts between Pol II and promoter DNA at distinct positions from −50 to +4 on the AdMLP. Moreover, the disruption of cross-linking correlates with the ability of an ncRNA to repress transcription, the ncRNAs that repress transcription inhibit cross-linking, whereas the ncRNAs that bind Pol II but do not repress transcription do not inhibit cross-linking.
Fig. 2.
B2 RNA and Alu RNA block cross-linking of Pol II to promoter DNA. Experiments were performed with AdMLP DNAs that contained a photoactivatable cross-linker between the positions specified. ncRNAs (4 nM) were added to reactions before the assembly of closed complexes. Products were resolved by SDS/PAGE and visualized by Phosphorimagery. Bands containing DNA cross-linked to Rpb1 or Rpb2 are indicated.
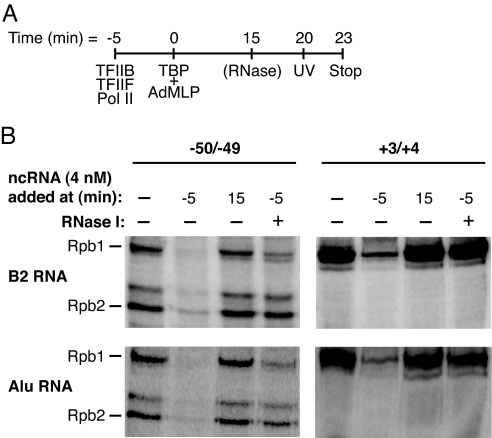
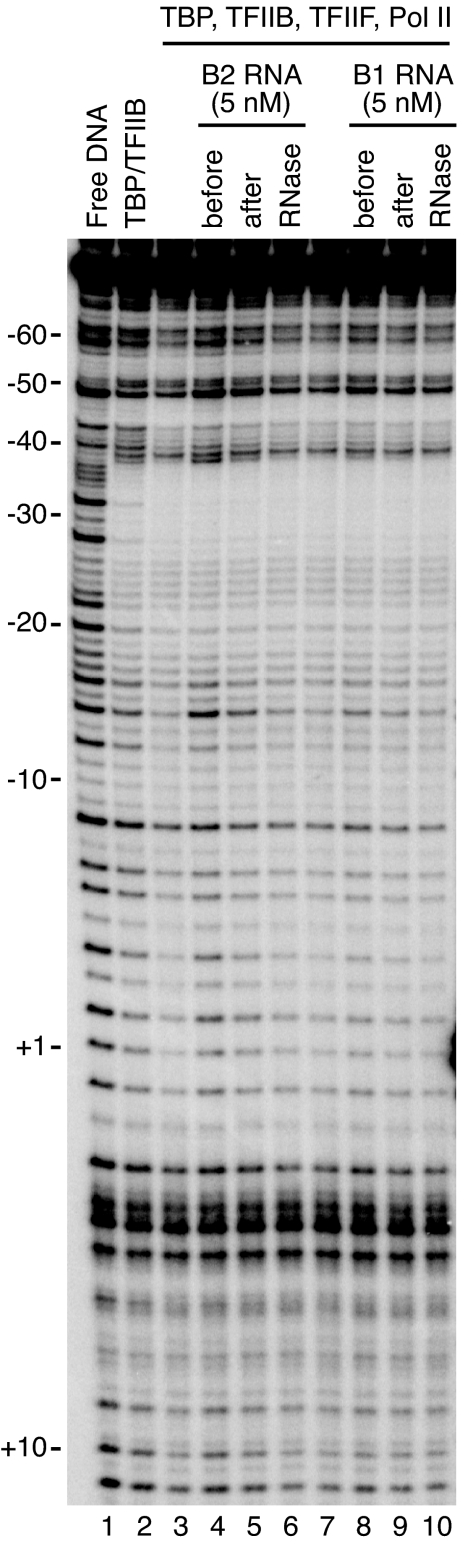
In previous work (4, 8) we showed that B2 and Alu RNAs inhibit transcription only when added to reactions before the assembly of complexes on the promoter; when ncRNAs were added to promoter-bound complexes containing Pol II and the general factors, inhibition was not observed. Additionally, we found that transcriptional repression by B2 RNA is reversible; when complexes were assembled in the presence of B2 RNA, then treated with RNase I to remove the B2 RNA, Pol II transcriptional activity was restored (8). We next tested whether inhibition of Pol II/DNA cross-linking correlated with these aspects of transcriptional repression. The schematic in Fig. 3A shows how reactions were assembled, with the inhibitory ncRNAs added at either the −5-min or 15-min time point (before or after closed complex formation, respectively). As shown in Fig. 3B, when B2 RNA (Upper) or Alu RNA (Lower) was added to reactions after closed complex had formed (15-min point), neither RNA affected cross-linking of Rpb1 or Rpb2 to DNA. Inhibition of cross-linking when the ncRNAs were added before closed complex formation (−5-min point) is shown as a positive control. Moreover, this inhibition is reversible; when either ncRNA was added to reactions before closed complex formation, then reactions were later treated with RNase I, cross-linking was restored. We conclude that the ability of B2 and Alu RNAs to inhibit specific cross-links between Pol II and the promoter correlates with their ability to repress transcription; both can occur only when the ncRNAs are added to reactions before the formation of closed complexes and both are reversible by RNase treatment.
Fig. 3.
B2 and Alu RNAs inhibit Pol II/promoter cross-links only under conditions in which they repress transcription. (A) A schematic showing the time course of assembly of reactions. ncRNAs were added at −5 min or 15 min. (B) Inhibition of cross-linking by B2 and Alu RNAs is reversible and only occurs when they are added to reactions before the formation of closed complexes. B2 RNA (Upper) and Alu RNA (Lower) were added to reactions at the points designated. Where indicated, reactions received 10 units of RNase I.
B2 RNA and Alu RNA Significantly Alter the Conformation of Complexes at the Promoter.
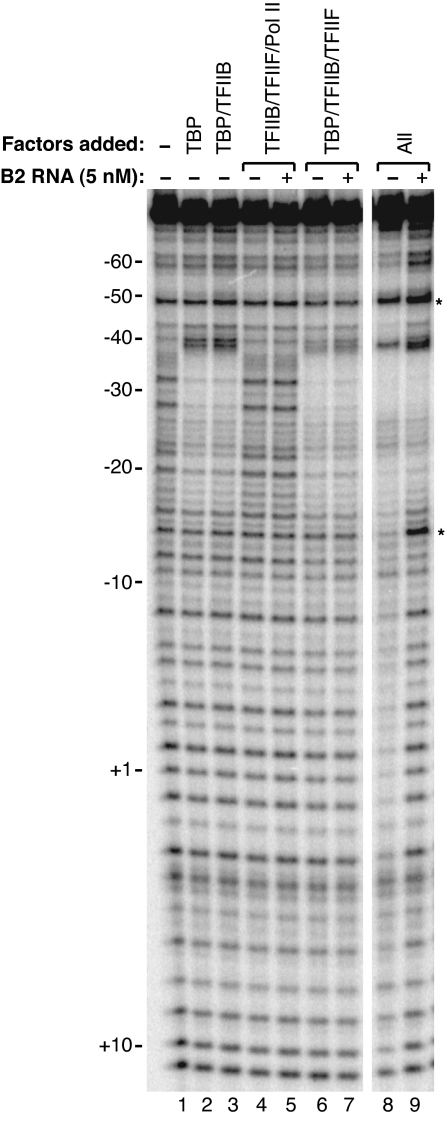
The cross-linking data show that B2 RNA and Alu RNA prevent Pol II from contacting the DNA at several positions. To obtain a broader view of the extent to which the ncRNAs disrupt protein–DNA contacts, we used DNase I footprinting (Fig. 4). We began by establishing conditions to observe a footprint of closed complexes on the AdMLP that contained TBP, TFIIB, TFIIF, and Pol II. TBP alone and the combination of TBP/TFIIB protected a relatively small region from −38 to −17 (Fig. 4, lanes 2 and 3 compared with lane 1). Including Pol II/TFIIF in the reactions resulted in additional protections extending from approximately −60 to +10 (Fig. 4, compare lanes 8 and lane 3). Addition of Pol II/TFIIF also enhanced the level of protection observed in the TATA box region. The extended footprint observed with all 4 factors required Pol II (Fig. 4, compare lanes 6 and 8), and when TBP was omitted, the remaining 3 factors caused no protection of the promoter DNA (Fig. 4, lane 4). The bands in several lanes were quantitated, and their intensities were plotted versus position on the gel (Fig. S1).
Fig. 4.
B2 RNA eliminates Pol II-specific protection from DNase I that is normally observed in closed complexes. TBP, TFIIB, TFIIF, Pol II, and B2 RNA (5 nM) were added to reactions as designated. The asterisks indicate bands that were enhanced in the presence of B2 RNA.
When B2 RNA was added to reactions before assembly of closed complexes, it dramatically changed the footprint (Fig. 4, compare lanes 9 and 8; these footprints are also compared in the middle plot of Fig. S1). In the presence of B2 RNA and all 4 factors, the footprint looked surprisingly similar to that observed with just TBP and TFIIB (Fig. 4, compare lanes 9 and 3; these footprints are also compared in the right plot of Fig. S1). The regions normally protected by Pol II/TFIIF (−60 to −38 and −18 to +10) were not protected when complexes were assembled in the presence of B2 RNA. Importantly, B2 RNA also caused enhanced DNase I cleavage at 2 positions (−48 and −14) and did not completely eliminate protection at positions −20 and −8. In addition, the enhanced protection of the TATA box region caused by Pol II/TFIIF is maintained in the presence of B2 RNA. These features of the footprint not only depended on the presence of B2 RNA, but required Pol II (Fig. 4, lane 7 versus lane 9) and TBP (Fig. 4, lane 5 versus lane 9), indicating that all 3 of these factors are present in the complex footprinted in Fig. 4, lane 9. We conclude that complexes assembled in the presence of B2 RNA contain all of the factors and have an altered conformation caused by the presence of the ncRNA, which allows increased accessibility of DNase I to the promoter in regions upstream and downstream of the TATA box. This increased accessibility to DNase I is likely the result of Pol II not properly contacting promoter DNA, which is consistent with the UV cross-linking results (Figs. 2 and 3).
We next tested whether the effect of B2 RNA on the DNase I footprint of a closed complex correlates with transcriptional repression. We performed additional experiments comparing the effect of B2 RNA with that of B1 RNA. In both cases, the ncRNAs were added to reactions either before or after closed complexes had formed, and in some reactions, complexes were treated with RNase I before footprinting. B2 RNA only affected the DNase I footprint of a complex under conditions in which transcriptional repression is known to occur (Fig. 5). The footprint of a closed complex is shown in Fig. 5, lane 3 and the effect of B2 RNA, when added to reactions before complexes form, is shown in Fig. 5, lane 4 (see Fig. S2A for a plot comparing these footprints). These data are similar to those described for Fig. 4 in which B2 RNA causes regions upstream and downstream of the TATA box to be susceptible to DNase I cleavage, while simultaneously causing enhancements at positions −48 and −14. By contrast, when B2 RNA was added to reactions after closed complexes had formed on the promoter, very little change in the footprint of the closed complex was observed (Fig. 5, lane 5 versus lane 3, and the middle plot of Fig. S2A). A similar result was obtained when complexes containing B2 RNA were treated with RNase I: this footprint looked nearly identical to that of the closed complex formed in the absence of B2 RNA (Fig. 5, lane 6 versus lane 3). Last, including B1 RNA in reactions did not substantially change the footprint under any of the conditions tested (Fig. 5, compare lanes 3 and 7 to lanes 8–10; the plot on the right in Fig. S2A compares the footprints in Fig. 5, lanes 3 and 8). We also performed DNase I footprinting experiments on complexes assembled in the presence of Alu RNA or scAlu RNA. As shown in Fig. S2B, the effects of Alu RNA and scAlu RNA mirror those of B2 RNA and B1 RNA, respectively. We conclude that the arrangement of protein–DNA contacts that occurs under conditions where B2 RNA or Alu RNA inhibit transcription results in transcriptional repression; this altered arrangement does not occur with an ncRNA that does not repress transcription.
Fig. 5.
B2 RNA reversibly alters the conformation of complexes assembled on DNA only under conditions in which it represses transcription. B2 RNA and B1 RNA were added to reactions before or after closed complex formation as indicated. When included, reactions received 10 units of RNase I.
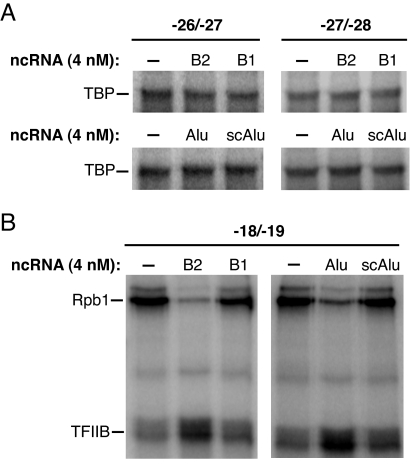
The DNase I footprinting data show that when complexes are assembled in the presence of B2 or Alu RNA, the ncRNA blocks the interaction of Pol II with promoter DNA; however, the protection over the TATA box region remains unchanged, indicating that TBP and perhaps TFIIB maintain interactions with promoter DNA in the presence of B2 and Alu RNAs. We directly probed for the effect of these RNAs on promoter/TBP and promoter/TFIIB contacts within inhibited complexes using site-specific UV cross-linking (17). As shown in Fig. 6A, the extent of TBP/promoter cross-linking at 2 positions in the TATA box was unchanged in complexes assembled in the presence of inhibitory RNAs (B2 RNA or Alu RNA) and noninhibitory RNAs (B1 RNA or scAlu RNA). Therefore, TBP is indeed present in inhibited complexes, and the manner in which it interacts with DNA is not affected by the ncRNAs. To monitor TFIIB/promoter interactions, we used a promoter that cross-links to either TFIIB or the Rpb1 subunit of Pol II (the photoactivatable group was between positions −18 and −19). Interestingly, B2 RNA and Alu RNA significantly enhanced cross-linking to TFIIB, while inhibiting cross-linking to Rpb1 (Fig. 6B). This effect was specific to the inhibitory ncRNAs because B1 RNA and scAlu RNA did not affect either the TFIIB or Rpb1 cross-link. We conclude that complexes assembled in the presence of repressor ncRNAs have an altered conformation that enhances TFIIB/promoter contacts while disrupting Pol II/promoter contacts.
Fig. 6.
Complexes assembled with B2 RNA and Alu RNA contain TBP and TFIIB and have an altered conformation. (A) ncRNAs do not affect site-specific TBP/promoter cross-linking. Single photoactivatable cross-linkers were located between the positions specified. ncRNAs (4 nM) were added to reactions before the assembly of closed complexes. Bands containing DNA cross-linked to TBP are indicated. (B) Inhibitory ncRNAs enhance TFIIB/promoter cross-linking and inhibit Rpb1/promoter cross-linking. The ncRNAs (4 nM) were added to reactions as indicated. Bands containing DNA cross-linked to TFIIB and Rpb1 are shown.
Discussion
Here, we investigated the mechanism by which mouse B2 RNA and human Alu RNA repress Pol II transcription. We first used a premelted template to bypass open complex formation and found that B2 and Alu RNAs still repressed transcription. Cross-linking and DNase I footprinting revealed that B2 and Alu RNAs change the molecular structure of the complex that assembles on a promoter; namely, they prevent Pol II from establishing many of its contacts with the DNA. Disruption of these contacts correlates tightly with transcriptional repression. Moreover, Pol II remains associated with promoter DNA in the presence of B2 and Alu RNAs, despite the lack of polymerase–DNA contacts. We conclude that B2 RNA and Alu RNA block the formation of closed complexes, thus resulting in transcriptional repression, as depicted in the model in Fig. 7.
Fig. 7.
Model depicting inhibitory ncRNAs blocking the formation of closed complexes by preventing the polymerase from properly engaging the DNA. See Discussion for a complete description. Abbreviations are as described in Figure 1A.
Closed complexes cannot properly form because B2 RNA and Alu RNA block interactions between Pol II and the promoter, which are essential for transcriptional activity. Both RNAs inhibited cross-linking between the 2 largest subunits of Pol II at all positions in the promoter tested. Both RNAs also largely eliminated the protection of promoter DNA from DNase I that is normally caused by the presence of Pol II/TFIIF in closed complexes, leading to a footprint that looked strikingly similar to that of TBP/TFIIB bound to the promoter. By contrast, B1 RNA and scAlu RNA, ncRNAs that in vitro bind Pol II but do not repress transcription, did not appreciably affect either cross-linking of Pol II to DNA or the DNase I footprint. Hence, the ability to inhibit the interaction of Pol II with promoter DNA is specific for repressor ncRNAs.
Although closed complexes cannot form in the presence of B2 and Alu RNAs, the inhibited complexes that do form (RP·ncRNA) appear to contain, in addition to the ncRNA, all of the protein factors and promoter DNA despite the lack of polymerase/promoter contacts. In the DNase I footprinting experiments, B2 and Alu RNA caused enhancements of bands at −48 and −14, and these enhancements required both TBP and Pol II, indicating that the ncRNA and both protein components are present in the inactive complexes. In addition, TBP/promoter and TFIIB/promoter cross-links were maintained in the presence of B2 or Alu RNA. Moreover, our previous EMSA studies showed that observing promoter-bound complexes containing B2 RNA or Alu RNA required Pol II and each general factor (4, 8). It is likely that Pol II is held in the inhibited complexes via its contacts with promoter-bound TBP/TFIIB. We found that B2 RNA blocked the assembly of artificial elongation complexes containing Pol II bound to an RNA:DNA hybrid (18). In the studies presented here, B2 RNA did not block complexes from assembling on promoter DNA despite the lack of promoter/Pol II contacts. This contrast can be explained by the presence of GTFs, which provide a network of interactions that hold inhibited complexes on the promoter in the absence of Pol II–DNA contacts. Such a network of contacts was not present during the assembly of the artificial elongation complexes (18). Interestingly, Escherichia coli 6S RNA has been shown to block bacterial RNA polymerase from binding promoter DNA (19). Here, we found that binding of an inhibitory ncRNA to Pol II also prevented interaction with promoter DNA; however, in the eukaryotic system the interactions between Pol II and the GTFs allowed complexes that contained promoter DNA to form. The single-stranded region of 6S RNA can engage the active site of bacterial RNA polymerase and serve as a template for synthesis of short RNA transcripts, which functions to derepress transcription (19). Future studies will likely reveal whether an analogous regulatory mechanism can occur with B2 and Alu RNAs.
Previously, we showed that B2 RNA and Alu RNA were unable to repress transcription when added to complexes that were preformed on the AdMLP contained on negatively-supercoiled DNA (4, 18). The complexes formed under these conditions were likely open complexes because they were transcriptionally active. The results presented here show that closed complexes are also resistant to the effects of B2 RNA and Alu RNA; addition of either RNA to closed complexes did not affect cross-linking or DNase I footprinting. Moreover, our data show that the ncRNA is not able to invade closed complexes once Pol II has engaged the DNA. In addition, once closed complexes form they are kinetically stable; if the reverse reaction occurred to an appreciable extent, B2 or Alu RNA would have blocked Pol II from re-engaging the promoter.
That B2 RNA and Alu RNA have apparently identical mechanisms of repression is somewhat surprising, given that these 2 ncRNAs are not similar in sequence or overall secondary structure (4, 18). The regions of B2 RNA and Alu RNA that bind to Pol II (Pol II binding domains) and those that repress transcription (repression domains) are distinct from one another (4, 18). For example, the Pol II binding domain of B2 RNA can bind Pol II and assemble into complexes at promoters, yet it does not repress transcription (18). Similarly, B1 RNA and scAlu RNA bind tightly to Pol II, but lack repression domains (4) and, as shown here, these ncRNAs do not interfere with Pol II/DNA contacts. Therefore, our data support a model in which it is the repression domains of B2 RNA and Alu RNA that exclude polymerase–promoter contacts. A crystal structure of a synthetic RNA aptamer that binds yeast Pol II and represses transcription shows that a region of the aptamer forms a double stem-loop structure in the DNA binding cleft of Pol II, which would preclude the template DNA strand from entering the cleft (20). The data presented here are consistent with a model whereby the repression domains of B2 and Alu RNAs block transcription by interacting with the DNA cleft of Pol II in a manner similar to the synthetic aptamer. Our data, however, also indicate that B2 and Alu RNAs cause a loss of Pol II contacts with DNA upstream of the TATA box, which is thought to be outside of the DNA binding cleft (21). Future studies will be needed to determine both where on the polymerase the Pol II binding domains of B2 RNA and Alu RNA dock and whether their repression domains indeed contact the DNA binding cleft.
Finding that B2 and Alu RNAs allow the formation of complexes on a promoter despite preventing the polymerase from engaging the DNA raises the possibility that such complexes form in cells and provides an intriguing means by which to regulate gene-specific transcription. We have found in heat-shocked cells that B2 RNA or Alu RNA, along with Pol II, occupy the promoters of repressed genes (4). Given the mechanism of repression described here, it is likely that Pol II has not engaged the DNA at these promoters. Moreover, in vitro removing the ncRNAs from complexes by RNase treatment restored contacts between Pol II and the promoter, thereby making these complexes transcriptionally active. A new mechanism for controlling gene-specific transcription in cells might exist whereby a factor that removes an ncRNA repressor would activate a prerecruited Pol II. Future studies will determine whether this type of regulation occurs and how widespread it may be in mammalian cells.
Materials and Methods
Plasmid Construction and RNA Preparation.
Construction of pUC-T7-B2, pUC-T7-B1, pUC-T7-Alu, and pUC-T7-scAlu, which encode B2 RNA, B1 RNA, Alu RNA, and scAlu RNA, respectively, was as described (3, 4). The ncRNAs were made by using in vitro transcription by T7 RNA polymerase as described (3).
In Vitro Transcription by Pol II.
Recombinant human TBP, TFIIB, TFIIF, and native human Pol II were prepared as described (22). DNA templates consisted of either negatively-supercoiled plasmid containing the AdMLP (−53 to +10) fused to an 90-bp G-less cassette or an 88-bp linear DNA containing the AdMLP with a mismatched region from −9 to +3 to create a transcription bubble. Reactions (20 μL) were assembled as described (18). Briefly, factors were used at the following final concentrations: 3.5 nM TBP, 10 nM TFIIB, 2 nM TFIIF, 1–3 nM Pol II, and 1 nM DNA template. DNA templates were preincubated with TBP at 30 °C for 4 min. TFIIB, TFIIF, Pol II, and ncRNA (when included) were incubated together in a separate tube at 30 °C for 4 min. The contents of these 2 tubes were mixed and incubated at 30 °C for 20 min to allow complexes to form, and then nucleotides were added to initiate transcription. Reactions were stopped after 20 min and RNA was resolved by denaturing PAGE.
Derivatized Promoter DNA Fragments.
The promoters used in Fig. 2 contained the AdMLP from −59 to +24 with a single phosphorothioate linkage. The modified linkages were contained within 5′ end-labeled PCR primers such that the products contained phosphorothioates between the following positions: −50 and −49 (nontemplate strand), −36 and −35 (nontemplate strand), −6 and −5 (template strand), +1 and +2 (template strand), and + 3 and +4 (template strand). The promoters used in Fig. 6 were assembled from annealed oligonucleotides (Invitrogen) containing single phosphorothioates between positions −27 and −28 (template strand), −26 and −27 (nontemplate strand), and −18/−19 (nontemplate strand). The phosphorothioates were derivatized with azidophenacyl bromide (AzBr) by incubating ≈500 ng of each DNA with 6 mM AzBr in a buffer containing 57% methanol and 35 mM potassium phosphate (pH 7.2) overnight in the dark at room temperature (23). Derivatized DNA was ethanol-precipitated; the efficiency of the derivatization reactions was 90% or more. EMSAs established that the derivatized promoter DNA fragments formed complexes with Pol II and the GTFs similarly to the underivatized promoter.
Site-Specific Protein–DNA Photocross-Linking.
Buffer conditions and concentrations of TFs were identical to those used for in vitro transcription, except DTT was omitted and the DNA template was 2 nM. Complexes (with or without ncRNAs) were formed at 30 °C in 20-μL reactions contained in polystyrene microcentrifuge tubes. The tubes were placed inside 13 × 100-mm borosilicate glass culture tubes to eliminate radiation with wavelengths <290 nm, then were UV-irradiated for 3 min at room temperature (≈3,000 μW/cm2) using a Stratagene UV Stratalinker 1800 equipped with 6 312-nm UV bulbs. After UV irradiation, bands were resolved by 5% SDS/PAGE and visualized with Phosphorimagery.
DNase I Footprinting.
DNase I footprinting was performed with a PCR-generated DNA fragment containing the AdMLP from −78 to +24 and a 32P-label on the 5′ end of the template strand. Buffer conditions and concentrations of TFs were identical to those used for in vitro transcription, except the DNA template was 0.5 nM. After forming complexes (with or without ncRNA), DNase I footprinting was performed by adding 2 μL of a solution containing 0.03 unit/μL of DNase I and 10 mM CaCl2 for 1 min at 30 °C. The reactions were stopped and processed as described (24).
Supplementary Material
Acknowledgments.
This work was supported by Public Health Service Grant R01 GM068414 from the National Institute of General Medical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810738106/DCSupplemental.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44:3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 4.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- 6.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 7.Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 9.Maraia RJ, Driscoll CT, Bilyeu T, Hsu K, Darlington GJ. Multiple dispersed loci produce small cytoplasmic Alu RNA. Mol Cell Biol. 1993;13:4233–4241. doi: 10.1128/mcb.13.7.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Carey M, Gralla JD. Polymerase II promoter activation: Closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 11.Holstege FCP, Fiedler U, Timmers HTM. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 14.Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 15.Luse DS, Jacob GA. Abortive initiation by RNA polymerase II in vitro at the adenovirus major late promoter. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 16.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 17.Kim TK, et al. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13:583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- 20.Kettenberger H, et al. Structure of an RNA polymerase II–RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat Struct Mol Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 21.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: A model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 22.Weaver JR, Kugel JF, Goodrich JA. The sequence at specific positions in the early transcribed region sets the rate of transcript synthesis by RNA polymerase II in vitro. J Biol Chem. 2005;280:39860–39869. doi: 10.1074/jbc.M509376200. [DOI] [PubMed] [Google Scholar]
- 23.Mayer AN, Barany F. Photoaffinity cross-linking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene. 1995;153:1–8. doi: 10.1016/0378-1119(94)00752-e. [DOI] [PubMed] [Google Scholar]
- 24.Galasinski SK, Lively TN, Grebe de Barron A, Goodrich JA. Acetyl-CoA stimulates RNA polymerase II transcription and promoter binding by TFIID in the absence of histones. Mol Cell Biol. 2000;20:1923–1930. doi: 10.1128/mcb.20.6.1923-1930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.