Abstract
The hereditary hearing–vision loss disease Usher syndrome (USH) is caused by defects in several proteins, most of which form an integrated protein network called Usher interactome. Harmonin/Ush1C is a master scaffold in the assembly of the Usher protein complexes, because harmonin is known to bind to every protein in the Usher interactome. However, the biochemical and structural mechanism governing the Usher protein complex formation is largely unclear. Here, we report that the highly-conserved N-terminal fragment of harmonin (N-domain) immediately preceding its PDZ1 adopts an autonomously-folded domain. We discovered that the N-domain specifically binds to a short internal peptide fragment of the cadherin 23 cytoplasmic domain. The structures of the harmonin N-domain alone and in complex with the cadherin 23 internal peptide fragment uncovered the detailed binding mechanism of this interaction between harmonin and cadherin 23. We further elucidated the harmonin PDZ domain-mediated cadherin 23 binding by solving the structure of the second harmonin PDZ domain in complex with the cadherin 23 carboxyl tail. The multidentate binding mode between harmonin and cadherin 23 provides a structural and biochemical basis for the harmonin-mediated assembly of stable tip link complex in the auditory hair cells.
Keywords: PDZ domains, scaffold protein, Usher syndrome
Usher syndrome (USH) is the most common form of hereditary hearing–vision loss in humans (1–3). Hearing loss in the USH patients originates from defects of sensory hair cells in ears, whereas blindness is caused by retinitis pigmentosa (RP). USH1 is the most severe form, characterized by severe to profound congenital hearing loss and prepubertal onset of RP. USH is clinically and genetically heterogeneous. To date, 5 genes (namely USH1B, USH1C, USH1D, USH1F, and USH1G) and 2 loci are identified for USH1, 3 genes for USH2 (USH2A, USH2C, and USH2D), and 1 gene (USH3A) and 1 locus for USH3 (1, 4, 5). Accumulating data indicate that the majority, if not all, of the Usher proteins form an integrated network of protein complexes (1, 2). In this network, the USH1C product harmonin functions as the mast scaffold in the assembly of the Usher protein complexes, because harmonin is capable of directly binding to all known Usher proteins (Fig. S1). However, the biochemical and structural mechanisms governing the Usher protein complex formation are largely unclear.
Hair cells are highly-specialized cells with hundreds of stereocilia projected from the apical surface of the cell body, and they are uniformly oriented. They are connected by different types of extracellular links. Cadherin 23 and Protocadherin 15, 2 adhesion molecules and products of USH1D and USH1E, form the tip link located at the extreme distal end of stereocilia (6, 7). The molecular mechanisms governing the ankle link and the basal link assemblies are not well characterized, but there is evidence showing several USH gene products including cadherin 23, VlgR, and Usherin are crucial for their formation (8–10). Studies using the animal models of USH showed that mutation of any one of the Usher genes affects the development of stereocilia at different degrees (11–14).
Harmonins can be divided into 3 isoforms, a, b, and c (15). Each isoform contains 2 N-terminal PDZ domains followed by 1 or 2 coiled-coil regions (Fig. S1). Harmonin a and b each contains an additional PDZ domain at its C-terminal end. Harmonin b also contains a F-actin-binding PST domain preceding the C-terminal PDZ domain (10). In the Usher proteins network, Cadherin 23, Protocadherin 15, Sans, VlgR, and Ush2C each contains a type I PDZ binding motif and binds to either the first or the second PDZ domains of harmonin (1, 6, 16, 17). The first PDZ domain of harmonin has been reported to bind to protein modules other than PDZ binding motif. These noncanonical harmonin PDZ1 binding domains include the globular tail domain of Myosin VIIa and the SAM domain from Sans (17). Such noncanonical PDZ/target interactions need to be verified because of the unprecedented PDZ domain-mediated target binding modes (18).
In this article, we discovered that the highly-conserved N-terminal fragment of harmonin adopts an autonomously-folded structure (termed the N-domain). We further discovered that the N-domain specifically binds to an internal peptide fragment in the cytoplasmic domain of cadherin 23. The molecular basis governing this interaction was elucidated by solving the structure of the complex. Together with the structure of harmonin PDZ2 in complex with the carboxyl tail of cadherin 23, we demonstrate that harmonin and cadherin 23 can form stable complex structures required at the functional tip links of hair cells via a multidentate binding mode. Our findings significantly expanded our knowledge on the action mode of harmonin, the master scaffold of the USH protein complex.
Results
Harmonin N-Terminal Domain Adopts an α-Helical Fold.
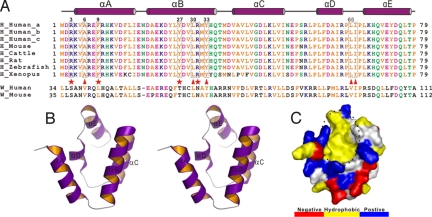
Current molecular analysis revealed that all 5 USH1 proteins are integrated into a protein network via binding to the PDZ domains of harmonin (16) (summarized in Fig. S1). As our first step in elucidating the molecular basis of harmonin-organized USH protein network, we carried out a sequence alignment analysis of harmonins from different species. Interestingly, we found that harmonin contains a highly-conserved, previously-uncharacterized, ≈90-residue peptide fragment preceding its first PDZ domain (Fig. 1A), indicating the potential functional importance of this region. The recombinant N-terminal region (residues 1–97, referred hereafter as the N-domain) was eluted at a volume indicative of a stable monomer on an analytical gel filtration chromatography, and the protein adopted a predominant α-helical structure as evaluated by CD spectroscopy. Additionally, the well-dispersed and homogeneous 1H-15N HSQC spectrum further indicated that the N-domain is well-folded (Fig. 2E). The structure of the N-domain was determined to high resolution by NMR spectroscopy. The domain contains 5 α-helices and adopts a compact 5-helix-bundle structure (Fig. 1B and Table S1). In the N-domain helix bundle, the αA- and αB-helices form a V-shaped helix-hairpin with a solvent-exposed cleft on one side of the hairpin and a hydrophobic core packing face (with αC and αD) on the other side (Fig. 1B). Interestingly, the solvent-exposed open cleft of the αA/αB-helix hairpin is hydrophobic, and the residues forming this exposed hydrophobic surface are highly conserved in harmonins (Fig. 1 A and C). We reasoned that this N-domain may function as a previously-uncharacterized protein–protein interaction-domain (see below for details). The αC- and αD-helices also form a V-shaped helix hairpin and packed with the αA/αB-helix hairpin to form the core of the bundle. The open side of the αC/αD-helix hairpin is capped by the αE-helix. A structural homology comparison using Dali (http://ekhidna.biocenter.helsinki.fi/dali_server) revealed that the harmonin N-domain represents a somewhat novel fold with no known protein structures with the same topology, although the arrangement of the first 4 helices (αA–αD) of the N-domain resembles the left-handed 4-helical bundle structures of the Sin3 PAH domain [Z score of 7.7; Protein Data Bank (PDB) entries for the PAH domain are 2CZY and 2RMR].
Fig. 1.
Harmonin contains a novel N-terminal domain. (A) Structure-based sequence alignment of the N-domain from the different harmonin family members. In this alignment, the conserved hydrophobic residues are shown in orange, negatively-charged residues are in magenta, positively-charged residues are in blue, and the rest of the highly-conserved residues are in green. The residues that are critical for binding to the cadherin 23 peptide are boxed and highlighted by red stars or triangles. For comparison, the N-terminal region of whirlin from human and mouse are also included. (B) Stereoview of a representative NMR structure of the harmonin N-domain drawn in the ribbon diagram. (C) Surface representation showing the existence of a unique exposed hydrophobic pocket in the N-domain (highlighted by a black circle), which is hypothesized to serve as the binding site for potential target proteins. In this presentation, the hydrophobic amino acid residues are drawn in yellow, the positively-charged residues are blue, the negatively-charged residues are red, and the uncharged polar residues are gray.
Fig. 2.
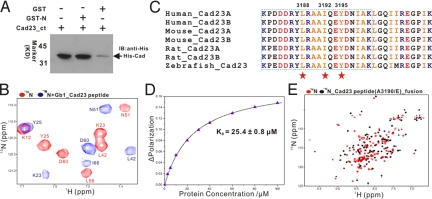
Harmonin N-domain binds to the cytoplasmic domain of cadherin 23. (A) Western blot analysis showing that GST-fused harmonin N-domain binds to the cytoplasmic domain of cadherin 23. Lane 1 represents cadherin 23 input. (B) Superposition plot of the 1H-15N HSQC spectrum of the free N-domain and the protein in the presence of a saturating amount of GB1-tagged cadherin 23 peptide. For clarity, only a selected region of each spectrum is shown. The chemical shift assignments for the free- and cadherin 23 peptide bound-forms of the N-domain are labeled. (C) Amino acid sequence alignment of the 31-residue harmonin N-domain-binding fragment of cadherin 23. The crucial residues involved in binding to the harmonin N-domain are highlighted by red stars. The minimal 20-residue harmonin N-domain binding fragment of cadherin 23 are boxed. (D) Fluorescence-based measurement of the binding affinity of the harmonin N-domain toward the cadherin 23 peptide. (E) Overlay plot of the 1H-15N HSQC spectra of the free N-domain and the protein fused with a cadherin 23 (A3190E) peptide at its C terminus.
The N-Domain Binds to an Internal Peptide Fragment of the Cadherin 23 Intracellular Domain.
As the cytoplasmic tail of cadherin 23 was previously shown to a bind to harmonin both in vivo and in vitro, and these 2 proteins are stably associated with each other in the stereocilia tips of the mature inner ear hair cells (6, 10), we wondered whether the harmonin N-domain might directly bind to cadherin 23 with a region other than the previously-identified C-terminal PDZ-binding motif. We used a GST-fusion protein pull-down assay to test this hypothesis. We found that the GST-fused N-domain can specifically bind to the recombinant cadherin 23 containing its entire cytoplasmic domain (Fig. 2A). The binding of the cadherin 23 cytoplasmic domain to the harmonin N-domain was further confirmed by titrating the 15N-labeled N-domain with unlabeled cadherin 23 by NMR spectroscopy (Fig. S2). To define the exact N-domain binding region of cadherin 23, we purified several fragments of the cadherin 23 cytoplasmic domain in their GB1- or Trx-fused forms (Fig. S2A). These purified cadherin 23 fragments were assayed for their binding to the N-domain by NMR-based titration approach (Fig. S2 B–D). We found that a 31-residue peptide fragment corresponding to amino acid residues 3181–3211 in the middle of cadherin 23 cytoplasmic domain was sufficient to bind to the harmonin N-domain (Fig. 2 B and C). To further narrow down the minimum N-domain binding region of cadherin 23, we combined point mutation and truncation approaches. Deletions up to 11 residues at the C-terminal end of the 31-residue N-domain-binding cadherin 23 fragment had no detectable changes to their binding; therefore, we narrowed the N-domain binding region down to the N-terminal 20 residues of the cadherin 23 fragment (Fig. 2C). Individual point mutations of the conserved hydrophobic residues within this 20-residue cadherin 23 fragment, including Leu-3188, Ile-3192, and Tyr-3195 to a Gln, abolished cadherin 23 binding to the N-domain, further supporting the specific interaction between the harmonin N-domain and cadherin 23. Whereas substitution of Ala-3190 or Leu-3201 with Gln had no obvious impact on its binding to the N-domain (Fig. S3). Finally, we used a 20-residue synthetic peptide corresponding to amino acid residues 3181–3200 (KPDDDRYLRAAIQEYDNIAK) of cadherin 23 (referred to as the Cad23 peptide) for further characterization of the interaction between cadherin 23 and the harmonin N-domain. Fluorescence titration assay showed that this Cad23 peptide binds to the harmonin N-domain with a KD of 25 μM (Fig. 2D), an affinity comparable to the harmonin PDZ/cadherin 23 carboxyl tail peptide complex (see below). The interaction between this 20-residue peptide and the N-domain was further verified by NMR spectroscopy, and the pattern of the 15N-labeled N-domain saturated with the cadherin 23 peptide is essentially identical to the N-domain bound to the 31-residue cadherin 23 fragment shown in Fig. 2B. Taken together, all of the above biochemical and spectroscopic data clearly indicate that the harmonin N-domain functions as a protein–protein interaction module and specifically interacts with a completely-conserved internal peptide sequence from the cytoplasmic domain of cadherin 23.
Structure of the N-Domain in Complex with the Cad23 Peptide.
To elucidate the molecular mechanism governing this harmonin/cadherin 23 interaction, we sought to determine the structure of the N-domain and the Cad23 peptide complex by NMR spectroscopy. The 1H-15N HSQC spectra of the N-domain in complex with the Cad23 peptide at protein concentrations >0.3 mM were poor in homogeneity, most likely because of partial aggregation of the complex and the unfavorable chemical exchange between the free and the Cad23 peptide-bound forms of the N-domain. To overcome this obstacle, we first fused the Cad23 peptide to the C terminus of the N-domain by using a flexible linker. Disappointedly, the N-domain–Cad23 peptide fusion protein precipitated at concentrations >0.2 mM, and this poor sample behavior prevented us from solving the complex structure by NMR. We suspected that the concentration-dependent aggregation of the N-domain–Cad23 peptide fusion protein is caused by nonspecific hydrophobic interactions, as we predicted that the hydrophobic Cad23 peptide would bind to the N-domain in an α-helical conformation and the complex would contain several solvent-exposed hydrophobic residues. Based on this prediction, we substituted Ala-3190 at the Cad23 peptide, which was shown to be not required for the N-domain binding (Fig. S3B), with Glu. The N-domain–Cad23(A3190E) fusion protein was highly soluble and displayed an excellent 1H-15N HSQC spectrum suitable for NMR-based structure determination (Fig. 2E). The HSQC spectrum of the N-domain–Cad23(A3190E) fusion protein is distinctly different from that of the free N-domain (Fig. 2E), further supporting specific interaction between the N-domain and cadherin 23. Importantly, the HSQC spectrum of the N-domain–Cad23(A3190E) fusion protein is highly similar to that of the free N-domain saturated with the Cad23 peptide (Fig. 2 B and E), indicating that the fusion of the Cad23 peptide to N-domain did not alter the overall interaction mode between the Cad23 peptide and the N-domain.
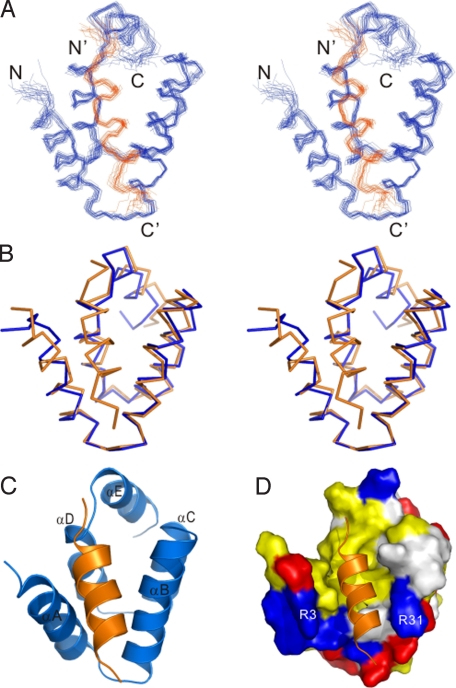
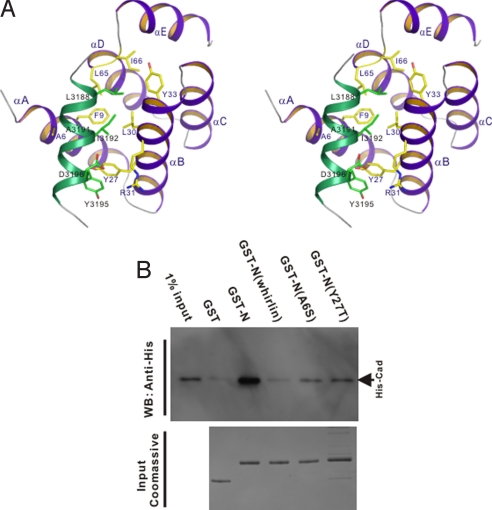
The high-resolution solution structure of the N-domain–Cad23(A3190E) fusion complex was solved by NMR spectroscopy (Fig. 3 and Table S1). In the complex, the Cad23 peptide forms an amphipathic α-helix and binds to the exposed hydrophobic pocket at the open side of the αA/αB-helix hairpin of the harmonin N-domain (Fig. 3 C and D). The binding of the Cad23 peptide does not induce large conformational changes to the N-domain (a rmsd value of 1.43 Å comparing the free- and the Cad23 peptide-bound forms of the N-domain; Fig. 3B). The N-domain/Cad23 peptide interface covers a total of ≈680-Å2 surface area. The hydrophobic side chains of Leu-3188, Ala-3191, and Ile-3192 from the Cad23 peptide pack into the N-domain hydrophobic pocket formed by the side chains of Ala-6, Phe-9, Leu-30, Tyr-33, Leu-65, and Ile-66. Specifically, the side chains of Leu-3188 and Ala-3191 are deeply embedded in the hydrophobic pocket and completely occluded from the solvent; the aromatic side chain of Tyr-3195 stacks with the aromatic side chain of harmonin Tyr-27 to form a pair of π–π interactions, explaining the absolute requirement of these 3 residues for the Cad23 peptide to bind to the harmonin N-domain. In addition, the negatively-charged side chain of Asp-3196 forms a pair of salt bridge with the positively-charged Arg-31 from αB of the N-domain (Figs. 3D and 4A). Again, we used the GST–fusion protein pull-down assay to verify the roles of selected residues from the N-domain in binding to cadherin 23. As shown in Fig. 4B, the N-domain mutants with Ala-6 substituted by Ser or Tyr-27 by Thr were largely defective in binding to cadherin 23, confirming the critical roles of these residues in binding to cadherin 23. As a control, we checked by recording their HSQC spectra that the A6S and Y27T mutants of the N-domain adopt the same overall structure as the wild-type protein.
Fig. 3.
The structure of the N-domain and the Cad23 peptide complex. (A) Stereoview showing the backbones of 20 superimposed NMR structures of the harmonin N-domain in complex with the Cad23 peptide. The Cad23 peptides are drawn in orange. (B) Stereoview showing the comparison of the conformations of the free N-domain (blue) and the N-domain in complex with the Cad23 peptide (orange). (C) Ribbon diagram of a representative NMR structure of the N-domain/Cad23 peptide complex. (D) Surface representation of the harmonin N-domain/Cad23 peptide complex. N-domain is shown in surface representation using the same coloring scheme as in Fig. 1C, and the Cad23 peptide is drawn in the ribbon model.
Fig. 4.
Molecular details of the interaction between the N-domain and the Cad23 peptide. (A) Stereoview showing the detailed interactions of the Cad23 peptide with the residues from the harmonin N-domain. (B) Western blot analysis showing the interactions between cadherin 23 with 2 harmonin N-domain mutants. The figure also shows that the corresponding N-domain from whirlin does not bind to cadherin 23.
The Second PDZ Domain of Harmonin Specifically Interacts with Cadherin 23 C-Terminal Peptide.
Cadherin 23 has a canonical type I PDZ binding motif at its extreme C terminus (−ITEL, referred to as Cad23–PBM). It was previously reported that the Cad23–PBM is able to interact with the second PDZ domains of harmonin (6). However, the structural basis of this interaction is not known. We first quantified the interaction of harmonin PDZ2 with a synthetic peptide comprising the last 9 residues of cadherin 23 (ETPLEITEL) by using fluorescence titration experiments. We found that PDZ2 of harmonin specifically binds to the Cad23–PBM peptide (KD ≈10 μM; Fig. S4B). It is important to note that the affinities of the PDZ2/Cad23 PBM complex and the N-domain/Cad23 internal peptide complex are of similar values, even though both interactions are moderate in strength. Although harmonin PDZ1 is also predicted to be a type I PDZ domain, it essentially does not bind to the Cad23–PBM peptide (KD >200 μM; Fig. S4B). As expected, no interaction could be observed between harmonin PDZ3 (a type II PDZ) and the Cad23–PBM peptide (6). To uncover the structural basis governing the specific interaction between harmonin PDZ2 and the Cad23–PBM peptide, we solved their complex structure by NMR spectroscopy (Fig. 5A and Table S1). Harmonin PDZ2 adopts a canonical PDZ domain fold consisting of a partially open β-barrel with 6 β-strands (βA to βF) and 2 helices (αA and αB) capping the opening sides of the β-barrel. An additional short α-helix is found after the final βF. Cad23–PBM binds to the αB/βB-groove of PDZ2 by augmenting the βB-strand of the PDZ domain in an antiparallel manner (Fig. 5A). The side chain of Leu(0) of Cad23–PBM inserts into the hydrophobic pocket at the end of the αB/βB groove. The carboxyl group of the peptide forms hydrogen bonds with the backbone amides of Leu-222, Gly-223, and Cys-224 (the so-called GLGF motif in PDZ domains) of PDZ2. The negatively-charged carboxylate is further stabilized by Lys-279 at the αB9 position of PDZ2. Therefore, Lys-279 plays an equivalent role to the positively-charged Arg or Lys at the beginning of the βA/βB loop in most PDZ domains, which is missing in harmonin PDZ2. As a type I PDZ ligand, the side chain hydroxyl group of Thr(−2) forms a strong hydrogen bond with the N-3 nitrogen of the imidazole ring of His-271 at the αB1 position. The methyl group of Thr(−2) contacts with the hydrophobic side chain of Val-275 1 helical turn above His-271 (Fig. 5A and Fig. S4A). The negatively-charged Glu(−1) of Cad23–PBM interacts with Lys-242 in the βC/αA-loop of PDZ domain. Finally, the harmonin PDZ2 and the Cad23–PBM complex are further stabilized by the charge–charge interaction between the side chains of Glu(−4) of the Cad23–PBM and Lys-272 of PDZ2. In the harmonin PDZ2/Cad23–PBM complex, Lys-242 in the βC/αA-loop of PDZ2 of the Cad23–PBM and Lys-272 of PDZ2 interacts with the negatively-charged Glu(−1) and Glu(−4) of Cad23–PBM, respectively (Fig. 5A). The corresponding residues in these 2 positions in PDZ1 are Ile-118 and Glu-148. This difference likely explains the weaker affinity between harmonin PDZ1 and Cad23 PBM, even though both PDZ1 and PDZ2 belong to the type I PDZ domain.
Fig. 5.
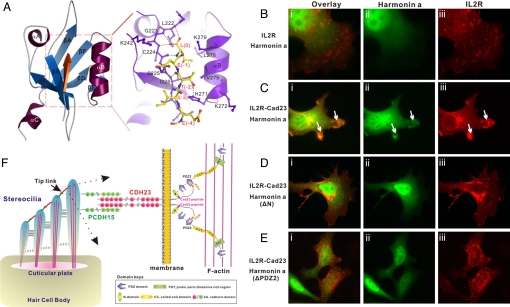
The multidentate interaction between harmonin and cadherin 23. (A) Specific interaction between harmonin PDZ2 and Cad23–PBM revealed by the NMR structure of the PDZ domain/peptide complex. Cad23-PBM is shown in red. The boxed region represents the Cad23–PBM-binding groove of the PDZ domain, and the detailed interactions between Cad23–PBM and the PDZ2 domain are drawn in the explicit atomic model. (Bi–Eiii) Both the harmonin N-domain/cadherin 23 internal peptide and the PDZ2/Cad23–PBM interactions are required for colocalization of harmonin and cadherin 23 in HeLa cells. (Bi–Biii) IL2R and harmonin did not colocalize with each other when the 2 proteins were overexpressed in HeLa cells. (Ci–Ciii) IL2R-cadherin 23 cytoplasmic domain chimera and harmonin colocalized with each other in the edges of cells (arrows). (Di–Eiii) Deletion of either the N-domain (Di–Diii) or PDZ2 (Ei–Eiii) of harmonin compromised the colocalization of harmonin and cadherin 23 in HeLa cells. (F) A schematic diagram depicting a model showing the synergistic, multidentate interaction between dimerized cadherin 23 and polyvalent harmonin in the tip link of hair cells. In this mode, harmonin via its N-domain and PDZ2 interacts with an internal peptide fragment and the C-terminal PBM of the cytoplasmic domain of cadherin 23. This multidendate interaction between harmonin and cadherin 23, together with the dimerization of cadherin 23, greatly enhances both specificity and stability of the assembly of the tip link complex in hair cells. Harmonin can further connect the tip link complex to the actin cytoskeleton via its PST module.
Cellular Colocalization Requires the Multidentate Interaction Between Harmonin and Cadherin 23.
Harmonin and cadherin 23 are known to colocalize at the tip of stereocillia in hair cells and in heterologous cells when overexpressed (10). We used a heterologous cell culture system to evaluate the role of the PDZ2/Cad23–PBM and the N-domain/Cad23 internal peptide interactions between harmonin and cadherin 23 in their cellular localizations. In this assay, the entire cytoplasmic domain of cadherin 23 was fused to the IL-2 receptor (IL2R) containing its transmembrane and extracellular domains. This IL2R–Cad23 chimera protein was used to evaluate interactions between harmonin and cadherin 23. Consistent with an earlier report (10), harmonin colocalized with IL2R–Cad23 chimera at the cell peripheries (Fig. 5C) when the 2 proteins were coexpressed. This colocalization was completely absent if IL2R lacking the cadherin 23 cytoplasmic domain was coexpressed with harmonin (Fig. 5B). Disruption of the either binding sites by deleting the N-domain or PDZ2 of harmonin compromised the colocalization of the 2 proteins (Fig. 5 D and E), indicating that both the PDZ2/Cad23–PBM and the N-domain/Cad23 internal peptide interactions are required for the colocalization of harmonin and cadherin 23 in heterologous cells.
Discussion
In this work, we discovered that the master Usher complex scaffold harmonin contains a well-folded protein–protein interaction domain at its very N-terminal end. This N-terminal domain is highly conserved throughout the evolution and is not affected by the alternative splicing of harmonins (Fig. 1A), implicating indispensable functions of this previously-uncharacterized protein module. We further discovered that a short internal peptide fragment (≈15 residues) of cadherin 23 specifically binds to the harmonin N-domain. Reciprocally, the harmonin N-domain-binding sequence of cadherin 23 is absolutely conserved throughout the evolution (Fig. 2C), supporting the functional importance of the interaction between the harmonin N-domain and cadherin 23. Before this study, the N-domain together with the first PDZ domain of harmonin were collectively defined as PDZ1. A number of Usher proteins were identified to interact with this combined N-PDZ1 tandem, and these N-PDZ1-interacting proteins include the cytoplasmic domain of cadherin 23 (6), SANS (17), the Myth4-FERM domain of myosin VII (17), and the carboxyl PDZ binding motifs of the rest Usher proteins (1, 6, 16) (Fig. S1). Among these interactions, the bindings of the internal peptide fragment of cadherin 23, the SAM domain of SANS, and the Myth4-FERM domain of myosin VII to harmonin N-PDZ1 are particularly noteworthy, because these interactions involve binding sequences other than the canonical PDZ domain binding motifs that are typically located at the extreme carboxyl termini of PDZ targets and these binding sequences do not share obvious homology. Here, we demonstrated that the interaction between the internal peptide fragment of cadherin 23 and harmonin is mediated by the N-domain instead of the PDZ1 domain of harmonin. We conclude that the highly-conserved harmonin N-domain represents a protein–protein interaction module capable of binding to other Usher proteins, including cadherin 23 and SANS.
Whirlin is highly homologous to harmonin in their domain organizations including the ≈100-residue N-terminal domain preceding its PDZ1 (Fig. S5). Mutation of whirlin gene at first is known to cause nonsyndromic profound type of recessive hearing loss (DFNB31) because of developmental defects of stereocilia of hair cells (19). Recently, whirlin was mapped to be component of the USH2 proteins (namely USH2D) (5). Additionally, whirlin has been shown to localize at the tips of stereocilia of hair cells (9). Amino acid sequence alignment analysis showed that the whirlin N-domain can be nicely aligned with the harmonin N-domain (Fig. 1A), although the amino acid sequence identity between whirlin and harmonin is lower than that among harmonins from different species. Thus, it is natural to ask whether the whirlin N-domain might also function as a protein–protein interaction domain in binding to cadherin 23. We purified GST-fused whirlin N-domain and assayed its potential interaction with the cytoplasmic domain of cadherin 23 and found that no interaction could be detected between these 2 proteins (Fig. 4B), indicating specific interaction between harmonin and cadherin 23. Several residues in the whirlin N-domain corresponding to those critical for the harmonin N-domain binding to cadherin 23 (Fig. 1A, highlighted by stars) are different between whirlin and harmonin, and these differences likely account for the distinct cadherin 23 binding properties of the 2 scaffold proteins in the Usher interactome. For example, Tyr-27 of harmonin N-domain, which is critical in packing with Tyr-3196 of cadherin 23, corresponds to a Thr residue in the whirlin N-domain (Figs. 1A and 4A). Substitution of Tyr-27 with a Thr (Thr-59 in human whirlin) abolished the cadherin 23 internal fragment binding capacity of the haromnin N-domain (Fig. 4B).
In addition to direct binding between the harmonin N-domain and cadherin internal peptide sequence, interaction between harmonin and cadherin 23 involves the harmonin PDZ2 and the carboxyl PDZ binding motif of cadherin 23 (ref. 6 and this study). We showed that the cadherin 23 carboxyl PDZ binding motif has negligible interactions with either PDZ1 or PDZ3 of harmonin. Therefore, the formation of the harmonin/cadherin 23 complex is mediated by 2 pairs of nonoverlapping interactions between the 2 proteins each with a moderate binding affinity (with a KD of ≈10–20 μM; Fig. 5F). The bidentate interaction nature between harmonin and cadherin 23 entails the formation of tight harmonin/cadherin 23 complex, as one can envision from the thermodynamic principle of the interaction. At the tip link, cadherin 23 forms a dimer unit in interacting with protocadherin 15 (6, 7). The interaction between dimerized cadherin 23 with harmonin (which also has potential to form dimer or even multimer via its coiled-coil domains) would further increase the interaction avidity between the 2 proteins (Fig. 5F). We therefore predict that harmonin can play a critical role in clustering cadherin 23 in a highly-restricted region (i.e., the tip link) in stereocilia of hair cells. In addition, the C-terminal PST domain could serve as an anchoring site for connecting the tip link complexes with the actin cytoskeletals (Fig. 5F).
In summary, we have identified a protein–protein interaction domain in the very N-terminal end of the master Usher protein complex scaffold harmonin. The harmonin N-domain binds to an absolutely-conserved, short internal peptide fragment of cadherin 23. The 3D structures of the harmonin N-domain alone and in the complex with the cadherin 23 fragment solved in this work elucidated the specific interaction mechanism between this previously-uncharacterized interaction mode between these 2 Usher proteins. We have also characterized the interaction between harmonin PDZ2 and the cadherin 23 carboxyl tail peptide in detail by solving the PDZ2/peptide complex structure. It is likely that the multidentate interaction between harmonin and cadherin 23 plays an important role in the assembly of stable tip link complex formed by cadherin 23/protocadherin15 in hair cells.
Materials and Methods
Protein Preparation.
The coding sequence of the harmonin N-domain was PCR-amplified from human Ush1C and cloned into a pET32a vector. The His6-tagged N-domains expressed in Escherichia coli cells were purified by using a Ni2+-nitrilotriacetic acid agarose column followed by size-exclusion chromatography. For in vitro biochemical analysis, the wild-type N-domain and its mutants were expressed as GST fusion proteins and purified by GSH-Sepharose affinity chromatography. Cadherin 23 and its various mutants were expressed as His6-tagged proteins and purified by Ni2+-nitrilotriacetic acid agarose affinity chromatography.
NMR Spectroscopy.
The protein samples for NMR studies were concentrated to ≈0.2 mM for titration experiments and ≈0.6 mM for structural determinations in 100 mM potassium phosphate at pH 6.5. NMR spectra were acquired at 30 °C on Varian Inova 500 or 750-MHz spectrometers. Backbone and side-chain resonance assignments were achieved by combination of the standard heteronuclear correlation experiments and 2D 1H NOESY experiments (20, 21). Approximate interproton distance restraints were derived from 2D 1H-NOESY, 3D 15N-separated NOESY, and 13C-separated NOESY spectra. Structures were calculated with the program CNS (22). The figures were prepared with the programs MOLSCRIPT (23), PyMOL (http://pymol.sourceforge.net), and MOLMOL (24).
GST Pull-Down Assay.
Direct interactions between cadherin 23 and various GST-fused proteins were assayed in PBS. Equal molar amounts of cadherin 23 and one of the GST fusion proteins (≈0.6 nmol each) were mixed in 100 μL of the assay buffer. The GST fusion protein/cadherin 23 complexes were pelleted by 15 μL of fresh GSH-Sepharose beads. The pellets were washed 3 times with 0.5 mL of the assay buffer and subsequently boiled with 15 μL of 2× SDS/PAGE sample buffer. The precipitated cadherin 23 in the gel was visualized either by Coomassie blue staining or immuno-detection using anti-His antibody.
Fluorescence Assay.
Fluorescence assays were performed on a PerkinElmer LS-55 fluorimeter equipped with an automated polarizer at 20 °C. In the assay, FITC (Molecular Probes)-labeled peptide samples (≈1 μM) were titrated with binding partners in buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT, and 1 mM EDTA, and the KD values were obtained by fitting the titration curves with the classical 1-site binding model.
Cellular Localization.
The wild-type harmonin a was cloned into pEGFP-C2 vector. EGFP–harmonin a lacking the N-domain (EGFP–harmonin a ΔN) and EGFP–harmonin a lacking its PDZ2 (EGFP–harmonin a ΔPDZ2) mutants were created by using the standard PCR-based method. HeLa cells were transiently transfected with 0.5 μg for each plasmid per well by using a Lipofectamine PLUS kit (Invitrogen), and cells were cultured for 24 h before fixation. The cells were imaged with a Nikon Eclipse TE2000 inverted fluorescence microscope.
Supplementary Material
Acknowledgments.
This work was supported by Research Grants Council of Hong Kong HKUST6442/06M, 663407, 663808, CA07/08.SC01, and AoE/B-15/01-II (to M.Z.). The NMR spectrometers used in this work were purchased with funds donated to the Biotechnology Research Institute HKUST, Hong Kong by the Hong Kong Jockey Club.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2KBQ, 2KBR, and 2KBS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901819106/DCSupplemental.
References
- 1.Reiners J, Wolfrum U. Molecular analysis of the supramolecular usher protein complex in the retina. Harmonin as the key protein of the Usher syndrome. Adv Exp Med Biol. 2006;572:349–353. doi: 10.1007/0-387-32442-9_49. [DOI] [PubMed] [Google Scholar]
- 2.El-Amraoui A, Petit C. Usher I syndrome: Unraveling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed ZM, Riazuddin S, Riazuddin S, Wilcox ER. The molecular genetics of Usher syndrome. Clin Genet. 2003;63:431–444. doi: 10.1034/j.1399-0004.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams DS. Usher syndrome: Animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008;48:433–441. doi: 10.1016/j.visres.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebermann I, et al. A novel gene for Usher syndrome type 2: Mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121:203–211. doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed] [Google Scholar]
- 6.Siemens J, et al. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci USA. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollner C, et al. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 2004;428:955–959. doi: 10.1038/nature02484. [DOI] [PubMed] [Google Scholar]
- 8.McGee J, et al. The very large G protein-coupled receptor VLGR1: A component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci. 2006;26:6543–6553. doi: 10.1523/JNEUROSCI.0693-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adato A, et al. Usherin, the defective protein in Usher syndrome type IIA, is likely to be a component of interstereocilia ankle links in the inner ear sensory cells. Hum Mol Genet. 2005;14:3921–3932. doi: 10.1093/hmg/ddi416. [DOI] [PubMed] [Google Scholar]
- 10.Boeda B, et al. Myosin VIIa, harmonin, and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit C. Usher syndrome: From genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- 12.Keats BJ, Corey DP. The usher syndromes. Am J Med Genet. 1999;89:158–166. [PubMed] [Google Scholar]
- 13.Friedman LM, Dror AA, Avraham KB. Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol. 2007;51:609–631. doi: 10.1387/ijdb.072365lf. [DOI] [PubMed] [Google Scholar]
- 14.Vrijens K, Van Laer L, Van Camp G. Human hereditary hearing impairment: Mouse models can help to solve the puzzle. Hum Genet. 2008;124:325–348. doi: 10.1007/s00439-008-0556-y. [DOI] [PubMed] [Google Scholar]
- 15.Verpy E, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 16.Reiners J, et al. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet. 2005;14:3933–3943. doi: 10.1093/hmg/ddi417. [DOI] [PubMed] [Google Scholar]
- 17.Adato A, et al. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet. 2005;14:347–356. doi: 10.1093/hmg/ddi031. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Wang W. Organization of signaling complexes by PDZ-domain scaffold proteins. Acc Chem Res. 2003;36:530–538. doi: 10.1021/ar020210b. [DOI] [PubMed] [Google Scholar]
- 19.Mburu P, et al. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- 20.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 21.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 22.Daniels DL, Cohen AR, Anderson JM, Brunger AT. Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat Struct Biol. 1998;5:317–325. doi: 10.1038/nsb0498-317. [DOI] [PubMed] [Google Scholar]
- 23.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 24.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.