Abstract
Bacterial pathogens sense host cues to activate expression of virulence genes. Most of these signals are sensed through histidine kinases (HKs), which comprise the main sensory mechanism in bacteria. The host stress hormones epinephrine (Epi) and norepinephrine are sensed through the QseC HK, which initiates a complex signaling cascade to regulate virulence gene expression in enterohemorrhagic Escherichia coli (EHEC). Epi signaling through QseC activates expression of the genes encoding the QseEF 2-component system. QseE is an HK, and QseF is a response regulator. Here, we show that QseE is a second bacterial adrenergic receptor that gauges the stress signals Epi, sulfate, and phosphate. The qseEF genes are organized within an unusual operonic structure, in that a gene is encoded between qseE and qseF. This gene was renamed qseG, and it was shown to encode an outer membrane (OM) protein. EHEC uses a type III secretion system (TTSS) to translocate effector proteins to the epithelial cells that rearrange the host cytoskeleton to form pedestal-like structures that cup the bacterium. QseE, QseG, and QseF are necessary for pedestal formation. Although QseE and QseF are involved in the transcriptional control of genes necessary for pedestal formation, QseG is necessary for translocation of effectors into epithelial cells. QseG is an OM protein necessary for translocation of TTSS effectors that also works in conjunction with a 2-component signaling system that senses host stress signals.
Keywords: enterohemorrhagic E. coli, epinephrine, interkingdom signaling, type III secretion
Bacteria and their hosts communicate with each other through hormone-like compounds. This communication is referred to as interkingdom cell-to-cell signaling (1). Many pathogens sense and respond to the host adrenergic signaling molecules epinephrine (Epi) and norepinephrine to recognize the host environment and promote the expression of virulence factors (1). These pathogens use the same histidine kinase (HK), QseC (2, 3), to recognize both host-derived adrenergic signals as well as a bacterial signal dubbed autoinducer-3 (AI-3) (1, 3, 4). Upon sensing any of these 3 signals, QseC augments its phosphorylation and subsequently phosphorylates a transcription factor, QseB (2), thereby relaying the presence of these signals to a complex regulatory cascade and leading to transcription of key virulence genes (1–5).
Enterohemorrhagic Escherichia coli (EHEC) causes hemorrhagic colitis and hemolytic uremic syndrome worldwide. EHEC colonizes the large intestine and adheres to epithelial cells forming attaching and effacing (AE) lesions, which efface the microvilli and reorganize the host cytoskeleton into a pedestal-like structure (6). Most of the genes necessary for AE lesion formation are encoded within the locus of enterocyte effacement (LEE). The LEE is composed of 41 genes organized in 5 major operons (named LEE1 to LEE5) and encodes all of the components of a type III secretion system (TTSSs)—an adhesin, intimin, and the translocated intimin receptor (Tir). EHEC secretes structural proteins of the TTSS, such as EspA, a filament that creates a sheath around the TTS needle, and EspBD, which creates a pore through the eukaryotic cell membrane. Tir is translocated through the TTSS into host cells, where it embeds itself in the membrane and acts as a receptor for Intimin. Tir also initiates a signaling cascade that leads to the recruitment of N-WASP and Arp2/3, leading to actin nucleation and the formation of the pedestal (6). EHEC's repertoire of virulence factors includes numerous TTS effectors, many of which are encoded outside the LEE but are translocated into host cells via the LEE TTSS (7, 8). One such effector is EspFu, which acts as a link between Tir and N-WASP and the Arp2/3 complex (9–11).
Most pathogens develop mechanisms to recognize when they have reached a certain niche. EHEC recognizes the AI-3 that is produced by the intestinal microbial flora (4, 12), as well as the host hormones Epi and norepinephrine (4). Norepinephrine is produced in the intestine by adrenergic neurons of the enteric nervous system, whereas Epi is a systemic hormone produced in the central nervous system and the adrenal medulla that reaches the intestine through the bloodstream (13, 14). These stress hormones affect many normal functions of the intestine, such as chloride and potassium secretion (13–15). Upon recognition of these signals, QseC autophosphorylates, then transfers a phosphate to QseB, which in turn activates the flagella regulon (2) and genes involved in AE lesion (3). This regulation is complex and involves many intermediary signaling proteins, such as QseA (16), and another 2-component system, QseEF, which is also involved in the transcriptional activation of espFu (17). QseE and QseF are a cognate pair, and QseE transfers its phosphate to QseF (18). QseF is a response regulator and contains a helix-turn-helix DNA-binding domain and a σ54 activation domain. It has also been reported that QseF can be phosphorylated by at least 4 noncognate sensors: BaeS, EnvZ, RstB, and UhpB (18). The qseEF genes are cotranscribed with a small gene, yfhG. YfhG contains a predicted secretion signal sequence and is predicted by in silico analysis (http://ca.expasy.org) to be a membrane protein. However, it lacks significant homology to other proteins and does not contain any predicted active sites or conserved domains. Membrane proteins are key players in allowing 2-component systems to recognize their cognate signals and communicate with other 2-component systems (19, 20). In this study we elucidated the role of YfhG (QseG) in pedestal formation and further characterized the QseEF system and its roles in responding to environmental signals and regulation of AE lesion formation.
Results
The qseEFG Operon.
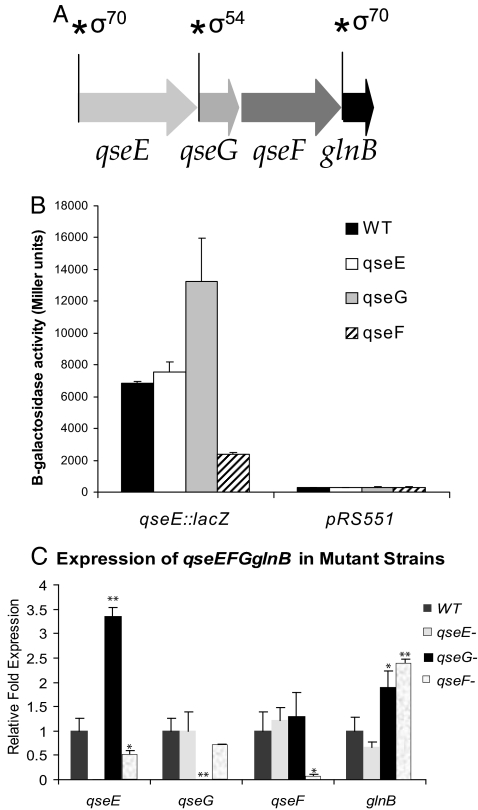
The qseEyfhGqseFglnB genes are cotranscribed in 1 operon (17). Because yfhG is cotranscribed with qseE and qseF, this gene was renamed qseG. Previous reports and in silico analysis revealed that the structure and regulation of this operon is complex, with at least 2 transcriptional start sites (TSSs) that have been mapped by primer extension, one upstream of glnB and another upstream of qseE. Both of these TSSs have σ70 promoters (21, 22). Additional in silico scans of the EHEC genome using a σ54 consensus sequence predicted a σ54 promoter upstream of qseG (www.promscan.uklinus.net/RpoN/data.html) (Fig. 1A).
Fig. 1.
Autoregulation of the qseEGFglnB operon. (A) The qseEGFglnB operon contains 2 σ70 promoters upstream of qseE and glnB, and a predicted σ54 upstream of qseG. (B) The qseE-lacZ fusions within WT and qseE, qseG, and qseF nonpolar mutants. (C) qRT-PCR analysis examining the expression of qseE, qseG, qseF, and glnB in WT and qseE, qseG, and qseF mutants. Error bars represent the standard deviation of 3 independent experiments. ∗, P < 0.05; ∗∗, P < 0.005.
Autoregulation is common within 2-component systems. Using a qseE-lacZ transcription fusion, we could not detect differences in transcription between WT and a qseE nonpolar mutant (Fig. 1B). Quantitative RT-PCR (qRT-PCR) for the qseE transcript and transcription of a qseE-lacZ transcription fusion showed that transcription of qseE was increased in the qseG mutant, suggesting that QseG exerts a repressive role in the expression of qseE (Fig. 1 B and C). Conversely, expression of qseE was decreased in a qseF mutant, suggesting that QseF activates the expression of qseE (Fig. 1 B and C). However, it is worth noting that QseF is a σ54-dependent response regulator, suggesting that QseF acts through an intermediary, yet unidentified transcription factor to activate the σ70 promoter upstream of qseE. These data indicate that the promoter upstream of qseE is activated through QseF. Transcription of this promoter was not affected in a qseE mutant, which can be explained by the previous observation that QseF is a promiscuous response regulator that can be phosphorylated by 4 other HKs in addition to QseE (18). Expression of qseE was repressed by QseG. In silico analyses using the Trans Term program (23) did not find any transcription terminators between qseF and qseG. The differential expression of qseE between qseF and qseG mutants could be due to posttranscriptional regulation.
Because there are 2 additional internal promoters in this operon, we investigated whether transcription autoregulation occurred through either one of these promoters. Transcription of both qseG and qseF was unaltered in any of the 3 mutants, suggesting that transcription of these 2 genes is not subject to autoregulation. In contrast, transcription of glnB was increased in the qseF and qseG mutants, suggesting that QseF and QseG act through the promoter upstream of glnB to repress its transcription. These data highlight the complexity of the autoregulatory circuit that governs the expression of the qseEGFglnB operon (Fig. 1C).
QseG Is an Outer Membrane Protein.
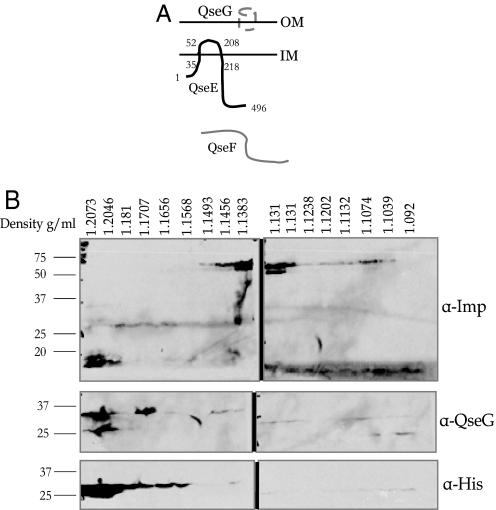
QseG is a 237-aa protein that separates on an SDS/PAGE gel at 27 kDa. The first 25 amino acids of QseG are predicted to be a signal sequence, and QseG contains lipid attachment sites (http://expasy.org/tools/scanprosite/). QseG shows homology to membrane proteins as well as alpha-helix proteins (http://expasy.org/tools/blast/). However, no additional domains or active sites were found in QseG based on in silico analysis. To determine the localization of QseG in EHEC, we isolated crude membrane fractions from an EHEC strain expressing a QseGMycHis fusion protein. These fractions were then separated over a sucrose density gradient. Density of each fraction was determined based on its refractive index. As seen in Fig. 2, 18 fractions ranging from 1.092 to 1.2073 g/mL were isolated. An anti-Imp antibody was used as a control for the sucrose fractions. This antibody recognizes an inner membrane protein of ≈55 kDa and the outer membrane (OM) protein, OmpA, at ≈19 kDa. When fractions were blotted with this antibody, OmpA appeared primarily in fractions 1 and 2 at 1.2076–1.2046 g/mL, whereas Imp appeared primarily in fractions 7–10 at 1.1493–1.1311 g/mL (Fig. 2). These weights correspond to the densities for the inner membranes and OMs from previously reported membrane fractionations (24, 25). When these fractions were probed by using both an anti-His antibody or an anti-QseG antibody, in both instances QseG appeared in the first 2–3 fractions (1.2073–1.18 g/mL), indicating that QseG localizes to the OM (Fig. 2).
Fig. 2.
Cellular localization of QseG. (A) Depiction of the localization of QseE, QseF, and QseG. (B) Membrane fractions showing that QseG localizes to the OM.
QseG Is Required for Pedestal Formation.
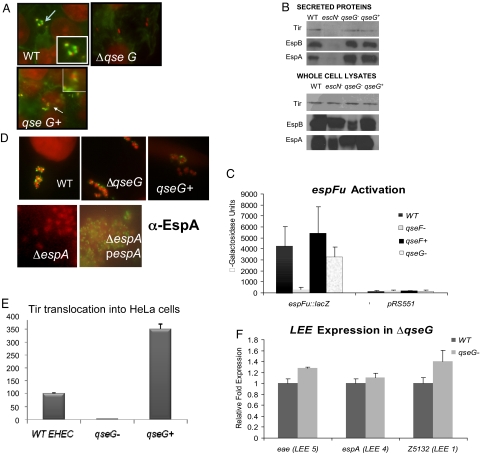
QseF is required for EHEC to form pedestals on epithelial cells because of its regulation of espFu (17). To investigate whether QseG is involved in the same process, we tested the ability of the qseG mutant to make pedestals (26). Actin was stained with FITC-labeled phalloidin, the bacteria and HeLa cell nuclei were stained with propidium iodide, and pedestals were visualized as brilliant patches of green underneath a red bacterium. The qseG mutant was unable to form pedestals, and pedestal formation was restored upon complementation (Fig. 3A). This mutant produced and secreted EspA, EspB, and Tir (Fig. 3B). QseG also did not activate expression of espFu (Fig. 3C) or the LEE genes (Fig. 3F). Because the qseG mutant is able to produce and secrete each one of the components necessary for assembly of the TTSS and its effectors, we investigated whether the secretion apparatus was properly assembled and whether effectors, while secreted into the media, were properly translocated into epithelial cells.
Fig. 3.
QseG is required for pedestal formation by EHEC. (A) FAS assays. Arrows indicate bacteria forming AE lesions. (Magnification: 100X.) (B) Western blots of whole-cell lysates (Top) and secreted proteins (Bottom) with anti-EspB, anti-EspA, and anti-Tir antisera. (C) Transcriptional regulation of espFu using an espFu-lacZ fusion. β-Galactosidase activity was measured in Miller units, and the triangle means the differences between qseG and WT were not statistically significant. (D) Detection of the EspA filament by using immunofluorescence with anti-EspA antibody and FITC-labeled secondary antibody. An EPEC espA mutant was used as a negative control, and the espA mutant expressing the EHEC EspA was used as a positive control. (Magnification: 100X.) (E) Translocation of Tir to host cells by using tir-cyaA fusions. WT levels of translocation were set at 100%. (F) Expression of the LEE genes by using qRT-PCR. Error bars in C, E, and F indicate the standard deviation of 3 independent experiments.
To visualize the TTSS in the ΔqseG strain, we used immunofluorescence. After infecting HeLa cells with WT and ΔqseG EHEC, we stained the EspA filament of the TTSS in green by using an anti-EspA antibody and an FITC-conjugated secondary antibody. The HeLa cell nuclei and the bacteria were stained in red with propidium iodide. The qseG mutant formed EspA filaments (Fig. 3D), demonstrating that it can assemble the TTSS. To further investigate why ΔqseG is unable to form pedestals, we used the cyaA gene reporter system developed by Sory and Cornelis (27). This system uses a fusion of a TTS effector and the calmodulin-dependent adenylate cyclase domain (cyaA) of Bordetella pertussis cytolysin. This toxin relies on calmodulin for activation, which is present in eukaryotic cells but not prokaryotic cells (28). Therefore, rises in the levels of cAMP in the host cell due to the activation of the adenylate cyclase toxin will only occur if the effector-cyaA fusion is translocated by the TTSS into host cells rather than simply secreted into the media. We used the tir-cyaA gene fusion (29) to monitor the translocation of Tir into host cells by WT EHEC, the ΔqseG EHEC, and the complemented strain. Levels of cAMP of HeLa cells infected with WT were set at 100%, and cAMP levels from cells infected with either the ΔqseG or complemented strains were expressed as a percentage of the levels from cells infected with WT. The ΔqseG did not translocate Tir (Fig. 3E), and translocation of Tir was restored upon complementation. The complemented strain translocated Tir 3.5-fold higher than the WT strain, suggesting that overexpression of qseG enhances Tir translocation. These results suggest that QseG does not transcriptionally regulate the LEE or espFu genes, nor is it involved in structural assembly of the TTSS. Instead, QseG is necessary for the translocation of effector molecules into epithelial cells.
QseE Senses Epi, Sulfate, and Phosphate Sources.
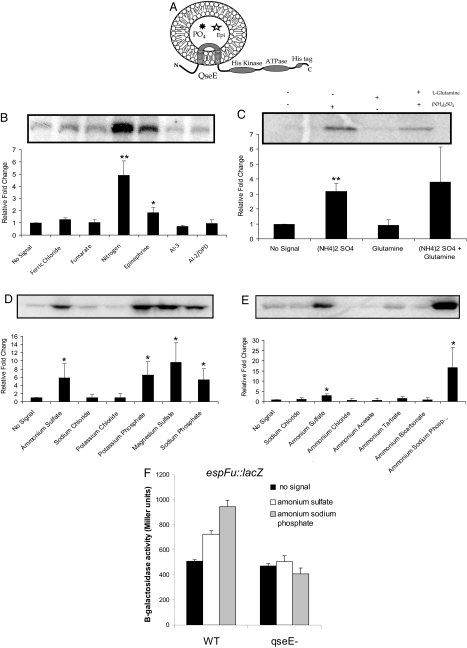
QseE is an HK that transfers a phosphate to QseF (18). To mimic the membrane environment in in vitro assays, liposomes have been used (2, 30). This system also provides information about domain orientation, given that the HK is inserted in an inside-out orientation. This orientation, as shown in Fig. 4A, allows different chemicals to be preloaded into the liposome and tested for their ability to enhance the HK's autophosphorylation (2, 30). To identify the physiological signals sensed by QseE, QseE was inserted into liposomes. Based on QseE's putative role in the EHEC AI-3/Epi/norepinephrine signaling cascade, we used sources of 50 μM Epi as well as 50 μM AI-3. In addition, we treated the liposomes with 100 μM 4,5-dihydroxy-2,3-pentanedione (Omm Scientific), the precursor to the AI-2 signal, as well as 20 mM fumarate, the known signal for DcuS (30). We also tested sources of iron (150 μM FeCl3) and nitrogen, signals known to be important in pathogenesis. As shown in Fig. 4B, QseE exhibited a low level of basal phosphorylation when no signals were added. However, QseE showed a robust response to nitrogen sources [15 mM glutamine and 15 mM (NH4)2SO4], and it significantly increased phosphorylation in response to Epi, but it showed no response to AI-3, AI-2, or iron (Fig. 4B). The phosphorylated band from autoradiography was confirmed to be QseE by Western blot analysis and mass spectrometry. These results suggested that QseE senses nitrogen sources and Epi but not quorum-sensing molecules.
Fig. 4.
QseE autophosphorylation in response to various agonists. (A) Schematic of QseE's orientation in the liposome. (B) QseE phosphorylation in response to nitrogen, iron, Epi, AI-3, AI-2, and fumarate. (C) QseE's response to ammonium sulfate and glutamine. (D) QseE's response to phosphate and sulfate sources. (E) QseE's response to ammonium sources containing phosphate or sulfate counter ions. (F) Transcription of espFu-lacZ in WT and qseE in the absence and presence of phosphate and sulfate. ∗, P < 0.05; ∗∗, P < 0.005.
To further investigate QseE's response to nitrogen sources, we preloaded QseE liposomes with glutamine or ammonium sulfate separately. This demonstrated that QseE was responding specifically to ammonium sulfate and not to glutamine (Fig. 4C). Considering the presence of glnB in this operon, it would be logical for QseE to be a nitrogen sensor. However, one needs to distinguish whether QseE was responding to ammonium (a true nitrogen source) or its counter ion sulfate. To this end, we conducted the autophosphorylation assays with numerous sources of phosphate or sulfate (Fig. 4D). QseE's autophosphorylation in each case was stimulated by the presence of either phosphate or sulfate sources but not by NaCl or KCl, which were used as controls. Although these experiments demonstrated that QseE autophosphorylation is stimulated by phosphate and sulfate sources, they did not rule out the possibility that QseE is a nitrogen sensor. To further elucidate QseE's role as a potential nitrogen sensor, we tested the ability of numerous ammonium compounds to activate QseE autophosphorylation. As observed in Fig. 4E, only the ammonium compounds containing a phosphate or sulfate counter ion were able to stimulate QseE autophosphorylation. These data indicate that QseE senses phosphate, sulfate, and Epi, but that it is not a nitrogen sensor. We have previously reported that epinephrine activates espFu expression (17). Given that QseE also senses sulfate and phosphate sources, we investigated whether phosphate and sulfate increased espFu expression. Congruent with the phosphorylation studies, phosphate and sulfate increased espFu expression (Fig. 4F). This activation was modest, which can again be explained by the fact that QseF can be phosphorylated by 4 other HKs in addition to QseE (18).
Discussion
Two-component signaling systems are major prokaryotic players in the recognition and transduction of environmental signals. Here, we report 2 genes encoding a 2-component system cotranscribed with a gene encoding an OM lipoprotein, QseG (Fig. 1). QseG is shown to be important for AE lesion formation (Fig. 3), and in contrast to QseE and QseF, it does not have a role in transcription regulation of the genes encoding the TTSS and its effectors. However, a ΔqseG strain is unable to translocate the effector Tir, which is essential for pedestal formation, into the host cell. Lipoproteins have been reported to play a role in expression of TTSSs (31), but QseG seems to play a role in translocation instead.
Although the genetics of many 2-component systems have been characterized, very few physiological signals that HKs recognize are known. It has been shown genetically that QseE and QseF are members of the EHEC Epi/norepinephrine/AI-3 signaling cascade, because they are regulated by both QseA and QseBC (17). Here, we show that QseE responds to Epi but not AI-3 (Fig. 4). This is logical, considering that QseEF plays a role in pedestal formation but not in flagellation or motility (17). EHEC has a low infectious dose, so it is hypothesized that the AI-3 it responds to upon entering the intestine is produced by the normal flora in the lumen (4). In the lumen it would be beneficial for EHEC to express flagellation genes for motility and not genes associated with AE lesion formation. Epi is systemic in the blood, and norepinephrine is released by adrenergic neurons in the intestine (13, 14). The sources of these compounds in the intestine suggest that they are present in highest concentration closest to the epithelial layer, the same location where EHEC begins the process of AE. Hence, the observation that QseE senses Epi but not AI-3 fits with the data showing that this 2-component system acts on pedestal formation but not flagellation. Although there are many parallels between QseE and QseC, QseE appears to be downstream in the signaling cascade from QseC (17), and these 2 molecules most likely function in tandem.
QseE also has a strong response to both phosphate and sulfate sources (Fig. 4). Little is known about sulfate levels in the intestine and how EHEC's sensing of sulfate might be involved in virulence. However, depletion in phosphate intestinal levels has been associated with increased stress and mortality by nosocomial infections (32). The ability of QseE to sense phosphate may allow EHEC to outcompete the overwhelmingly large population of microorganisms in gaining nutrients, and therefore successfully colonize the intestinal epithelium.
This study takes the first steps toward characterizing QseEFG. The data presented indicate that the regulation and function of the QseEF system and QseG are complex. Because QseEFG is also present in nonpathogenic strains of E. coli, it is likely that they have 2 distinct roles in nonpathogenic and pathogenic strains. It is possible that although QseEFG is involved in AE lesion formation in EHEC, its role in nonpathogenic E. coli involves metabolism or stress responses. These roles could coincide if they involve sensing environmental cues that allow EHEC to inhabit the intestine. Understanding how commensal E. coli genes have been diverted to also play a role in the regulation of virulence genes in EHEC is important in the study of pathogenic E. coli. This is increasingly relevant as 2-component systems are evaluated as drug targets (3, 33).
Materials and Methods
Strains and Plasmids, Recombinant DNA.
All bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli strains were grown in DMEM at 37 °C or in LB. Recombinant DNA and molecular biology techniques were performed as previously described (34). Primers used in qRT-PCR and cloning are listed in Table S2. Plasmid pNR30 was constructed by amplifying qseE from EHEC genomic DNA with primers qsepet21F and qseEpet21R and cloning into (BamHI and NotI) pET21 (Novagen). Plasmid pNR03 was created by amplifying qseG and cloning into (EcoRI and KpnI) pBadMycHisA (Invitrogen). NR03 was constructed by using λ-Red as previously described (35) using primers yfhGRedF and yfhGRedR. To create the nonpolar mutant, NR03, the chloramphenicol cassette was resolved by using pCP20 (35). NR03 was complemented with plasmid pNR03 to create strain NR05.
SDS/PAGE and Immunoblotting.
Secreted proteins were isolated from EHEC WT, CVD451, NR03, and NR05 as previously described (36). Whole-cell lysates were prepared from strains grown in DMEM to an OD600 of 1.0. SDS/PAGE and immunoblotting were completed as previously described (34). Protein concentration from whole-cell lysates was determined by using the Bradford assay (34). Preparations were probed by Western blot analysis using polyclonal antisera against EspA, EspB, Tir, or intimin.
AE Lesion and TTSS Assembly Tests.
Fluorescent actin staining (FAS) and the EspA immunofluorescent filament staining tests were performed as previously described (26, 37). As a negative control, we used an EPEC espA mutant, strain UMD872 (38), and as a positive control, we used UMD872 with plasmid pICC284 expressing the EHEC EspA protein from a pBADMycHis vector (39). Tir translocation assays were performed as previously described (29).
Phosphorylation of QseE-His in Liposomes.
QseE-His-loaded liposomes were prepared and tested for autophosphorylation in response to various signals as previously described (2) (details in SI Materials and Methods).
Real-Time qRT-PCR.
Cultures were grown in DMEM at 37 °C to an OD600 of 1.0. RNA was extracted from 3 replicates of each strain by using a RiboPure bacterial RNA isolation kit (Ambion) according to the manufacturer's instructions. Primers used in qRT-PCR analysis are listed in Table S2. The qRT-PCR analysis was conducted by using an Applied Biosystems ABI 7500 sequence detection system using a 1-step reaction as previously described (12). The rpoZ gene was used as an internal control. For details, please see SI Materials and Methods.
Membrane Preparation and Sucrose Density Gradient Centrifugation.
Membrane separation methodology was adapted from previously published methods for isolation of OMs from Gram-negative bacteria (24, 25, 40). For details, see SI Materials and Methods.
β-Galactosidase Assays.
Bacteria containing the lacZ fusions were grown in DMEM to an OD600 of 1.0 at 37 °C. These cultures were assayed for β-galactosidase activity as described previously (41). β-Galactosidase activity of the qse-lacZ in WT and qseE, qseF, and qseG mutants was assayed in DMEM. β-Galactosidase activity of the espFu-lacZ in WT and qseE mutant was assayed in minimal media in the absence and presence of 15 mM phosphate and sulfate.
Supplementary Material
Acknowledgments.
We thank J. R. Falck (University of Texas Southwestern Medical Center) for the AI-3; James Kaper (University of Maryland School of Medicine, Baltimore) for the EspB, Tir, and intimin antibodies and the cya fusions; Thomas Silhavy (Princeton University, Princeton) for the Imp antibody; Gad Frankel (Imperial College, London) for the EspA antibody and plasmid pICC284; and Jason Huntley for assistance with sucrose gradient centrifugation. This work was supported by National Institutes of Health Grant AI053067, the Ellison Medical Foundation, and the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811409106/DCSupplemental.
References
- 1.Hughes DT, Sperandio V. Inter-kingdom signaling: Communication between bacteria and host. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2007;44:271–278. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 7.Tobe T, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng W, et al. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campellone KG, Robbins D, Leong JM. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Garmendia J, et al. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol. 2004;6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 11.Cheng HC, Skehan BM, Campellone KG, Leong JM, Rosen MK. Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U) Nature. 2008;454:1009–1013. doi: 10.1038/nature07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furness JB. Types of neurons in the enteric nervous system. J Auton Nerv Syst. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 14.Purves D, et al. Neuroscience. New York: Sinaur Associates; 2001. [Google Scholar]
- 15.Horger S, Schultheiss G, Diener M. Segment-specific effects of epinephrine on ion transport in the colon of the rat. Am J Physiol. 1998;275:G1367–G1376. doi: 10.1152/ajpgi.1998.275.6.G1367. [DOI] [PubMed] [Google Scholar]
- 16.Sperandio V, Li CC, Kaper JB. Quorum-sensing Escherichia coli regulator A: A regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect Immun. 2002;70:3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reading NC, et al. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol. 2007;189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto K, et al. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 19.Eguchi Y, et al. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanie-Cornet MP, Cam K, Jacq A. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J Bacteriol. 2006;188:4264–4270. doi: 10.1128/JB.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B, Choi KY, Zalkin H. Regulation of Escherichia coli glnB, prsA, and speA by the purine repressor. J Bacteriol. 1993;175:3598–3606. doi: 10.1128/jb.175.11.3598-3606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Magasanik B. The glnB region of the Escherichia coli chromosome. J Bacteriol. 1993;175:7441–7449. doi: 10.1128/jb.175.22.7441-7449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido H. Isolation of outer membranes. Methods Enzymol. 1994;235:225–234. doi: 10.1016/0076-6879(94)35143-0. [DOI] [PubMed] [Google Scholar]
- 25.Osborn MJ, Gander JE, Parisi E, Carson J. Mechinism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 26.Knutton S, Baldwin T, Williams PH, McNeish AS. Actin accumulation at sites of bacterial adhesion to tissue culture cells: Basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 28.Wolff J, Cook GH, Goldhammer AR, Berkowitz SA. Calmodulin activates prokaryotic adenylate cyclase. Proc Natl Acad Sci USA. 1980;77:3841–3844. doi: 10.1073/pnas.77.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford JA, Kaper JB. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol Microbiol. 2002;46:855–868. doi: 10.1046/j.1365-2958.2002.03214.x. [DOI] [PubMed] [Google Scholar]
- 30.Janausch IG, Garcia-Moreno I, Unden G. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J Biol Chem. 2002;277:39809–39814. doi: 10.1074/jbc.M204482200. [DOI] [PubMed] [Google Scholar]
- 31.Fardini Y, et al. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica Serovar Enteritidis. Infect Immun. 2007;75:358–370. doi: 10.1128/IAI.00716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, Zaborina O, Holbrook C, Zaborin A, Alverdy J. Depletion of intestinal phosphate after operative injury activates the virulence of P aeruginosa causing lethal gut-derived sepsis. Surgery. 2008;144:189–197. doi: 10.1016/j.surg.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada A, et al. Targeting two-component signal transduction: A novel drug discovery system. Methods Enzymol. 2007;422:386–395. doi: 10.1016/S0076-6879(06)22019-6. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarvis KG, et al. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knutton S, et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenny B, Lai LC, Finlay BB, Donnenberg MS. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 39.Crepin VF, Shaw R, Abe CM, Knutton S, Frankel G. Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J Bacteriol. 2005;187:2881–2889. doi: 10.1128/JB.187.8.2881-2889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huntley JF, Conley PG, Hagman KE, Norgard MV. Characterization of Francisella tularensis outer membrane proteins. J Bacteriol. 2007;189:561–574. doi: 10.1128/JB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.