Abstract
The earliest Neotropical primate fossils complete enough for taxonomic assessment, Dolichocebus, Tremacebus, and Chilecebus, date to approximately 20 Ma. These have been interpreted as either closely related to extant forms or as extinct stem lineages. The former hypothesis of morphological stasis requires most living platyrrhine genera to have diverged before 20 Ma. To test this hypothesis, we collected new complete mitochondrial genomes from Aotus lemurinus, Saimiri sciureus, Saguinus oedipus, Ateles belzebuth, and Callicebus donacophilus. We combined these with published sequences from Cebus albifrons and other primates to infer the mitochondrial phylogeny. We found support for a cebid/atelid clade to the exclusion of the pitheciids. Then, using Bayesian methods and well-supported fossil calibration constraints, we estimated that the platyrrhine most recent common ancestor (MRCA) dates to 19.5 Ma, with all major lineages diverging by 14.3 Ma. Next, we estimated catarrhine divergence dates on the basis of platyrrhine divergence scenarios and found that only a platyrrhine MRCA less than 21 Ma is concordant with the catarrhine fossil record. Finally, we calculated that 33% more change in the rate of evolution is required for platyrrhine divergences consistent with the morphologic stasis hypothesis than for a more recent radiation. We conclude that Dolichocebus, Tremacebus, and Chilecebus are likely too old to be crown platyrrhines, suggesting they were part of an extinct early radiation. We note that the crown platyrrhine radiation was concomitant with the radiation of 2 South American xenarthran lineages and follows a global temperature peak and tectonic activity in the Andes.
Keywords: mitochondrial phylogeny, molecular clock, New World monkeys, primate evolution, platyrrhine origins
South America existed as an island continent from 80 to 3.5 Ma, resulting in a highly endemic mammalian fauna. Primates are thought to have joined this fauna sometime between 50 and 30 Ma (reviewed in ref. 1) by rafting or island hopping across the then-narrower Atlantic Ocean (2). The phylogenetic relationships and divergence times of the Neotropical primates (Platyrrhini) are not as well understood as those of their closest relatives, the Old World monkeys and apes (Catarrhini) (e.g., refs. 3 and 4). It has been suggested that the macroevolutionary pattern among platyrrhines is markedly different from that of catarrhines, with platyrrhine evolution characterized by a single ancient radiation with long-lived and morphologically conservative lineages, whereas catarrhines have experienced significant faunal turnover (5, 6). This “morphological stasis hypothesis” (MSH) is based on an interpretation of the platyrrhine fossil record that places the known fossil taxa, including the earliest, within the crown platyrrhine radiation. The “successive radiations hypothesis” (SRH) suggests that the earliest fossil taxa are outside the crown radiation and part of an earlier, extinct radiation (7, 8), thus making platyrrhine and catarrhine evolution more similar. If the MSH is correct, then the most recent common ancestor (MRCA) of crown platyrrhines must be ancient, and the major lineages must have diverged before the first appearance of their fossil relatives. Here we test the MSH with divergence date estimates and evolutionary rate comparisons inferred from complete heavy-strand protein-coding mitochondrial DNA sequences.
Recent molecular studies (9–14) have confirmed monophyly of the 3 major divisions within the platyrrhines: the pitheciids, consisting of the pitheciines and Callicebus; the atelids; and the cebids, consisting of the callitrichines, Cebus, Saimiri, and Aotus (Table 1). The relationships among these larger groups have proved difficult to resolve (9–14), although a recent study of rare genomic insertion events supports a basal division of pitheciids and a sister relationship between atelids and cebids (12). The earliest fossil platyrrhines are known from localities in Argentina, Chile, and Bolivia. Early primate-bearing sediments span the Oligocene–Miocene boundary from 26 to 20 Ma (15, 16). The oldest fossil taxa with diagnosable morphology, Dolichocebus, Tremacebus, and Chilecebus, are found in early Miocene layers (approximately 20 Ma). Proponents of the MSH consider Dolichocebus a relative of Saimiri (17), Tremacebus of Aotus (5), and Chilecebus of the cebines, Cebus and Saimiri (16). This requires the 3 major crown platyrrhine lineages to have diverged by 20 Ma. The SRH considers these taxa to be stem platyrrhines outside the crown radiation (7, 8). Under the SRH the radiation of extant platyrrhines is more recent, and similarities between modern taxa and the early Miocene taxa are convergent (7).
Table 1.
Sequences used in this study
| Group | Taxa | Common name | Accession no. |
|---|---|---|---|
| Haplorrhine | |||
| Anthropoid | |||
| Platyrrhine | |||
| Cebid | Aotus lemurinus* | Gray-bellied night monkey | FJ785421 |
| Cebus albifrons | White-fronted capuchin | AJ309866 | |
| Saimiri sciureus* | Common squirrel monkey | FJ785425 | |
| Saguinus oedipus* | Cottontop tamarin | FJ785424 | |
| Atelid | Ateles belzebuth* | White-fronted spider monkey | FJ785422 |
| Pitheciid | Callicebus donacophilus* | White-eared titi monkey | FJ785423 |
| Catarrhine | |||
| Cercopithecoid | Chlorocebus sabaeus | Green monkey | EF597503 |
| Papio hamadryas | Hamadryas baboon | NC_001992 | |
| Theropithecus gelada* | Gelada | FJ785426 | |
| Colobus guereza | Mantled guereza | AY863427 | |
| Trachypithecus obscurus | Dusky leaf monkey | NC_006900 | |
| Hominoid | Homo sapiens | Human | EF061150 |
| Pan troglodytes | Common chimpanzee | EU095335 | |
| Pongo pygmaeus | Bornean orangutan | NC_001646 | |
| Hylobates lar | White-handed gibbon | NC_002082 | |
| Tarsioid | Tarsius bancanus | Western tarsier | NC_002811 |
| Strepsirhine | |||
| Lemurid | Lemur catta | Ring-tailed lemur | AJ421451 |
| Lorisid | Nycticebus coucang | Slow loris | NC_002765 |
*Produced for this study.
Several studies have estimated platyrrhine divergence dates from nucleotide sequences, with findings of the MRCA of living platyrrhines ranging from 20 to 26 Ma (10, 11, 18, 19). Those studies finding a MRCA of approximately 20 Ma would seem to falsify the MSH, but the confidence intervals for these dates are large enough that an MRCA before the Miocene cannot be excluded (10, 18). By choosing mitochondrial genomes, we were able to increase resolution by using long DNA sequences and multiple well-constrained fossil calibration dates (3). In a comprehensive analysis using multiple fossil calibration schemes, we developed an exploratory approach to estimating the evolutionary rates and divergence times of the major platyrrhine lineages under both hypotheses. We applied the Bayesian method of Thorne et al. (20, 21) as implemented with multidivtime from the MULTIDISTRIBUTE package. This method accommodates uncertainty in fossil calibration points, which avoids potentially misleading confidence intervals associated with fixed calibration points (22). In addition, rates of evolution are allowed to vary between branches (i.e., no strict molecular clock is enforced), and variance and credibility intervals of node times and branch rates are calculated. In total more than 665 multidivtime analyses were performed.
Results and Discussion
Phylogenetic Inference.
We sequenced a complete mitochondrial genome from Aotus lemurinus, Saimiri sciureus, Saguinus oedipus, Ateles belzebuth, and Callicebus donacophilus and added them to a mitochondrial genome of Cebus albifrons taken from GenBank, thus representing each of the major platyrrhine lineages. We also sequenced a mitochondrial genome of Theropithecus gelada. This taxon was chosen because the fossil record of Theropithecus is well known, and therefore the timing of the Papio/Theropithecus split is well constrained. To these sequences we added a selection of available mitochondrial genomes from GenBank chosen to provide nodes temporally constrained by a good fossil record, to fill out major branches of the haplorhine tree, or to root the tree (Table 1).
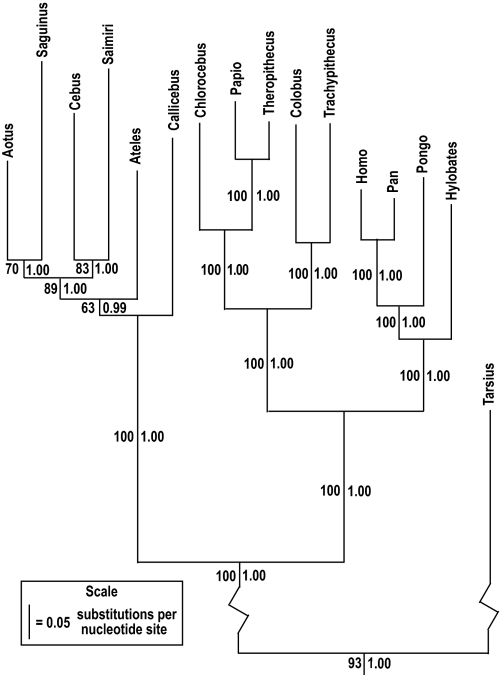
An alignment of the heavy-strand protein-coding genes (10,906 bp total) was used for phylogenetic inference using maximum likelihood and Bayesian methods as implemented in PAUP* (23) and MrBayes (24), respectively. In both cases the data were analyzed with the GTR + I + G model of sequence evolution (chosen by both Modeltest and MrModeltest). Both methods recovered the same topology (Fig. 1). Although the Bayesian clade credibility values are high across the tree, the maximum likelihood bootstrap values are low for all nodes within the platyrrhines. This is not surprising given the very short branch lengths inferred for all of the platyrrhine internodes (see branch lengths below and Fig. 1). Two of our inferred relationships are of note. First, we infer a cebid/atelid clade to the exclusion of the pitheciids. No sequence-based study, including this one, has resolved the platyrrhine family relationships convincingly (9–11, 13, 14). Our findings agree with the strong evidence provided by 5 shared Alu insertion events between cebids and atelids (12). Second, within the cebids, Aotus was found to be sister to Saguinus. Previous sequence-based studies have inferred every possible relationship between Aotus, the callitrichines, and the cebines with various levels of support (9–11, 13, 14). Our results differ from the Alu insertion data showing one shared event between Aotus and Saimiri to the exclusion of Saguinus (12). It remains unclear whether the inferred mitochondrial phylogeny within the cebids is reflective of the true population history of divergence. This is not problematic for our purpose. Even if the mitochondrial history differs from the population history, it must be the case that the populations diverged after the mitochondrial lineages diverged.
Fig. 1.
Topology and branch lengths used for divergence time estimates. Phylogeny was inferred through maximum likelihood (PAUP*) and Bayesian (MrBayes) analysis with Lemur and Nycticebus as outgroups. Maximum likelihood bootstrap percentages are left of the branches, and Bayesian clade credibility proportions are right. Maximum likelihood branch lengths were estimated using the F84 + G model of evolution (estbranches).
Branch Lengths.
Because of uncertainty in tree topology we have used 4 alternate hypotheses for molecular dating: (i) the best inferred topology (Fig. 1), (ii) pitheciids sister to atelids, (iii) Aotus, Saguinus, Cebus/Saimiri trichotomy, and (iv) Aotus sister to Cebus/Saimiri. The latter were constructed by maintaining all relationships shown in Fig. 1 except for those indicated. In all cases only Lemur was used to root the trees. First, maximum likelihood parameters under the F84 model of evolution with 5 discrete γ categories were calculated using baseml from the PAML package (25) for all 4 topologies. These parameters were then used to estimate branch lengths and their variance–covariance matrix using the estbranches program from the MULTIDISTRIBUTE package.
The estimated branch lengths for the best topology are depicted in Fig. 1. Of note is the long branch leading from the anthropoid MRCA to the platyrrhine MRCA. This branch is 64% longer than the branch leading to the catarrhine MRCA, suggesting that platyrrhines either diversified much more recently or that the ancestral rate of sequence evolution was faster leading to the platyrrhine MRCA than to the catarrhine MRCA. The mean branch from the anthropoid MRCA to tip amongst platyrrhines has 0.64 substitutions per site, compared with 0.60 substitutions per site amongst catarrhines, suggesting that platyrrhines have on average evolved faster than catarrhines. However, it has long been recognized that hominoids have evolved at a slower rate than cercopithecoids (3, 26, 27). The mean branch from the anthropoid MRCA to tip amongst hominoids has 0.54 substitutions per site, and among cercopithecoids has 0.64 substitutions per site. Thus, platyrrhines have, on average, undergone an equivalent amount of sequence evolution as have cercopithecoids, whereas hominoids have experienced less. This suggests that it is indeed a “hominoid slowdown” and not a cercopithecoid speed-up. Also of note are the extremely short internodes within platyrrhines. The 4 platyrrhine internodes represent the 4 shortest branches in the tree. This implies that the diversification of the living platyrrhines must represent a rapid radiation.
Divergence Dates with Best Available Constraints.
We estimated the times of divergence between all lineages using the best constraints available for all nodes in the tree for all 4 topologies. Because the test of the hypothesis depends on the Bayesian credibility intervals estimated for the platyrrhine nodes, we used the largest reasonable ranges when assigning constraints to conservatively estimate credibility intervals. Upper constraints (more recent) are logically established by the first appearance of a derived lineage in the fossil record. Lower constraints cannot be definitively set, so we have chosen to place lower bounds at or beyond what most authorities consider possible. When too little fossil information is available we have not set lower bounds.
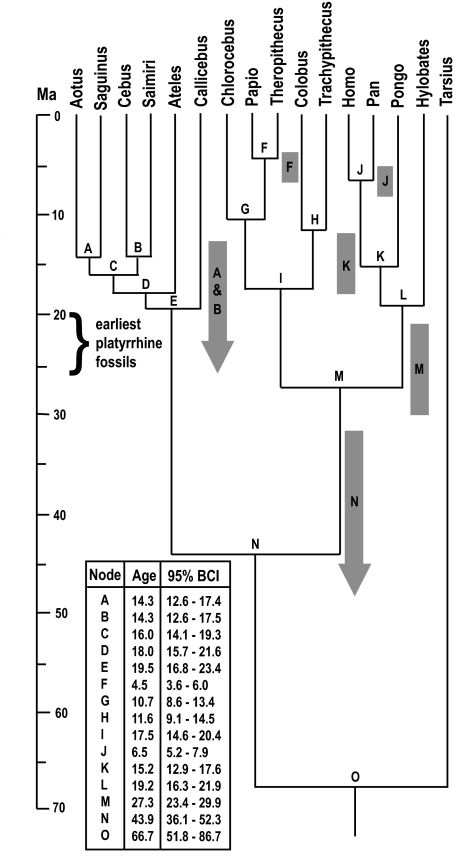
We set calibration constraints on 5 nodes within the catarrhines. The only constraints used within the platyrrhines were that the Cebus/Saimiri and the Aotus/Saguinus splits must have occurred by 12.5 Ma on the basis of the La Venta fauna (Table 2). The lack of lower bounds within the platyrrhines fully allows for the data to support the MSH. However, the estimated divergence dates for the best topology (Fig. 2) are clearly inconsistent with the MSH. The MRCA of the living platyrrhines is estimated to have lived 19.5 Ma (95% credibility interval 16.8–23.4 Ma), and the Aotus/Saguinus and Cebus/Saimiri splits are estimated at 14.3 (95% credibility interval 12.6–17.4) and 14.3 (95% credibility interval 12.6–17.5) Ma, respectively. Although the credibility interval of the platyrrhine MRCA encompasses the early Miocene fossils, the Cebus/Saimiri and Aotus/Saguinus splits are both far younger than the early Miocene fossils, suggesting that Dolichocebus and Tremacebus cannot be close relatives of Saimiri and Aotus, respectively, as proponents of the MSH purport (5, 17). The lower bound of credibility for the split between the cebids is 19.3 Ma, also making it unlikely that Chilecebus is a cebid (16). Overall, these divergence estimates suggest that if Dolichocebus, Tremacebus, or Chilecebus fall within extant diversity, they can only be pitheciids or stem members of the atelid/cebid clade.
Table 2.
Evolutionary rate calibration constraints
| Constraint | Divergence | Upper (Ma) | Lower (Ma) | Fossil (reference) | Age (Ma) |
|---|---|---|---|---|---|
| 1 | Homo/Pan | 5.0 | 8.0 | Ardipithecus (28) | 5.2 |
| Orrorin (29) | 6.0 | ||||
| Sahelanthropus (30, 31) | 6.0–7.0 | ||||
| 2 | Homo/Pongo | 12.5 | 18.0 | Sivapithecus (32) | ≈12.5 |
| 3 | Papio/Theropithecus | 3.5 | 6.5 | Theropithecus (33) | ≈3.5 |
| 4 | Hominoid/cercopithecoid | 21.0 | 30.0 | Morotopithecus (34) | >20.6 |
| Victoriapithecus (35) | ≈19.0 | ||||
| 5 | Platyrrhine/catarrhine | 31.5 | NA | Fayum catarrhines (36, 37) | 31.5 |
| 6 | Aotus/Saguinus | 12.5 | NA | Aotus dindensis (38) | ≈12.5 |
| 7 | Cebus/Saimiri | 12.5 | NA | Neosaimiri (39) | ≈12.5 |
Fig. 2.
Divergence time estimates with Bayesian credibility intervals (BCI) using the best available fossil constraints. Gray bars indicate constraint ranges for the nodes indicated on them. Arrows indicate that no lower constraint was used.
To test the effects of topology on the divergence dates, we performed the same analysis as above for each of the 3 alternative topologies. In no case did the lower bounds of the credibility intervals accommodate the MSH. Making pitheciids and atelids sister reduces the age estimate of the platyrrhine MRCA to 17.7 Ma (95% credibility interval 15.3–21.6), with the lower credibility bounds of the Aotus/Saguinus and Cebus/Saimiri splits being no older than 18.2 Ma. Moving Aotus within the cebids (Aotus, Saguinus, Cebus/Saimiri trichotomy and Aotus sister to Cebus/Saimiri) results in ages slightly older than those for the best topology; however, in no case are the lower-bounds of the cebid divergences old enough to accommodate Dolichocebus, Tremacebus, or Chilecebus within them. Although topology can be critical to molecular date estimates, the extremely short platyrrhine internodes mean that any reasonable rearrangement adds little length to the tree and consequently does not effect the dates greatly. In all subsequent analyses we use the best topology (Fig. 1).
Concordance with Catarrhine Fossil Record.
Next, we assessed concordance between divergence dates on the platyrrhine fossil record and the better-known catarrhine fossil record. Fossil estimates of divergence times are concordant when the divergence times independently predict each other given the observed branch lengths (40). This happens when the branch length per unit time between fossils is proportional to the branch length per unit time between each fossil and time 0 at the terminal branches. We first tested for concordance among the 4 catarrhine divergences used as constrained calibration nodes in the previous analysis. To determine the effects of the calibration constraints at each node on the others, we varied constraints on each node individually while leaving all other nodes unconstrained. We slid lower and upper constraints, separated by 1 million years (Myr), across a wide range of ages at 1-Myr intervals. We varied the Papio/Theropithecus split between 0 and 16 Ma, the Pan/Homo split between 1 and 20 Ma, the Pongo/Homo split between 6 and 30 Ma, and the MRCA of catarrhines between 10 and 40 Ma. In total 89 multidivtime analyses were performed, and the predicted times of divergence from the unconstrained nodes were recorded from each analysis. We found very strong agreement between the fossil estimates of divergence times given the estimated branch lengths (data not shown). Only calibration constraints consistent with fossil estimates for a node realistically predict the remaining 3 nodes. For example, a Pan/Homo split between 5 and 6 Ma predicts the Papio/Theropithecus split at 3.7 Ma, the Pongo/Homo split at 12.9 Ma, and the catarrhine MRCA at 23.1 Ma, whereas a Pan/Homo split between 9 and 10 Ma predicts the Papio/Theropithecus split at 6.2 Ma, the Pongo/Homo split at 21.7 Ma, and the catarrhine MRCA at 38.3 Ma. Thus, the 4 independent fossil estimations of catarrhine divergence times do not require much evolutionary rate variation given the observed branch lengths.
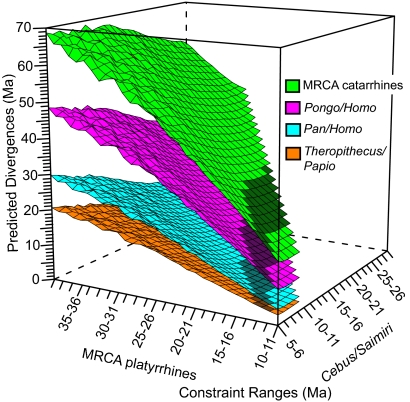
Because of the strong concordance between the 4 catarrhine nodes, we used these as tests to look for concordant platyrrhine divergence times. To find such times, we took a similar exploratory approach to that described above. However, here we varied 1-Myr constraint intervals on the Cebus/Saimiri split and the platyrrhine MRCA simultaneously. We varied the Cebus/Saimiri split between 5 and 30 Ma and the platyrrhine MRCA between 10 and 40 Ma in 1-Myr increments. We ran an analysis with every possible combination of constraints on these nodes. In total, 540 multidivtime analyses were performed (Fig. 3). This process was automated with a Perl script written for this project.
Fig. 3.
Surface plot of predicted catarrhine divergence times given various platyrrhine divergence scenarios. The figure summarizes the results of 540 multidivtime analyses. The constraints used for each analysis are given on the x and y axes, and the predicted catarrhine divergence times given these constraints are shown on the z axis. Shaded regions indicate platyrrhine calibrations that predict catarrhine divergences concordant with the catarrhine fossil record.
There were 47 platyrrhine constraint combinations that predicted divergence times for the Pan/Homo, Papio/Theropithecus, Pongo/Homo, and catarrhine MRCA within the conservative ranges inferred from the fossil record and described above (Fig. 3, shaded regions). The oldest the Cebus/Saimiri split can be to predict the 4 test nodes within the defined ranges is between 18 and 19 Ma, whereas the oldest platyrrhine MRCA is between 20 and 21 Ma. Platyrrhine constraint times consistent with the MSH predict divergence times for the catarrhine nodes that are older than the fossil record suggests. For example, a Cebus/Saimiri split between 22 and 23 Ma and a platyrrhine MRCA between 27 and 28 Ma predicts the Pan/Homo split at 10.1 Ma, the Papio/Theropithecus split at 6.6 Ma, the Pongo/Homo split at 22.8 Ma, and the catarrhine MRCA at 39.6 Ma.
Rates of Evolution.
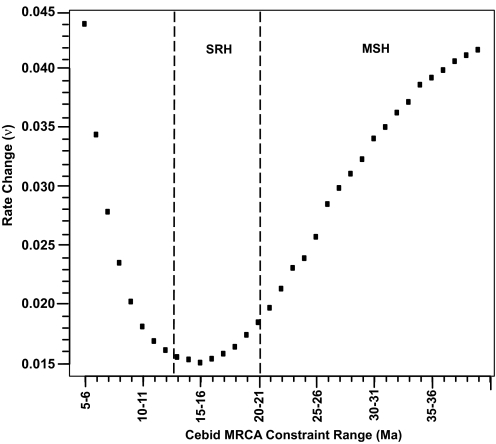
Because platyrrhine divergence times consistent with the MSH are not concordant with the catarrhine fossil record, if the MSH is correct then rates of evolution must have changed through time. We characterized the amount of evolutionary rate change required by various platyrrhine divergence times in 2 ways. First, we tracked changes in the autocorrelation parameter ν calculated with multidivtime under different platyrrhine divergence scenarios. This parameter characterizes the amount of rate variation among adjacent branches, with ν = 0 meaning no rate change (e.g., strict molecular clock) and increasing rate variation as ν increases (20, 21). For this analysis, we applied the constraints used in the first analysis for the 5 nodes outside platyrrhines. Within the platyrrhines we chose to apply constraints to the MRCA of cebids because Tremacebus, Dolichocebus, and Chilecebus are considered to be cebids by the MSH. Using an approach similar to that used in the concordance analysis, we slid lower and upper constraints separated by 1 Myr between 5 and 40 Ma in 1-Myr intervals. Thirty-five multidivtime analyses were performed, and ν was recorded for each (Fig. 4). It was found that a cebid MRCA between 15 and 16 Ma requires the least amount of evolutionary rate change (ν = 0.0151). For cebid MRCA divergence times consistent with the SRH, ν ranges from 0.0151 to 0.0175, whereas for times consistent with the MSH, the minimum ν is 0.0185.
Fig. 4.
Change in the rate of evolution between branches given various platyrrhine divergence scenarios. The figure gives the estimated autocorrelation parameter (ν) for 35 multidivtime runs. Five constraint intervals were constant for all runs (Table 2, constraints 1–5). The cebid MRCA was varied for each run, and the constraint intervals used are shown on the x axis. The dashed lines indicate the La Venta fossils at 12.5 Ma and the disputed early Miocene fossils at 20 Ma and demarcate the SRH and the MSH.
Finally, we estimated the average difference in evolutionary rates between adjacent branches under the MSH and the SRH. To do so we used multidivtime to estimate evolutionary rates along each branch using the best estimates of divergence times consistent with each hypothesis respectively. We then calculated the average rate change as the mean absolute difference in rate between adjacent branches. To calculate rates under the SRH we used the 5 catarrhine constraints used in the previous analyses (Table 2) and constrained the Cebus/Saimiri and Aotus/Saguinus divergences between 12.5 and 20 Ma. To calculate rates under the MSH we took a similar approach but constrained the Cebus/Saimiri and Aotus/Saguinus divergences older than 20 Ma. The average evolutionary rate change between adjacent branches given the SRH constraints is 0.0028 substitutions per site per Myr, whereas that for the MSH is 0.0031 substitutions per site per Myr. Thus, the MSH requires on average slightly greater change in evolutionary rates between adjacent branches; however, a paired t test shows this difference not to be significant (t = 0.815, df = 29, P = 0.211, one-tailed).
In the previous analysis, given the SRH constraints, the estimated divergence times are similar to those shown in Fig. 2, whereas the divergence times given the MSH constraints are very different. To minimize the amount or evolutionary rate change in the latter experiment, multidivtime pushed the divergence times for the nodes outside the platyrrhines back in time to account for the necessarily older platyrrhine divergences. For example, the divergence between platyrrhines and catarrhines is pushed back to 52.8 Ma (95% CI 45.6–62.6) and the divergence between tarsiers and anthropoids to 79.7 Ma (95% CI 63.8–106.2). These times are well beyond what most authorities consider possible. To make the rates comparison more realistic we reanalyzed the data by tightly constraining all nodes in the tree except for the platyrrhine nodes. To do so we constrained the nodes outside the platyrrhines to ±1 Myr of the best estimate of divergence times shown in Fig. 2. To these we added either the MSH or SRH platyrrhine constraints used in the previous analysis. We found that given the SRH the mean rate change between branches is 0.0027 substitutions per site per Myr, compared with 0.0036 substitutions per site per Myr given the MSH. Thus, given the divergence times for the taxa outside the platyrrhines shown in Fig. 2, the MSH requires on average 33% greater evolutionary rate change between adjacent branches than does the SRH. This difference was found to be significant with a paired t test (t = 1.735, df = 29, P = 0.047, one-tailed).
The reason for the greater amount of evolutionary rate change required by the MSH is clear from the inferred branch lengths. The branch leading from the anthropoid MRCA to the platyrrhine MRCA is much longer than that leading to the catarrhine MRCA (Fig. 1). However, the MSH requires the MRCA of platyrrhines be as old or older than the MRCA of catarrhines. Consequently, the MSH requires relatively fast evolution in the ancestral platyrrhine lineage. Because the root-to-tip branch lengths of the platyrrhines are on average as long as those of cercopithecoids (Fig. 1), the MSH then requires the rate of evolution to slow independently in many of the terminal platyrrhine lineages. This accounts for the greater amount of evolutionary rate change seen in the MSH compared with the SRH. Thus, accepting fossil interpretations consistent with the MSH requires accepting much greater disparity in evolutionary rates among branches (e.g., ref. 41).
Our analyses suggest that the radiation of crown platyrrhines was rapid and began approximately 20 Ma, long after primates reached South America. This suggests that the multiple fossil taxa present before 20 Ma are outside of the crown radiation and are therefore part of an earlier radiation. The cause of faunal turnover in South American primates is unclear. We note that amongst the South American-endemic xenarthrans (armadillos, sloths, and anteaters), both armadillos and sloths independently experienced radiations at approximately 20 Ma (42). The primate and xenarthran radiations in the early Miocene follow a peak in global temperature (43) and a major compressional episode in the Chilean Andes caused by an acceleration in the rate at which the Nazca and South American plates met (44). It remains to be seen whether climatic or tectonic changes played any role in the faunal turnover of South American primates or the simultaneous radiations among xenarthrans.
Conclusions
Using a phylogeny representing all major anthropoid clades, we have demonstrated that the MRCA of platyrrhines likely lived too recently for the early Miocene South American primate fauna to fall within extant diversity, indicating that Dolichocebus, Tremacebus, and Chilecebus are stem platyrrhines. This suggests that platyrrhine evolution is characterized by an extinct early radiation followed by the radiation of crown platyrrhines. Although platyrrhine evolution may involve morphological stasis from the time of La Venta approximately 12.5 Ma, this stasis is not likely to extend to the early Miocene.
Materials and Methods
Mitochondrial Genome Sequencing.
Total DNA was extracted from tissue, and mitochondrial genomes were sequenced with established methods that ensure true mitochondrial sequence and not nuclear insertions (3, 45). Sequencing and PCR primers are available upon request.
Phylogenetic Inference.
Heavy-strand protein-coding genes were extracted from complete genome sequences according to GenBank annotations. These genes were individually aligned with ClustalW 2.0.8 (46), and these alignments were concatenated. This process was automated with a Perl script written for this project. To infer the model of nucleotide change that best describes the data set, Modeltest 3.7 (47) and MrModeltest 2.2 were used and evaluated under the Aikake Information Criterion. Model and parameters chosen by Modeltest 3.7 were used for maximum likelihood phylogenetic inference implemented in PAUP* 4.0b10 (23). Topologic support was estimated with 660 bootstrap replicates. The model chosen by MrModeltest 2.2 was used for Bayesian phylogenetic inference as implemented by MrBayes 3.1.2 (24). For the Bayesian analysis, the data were partitioned by codon position, and model parameters were estimated independently for each partition. Four Markov Chain Monte Carlo (MCMC) simulations were run for 10,000,000 generations sampled every 1,000 generations after a burn-in period of 1,000,000 generations.
Molecular Dating.
The various divergence date and evolutionary rate analyses were performed as described above. Bayesian priors for multidivtime were chosen according to J. Thorne's recommendations in the multidivtime manual. Root-to-tip mean was set at 75 Ma, with a 75 Ma standard deviation. The evolutionary rate at the root was set at 0.12 substitutions per nucleotide site per Myr, with a standard deviation of 0.12. This was calculated as the median root-to-tip branch length divided by the root-to-tip mean prior. The autocorrelation parameter prior (brownmean) and its standard deviation were set to 0.026, such that brownmean multiplied by the root-to-tip mean prior (75 Ma) equals ≈2. After a burn-in period of 100,000 generations, MCMC chains were sampled every 100 generations until 10,000 samples were taken. To test the effect of the priors on the posteriors, several runs were performed with various prior choices. The results were extremely robust to changes in priors.
Acknowledgments.
We thank Alfred Rosenberger and Terry Harrison for helpful comments and suggestions; Heidi Reinholdt and Laura Gaydosh for help with laboratory work; and Anthony Di Fiore, Cathi Lehn, and Derek Wildman for providing samples. This research was supported by National Institutes of Health Grants R01-GM060760 and R24-GM65580 (to C.-B.S. and T.R.D.).
Footnotes
References
- 1.Flynn JJ, Wyss AR. Recent advances in South American mammalian paleontology. Trends Ecol Evol. 1998;13:449–454. doi: 10.1016/s0169-5347(98)01457-8. [DOI] [PubMed] [Google Scholar]
- 2.Hartwig WC. Patterns, puzzles and perspectives on platyrrhine origins. In: Corruccini RS, Ciochon RL, editors. Integrative Paths to the Past. Englewood Cliffs, NJ: Prentice-Hall; 1994. pp. 69–93. [Google Scholar]
- 3.Raaum RL, et al. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. J Hum Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Stewart CB, Disotell TR. Primate evolution—In and out of Africa. Curr Biol. 1998;8:R582–588. doi: 10.1016/s0960-9822(07)00367-3. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberger AL. Platyrrhine paleontology and systematics: The paradigm shifts. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 151–159. [Google Scholar]
- 6.Delson E, Rosenberger AL. Are there any anthropoid primate living fossils. In: Eldridge N, Stanley SM, editors. Living Fossils. New York: Springer Verlag; 1984. pp. 50–61. [Google Scholar]
- 7.Kay RF, et al. The anatomy of Dolichocebus gaimanensis, a stem platyrrhine monkey from Argentina. J Hum Evol. 2008;54:323–382. doi: 10.1016/j.jhevol.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Kay RF. The phyletic relationships of extant and fossil Pitheciinae (Platyrrhini, Anthropoidea) J Hum Evol. 1990;19:175–208. [Google Scholar]
- 9.Horovitz I, Zardoya R, Meyer A. Platyrrhine systematics: A simultaneous analysis of molecular and morphological data. Am J Phys Anthropol. 1998;106:261–281. doi: 10.1002/(SICI)1096-8644(199807)106:3<261::AID-AJPA1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Schrago CG. On the time scale of New World primate diversification. Am J Phys Anthropol. 2007;132:344–354. doi: 10.1002/ajpa.20459. [DOI] [PubMed] [Google Scholar]
- 11.Opazo JC, et al. Phylogenetic relationships and divergence times among New World monkeys (Platyrrhini, Primates) Mol Phylogenet Evol. 2006;40:274–280. doi: 10.1016/j.ympev.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Ray DA, et al. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Steiper ME, Ruvolo M. New World monkey phylogeny based on X-linked G6PD DNA sequences. Mol Phylogenet Evol. 2003;27:121–130. doi: 10.1016/s1055-7903(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 14.von Dornum M, Ruvolo M. Phylogenetic relationships of the New World monkeys (Primates, Platyrrhini) based on nuclear G6PD DNA sequences. Mol Phylogenet Evol. 1999;11:459–476. doi: 10.1006/mpev.1998.0582. [DOI] [PubMed] [Google Scholar]
- 15.Takai M, Anaya F, Shigehara N, Setoguchi T. New fossil materials of the earliest new world monkey, Branisella boliviana, and the problem of platyrrhine origins. Am J Phys Anthropol. 2000;111:263–281. doi: 10.1002/(SICI)1096-8644(200002)111:2<263::AID-AJPA10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Fleagle JG, Tejedor MF. Early platyrrhines of southern South America. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 161–173. [Google Scholar]
- 17.Rosenberger AL. Cranial anatomy and implications of Dolichocebus, a late Oligocene ceboid primate. Nature. 1979;279:416–418. doi: 10.1038/279416a0. [DOI] [PubMed] [Google Scholar]
- 18.Poux C, et al. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst Biol. 2006;55:228–244. doi: 10.1080/10635150500481390. [DOI] [PubMed] [Google Scholar]
- 19.Schneider H. The current status of the New World monkey phylogeny. Anais da Academia Brasileira de CiÍncias. 2000;72:165–172. doi: 10.1590/s0001-37652000000200005. [DOI] [PubMed] [Google Scholar]
- 20.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 21.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 22.Graur D, Martin W. Reading the entrails of chickens: Molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Swofford DL. Sunderland, MA: Sinauer Associates; 2004. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Version 4. [Google Scholar]
- 24.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 26.Goodman M. The role of immunochemical differences in the phyletic development of human behavior. Hum Biol. 1961;33:131–162. [PubMed] [Google Scholar]
- 27.Steiper ME, Young NM, Sukarna TY. Genomic data support the hominoid slowdown and an Early Oligocene estimate for the hominoid-cercopithecoid divergence. Proc Natl Acad Sci USA. 2004;101:17021–17026. doi: 10.1073/pnas.0407270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haile-Selassie Y. Late Miocene hominids from the Middle Awash, Ethiopia. Nature. 2001;412:178–181. doi: 10.1038/35084063. [DOI] [PubMed] [Google Scholar]
- 29.Senut B, et al. First hominid from the Miocene (Lukeino Formation, Kenya) Comptes Rendus De L Academie Des Sciences Serie Ii Fascicule a-Sciences De La Terre Et Des Planetes. 2001;332:137–144. [Google Scholar]
- 30.Vignaud P, et al. Geology and palaeontology of the Upper Miocene Toros-Menalla hominid locality, Chad. Nature. 2002;418:152–155. doi: 10.1038/nature00880. [DOI] [PubMed] [Google Scholar]
- 31.Brunet M, et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418:145–151. doi: 10.1038/nature00879. [DOI] [PubMed] [Google Scholar]
- 32.Kelley J. The hominoid radiation in Asia. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 369–384. [Google Scholar]
- 33.Leakey MG. Evolution of Theropithecus in the Turkana Basin. In: Jablonski NG, editor. Theropithecus: The Rise and Fall of a Primate Genus. Cambridge, UK: Cambridge Univ Press; 1993. pp. 85–123. [Google Scholar]
- 34.Young NM, MacLatchy L. The phylogenetic position of Morotopithecus. J Hum Evol. 2004;46:163–184. doi: 10.1016/j.jhevol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Benefit BR, McCrossin ML. The Victoriapithecidae, Cercopithecoidea. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 241–253. [Google Scholar]
- 36.Rasmussen DT. Early catarrhines of the African Eocene and Oligocene. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 203–220. [Google Scholar]
- 37.Seiffert ER. Revised age estimates for the later Paleogene mammal faunas of Egypt and Oman. Proc Natl Acad Sci USA. 2006;103:5000–5005. doi: 10.1073/pnas.0600689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setoguchi T, Rosenberger AL. A fossil owl monkey from La Venta, Colombia. Nature. 1987;326:692–694. doi: 10.1038/326692a0. [DOI] [PubMed] [Google Scholar]
- 39.Hartwig WC, Meldrum DJ. Miocene platyrrhines of the northern Neotropics. In: Hartwig WC, editor. The Primate Fossil Record. Cambridge, UK: Cambridge Univ Press; 2002. pp. 175–188. [Google Scholar]
- 40.Near TJ, Sanderson MJ. Assessing the quality of molecular divergence time estimates by fossil calibrations and fossil-based model selection. Philos Trans R Soc Lond B Biol Sci. 2004;359:1477–1483. doi: 10.1098/rstb.2004.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberger AL. Fossil New World monkeys dispute the molecular clock. J Hum Evol. 1984;13:737–742. [Google Scholar]
- 42.Delsuc F, Vizcaino SF, Douzery EJ. Influence of Tertiary paleoenvironmental changes on the diversification of South American mammals: A relaxed molecular clock study within xenarthrans. BMC Evol Biol. 2004;4:11. doi: 10.1186/1471-2148-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zachos J, et al. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 44.Sempere T, Marshall LG, Rivano S, Godoy E. Late Oligocene-Early Miocene compressional tectosedimentary episode and associated land-mammal faunas in the Andes of central Chile and adjacent Argentina (32–37∞s) Tectonophysics. 1994;229:251–264. [Google Scholar]
- 45.Sterner KN, et al. Mitochondrial data support an odd-nosed colobine clade. Mol Phylogenet Evol. 2006;40:1–7. doi: 10.1016/j.ympev.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]