Abstract
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) has recently emerged worldwide. The United States, in particular, is experiencing a serious epidemic of CA-MRSA that is almost entirely caused by an extraordinarily infectious strain named USA300. However, the molecular determinants underlying the pathogenic success of CA-MRSA are mostly unknown. To gain insight into the evolution of the exceptional potential of USA300 to cause disease, we compared the phylogeny and virulence of USA300 with that of closely related MRSA clones. We discovered that the sublineage from which USA300 evolved is characterized by a phenotype of high virulence that is clearly distinct from other MRSA strains. Namely, USA300 and its progenitor, USA500, had high virulence in animal infection models and the capacity to evade innate host defense mechanisms. Furthermore, our results indicate that increased virulence in the USA300/USA500 sublineage is attributable to differential expression of core genome-encoded virulence determinants, such as phenol-soluble modulins and α-toxin. Notably, the fact that the virulence phenotype of USA300 was already established in its progenitor indicates that acquisition of mobile genetic elements has played a limited role in the evolution of USA300 virulence and points to a possibly different role of those elements. Thus, our results highlight the importance of differential gene expression in the evolution of USA300 virulence. This finding calls for a profound revision of our notion about CA-MRSA pathogenesis at the molecular level and has important implications for design of therapeutics directed against CA-MRSA.
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) is a major public health problem in the United States (1). In contrast to hospital-associated (HA)-MRSA, CA-MRSA strains cause infections in healthy individuals without predisposing risk factors and outside of the hospital setting. Although CA-MRSA has emerged worldwide, it is epidemic in the United States (2, 3). The vast majority of CA-MRSA infections in the United States are caused by a recently emerged S. aureus clone known as pulsed-field type USA300 (USA300). USA300 infections are primarily those of skin and soft tissue, but the pathogen can cause severe invasive disease and spreads easily and sustainably among humans (3–5). CA-MRSA infections in other countries have not attained comparable levels, produce less severe disease phenotypes, and are commonly caused by clones unrelated to USA300 (2, 6, 7). Given the high transmissibility of USA300, it is possible that this clone could become a problem worldwide.
The molecular determinants underlying the success of USA300 as a pathogen are not understood. Based on genome comparisons and epidemiological evidence, it has been speculated that determinants encoded on mobile genetic elements (MGEs), such as the Panton-Valentine leukocidin (PVL), have a predominant impact on virulence (8–10). However, recent reports indicate that the contribution of these unique MGEs to CA-MRSA virulence may be comparatively minor (11–14). This view is supported by reports on CA-MRSA infections that are caused by PVL-negative strains (2, 6, 7). Thus, there likely exist alternative explanations for the basis and evolution of the enhanced capacity of USA300 to cause widespread disease.
Increases in disease severity or frequency are often linked to the emergence of distinct bacterial clones of high virulence (15, 16). Therefore, to understand the evolution of USA300 better, we performed a comprehensive analysis of the major MRSA subclones of the clonal complex (CC) 8 lineage from which USA300 arose (5, 10). We analyzed CC8 isolates obtained from different geographical regions around the world to determine presence and expression of key virulence factors and evaluated virulence in vitro using human leukocytes and in vivo using animal infection models. We demonstrate that the S. aureus sublineage that led to the emergence of USA300 is characterized by enhanced virulence, clearly distinguishing that sublineage from others within CC8. Furthermore, our analyses indicate that enhanced virulence of USA300 is mainly based on high expression of core genome-encoded virulence determinants rather than the acquisition of additional virulence genes via MGEs. These findings represent a paradigmatic shift of our notion about how the pathogenic potential of USA300 evolved and are important for ongoing efforts aimed to find therapeutics directed against CA-MRSA.
Results
Major MRSA Subclones of the CC8 Lineage.
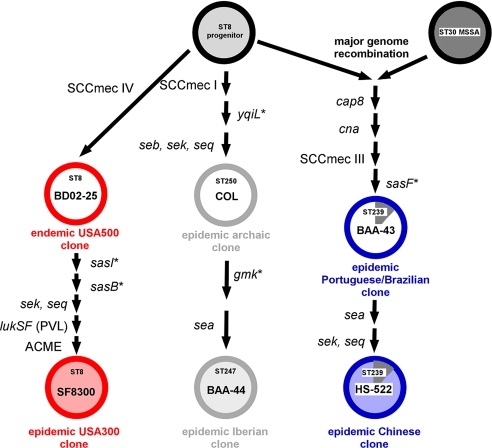
To investigate the evolution of USA300 virulence, we selected 6 S. aureus isolates representing all major epidemic clones of CC8 (17–19) (Fig. 1 and Table S1). First, to infer the genetic relation of these clones, we analyzed DNA sequence variations within 7 housekeeping and 7 surface protein-encoding gene fragments (Fig. 1 and Table S1). According to our analysis, CC8 is split into 3 distinct sublineages. Importantly, the data indicate that USA300 evolved from a USA500 clone. Both USA500 and USA300 are epidemic in hospital and community settings, predominantly in the United States. The second sublineage, comprising the Archaic MRSA clone and its contemporary successor, the Iberian MRSA clone, was widely disseminated in hospitals in the United Kingdom and other parts of Europe shortly after the introduction of β-lactamase–resistant penicillin into clinical use in 1961 (20). The third sublineage, comprising the Brazilian/Portuguese MRSA clone and its successor, the Chinese MRSA clone, is widespread in hospitals in South America and Europe and hospitals in Asia, respectively (17, 18). This sublineage evolved through a rare genetic event involving lateral transfer and homologous recombination with a ≈557-kb fragment from the chromosome of a strain belonging to CC30, another epidemic MRSA lineage (21) (Fig. 1).
Fig. 1.
Evolutionary relation of CC8 subclones. The tree architecture was inferred by analysis of 7 housekeeping and 7 surface protein genes (Table S1). Presence of virulence genes was determined by analytical PCR (Table S2). Strains analyzed in this study are shaded in gray, blue, and red, representing the 3 different CC8 sublineages, and are labeled with the specific strain designations. *Single-nucleotide polymorphisms in housekeeping or surface protein-encoding genes used to infer evolutionary relation (Table S1).
Sequential Acquisition of Virulence Genes by Contemporary CC8 Clones.
To gain information about the genetic basis for the pathogenic potential of the major CC8 clones, we assessed the presence of 43 core genome- or MGE-encoded virulence genes in representative isolates by PCR (Table S2). With specific regard to the evolution of the USA300 sublineage, this analysis led to the key observations that (i) USA500 MRSA does not contain any known enterotoxin genes typically encoded on prophages and pathogenicity islands and (ii) USA300 MRSA evolved from the USA500 background genome, subsequently acquiring multiple additional virulence determinants from MGEs, including the arginine catabolic mobile element (ACME), the staphylococcal pathogenicity island encoding enterotoxin K (sek) and enterotoxin Q (seq), and a prophage that contains the lukSF-PV genes encoding PVL (10, 12).
Significant Differences in Virulence Exist Among CC8 Clones.
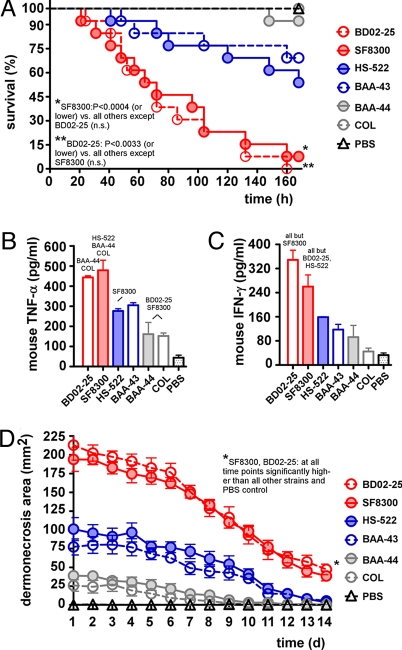
We next evaluated virulence of the 6 selected clones using murine models of bacteremia and skin infection, which are frequent manifestations of disease caused by CA-MRSA (2). USA500 and USA300 were significantly more virulent in the animal infection models compared with the other CC8 strains tested (Fig. 2 A and D). In the bacteremia model, we also measured levels of TNF-α and IFN-γ in blood as markers of inflammation at the time of death (Fig. 2 B and C). The levels of these cytokines correlated well with survival curves in the bacteremia model (compare Fig. 2A with Fig. 2 B and C), consistent with the idea that animals experienced bacterial sepsis. Notably, results for the USA300 and USA500 strains in the infection models were virtually identical, indicating that the acquisition of virulence determinants on MGEs by USA300 did not lead to a significantly enhanced virulence potential by comparison. This is especially noteworthy, given the absence of prophage- and pathogenicity island-encoded enterotoxins in USA500. The virulence of the other strains as assessed by the animal infection models was also similar within the respective sublineages. Notably, in vitro growth of the tested strains was virtually indistinguishable except for strain COL, which showed slightly slower in vitro growth (Fig. S1). This indicates that the presence of different staphylococcal cassette chromosome mec (SCCmec) elements (Fig. 1 and Table S1) does not have a strong impact on bacterial fitness in the analyzed strains, as suggested particularly for the SCCmec type IV element, which is smaller and size and does not lead to different persistence in a competitive infection model compared with an isogenic SCCmec-negative strain (12). Finally, distribution of the 43 virulence genes tested by PCR (Table S2) could not explain the virulence patterns of CC8 subclones detected in the animal infection models simply by gene presence. Thus, our results emphasize the importance of genetic factors intrinsic to the CC8 genotype rather than specific virulence factors acquired via horizontal gene transfer from prophages or pathogenicity islands and suggest that virulence within CC8 is determined to a considerable extent by differential gene expression.
Fig. 2.
Virulence assessment of CC8 subclones by animal infection models. (A) Bacteremia model. CD1 Swiss female mice were infected with 108 cfus of the indicated MRSA strains. Survival curves were compared using log-rank (Mantel-Cox) tests. Concentration of TNF-α (B) and IFN-γ (C) in mouse blood at death. Strains for which statistically significant differences were achieved are named above bars. Compared with PBS, values were significantly different for BD02-25, SF8300, HS-522, and BAA43 (TNF-α) and for BD02-25 and SF8300 (IFN-γ). n.s., not significant. (D) Abscess model. As a control, SKH1-hrBR hairless mice were infected with 107 cfus of the indicated MRSA strains, and abscess or dermonecrosis areas were measured each day.
Interaction with Components of Innate Host Defense.
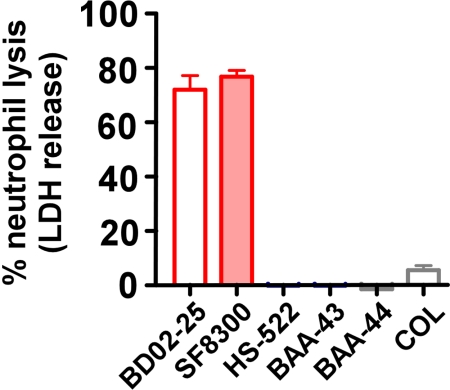
Virulence of S. aureus is largely dependent on the interaction with innate host defense. Neutrophils are the most prominent cellular component of human host defense against S. aureus. Host antimicrobial peptides (AMPs) are a major component of innate host defense in neutrophil phagosomes and on epithelial surfaces (22). To evaluate whether the CC8 strains have differential capacity to interact with these key components of innate host defense, we measured the ability of S. aureus exoproteins to cause lysis of human neutrophils and tested resistance to AMPs. Culture filtrates from USA300 and USA500 caused significant lysis of human neutrophils (70–80%) (Fig. 3). This finding is in pronounced contrast to that observed for all other strains (<10% at most). Additionally, the USA500 and USA300 strains had higher resistance to dermcidin and indolicidin, 2 AMPs of human/mammalian origin, whereas resistance to other AMPs was almost unchanged (Table 1). In summary, these findings indicate that the USA300/USA500 sublineage has increased capacity to evade mechanisms of innate host defense.
Fig. 3.
Neutrophil lysis by CC8 subclones. Lysis of human neutrophils by culture filtrates from CC8 strains was determined by release of lactate dehydrogenase (LDH). Values were significantly different for all comparisons except BD02-25 vs. SF8300 and all among HS-522, BAA-43, BAA-43, and COL.
Table 1.
Minimum inhibitory concentrations (mg/mL) of different strains to AMPs
| AMPs | BD02-25 | SF8300 | HS-522 | BAA-43 | BAA-44 | COL |
|---|---|---|---|---|---|---|
| Dermcidin-1 | 1.28 | 1.28 | 0.48 | 0.48 | 0.64 | 0.48 |
| Indolicidin | 0.05 | 0.06 | 0.01 | 0.04 | 0.03 | 0.01 |
| Nisin | 0.004 | 0.004 | 0.002 | 0.002 | 0.004 | 0.008 |
| Gramicidin | 2.56 | 2.56 | 1.92 | 1.92 | 2.56 | 1.92 |
| Melittin | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 |
Expression of key Virulence Genes.
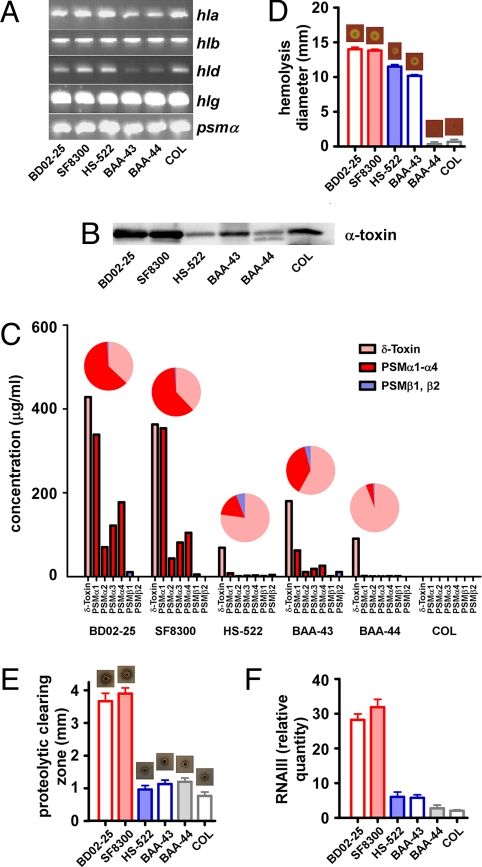
To test the hypothesis that differential expression of virulence determinants dictates pathogenic potential of MRSA sublineages within CC8, we evaluated presence and expression of cytolytic toxins, molecules with a key impact on immune evasion and virulence of S. aureus (23, 24). We first confirmed that the genes encoding cytolysins were present in all strains investigated (Fig. 4A and Table S2).
Fig. 4.
Expression of virulence determinants in CC8 subclones. (A) Presence of cytolysin genes by analytical PCR. (B) Western blot of α-toxin production. (C) PSM production. PSM production in culture filtrates was determined by RP-HPLC/electrospray ionization-MS. Relative production of δ-toxin (pink), other α-type (red), and β-type (blue) PSMs is shown in pie diagrams. (D) Hemolysis. Lysis of red blood cells was determined by dropping equal amounts of cells on sheep blood agar plates, incubating at 37 °C, and measuring clearing zones after 24 h. Values were significantly different for all comparisons except BD02-25 vs. SF8300 and BAA-44 vs. COL. (E) Proteolysis. Proteolysis of supernatants concentrated by lyophilization was measured by an agar diffusion assay. (F) RNAIII expression by qRT-PCR. (E and F) Values were significantly different for all comparisons except BD02-25 vs. SF8300 and all among HS-522, BAA-43, BAA-43, and COL.
Next, we determined expression of α-toxin and phenol-soluble modulins (PSMs), S. aureus cytolytic toxins that kill white blood cells and strongly influence virulence of CA-MRSA (25, 26). Among PSMs, those of the α-type have, by far, the greatest cytolytic potential toward human neutrophils (25). Importantly, α-type PSMs are largely responsible for neutrophil lysis, which is caused by secreted factors of CA-MRSA, including USA300 (25). We detected higher production of α-toxin (Fig. 4B) and α-type PSMs (Fig. 4C) in the USA300/USA500 sublineage than in the other strains. Furthermore, relative production of α-type PSMs in comparison to other PSMs with a lower cytolytic potential was increased in USA300 and USA500 (Fig. 4C). These findings indicate that the observed increased potential of USA500 and USA300 to lyse neutrophils is largely attributable to increased expression of α-type PSMs.
In addition, we determined the capacity of the strains to lyse erythrocytes, another target cell of cytolytic toxins (Fig. 4D). Compared with USA500 and USA300, capacities of the Archaic and Iberian clones to lyse red blood cells were considerably reduced. However, we did not detect significant differences between the 2 other sublineages. Nevertheless, these results revealed differences in hemolytic capacities among strains containing identical hemolysin genes.
S. aureus-secreted proteases have a key role in virulence as a means to acquire nutrients from host tissues and protect against AMPs. We found that proteolytic capacity of USA300 and USA500 culture filtrates was significantly increased compared with all other strains (Fig. 4E). Most likely, the differences in AMP resistance that we observed (Table 1) are, in part, attributable to differential expression of secreted proteases and differential susceptibility of AMPs to proteolytic digestion. That is, degradation of dermcidin but not melittin correlated with increased proteolysis (compare Fig. 4E and Fig. S2).
Thus, production of virulence determinants that are present in the representative CC8 genomes tested here, particularly those previously implicated in the virulence of CA-MRSA (25, 26), correlated with virulence observed in the animal infection models. These findings underscore the importance of differential gene expression for the evolution of virulence within the CC8 complex.
PSMs.
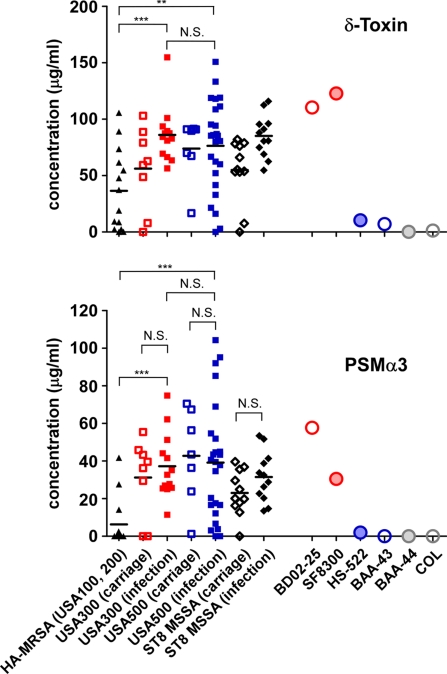
Because PSMs have a demonstrated crucial role in determining CA-MRSA infection (25), we analyzed expression of PSMs in more detail. We previously reported that selected CA-MRSA strains have increased expression of PSMs compared with representative HA-MRSA (25). Although appropriate for analysis of the most prominent clones, the number of isolates analyzed was limited. Therefore, we measured PSMα3 and δ-toxin expression, exemplifying relative expression of all PSMs (27), in a large subset of S. aureus strains (n = 88) from the same geographical origin (San Francisco) (Fig. 5). The strains were from individuals with carriage, hospital, or community infections and MRSA or methicillin-susceptible Staphylococcus aureus (MSSA). Average PSM production was significantly higher in USA300 and USA500 clinical isolates compared with that from the major HA-MRSA lineages, USA100 and USA200. These results confirm that CA-MRSA strains have increased PSM production compared with the most prominent HA-MRSA strains. Consistent with the results obtained using representative USA500 and USA300 isolates (Fig. 4C), there was no significant difference in average PSM production between a larger collection of USA300 and USA500 strains. These data indicate that high PSM expression was established a priori in USA500, the USA300 progenitor. Furthermore, average PSM expression in USA300 and USA500 was not significantly different from that in a mixture of CC8 MSSA clinical isolates (Fig. 5). This finding confirms that methicillin resistance does not have an impact on virulence, as shown recently in the USA300 and USA400 clones (12, 13, 28) and is consistent with reports on comparable clinical outcomes observed in CA-MRSA and CA-MSSA strains (29–31). Finally, we did not detect significant differences in PSMα3 expression between carriage vs. infection isolates, indicating that high expression of this key virulence determinant is not selected for in strains causing disease but is a general characteristic of the USA300/USA500 sublineage.
Fig. 5.
PSM production in HA-MRSA, CA-MRSA, and ST8 MSSA strain collections. Strains collected at hospitals in the San Francisco area were analyzed by RP-HPLC/electrospray ionization-MS for PSM production. Carriage isolates are from nonhospitalized homeless youth and urban poor in the same area. PSMα3 and δ-toxin levels are shown. Horizontal bars depict the mean. Statistical analysis is by unpaired t tests or one-way ANOVA for comparison of USA300, USA500, and MSSA carriage or infection isolates, respectively, which showed no statistically significant differences. N.S., not significant. ∗∗P < 0.01, ∗∗∗P < 0.001.
Role of the Global Virulence Regulator agr.
Differential expression of a series of virulence determinants as observed here for USA300 and USA500 is often caused by changes in activity of global regulatory systems in charge of virulence control. PSMs, α-toxin, and secreted proteases are classic examples of virulence determinants that are under control of the pivotal virulence regulator agr (25, 32, 33). To evaluate the hypothesis that agr expression contributes to the differential expression of virulence determinants in the strains tested, we measured expression of RNAIII, the major intracellular effector of agr (34). Activity of agr was significantly increased in USA300 and USA500 (Fig. 4F). This result indicates that the enhanced virulence potential of these strains may, at least in part, be attributable to high expression of this regulatory system.
On the other hand, several phenotypes, such as neutrophil lysis, hemolysis, and proteolytic capacity, did not completely correlate with RNAIII expression (Figs. 3 and 4 D, E, and F). Furthermore, there was a greater difference in production of PSMα3 (5.93-fold increased for USA300 vs. USA100/USA200 infection isolates; 6.23-fold increased for USA500 infection isolates) than in that of δ-toxin (2.35- and 2.09-fold, respectively) between USA300 and USA500 strains vs. USA100/USA200. Although both the psmα operon and the RNAIII-embedded δ-toxin gene hld are under direct control of the AgrA response regulator protein (27, 35), we previously found that the intergenic region in front of the psmα operon is subject to additional regulatory influences on psmα expression (27). The findings presented here are in accordance with these previous results and indicate that although agr activity plays a major role in defining the exceptional virulence potential of the USA300/USA500 sublineage, regulatory influences other than agr also have an impact on expression of key virulence determinants in these strains.
Discussion
Bacterial strains that cause epidemics, such as USA300, commonly combine extraordinary virulence with efficient colonization and host-to-host transmissibility. Whereas the analysis of colonization capacity and transmissibility helps to explain a pathogen's persistence and spread, assessment of the virulence potential allows predictions on the severity of disease that a pathogen may cause. Distinction between these phenotypes is crucial, because the underlying molecular factors may be entirely different, especially in the case of S. aureus, which may colonize humans in an asymptomatic fashion. Notably, the analysis of virulence potential is a key prerequisite for endeavors to find anti–CA-MRSA therapeutics, particularly those that use modern target-oriented drug development (36). Therefore, in an effort to establish a scientific basis for drug development against CA-MRSA, we focused on the evolution of virulence within CC8. Analysis of representative strains demonstrated that virulence was considerably increased in the lineage that comprises USA300 compared with the other CC8 MRSA lineages. This is consistent with the reported potential of USA300 to cause severe and widespread disease (37, 38).
MGEs, such as plasmids, prophages, transposons, and pathogenicity islands, often contribute to bacterial virulence, especially in S. aureus (39). However, our results indicate that for the CC8 lineage, particularly for USA300, differential expression of core genome-encoded virulence factors rather than MGEs may have a more profound impact on the evolution of virulence. This is based on our observations that (i) high virulence potential of USA300 was established in its progenitor strain, USA500 (i.e., before the acquisition of additional virulence determinants on MGEs); (ii) virulence and virulence factor expression were comparable among clones within the specific CC8 lineages; and (iii) differential distribution of virulence genes among CC8 strains tested failed to explain the observed differences in virulence merely by gene presence. Our results are in accordance with experimental infection studies that indicate there is little or no contribution of MGEs to USA300 virulence (11–14, 26), particularly compared with the dramatic influence of the core genome-encoded α-toxin and PSMs (25, 26).
This leaves the question of why we do not see severe infections with USA500 as often as with USA300. In that regard, it has been speculated that MGEs may contribute to USA300 transmission rather than virulence, which is, in part, based on the putative involvement of the ACME element in pH homeostasis (2, 10). Although this idea remains to be evaluated, it might explain the more pronounced spread of USA300, which would also cause a higher frequency of infections by USA300 compared with USA500. Furthermore, we cannot rule out the possibility that the toxin genes that are encoded on MGEs and by which USA300 differs from USA500, namely, the genes encoding PVL and the enterotoxins K and Q, have a yet unidentified role in USA300 virulence that is not detectable in mouse infection models. However, recent epidemiological evidence shows that CA-MRSA infections by strains other than USA300 may also be caused by PVL-negative strains (2, 6, 7). This supports the notion that PVL has a much less significant role in CA-MRSA virulence than previously assumed.
Our results suggest that the global virulence and quorum-sensing regulator agr has an important yet not exclusive role in defining the virulence gene expression pattern resulting in the increased virulence potential of USA300. Support for a key function of agr in defining CA-MRSA virulence has come from a recent study showing that the in vivo gene expression pattern of USA300 is indicative of a highly active agr system (40). Furthermore, Montgomery et al. (41) suggested that higher agr activity of USA300 may have caused the substitution of the early USA400 CA-MRSA clone by USA300. However, results from our previous studies indicate that agr activity is not likely involved in the differential virulence potential of these 2 more distant CA-MRSA clones (25, 28).
The increasing burden of CA-MRSA, including the ongoing epidemic in the United States, underscores the need to find innovative therapeutics for MRSA disease. Although CA-MRSA isolates are typically susceptible to many non–β-lactam antibiotics, there is recent emergence of multidrug-resistant CA-MRSA (42), thus confounding the current serious public health problem. One major focus in the development of antistaphylococcal therapeutics is the design of agents that neutralize virulence determinants (43). For these approaches to be successful, an in-depth evaluation of the target virulence factors of S. aureus is critical. Our findings have important implications for strategies directed to find innovative anti–CA-MRSA therapeutics because they suggest that virulence determinants of the core genome, such as α-toxin and PSMs, are more appropriate for target-oriented drug development than MGEs, on which many of these endeavors are currently being focused. In addition, a therapeutic that targets core genome-encoded virulence determinants would have much broader applicability compared with one aimed at neutralizing virulence determinants limited to CA-MRSA strains.
In summary, our study establishes that expression of core genome-encoded virulence genes plays a more crucial role in CA-MRSA virulence than factors present in MGEs. These findings represent a substantial change in our notion of how CA-MRSA virulence evolved and have important implications for drug development against this leading human pathogen.
Methods
Bacterial Strains, Growth Conditions, and Basic Molecular Biology Methods.
MRSA isolates of sequence types USA100 (ST 5), USA200 (ST 36), USA300 (ST 8), USA500 (ST 8), USA1100 (ST 30), and USA1000 (ST 59) were isolated from patients at San Francisco hospitals or from nonhospitalized individuals in the San Francisco area. BAA-44 (ST 247) and BAA-43 (ST 239) were obtained from American Type Culture Collection (ATCC). Six ST 239 clones (including HS-522) were randomly selected from unique patient clinical specimens of the ST 239 sequence type, which accounted for ≈40% of MRSA isolates, at Huashan Hospital, Shanghai, China between January 2006 and December 2006. Control isolates for PCR-based assays were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) and included the 5 reference strains with fully sequenced genomes—N315, Mu50, MW2, COL, and NCTC8325. Bacteria were grown in tryptic soy broth unless otherwise noted. Cultures were incubated at 37 °C with shaking at 200 rpm. DNA manipulation was performed using standard procedures. PCR-based assays for virulence and housekeeping genes were performed using the primers listed in Table S3. Primers for DNA amplifications were purchased from Sigma Genosys. PCR reactions were performed with Ready-To-Go PCR Beads (Amersham Biosciences) as recommended by the manufacturer. DNA was sequenced using Big Dye Terminator cycle sequencing (version 3.0) on an ABI3700 sequencer (Applied Biosystems). Nucleotide sequences were analyzed using the program Vector NTI Suite (InforMax).
Mouse Bacteremia and Skin Abscess Models.
Outbred immunocompetent CD1 Swiss female mice were used for the bacteremia model, and outbred immunocompetent Crl:SKH1-hrBR hairless mice (Charles River Laboratories) were used for the abscess model. All mice were between 4 and 6 weeks of age at the time of use. S. aureus strains were grown to midexponential phase, washed once with sterile PBS, and then resuspended in PBS at 1 × 108 cfus/100 μL (bacteremia model) or 1 × 107 cfus/50 μL (abscess model) as described (5). For the bacteremia model, we injected each mouse with 108 cfus of live S. aureus in 0.1 mL of sterile saline into the retro-orbital vein. Control animals received sterile saline only. After inoculation, mouse health and disease advancement were monitored every 3 h for the first 24 h and then every 8 h for up to 72 h. We euthanized the mice immediately if they showed signs of respiratory distress, mobility loss, or inability to eat and drink. All surviving animals were euthanized at 72 h. At the time of death, serum samples were harvested from test animals for the measurement of cytokine expression with commercial ELISA kits (R&D Systems) according to the manufacturer's instructions. For the abscess model, mice were anesthetized with isoflurane and inoculated with 50 μL of PBS containing 107 cfus of live S. aureus or saline alone in the right flank by s.c. injection. Test animals were examined at 24-h intervals for a total of 14 days; we measured skin lesion dimensions daily with a caliper. We applied length (L) and width (W) values to calculate the area of lesions with the formula L × W. All animals were euthanized after completion of the entire procedure. Animal studies were approved by the Animal Care and Use Committee, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases.
Further detailed protocols are reported in SI Methods.
Supplementary Material
Acknowledgments.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (M.O., F.R.D.); a grant from the Shanghai Medical Key Discipline (to M.L. and Y.L.); Microbial Pathogenesis and Host Defense Postdoctoral Fellowship 5T32AI060537–02 (to B.A.D.); and U.S. Public Health Service Grant NIAID R01 AI070289 (to H.F.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900743106/DCSupplemental.
References
- 1.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J Am Med Assoc. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran GJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 4.Chambers HF. Community-associated MRSA—Resistance and virulence converge. N Engl J Med. 2005;352:1485–1487. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otter JA, French GL. The emergence of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital, 2000–2006. Clin Microbiol Infect. 2008;14:670–676. doi: 10.1111/j.1469-0691.2008.02017.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang K, McClure JA, Elsayed S, Tan J, Conly JM. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. J Infect Dis. 2008;197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 8.Gillet Y, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 9.Vandenesch F, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 11.Diep BA, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diep BA, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 13.Voyich JM, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 14.Wardenburg JB, Palazzolo-Ballance AM, Otto M, Schneewind O, Deleo FR. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J Infect Dis. 2008;198:1166–1170. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musser JM. Molecular population genetic analysis of emerged bacterial pathogens: Selected insights. Emerg Infect Dis. 1996;2:1–17. doi: 10.3201/eid0201.960101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beres SB, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright MC, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira DC, Tomasz A, de Lencastre H. Secrets of success of a human pathogen: Molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002;2:180–189. doi: 10.1016/s1473-3099(02)00227-x. [DOI] [PubMed] [Google Scholar]
- 19.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–369. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jevons MP. “Celbenin”-resistant staphylococci. Br Med J. 1961;1:124–125. [Google Scholar]
- 21.Robinson DA, Enright MC. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol. 2004;186:1060–1064. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 23.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 24.Prevost G, Mourey L, Colin DA, Menestrina G. Staphylococcal pore-forming toxins. Curr Top Microbiol Immunol. 2001;257:53–83. doi: 10.1007/978-3-642-56508-3_4. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 26.Wardenburg JB, Bae T, Otto M, DeLeo FR, Schneewind O. Poring over pores: Alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 27.Queck SY, et al. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voyich JM, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 29.Miller LG, et al. Clinical and epidemiologic characteristics cannot distinguish community-associated methicillin-resistant Staphylococcus aureus infection from methicillin-susceptible S. aureus infection: A prospective investigation. Clin Infect Dis. 2007;44:471–482. doi: 10.1086/511033. [DOI] [PubMed] [Google Scholar]
- 30.Miller LG, et al. A prospective investigation of outcomes after hospital discharge for endemic, community-acquired methicillin-resistant and -susceptible Staphylococcus aureus skin infection. Clin Infect Dis. 2007;44:483–492. doi: 10.1086/511041. [DOI] [PubMed] [Google Scholar]
- 31.Wang JL, et al. Comparison of both clinical features and mortality risk associated with bacteremia due to community-acquired methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. Clin Infect Dis. 2008;46:799–806. doi: 10.1086/527389. [DOI] [PubMed] [Google Scholar]
- 32.Novick RP, Muir TW. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr Opin Microbiol. 1999;2:40–45. doi: 10.1016/s1369-5274(99)80007-1. [DOI] [PubMed] [Google Scholar]
- 33.Recsei P, et al. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 34.Novick RP, et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol. 2004;186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alksne LE, Projan SJ. Bacterial virulence as a target for antimicrobial chemotherapy. Curr Opin Biotechnol. 2000;11:625–636. doi: 10.1016/s0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- 37.Miller LG, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 38.Francis JS, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 39.Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3:585–594. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 40.Loughman JA, Fritz SA, Storch GA, Hunstad DA. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis. 2008;199:294–301. doi: 10.1086/595982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery CP, et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 42.Diep BA, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008;148:249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- 43.Otto M. Antibodies to block staph virulence. Chem Biol. 2007;14:1093–1094. doi: 10.1016/j.chembiol.2007.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.