Abstract
We found that Arabidopsis AtTDX, a heat-stable and plant-specific thioredoxin (Trx)-like protein, exhibits multiple functions, acting as a disulfide reductase, foldase chaperone, and holdase chaperone. The activity of AtTDX, which contains 3 tetratricopeptide repeat (TPR) domains and a Trx motif, depends on its oligomeric status. The disulfide reductase and foldase chaperone functions predominate when AtTDX occurs in the low molecular weight (LMW) form, whereas the holdase chaperone function predominates in the high molecular weight (HMW) complexes. Because deletion of the TPR domains results in a significant enhancement of AtTDX disulfide reductase activity and complete loss of the holdase chaperone function, our data suggest that the TPR domains of AtTDX block the active site of Trx and play a critical role in promoting the holdase chaperone function. The oligomerization status of AtTDX is reversibly regulated by heat shock, which causes a transition from LMW to HMW complexes with concomitant functional switching from a disulfide reductase and foldase chaperone to a holdase chaperone. Overexpression of AtTDX in Arabidopsis conferred enhanced heat shock resistance to plants, primarily via its holdase chaperone activity.
Keywords: disulfide reductase, foldase chaperone, holdase chaperone, functional switching, Yedox
The ability of thioredoxins (Trxs) to recognize and reduce target proteins containing disulfide bonds is their primary role in redox regulation. The reduction mechanism of Trx is composed of 2 steps centered around its active-site Cys residue. An active Cys of Trx first attacks the disulfide bridge of oxidized substrates, producing a mixed disulfide bond between the substrate and active Cys residue of Trx. Then, the other Cys attacks this disulfide bond, releasing a reduced substrate and an oxidized Trx. The oxidized Trx is usually reduced by Trx reductase and NADPH to complete the catalytic cycle (1). In addition, Trxs control several redox-independent cellular processes, such as the assembly of T7-DNA polymerase and filamentous phages (2, 3) by unknown reaction mechanisms.
Whereas 2 or 3 Trx isotypes exist in mammals (4, 5), yeast (6, 7), and Escherichia. coli (8), plants contain 2 large Trx families, namely the Trx and Trx-like proteins (9). Members of the Trx family have a distinct Trx motif, whereas the Trx-like protein family has a second functional domain in addition to the Trx motif (9). Although >20 different types of Trx genes have been identified in the Arabidopsis genome, there are only a few Trx-like proteins, including the PKC-interacting cousin of Trx, glutaredoxin-related proteins, and protein disulfide isomerase (PDI), whose functions remain to be elucidated (10–13). Among the proteins containing the Trx fold, 2-Cys peroxiredoxins (Prxs) have been shown to alter their conformation from low molecular weight (LMW) to high molecular weight (HMW) complexes in response to heat shock and radical stresses (14, 15). The conformational change is accompanied by functional switching from a peroxidase to a molecular chaperone (14). Considering these observations, we screened for heat shock-induced HMW complex proteins that have a similar regulation mode as the 2 Cys Prxs from heat-treated Arabidopsis suspension cells. In the screen, we identified a plant-specific Trx-like protein containing 3 tetratricopeptide repeat (TPR) domains and a Trx motif, which we designated AtTDX (GenBank accession no. At3g17880). Although AtTDX has been shown to functionally complement Δtrx1/2 mutant yeast (16), its biochemical nature and physiological function in plants remain obscure and warrant further study. The TPR units are particularly critical for protein–protein interaction and formation of multiprotein complexes (17), which are characteristic properties of molecular chaperones. Chaperones are largely classified into 2 groups: foldase chaperones, which support the folding of denatured proteins to their native state, and holdase chaperones, which bind to folding intermediates, thereby preventing their nonspecific aggregation (18).
Given that the E. coli Trx (19) and PDI (20), which contain a Trx motif and 2 Cys Prxs (14) with a Trx fold, exhibit chaperone functions, we explored the functions of AtTDX from Arabidopsis in the present study. We demonstrated in a plant system that an Arabidopsis Trx-like protein, AtTDX, displayed multiple functions that depended on its oligomeric status, alternatively acting as a disulfide reductase, foldase chaperone, and holdase chaperone. Furthermore, we showed that the holdase chaperone function of AtTDX significantly enhanced heat shock resistance in Arabidopsis.
Results
Multiple Functions of AtTDX.
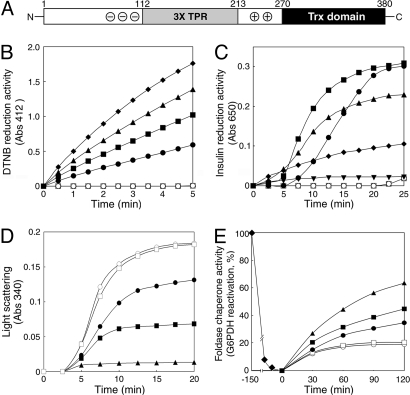
Using size exclusion chromatography (SEC) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analyses, we screened heat-treated Arabidopsis suspension cells for heat-induced HMW complex proteins that are regulated in a similar fashion to the 2 Cys Prxs (14) and isolated a plant-specific Trx-like protein, AtTDX. The AtTDX protein consists of 380 amino acids and is encoded by a single copy gene on Arabidopsis chromosome 3 (16) (Fig. S1). It contains a bipartite molecular structure, with 3 TPR domains in its N-terminal region (between residues 112 and 213), which are flanked by acidic residues on one side and by basic residues on the other, and a Trx motif in its C terminus (Fig. 1A). Given that disulfide reductase activity is a characteristic of Trxs, we purified bacterially-expressed recombinant AtTDX to homogeneity (Fig. S2) and examined its ability to reduce 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) in the presence of NADPH and Trx reductase as reductants. We observed that DTNB reduction increased linearly with increasing amounts of AtTDX (Fig. 1B), but that replacing DTNB with insulin as a protein substrate resulted in a more complex interaction. In the presence of 0.5 mM DTT, insulin reduction activity increased in a concentration-dependent manner up to 10 μM AtTDX but, surprisingly, decreased significantly with AtTDX concentrations of >20 μM (Fig. 1C). The results shown in Fig. 1 B and C strongly suggest that at higher molar ratios of AtTDX to substrate the reduced and partially denatured insulin molecules produced by its disulfide reductase activity are immediately trapped into soluble complexes by AtTDX (Fig. S3), which is typical of holdase chaperone activity.
Fig. 1.
Multiple functions of AtTDX as a disulfide reductase, a foldase chaperone, and a holdase chaperone. (A) A scheme of the bipartite AtTDX protein structure. The N-terminal TPR domain is flanked by acidic (⊝) and basic (⊕) residues and the C terminus bears a Trx motif. (B and C) Disulfide reduction activity of AtTDX. Reduction of disulfide bonds using either 5 mM DTNB in the presence of NADPH and Trx reductase (B) or 30 μM insulin in the presence of 0.5 mM DTT (C) was measured by absorbance changes at A412 (B) and A650 (C), respectively. Concentrations of AtTDX used were 5 μM (●), 10 μM (■), 20 μM (▴), 30 μM (□), and 60 μM (▾). (D) Holdase chaperone activity of AtTDX. Thermal aggregation of 1.8 μM MDH was examined, with the molar ratios of AtTDX to MDH set at 1:1 (●), 3:1 (■), and 6:1 (▴) at 43 °C. (E) Foldase chaperone activity of AtTDX. The Cys-free form of G6PDH (10 μM) was denatured in 6 M urea for 2.5 h (□) and then refolded in a renaturation buffer in the presence of 5 μM (●), 10 μM (■), or 30 μM (▴) AtTDX. Activity of native G6PDH was set to 100%. Reactions using 30 μM ovalbumin (□) instead of AtTDX or lacking both AtTDX and ovalbumin (○) were used as controls.
To test this hypothesis, holdase chaperone activity of AtTDX was measured by using malate dehydrogenase (MDH) as a substrate. Incubation of MDH with increasing amounts of AtTDX prevented thermal aggregation, and aggregation was completely blocked at a subunit molar ratio of 1 MDH/6 AtTDX (Fig. 1D). In addition, given that E. coli Trx exhibits a foldase chaperone function (19), we also examined the foldase chaperone activity of AtTDX by using glucose-6-phosphate dehydrogenase (G6PDH). To distinguish the foldase chaperone function of AtTDX from its disulfide reductase function, the Cys-free form of G6PDH was used (21). Using chemically-denatured G6PDH, we found that the foldase chaperone activity attained up to 63% of native G6PDH activity within 2 h at 30 μM AtTDX, whereas 18% was renatured in the absence of AtTDX (Fig. 1E). However, when we replaced AtTDX with 30 μM ovalbumin, neither holdase nor foldase chaperone activity was detected, suggesting that the multiple activities were derived from the specific function of AtTDX.
Recombinant AtTDX Forms Discretely-Sized Multiple Oligomers.
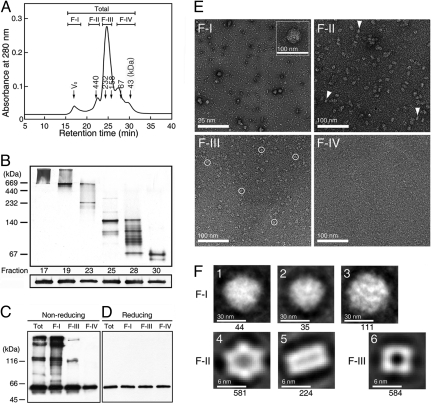
HMW complex formation is a well-conserved feature of holdase chaperones (22). Because AtTDX was isolated as a heat-stable HMW protein from Arabidopsis suspension cells and exhibited holdase chaperone function (Fig. 1D), we examined the oligomerization status of recombinant AtTDX. Purified AtTDX could be separated by SEC into several distinct peaks containing monomer, dimer, oligomer, and HMW complexes (Fig. 2A). The polymeric status was confirmed by using 10% native PAGE followed by silver staining. Protein fractions separated by SEC ranged from 50 to >1,000 kDa in size, as determined by native PAGE (Fig. 2B Upper). However, all of the fractions ran as a single protein band with an apparent molecular mass of ≈53 kDa in SDS/PAGE (Fig. 2B Lower), although the theoretical mass of AtTDX is 42,847 Da. The result suggests that multiple complexes of AtTDX are homopolymers consisting of variable numbers of monomer. In addition, we subjected SEC-separated proteins to reducing and nonreducing SDS/PAGE and Western blot analysis with anti-AtTDX antibody, after verifying the immuno-specificity of the antibody (Fig. S4). In nonreducing conditions, F-I and F-II proteins were present as SDS-resistant oligomers even at high SDS concentrations (Fig. 2C). However, the addition of β-mercaptoethanol completely dissociated the multimeric proteins into monomers (Fig. 2D), suggesting that the HMW complexes of AtTDX are associated not only by hydrophobic interactions, but also by disulfide bonds.
Fig. 2.
Oligomeric status of AtTDX analyzed by chromatographic, electrophoretic, and microscopic techniques. (A) SEC was performed by using a Superdex 200 HR column, and the fractions were divided into 4 classes (F-I–F-IV). Molecular masses of known standards run on the same column are indicated in the chromatogram. (B) Each fraction of SEC in A was separated on 10% native PAGE (Upper) or 12% reducing SDS/PAGE (Lower) gels, followed by Western blot analysis with an anti-AtTDX antibody. (C and D) All protein fractions in A were separated by nonreducing (C) and reducing (D) SDS/PAGE gels, followed by silver staining. (E) Electron micrographs of the F-I to F-IV proteins in A were analyzed by negative staining with 2% uranyl acetate. The scale bars represent 25 nm (F-I) and 100 nm (F-II–F-IV). In the inset of F-I, spherical HMW complexes are enlarged. (F) Multivariate statistical analysis of AtTDX complexes. In F-I, the spherical complexes are grouped according to the similarity of features based on eigenvector-eigenvalues and have diameters of ≈45.6 (1), ≈42.6 (2), and ≈51.7 (3) nm (3). In F-II, an end-on view of hexameric AtTDX (4) (diameter ≈10.4 nm) and the side perspective of AtTDX, forming a double-layered dodecameric form with a dyad across the equatorial plane (5) are shown, and the latter structures are indicated by triangles in E, F-II. Tetramers with diameters of ≈7.7 nm are represented in F-III (6) and indicated by circles in E, F-III. The number of particles in each class is indicated beneath each panel.
To further analyze the oligomeric status of AtTDX, we examined the peak fractions of SEC by transmission EM. The electron micrographs of negatively-stained AtTDX revealed several unique configurations: a spherically-shaped globular structure, a single- or double-layered ring structure, and an irregularly-shaped small particle (Fig. 2E). When we subjected the globular-shaped HMW complexes observed in F-I to rotational and translational alignment, they could be classified into 3 groups based on eigenvector–eigenvalue data (Fig. 2F 1–3). The diameter of the projected images ranged from 42 to 52 nm. To define the protein structures of F-II, a total of 581 end-on views and 224 side-on views of well-stained particles were translationally and rotationally aligned and subjected to multivariate statistical analysis (23). Image processing of F-II oligomers yielded 2 basic views depending on the orientation of the grid: ring-shaped hexameric structures with an end-on orientation, and double-layered dodecameric structures with a side-on orientation, forming a dyadic structure across the equatorial planes (Fig. 2F, 4 and 5). Although most of the F-III protein formed rectangular tetramers (Fig. 2F, 6), the monomers and dimers in the F-IV fraction (Fig. 2E) did not produce a regular structure.
Multiple Functions of AtTDX Are Associated with its Oligomeric Status.
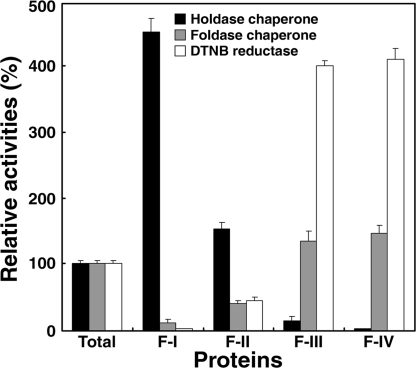
To explore the relationship between the oligomeric status of AtTDX and its function, we examined the 3 defined functions of AtTDX in the fractions isolated from the SEC column (Fig. S5). The SDS-resistant form of HMW complexes (Fig. 2C, F-I) exhibited a 4- to 5-fold higher holdase chaperone activity than that of the total protein, which had negligible DTNB reduction and foldase chaperone activities (Fig. 3, F-I). In contrast, the F-II fraction displayed a ≈1.5-fold higher holdase chaperone activity and ≈50% lower DTNB reduction and foldase chaperone activities than those of the total protein. When we compared the activities of LMW fractions with those of the total protein, F-III and F-IV showed significantly lower holdase chaperone activity (<10% of the activity in the total protein) with ≈1.3- and 4-fold higher foldase chaperone and disulfide reductase activities, respectively. From these results, we conclude that the function of AtTDX is determined by its oligomerization status. Multimerization of AtTDX enhances its holdase chaperone activity, whereas dissociation promotes its disulfide reductase and foldase chaperone functions.
Fig. 3.
Multiple functions of AtTDX are associated with its oligomeric status. Holdase chaperone, foldase chaperone, and DTNB reductase activities were measured on equal amounts of protein fractions obtained from SEC (F-I–F-IV of Fig. S5). The activities in each fraction were compared with those in the total protein (Total), whose activities were set to 100%.
The domains of AtTDX responsible for its 3 functions were identified by preparing a number of truncated constructs, expressing them in E. coli, and measuring the activities for each mutant (Fig. S6). The 40-380 mutant protein, which contains both the TPR and Trx domains behaved similarly to native AtTDX, whereas the 1-111 polypeptide lacking both of these domains did not show any activity. Mutant proteins containing the TPR domain but not the Trx domain (residues 1–266 and 40–266) displayed a slightly lower level of holdase chaperone activity than native AtTDX, and the holdase chaperone activity was found to correlate with oligomeric status, as determined by Western blot analysis on native, nonreducing, and reducing SDS/PAGE gels (Fig. S7). In addition, the protein containing only the Trx motif (residues 270–380) or the TPR-deficient mutant protein (Δtpr) exhibited a 4-fold higher DTNB reduction activity than native AtTDX with no holdase chaperone activity. These results strongly suggest that the TPR domains of AtTDX are required for its holdase chaperone function and that the domains may block the Trx motif from acting as a disulfide reductase. Moreover, the Cys mutant proteins (C304S, C307S, and C304/307S) did not exhibit disulfide reductase activity, but did display strong holdase chaperone activity, suggesting that the additional Cys in position 350 (Fig. S1) might contribute to the formation of the SDS-resistant but mercaptoethanol-sensitive oligomeric conformation of AtTDX (Fig. S7).
Heat Shock-Mediated Switching of AtTDX Oligomerization and Its Effect on in Vitro Protein Function.
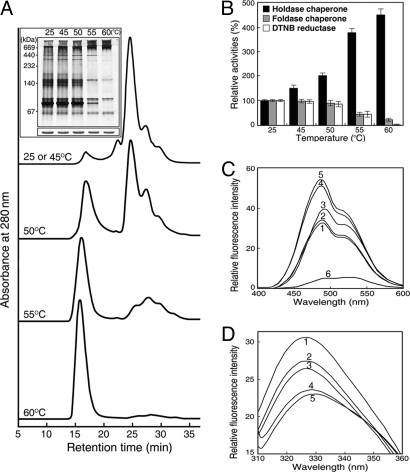
We analyzed the effect of heat shock treatment on AtTDX oligomerization status. The concentration of HMW complexes of AtTDX increased in SEC-separated proteins as incubation temperatures rose, with a concomitant decrease in LMW protein species (Fig. 4A). The heat shock-mediated shift in oligomeric status was confirmed by native PAGE followed by silver staining (Fig. 4A Inset). The results suggest that LMW proteins of AtTDX assemble into HMW complexes during heat shock.
Fig. 4.
Heat shock-dependent switching of AtTDX oligomerization and in vitro activity. (A) Changes in AtTDX oligomeric status in response to various heat shock treatments for 30 min, as analyzed by SEC. (Inset) The oligomeric status of AtTDX was confirmed by analysis on a 10% native PAGE gel (Upper) or a 12% SDS/PAGE gel (Lower), followed by silver staining. (B) Holdase chaperone, foldase chaperone, and DTNB reductase activities of heat-treated AtTDX were compared with those of native AtTDX incubated at 25 °C, which were set to 100%. (C and D) Heat shock-dependent changes of AtTDX hydrophobicity were measured by bis-ANS binding (C) and tryptophan fluorescence (D). AtTDX was incubated at 25 °C (line 1), 45 °C (line 2), 50 °C (line 3), 55 °C (line 4), and 60 °C (line 5) for 30 min. The fluorescence spectrum of 10 μM bis-ANS incubated alone at 60 °C is shown by line 6.
In addition to changes in oligomeric status after heat shock, holdase chaperone function of AtTDX significantly increased. A temperature shift from 25 °C to 60 °C caused a ≈4.5-fold increase in holdase chaperone activity, whereas the DTNB reduction and foldase chaperone activities decreased with rising temperatures (Fig. 4B). As a measure of oligomer changes, binding of 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (bis-ANS) was analyzed in heat-treated AtTDX. The fluorescence intensity of bis-ANS significantly increased at higher temperatures, which indicates that more hydrophobic patches are exposed on AtTDX at elevated temperatures (Fig. 4C). The heat shock-dependent oligomeric changes of AtTDX were also measured by using tryptophan fluorescence. The fluorescence decreased and the maximal emission was shifted to longer wavelengths with increasing temperatures (Fig. 4D). These results suggest that heat shock-mediated oligomeric changes of AtTDX are closely associated with its functional switching.
The Holdase Chaperone Function of AtTDX Enhances Thermo-Tolerance in Arabidopsis.
To understand the physiological function of AtTDX in vivo, we investigated the changes in AtTDX oligomeric status after heat shock treatment at 40 °C for various lengths of time by using Arabidopsis suspension cells. The oligomeric status of cytosolic AtTDX was analyzed by Western blot analysis with the use of an anti-AtTDX antibody that specifically reacted with AtTDX (Fig. S4) on a native PAGE gel (Fig. 5A Upper). The concentration of HMW AtTDX complexes rose after heat shock with a concomitant decrease in LMW species (Fig. 5A Upper, lanes 1–4). When the heat-treated cells were restored to their optimal temperature (22 °C), the concentration of HMW AtTDX complexes slowly decreased and returned to its preheat shock level (Fig. 5A Upper, lanes 4–7). However, the protein levels of AtTDX in all samples analyzed by Western blot on a reducing SDS/PAGE gel were nearly unchanged (Fig. 5A Lower). Considering that the multiple functions of AtTDX are determined by its oligomeric status (Fig. 3), it is possible that heat shock causes AtTDX to switch functions in vivo from a disulfide reductase and a foldase chaperone to a holdase chaperone, in a reversible manner.
Fig. 5.
The holdase chaperone function of AtTDX enhances heat shock recovery in Arabidopsis. (A) Heat shock-dependent reversible changes of AtTDX oligomeric status in Arabidopsis suspension cells. Suspension cells cultured at 22 °C were heat-stressed at 40 °C for 10, 30, or 60 min. After 60 min of heat treatment (lane 4), the cells were transferred to their optimal temperature and incubated again for 3, 6, or 12 h. Cytosolic proteins were extracted from the treated cells and equal amounts (50 μg) were loaded onto the lanes of a native PAGE gel (Upper) and a reducing SDS/PAGE gel (Lower), followed by Western blot analysis with an anti-AtTDX-antibody. (B) (Upper) Expression levels of AtTDX in WT Arabidopsis and representative T3 lines of transgenic Arabidopsis overexpressing AtTDX (OE), the AtTDX Cys-mutant (C304/307S), or the mutant suppressing the endogenous AtTDX protein (Sup) were analyzed by Western blotting on a SDS/PAGE gel. (Lower) Arabidopsis plants grown at 22 °C for 4 weeks are shown. (C) Comparison of heat shock tolerance between WT and transgenic Arabidopsis seedlings. (Upper) A scheme of heat shock treatment and recovery of the various Arabidopsis seedlings is depicted. (Lower) Comparison of plants on the final day of recovery after heat shock is shown.
We further investigated the physiological consequences of heat-shock-mediated oligomeric changes of AtTDX in transgenic Arabidopsis. Several lines of Arabidopsis T3 homozygotes, including WT lines, AtTDX overexpression lines, AtTDX suppression lines, and Cys-mutant (C304/307S) AtTDX overexpression lines that only have the holdase chaperone function, were analyzed. Expression levels of AtTDX in transgenic plants were measured by Western blot analysis (Fig. 5B Upper). The overexpression and suppression lines showed no phenotypic difference from the WT under normal growing conditions (Fig. 5B Lower) and, moreover, were similarly damaged by the heat stress incurred at 38 °C for 5 days. However, when the heat-stressed plants were returned to their optimal temperature (Fig. 5C), the transgenic lines overexpressing the native or C304/307S mutant AtTDX rapidly recovered during the poststress recovery period. The observation is in good agreement with the results of Arabidopsis stromal HSP70 (24). In contrast, almost all of the WT plants and the suppression lines were unable to recover from the damage caused by heat shock (Fig. 5C Lower). The inability of the WT and the suppression lines to recover from heat stress was verified by measuring the chlorophyll content and photosynthetic yield during the recovery period (Fig. S8). Whereas the heat shock-damaged plants of WT and AtTDX suppression lines of Arabidopsis failed to recover chlorophyll content and quantum yield during the recovery period, the parameters were fully restored in the Arabidopsis lines overexpressing native AtTDX and C304/307S AtTDX. These results suggest that the holdase chaperone function of AtTDX plays a major role in the protection of Arabidopsis from heat shock during the heat stress and/or recovery stage.
Discussion
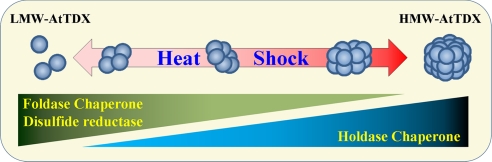
Although heat tolerance and rapid adaptation to daily temperature fluctuations are very important for the survival of sessile plants, the critical factors that confer heat resistance in higher plants are poorly understood (25). We have shown here that the heat-stable and multifunctional AtTDX plays a pivotal role in heat shock resistance in Arabidopsis during the poststress period, primarily via its holdase chaperone function. In particular, the multiple functions of AtTDX are closely associated with discretely-sized oligomeric states that reversibly switch from LMW to HMW complexes in response to heat shock. Our results are summarized in a comprehensive model in Fig. 6. The apparent functions of AtTDX are regulated by molar ratios of AtTDX to its substrate proteins (Fig. 1). PDI is similar: In the presence of higher molar ratios of PDI to substrate, PDI shows a strong holdase chaperone activity, whereas low PDI/substrate ratios result in an antichaperone activity (26, 27). Oligomerization-dependent functional changes have also been observed in human glyceraldehyde-3-phosphate dehydrogenase (G3PD). Depending on its oligomeric status, G3PD changes its function from a dehydrogenase to a nuclear uracil-DNA glycosylase (28). The TPR domain of AtTDX was shown to interact with yeast Ssb2, a cytosolic HSP70, in an oxidative stress-dependent manner, and point mutations of the Cys residues enhance the interaction (16). Similarly, we demonstrated that the AtTDX Cys mutant significantly increased its holdase chaperone activity and lost its disulfide reductase and foldase chaperone activities.
Fig. 6.
A representative model of changes in oligomeric status and function of AtTDX in response to heat shock. When plant cells are exposed to heat shock, LMW AtTDX reversibly change to HMW complexes, in parallel with functional switching from disulfide reductase and foldase chaperone activity to holdase chaperone activity. LMW AtTDX predominantly functions as a disulfide reductase and foldase chaperone, whereas the HMW complex protein acts as a holdase chaperone.
Molecular analyses of T3-transgenic Arabidopsis showed that plants overexpressing AtTDX recovered much faster from heat shock during the recovery phase than either WT or suppression lines (Fig. 5 and Fig. S8). The heat shock-resistant property of plants overexpressing AtTDX is likely caused by the formation of a stable complex between AtTDX and heat shock-induced partially-denatured cellular proteins and their ability to refold these proteins during the recovery period. Likewise, yeast HSP104 increases the survival of cells at high temperatures by facilitating reactivation of stress-induced protein aggregation (29).
In conclusion, our studies reveal the novel functions of a plant-specific AtTDX and demonstrate the physiological roles of AtTDX in response to heat shock. Thus, the data from this study provide important clues for understanding the molecular mechanisms that regulate plant defense signaling under normal and stress conditions.
Materials and Methods
Materials.
The NADH-dependent form of porcine heart mitochondrial MDH, the Cys-free form of Leuconostoc mesenteroides G6PDH, insulin, DTT, bis-ANS, and H2O2 were purchased from Sigma. Molecular markers for native PAGE and SEC, the Superdex 200 HR 10/30 column for fast performance liquid chromatography (FPLC), and ovalbumin were purchased from Amersham Biosciences.
Isolation of Heat-Stable, HMW Complex Proteins from Arabidopsis Suspension Cells.
Cytosolic extracts prepared from an Arabidopsis suspension cell culture (Arabidopsis thaliana L. Heynh, Ecotype Columbia) were heat-treated at 70 °C for 30 min and ultracentrifuged at 134,600 × g for 15 min. The supernatants were subjected to SEC by using a Superdex 200 HR column, and HMW complexes eluted from the void volume of SEC were collected and identified by 2D PAGE and MALDI-TOF analyses, as described (30). Among the proteins identified, AtTDX was chosen to be a target in this study.
Cloning of the AtTDX Gene, Protein Expression in E. coli, and Preparation of an Anti-AtTDX Antibody.
AtTDX was cloned from an Arabidopsis cDNA library by PCR. Various AtTDX DNA constructs were subcloned into the pET-28a vector (Novagen) and transformed into E. coli BL21(DE3). His-tagged AtTDX proteins were purified by using Ni-NTA agarose and cleaved by thrombin. Purified AtTDX dialyzed into 20 mM Hepes-KOH (pH 8.0) was used for biochemical analyses and preparation of a polyclonal antibody by injecting the protein 4 times into rabbits.
Determination of Disulfide Reductase, Foldase Chaperone, and Holdase Chaperone Activities.
Disulfide reductase activity of AtTDX was measured by using either DTNB as a substrate in the presence of NADPH and Trx reductase (31), or insulin with 0.5 mM DTT as a reductant (1). Foldase chaperone activity was analyzed by using 6 M urea-denatured G6PDH as a substrate as described (21). Briefly, denatured G6PDH (10 μM) was diluted 100-fold in a 100 mM sodium phosphate buffer (pH 7.2), and the refolding activity of AtTDX was monitored by assaying G6PDH activity. Holdase chaperone activity was measured by using MDH as a substrate. The substrate was incubated in a 50 mM Hepes-KOH (pH 8.0) buffer at 45 °C with various concentrations of AtTDX. Thermal aggregation of the substrate was determined by monitoring the turbidity increase at A340 as described (14, 32).
Fluorescence Measurements.
Exposure of hydrophobic domains was analyzed by measuring bis-ANS fluorescence with a SFM25 spectrofluorometer (Kontron). The excitation wavelength was set at 380 nm, and the emission was scanned between 400 and 600 nm (33). For the analysis of intrinsic tryptophan fluorescence, AtTDX (100 μg/mL) incubated at various temperatures for 30 min was scanned after setting the excitation wavelength to 295 nm.
SEC, PAGE, and Western Blot Analyses.
SEC was performed at 25 °C by FPLC using a Superdex 200 HR column equilibrated with a 50 mM Hepes-KOH (pH 8.0) buffer containing 100 mM NaCl. SDS/PAGE under reducing or nonreducing conditions, native PAGE, and Western blot analysis were performed as described (34).
EM and Image Processing.
Different MW fractions of AtTDX were applied to glow-discharged carbon-coated copper grids. After allowing the protein to absorb for 1–2 min, the grids were rinsed on droplets of deionized water and stained with 1% (wt/vol) uranyl acetate. Electron micrographs were recorded with an FEI Technai 12 microscope at an acceleration voltage of 120 kV and a magnification of 21 K and 52 K as described (35).
Oigomerization Changes of AtTDX After Heat Shock in Vivo.
Oligomerization changes of AtTDX in vivo were analyzed by using Arabidopsis suspension cells. The cells were grown in Jouanneau and Péaud-Lenoel's medium containing 1.5% (wt/vol) sucrose under continuous white light (40 μE·m−2·S−1) at 22 °C while shaking at 1.13 × g. For heat shock treatment, 4-day-old cultures were incubated at 40 °C for various lengths of time and then transferred to 22 °C for 5 days. Total proteins from the cells were extracted in buffer [50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, and 1 mM PMSF]. Oligomerization changes were analyzed by Western blotting on a native PAGE gel.
Preparation of Transgenic Arabidopsis Lines and Heat Shock Treatment.
For the construction of transgenic Arabidopsis plants overexpressing the native AtTDX or the AtTDX C304/307S mutant, native or mutant cDNAs, respectively, were ligated into pBI121 under control of the cauliflower mosaic virus 35S promoter. Plasmids were individually transformed into Agrobacterium tumefaciens GV3101 and introduced into Arabidopsis by using the floral dip technique (36). Expression levels of AtTDX were observed by Northern and Western blot analyses. As a result, 7 transgenic Arabidopsis lines overexpressing AtTDX were obtained and 11 suppression lines were obtained simply by cosuppression when generating overexpressors. Heat shock resistance of representative lines was tested by using 4-week-old Arabidopsis plants. All of the data presented are averages of at least 3 independent measurements.
For additional methods see SI Text.
Supplementary Material
Acknowledgments.
This work was supported by Ministry of Education, Science, and Technology/Korea Science and Engineering Foundation Grant R15-2003-012-01001-0 for the Environmental Biotechnology National Core Research Center, World Class University Program Grant R32-10148, Crop Functional Genomic Frontier Grant CG3313-1 (to K.O.L.), Medical Research Center Grant R13-2005-012-01003–0 (to K.R.K.), and Korea Research Foundation Grant KRF-2007-359-C00022 (to J.R.L.). H.B.C. and Y.J.J. were supported by a scholarship from the BK21 Program of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811231106/DCSupplemental.
References
- 1.Dyson HJ, et al. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: Structural and functional characterization of mutants of Asp-26 and Lys-57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- 2.Feng JN, et al. A permeabilized cell system that assembles filamentous bacteriophage. Proc Natl Acad Sci USA. 1997;94:4068–4073. doi: 10.1073/pnas.94.8.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamdan SM, et al. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci USA. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spyrou G, et al. Cloning and expression of a novel mammalian thioredoxin. J Biol Chem. 1997;272:2936–2941. doi: 10.1074/jbc.272.5.2936. [DOI] [PubMed] [Google Scholar]
- 5.Wollman EE, et al. Cloning and expression of a cDNA for human thioredoxin. J Biol Chem. 1988;263:15506–15512. [PubMed] [Google Scholar]
- 6.Gan ZR. Yeast thioredoxin genes. J Biol Chem. 1991;266:1692–1696. [PubMed] [Google Scholar]
- 7.Pedrajas JR, et al. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J Biol Chem. 1999;274:6366–6373. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- 8.Caldas T, et al. The Escherichia coli thioredoxin homolog YbbN/Trxsc is a chaperone and a weak protein oxidoreductase. Biochem Biophys Res Commun. 2006;343:780–786. doi: 10.1016/j.bbrc.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Meyer Y, et al. Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol. 2002;347:394–402. doi: 10.1016/s0076-6879(02)47039-5. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez-Marcos JF, et al. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houston NL, et al. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte S, et al. Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J Biol Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 14.Jang HH, et al. Two enzymes in one, two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Meissner U, et al. Formation, TEM study, and 3D reconstruction of the human erythrocyte peroxiredoxin-2 dodecahedral higher-order assembly. Micron. 2007;38:29–39. doi: 10.1016/j.micron.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Vignols F, et al. Redox control of Hsp70-Co-chaperone interaction revealed by expression of a thioredoxin-like Arabidopsis protein. J Biol Chem. 2003;278:4516–4523. doi: 10.1074/jbc.M210080200. [DOI] [PubMed] [Google Scholar]
- 17.Blatch GL, Lassle M. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Beissinger M, Buchner J. How chaperones fold proteins. Biol Chem. 1998;379:245–259. [PubMed] [Google Scholar]
- 19.Kern R, et al. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. Biochem J. 2003;371:965–972. doi: 10.1042/BJ20030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai B, et al. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 21.Hansen JE, Gafni A. Thermal switching between enhanced and arrested reactivation of bacterial glucose-6-phosphate dehydrogenase assisted by GroEL in the absence of ATP. J Biol Chem. 1993;268:21632–21636. [PubMed] [Google Scholar]
- 22.Haslbeck M, et al. A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J Mol Biol. 2004;343:445–455. doi: 10.1016/j.jmb.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 23.van Heel M, Frank J. Use of multivariate statistics in analyzing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 24.Su PH, Li HM. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008;146:1231–1241. doi: 10.1104/pp.107.114496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Queitsch C, et al. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puig A, Gilbert HF. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J Biol Chem. 1994;269:7764–7771. [PubMed] [Google Scholar]
- 27.Zhao TJ, et al. Catalysis of creatine kinase refolding by protein disulfide isomerase involves disulfide cross-link and dimer to tetramer switch. J Biol Chem. 2005;280:13470–13476. doi: 10.1074/jbc.M413882200. [DOI] [PubMed] [Google Scholar]
- 28.Meyer-Siegler K, et al. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsell DA, et al. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 30.Hajheidari M, et al. Proteomics uncovers a role for redox in drought tolerance in wheat. J Proteome Res. 2007;6:1451–1460. doi: 10.1021/pr060570j. [DOI] [PubMed] [Google Scholar]
- 31.Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, et al. Oligomerization, chaperone activity, and nuclear localization of p26, a small heat shock protein from Artemia franciscana. J Biol Chem. 2004;279:39999–40006. doi: 10.1074/jbc.M406999200. [DOI] [PubMed] [Google Scholar]
- 33.Sharma KK, et al. Interaction of 1,1′-bi(4-anilino)naphthalene-5,5′-disulfonic acid with α-crystallin. J Biol Chem. 1998;273:8965–8970. doi: 10.1074/jbc.273.15.8965. [DOI] [PubMed] [Google Scholar]
- 34.Moon JC, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 35.Park SC, et al. Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Mol Cells. 2006;21:129–134. [PubMed] [Google Scholar]
- 36.Clough SJ. Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol Biol. 2005;286:91–102. doi: 10.1385/1-59259-827-7:091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.