Abstract
RhoGDI2 is a suppressor of metastasis in human bladder cancer. Although diminished RhoGDI2 expression in tumors is associated with decreased patient survival, normal expression in some metastatic tumors led us to wonder whether other mechanisms regulate RhoGDI2 function. Protein interaction analysis identified Src as a novel RhoGDI2 interaction partner. Gene expression profiling and immunohistochemistry of human tumors revealed that Src levels diminish as a function of bladder cancer stage. In addition, diminished Src levels and RhoGDI2 levels appear mutually exclusive in individual tumors, indicating that both genes are likely involved in the same signaling pathway leading to metastasis suppression. Studies confirmed that activated Src kinase binds and phosphorylates RhoGDI2 in vitro and vivo. Mutagenesis revealed that Tyr-153 and, to a lesser degree, Tyr-24 were the primary Src phosphorylation sites. Phosphorylation decreased the amount of Rac1 in RhoGDI2 complexes and increased RhoGDI2 association with cell membranes. Stable expression of phosphomimetic Tyr-153 RhoGDI2 in metastatic human bladder cancer cell lines had no effect on primary tumor growth but suppressed metastasis more potently than WT RhoGDI2. These data suggest that phosphorylation by Src enhances RhoGDI2 metastasis suppression and that loss of Src relieves metastasis suppression in tumor cells that maintain RhoGDI2 expression. Our findings also suggest caution in using Src inhibitors in the hope of delaying progression in patients with bladder cancer.
Keywords: bladder neoplasms, guanine nucleotide dissociation inhibitors, neoplasm metastasis, src-family kinases
Bladder cancer is common in the United States (1). Most deaths from this disease are due to metastases, commonly to lung and liver. RhoGDI2 (LyGDI/D4GDI) has been shown to be a metastasis suppressor in animal models of bladder cancer and to be lost in many metastatic human tumors (2, 3). RhoGDI2 belongs to a family of related proteins that includes RhoGDI1 and RhoGDI3 (reviewed in ref. 4). RhoGDI1 is the most widely expressed and the most intensely studied. Previously, it was thought that RhoGDI2 expression was confined to cells of hematopoietic origin, but our studies showed it is expressed in a wider range of tissues and cell types (5). Much less is known about RhoGDI3. Its expression is highest in brain, lung, and testis and, in contrast to RhoGDI1 and RhoGDI2, it is not found in the cytoplasm, but is associated with vesicular membranes. RhoGDIs are thought to inhibit the activation of small GTPases of the Rho family by sequestering the GTPases in the cytosol and blocking activation by guanine nucleotide exchange factors (GEFs) (4). Previous work identified RhoA, Rac1, and Rac2 as targets of RhoGDI2-mediated inactivation (6, 7), suggesting that RhoGDI2's function overlaps that of RhoGDI1.
Although RhoGDI2 expression is reduced in many metastatic bladder cancers, ≈35% of patients with moderate or high levels of RhoGDI2 protein still developed metastatic disease. Although factors unrelated to the RhoGDI2 pathway could mediate lung metastasis in bladder cancer, we also considered the possibility that RhoGDI2 might be regulated by means other than expression. To explore this hypothesis, we carried out a mass spectrometry-based protein interaction screen for binding partners of RhoGDI2. Here, we show that Src associates with RhoGDI2 and enhances the ability of RhoGDI2 to suppress lung colonization in xenograft models of human bladder cancer.
Results
Mass Spectroscopic Identification of RhoGDI2 Immunocomplex Partners.
To identify interaction partners of RhoGDI2, metastatic UMUC3 human bladder carcinoma cells with low endogenous RhoGDI2 protein levels were transfected with pFLAG-CMV4-RhoGDI2 or pFLAG-CMV4 as a control. Eluates from 4 separate precipitations as well as 2 pulldowns from control cells were analyzed by mass spectrometry. FLAG immunoprecipitation and elution revealed 298 protein matches (Table S1). In addition to RhoGDI2 itself, known RhoGDI2-binding proteins in the immunoprecipitates included RhoA, Rac1, and Vav1, which supported the validity of the assay in finding proteins in complex with RhoGDI2. Interestingly, Src was also found to coprecipitate with RhoGDI2. Recent reports identified RhoGDI1 (8, 9) and RhoGDI2 (10) as tyrosine-phosphorylated proteins in cancer cells. Furthermore, Src regulates RhoGDI1 function through phosphorylation on a tyrosine that is conserved in RhoGDI2 (11). We therefore selected Src for further study.
Src Expression Decreases as a Function of Tumor Stage in Human Bladder Cancer.
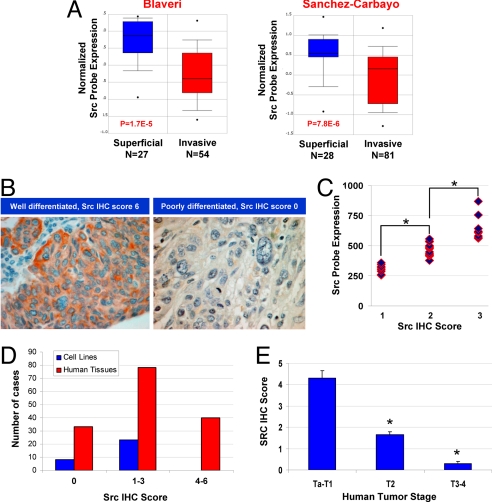
We next sought to determine Src's possible relevance to this system by examining the relationship between Src expression and tumor stage. Examination of publicly available data at www.oncomine.org revealed 2 studies in which bladder cancer gene expression was examined as a function of stage. Both studies showed statistically significant reduction of Src expression with higher stage (Fig. 1A). To evaluate the relationship of Src protein to human tumor stage, we developed 2 tissue microarrays (TMAs): one of human bladder cell lines and one of human bladder cancer tissues. We also analyzed gene expression data from tumor lines by using the same Affymetrix microarray used in one of the human studies shown in Fig. 1A. Immunohistochemical analysis and scoring of the cell lines (Fig. 1B) showed that Src protein levels correlated with Src message (Fig. 1C). Furthermore, Src staining of the cell lines was lower than for the tumor tissues (Fig. 1D), consistent with the finding that all but one (RT-4) of these cell lines are derived from highly aggressive cancers. Finally, immunohistochemistry (IHC) analysis of 151 primary tumors showed the reduction of Src expression correlated with tumor stage (Fig. 1E). Taken together, these studies indicate that Src is commonly lost in aggressive bladder cancers.
Fig. 1.
Src expression in human bladder cancer. (A) Src expression in human bladder cancer as a function of tumor stage in 2 independent studies (31, 32) culled from the Oncomine database (www.oncomine.org). Number of tumors in each stage group and statistical significance of differences in expression levels as a function of stage are shown. The Sanchez-Carbayo study (Right) used an HU-133A Affymetrix array probe set at 213324_at, whereas the Blaveri study (Left) used a custom 10,368-element cDNA microarray. (B) IHC evaluation of Src in human urothelial cancer TMA using GD11 antibody. Examples shown demonstrate scoring of Src staining. (C) Mean Src expression level by IHC in the TMA compared with the level as evaluated by HU-133A Affymetrix array (probe set is 213324_at) on 31 cell lines. *, P < 0.05 by t test. (D) Mean Src expression level by IHC in the TMA on cell lines and human tumors (n = 151). (E) Mean Src expression level by IHC in the human tumor TMA as a function of tumor stage (n = 151). Bars are SE. *, P < 0.05 by t test for group compared to Ta-T1.
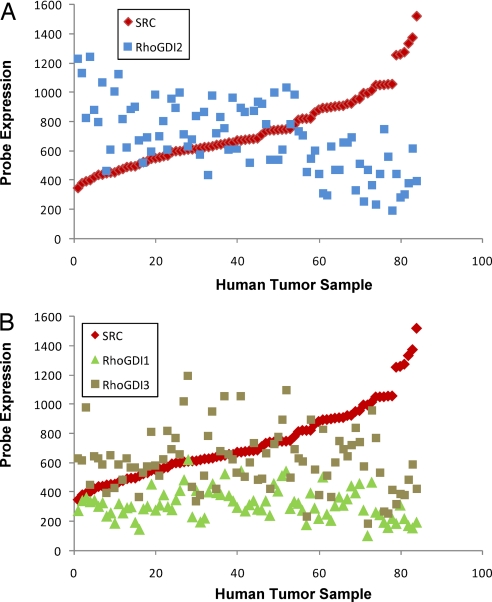
To determine whether loss of Src and RhoGDI2 showed any correlation, we evaluated the expression of both genes in 85 human tumors by microarray. Interestingly, expression of these genes showed a significant inverse relationship (Fig. 2A), which was not the case with RhoGDI1 or RhoGDI3 (Fig. 2B). This observation suggests that reduced expression of Src and RhoGDI2 is mutually exclusive, indicating that both genes are likely to be involved in the same signaling pathway leading to metastasis suppression.
Fig. 2.
The relationship of Src RNA expression to that of RhoGDI family genes in human bladder cancer. (A) Src and RhoGDI2 expression in human bladder cancer in 85 human bladder tumors profiled by the HU-133A Affymetrix array (12, 25). (B) Src and RhoGDI1 and RhoGDI3 expression in human bladder cancer in 85 human bladder tumors profiled by the HU-133A Affymetrix array (12, 25).
RhoGDI2 Is Bound and Phosphorylated in Vitro and in Vivo by Src.
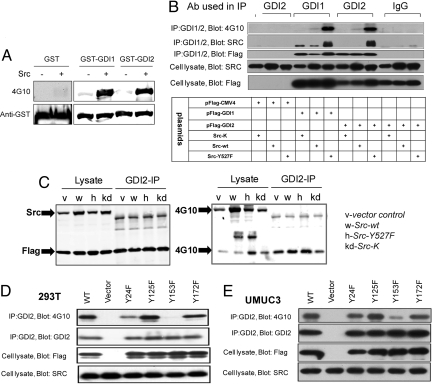
We next investigated whether Src phosphorylates RhoGDI2 in vitro. A kinase assay with purified active Src showed strong phosphorylation of RhoGDI2 (Fig. 3A). We then evaluated whether Src phosphorylates RhoGDI2 in cells by using 293T cells cotransfected with FLAG-tagged RhoGDI1 or RhoGDI2 together with inactive Src-K−, WT Src, or constitutively active Src (Y527F). RhoGDI1 or RhoGDI2 was immunoprecipitated and then probed for FLAG, Src, and phosphotyrosine. Control immunoprecipitations using nonimmune rabbit IgG were also analyzed (Fig. 3B). RhoGDI1, included as a positive control, showed substantial association and phosphorylation by Src kinase, consistent with published data (11). We observed that RhoGDI2 complexed weakly with WT Src but more strongly with constitutively active Src, at levels comparable to RhoGDI1. RhoGDI2 phosphorylation on tyrosine showed a similar pattern. Neither association nor phosphorylation was observed with inactive Src-K−. RhoGDI2 phosphorylation on tyrosine showed a similar pattern in human metastatic prostate cancer cells PC-3 (Fig. 3C).
Fig. 3.
RhoGDI phosphorylation and complex formation with Src. (A) In vitro kinase assay with Src and RhoGDIs 1 and 2 carried out as described in Materials and Methods. (B) 293T cells were cotransfected with FLAG-tagged RhoGDI1 and RhoGDI2 and different HA-tagged Src plasmids (inactive Src-K−, WT Src, and constitutively active Src-Y527F) as indicated. Shown is RhoGDI containing immunoprecipitated complexes probed with 4G10 phosphotyrosine, Src, and FLAG antibody as described in Materials and Methods. The IgG IP control represents a nonspecific rabbit IgG pulldown. (C) PC3 prostate cancer cells cotransfected with FLAG-RhoGDI2 and Src. The cells were lysed in RIPA buffer, and immunoprecipitations were carried out with anti-RhoGDI2, rabbit IgG as control. The recoveries of RhoGDI2 and Src were examined by Western blotting using anti-FLAG and anti-Src. The phosphorylations of RhoGDI2 and Src were detected by 4G10 (P-Tyr) antibody blotting. (D) 293T and (E) UMUC3 cells cotransfected with active Src Y527F (or empty vector) and the indicated mutant versions of FLAG-tagged RhoGDI2. (Third and Bottom) Equal amounts of each construct were overexpressed and immunoprecipitated. Western blots of immunoprecipitated RhoGDI2 constructs with 4G10 phosphotyrosine antibody indicated that RhoGDI2 is phosphorylated by Src at Tyr-153, to a lesser extent at Tyr-24, but not at Tyr-125 and Tyr-172.
The NetPhos 2.0 program (33) predicted that Tyr-24, Tyr-125, Tyr-153, and Tyr-172 in RhoGDI2 were potential sites for Src kinase. To evaluate these sites, we mutated each one individually to phenylalanine (F) and then assessed phosphorylation when cotransfected with constitutively active SrcY527F. In both 293T (Fig. 3D) and UMUC3 (Fig. 3E) cells, mutation of Tyr-153 nearly eliminated RhoGDI2 phosphorylation by Src kinase. The Y24 mutant also showed a partial reduction in phosphorylation, which was significant in 293T but noticeably less in UMUC3 human bladder cancer cells. In contrast, mutation of either Tyr-125 or Tyr-172 had no effect in either cell line. We conclude that Y153 is the major site of phosphorylation. Interestingly, when using the Scansite program (http://scansite.mit.edu) at moderate stringency, EGFR, PDGFRB, FGR, SHC1, CRK, LCK, and ITK are also predicted to phosphorylate and/or bind at Y153.
Tyrosine Phosphorylation of RhoGDI2 Alters Its Complex Formation with Rac1.
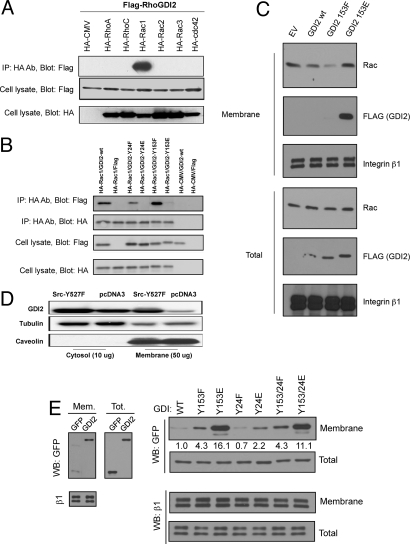
To identify which Rho GTPases associate with RhoGDI2, we individually coexpressed HA-tagged Rac1, Rac2, Rac3, RhoA, RhoC, and Cdc42 with FLAG-tagged RhoGDI2 in 293T cells. Immunoprecipitation with anti-HA antibody and Western blotting for FLAG detected RhoGDI2 only in the Rac1 precipitate (Fig. 4A). Thus, Rac1 seems to be the major target for RhoGDI2. We next evaluated the effect of Src-mediated phosphorylation on RhoGDI2's ability to complex with Rac1. We generated RhoGDI2 Y153 and Y24 mutants in which tyrosines were replaced with glutamic acid (E) to mimic phosphorylation or with phenylalanine (F) to prevent phosphorylation. These mutants were cotransfected with HA-tagged Rac1, cell lysates were precipitated with HA antibody, and the precipitates were probed for Rac1 and RhoGDI2. RhoGDI2 glutamic acid mutants all exhibited decreased co-IP with Rac1, whereas the corresponding phenylalanine residue mutants showed a modest increase relative to WT RhoGDI2 (Fig. 4B). These data suggest that RhoGDI2 phosphorylation by Src decreases its association with Rac1 in complex.
Fig. 4.
Rho family GTPases in complex with RhoGDI2. (A) HA-tagged Rho proteins Rac1, Rac 2, Rac 3, RhoA, RhoC, and Cdc42 were pulled down from 293T cell lysates by HA antibody. Pulldowns were probed with FLAG antibody to detect the RhoGDI2 presence. Representative results from 1 of 3 experiments show that only Rac1 is found in complex with RhoGDI2. (B) RhoGDI2 phosphomimetic mutants in complex with Rac1. HA-tagged Rac1 was pulled down from 293T cell lysates by HA antibody. Pulldowns were probed for the presence of both Rac1 (Second) and associated RhoGDI2 (Top). (C) RhoGDI2 and Rac1 in UMUC3 cell fractions. Cell fractions were prepared as described in Materials and Methods from UMUC3 cells transiently transfected and expressing FLAG-tagged WT RhoGDI2 or GDI2-Y153F or GDI2-Y153E mutants and fractions probed for FLAG (RhoGDI2), Rac1, and integrin β1. (D) Membrane versus cytosol fractions of RhoGDI2 as a function of activated Src. Fractions were prepared as described in Materials and Methods by ultracentrifugation from RhoGDI2, SrcY527F-expressing, or its vector in 293T cells. The membrane fractions versus cytosol fractions were prepared by ultracentrifuge from SrcY527F-expressing and its vector in 293T cells, and then the fractions were probed for RhoGDI2, tubulin, and caveolin. (E) Membrane targeting of RhoGDI2-GFP and mutants. (Left) UMUC3 cells transfected with GFP alone or RhoGDI2-GFP were fractionated as above and analyzed by Western blotting with anti-GFP. Antibody to the integrin β1 subunit was used as a loading control. (Right) UMUC3 cells transfected with GFP fusions of the indicated mutants were fractionated and analyzed by Western blotting in the same way. Values represent membrane/total signal for GFP relative to WT RhoGDI. Two experiments gave similar results.
To test the effects of Y153 phosphorylation on Rac1, membrane targeting of Rac1 was assayed in UMUC3 cells expressing FLAG-tagged RhoGDI2 WT, Y153F, or Y153E mutants. Membrane fractions of these cells were probed for Rac1, with integrin β1 levels used for normalization. WT RhoGDI2 moderately decreased Rac1 membrane targeting (Fig. 4C). This effect was slightly more pronounced in the Y153F mutation and was abrogated by the Y153E mutation. Taken together, these results suggest that a negative charge at residue 153 decreases RhoGDI2 complex formation with Rac1 and prevents its inhibition of Rac1 membrane targeting.
Activated Src Enhances RhoGDI2 Membrane Localization.
Src phosphorylation of RhoGDI1 induces membrane association (11). To determine whether Src also alters the intracellular localization of RhoGDI2, we fractionated 293T cells and evaluated the presence of endogenous RhoGDI2 in the cytosol and membranes with or without transfection of active Src. Src caused a large increase in RhoGDI2 in membrane fractions (Fig. 4D, lane 3 vs. lane 4). To further explore this effect, we expressed WT or mutant forms of RhoGDI2, then evaluated the levels of these constructs in the cytosolic and membrane fractions. WT GFP-GDI2 showed weak but detectable membrane association compared with GFP (Fig. 4E Left). Interestingly, the 153E phosphomimetic mutation caused a large increase in RhoGDI2 in the membrane fraction (Fig. 4E Right). By contrast, the phosphomimetic mutation at Y24 had little effect. Curiously, mutation of Y153 to F also modestly increased membrane association, although much less than Y153E. We conclude that tyrosine phosphorylation of RhoGDI2 on Y153 but not Y24 mimics Src by inducing association with membranes. Results with Y153F suggest that this tyrosine hydroxyl group may be important for preventing membrane binding, but this hypothesis requires further investigation.
The Y153E RhoGDI2 Mutation Enhances Suppression of Lung Colonization.
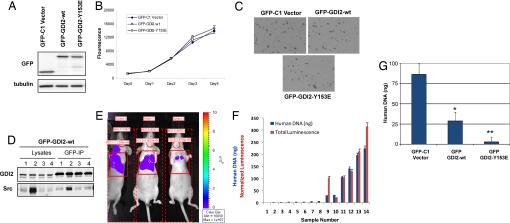
UMUC3 cells were transfected with GFP fusions of WT or Y153E RhoGDI2, or with GFP-C1 as a control. Stable cell lines were subjected to antibiotic selection and then FACS-sorted to obtain equivalent transgene expression. Western blotting for GFP verified that these lines had similar transgene levels (Fig. 5A). These cell lines were then evaluated in vitro and in vivo. All of the lines showed similar rates of monolayer growth in vitro (Fig. 5B) and similar colony formation in soft agar (Fig. 5C). No differences in tumor size were noted when these cells were injected s.c. into mice, consistent with prior data with WT RhoGDI2 (2, 12, 13). We also used GFP-GDI2-WT excised s.c. tumor tissue to evaluate whether Src and RhoGDI2 coprecipitate. Although total Src levels were quite variable between tumors, some s.c. tumors demonstrated coimmunoprecipitation of Src with RhoGDI2 (Fig. 5D).
Fig. 5.
Biological consequences of RhoGDI2 mutations in UMUC3 bladder cancer cells. (A) Expression levels of RhoGDI2-GFP fusion proteins in stably transfected cells. (Upper) Similar GFP fusion protein levels on stable cell lines of GFP-C1, GFP-GDI2-WT, and GFP-GDI2-Y153E. (Lower) The similar total protein loading indicated by α-tubulin. (B) Growth curves in monolayer culture of GFP-C1 vector, GFP-GDI2-WT, GFP-GDI2-Y153E stable cells. Cells were plated at 105 cells per well in 6-well plates for various times, and cell numbers were estimated by using an Alamar Blue assay. Fluorescence emission was assessed on days 0, 1, 2, 3, and 4 from plating. Each bar indicates the mean and SD of values obtained from triplicate samples. (C) Colony formation in agar was carried out in 0.4% agar/media containing 5% serum and evaluated at 13 days. (D) The s.c. tumors generated from inoculation of 106 GFP-GDI2-WT cells into nude mice were lysed in RIPA buffer. A total of 1 mg of total protein was immunoprecipitated with anti-GFP agarose. The recovery of RhoGDI2 was examined by anti-RhoGDI2 blotting. The Src in the complex was detected by anti-Src blotting. (E) Representative bioluminescent imaging using IVIS Imaging Systems (Caliper Life Sciences) of nude mice inoculated in the tail vein with UMUC3 cells stably expressing luciferase. (F) Quantitative (normalized to fit same scale as DNA quantitation) bioluminescent imaging compared with lung DNA assessment (14) of murine lungs following inoculation of cells in E in 14 mice. Mean human DNA amount (ng) in 2 mg of mouse lung DNA is shown. Error bar is SD. (G) Quantitative lung DNA assessment (14) for cells in A at 4 weeks after tail vein inoculation. Mean human DNA amount (ng) in 2 mg of mouse lung DNA is shown. Error bar is SD. A total of 20 mice were injected per group. Experiments were repeated twice with similar results. *, P < 0.001 by Student's t test compared to GFP-C1 vector. **, P < 0.0001 by Student's t test compared to GFP-GDI2-WT.
To reduce the time required and improve the quantitative evaluation of lung metastasis by human xenografts in murine hosts, we developed a DNA PCR assay (14). Using this assay, the amount of human DNA in the lung following tail-vein inoculation of cancer cells was found to correlate with surrogates of murine survival (14). Here, we sought to adapt this assay to determine the effect of RhoGDI2 on lung colonization, defined as the growth of human tumors in the lung to a sufficient size for visible detection by using imaging. To accomplish this, we evaluated the relationship of human DNA levels in the lung with tumor bioluminescence in this organ (13) (Fig. 5E). Using lungs harvested from 2 to 8 weeks after tail-vein inoculation with UMUC3 cells stably expressing luciferase, we generated a curve comparing human DNA levels to intensity of bioluminescence (13) (Fig. 5F). Results suggest that PCR signals >25 ng of DNA reflect lung colonization imaged by this modality. Following tail-vein inoculation, PCR analysis of lungs at 4 weeks indicated that vector-transfected UMUC3 cells led to lung colonization. In contrast, GFP-GDI2 suppressed lung colonization, consistent with previous results (2, 12, 13). Strikingly, GFP-GDI2-Y153E was associated with a nearly absent lung tumor burden compared with that of GFP-GDI2 (Fig. 5D).
Discussion
RhoGDI2 has been shown to be a metastasis suppressor in different cancers, including bladder cancer, breast cancer, and leukemia (5, 15, 16). Several reports have demonstrated that RhoGDI2 is phosphorylated in response to cell addition of phorbol esters or stimulation through the T cell receptor (17–19), but these reports did not identify which kinase directly or indirectly affects RhoGDI2 phosphorylation nor at what residues RhoGDI2 is phosphorylated. Furthermore, they did not evaluate the relevance of such changes on RhoGDI2 functions.
Using a mass spectrometry-based protein interaction screen, we identified Src in RhoGDI2 immunoprecipitates from tumor cells. Interestingly, in a small study 15 years ago (21), Src expression and activity were shown to decrease with bladder tumor stage and aggressiveness. We confirmed and extended this result by evaluating message levels in a large number of patients and by immunohistochemical analysis of Src in human bladder tumors. Src expression in bladder tumors decreased in tumors with increasing stage, and this change was observed in carcinomas rather than stroma. Although this relationship is unusual compared with other carcinomas, it is consistent with a role for Src as a tumor or metastasis suppressor in bladder cancer. This idea is further supported by the reciprocal relationship of expression of these 2 genes, consistent with involvement in the same molecular pathway of metastasis suppression.
Further evaluation revealed that RhoGDI2 was phosphorylated by Src both in vitro and in cells, and that in bladder cancer cells the major site of phosphorylation is Tyr-153, with minor phosphorylation at Tyr-24. This result is interesting because work from the Bokoch laboratory (4, 11, 20) has shown phosphorylation of RhoGDI1 by Src on the corresponding residue, Tyr-156. Interestingly, this phosphorylation modulates RhoGDI1-Rho GTPase complex formation (11). Although RhoA, Rac1, and Rac2 were reported to bind RhoGDI2 in vitro (4), our data indicated that only Rac1 could be detected in RhoGDI2 immunoprecipitates. We also showed that mutation of RhoGDI2 Y153 to glutamic acid strongly decreased the association with Rac1, whereas mutation to Phe somewhat increased binding. These results suggest that phosphorylation by Src on this site may regulate its binding to Rac1. The other Tyr-to-Glu RhoGDI2 mutants also associated weakly with Rac1, but whether they are physiological sites for other kinases is currently unknown. Although Tyr-125 and Tyr-172 are not Src targets, these sites, if phosphorylated by other kinases, might also alter RhoGDI2 association with Rac1.
Interestingly, we found more RhoGDI2 in the membrane fraction of cells when cotransfected with constitutively active SrcY527F or when the RhoGDI2 Y153E mutant was analyzed. RhoGDI1 behaved similarly when phosphorylated by Src (11). Mutation of the Y24 site had only a weak effect, again suggesting that Y153 is the critical residue. It is possible that Src is the membrane-binding site, because phosphorylated RhoGDI2 bound well to Src kinase. However, the 153E mutant would not be predicted to form a good binding site for the Src SH2 domain and instead suggests that phosphorylation promotes membrane association, either through a charge interaction or a conformational change that reveals a cryptic binding site. The membrane anchor(s) for phosphorylated RhoGDI1 and RhoGDI2 will be an important question for future research. In our prior animal experiments, WT RhoGDI2 was consistently shown to suppress experimental metastasis (2). Interestingly, the phosphomimetic GFP-GDI2-Y153E was a more potent suppressor of lung metastasis than its WT counterpart. This result therefore suggests that membrane association may be a component of the tumor suppression pathway.
In summary, our data define a novel mechanism for how loss of Src expression and activity (21) in bladder cancer as a function of tumor progression conspires to reduce the potency of proteins, such as RhoGDI2, that guard against metastasis. Furthermore, given the ongoing development of Src family kinase inhibitors for cancer therapy, our data suggest cautious use aimed at preventing or reducing bladder cancer progression.
Materials and Methods
Cell Lines, Plasmid Constructs, and Transfection.
293T cells were maintained in 10% FBS DMEM (Sigma). UMUC3 and other human bladder cancer cell lines were maintained and expression profiled by Affymetrix HU-133A as described previously (12). NetPhos 2.0 was used to predict RhoGDI2 tyrosine phosphorylation sites (22). Predicted phosphorylation sites of RhoGDI2 were mutated and subcloned from full-length RhoGDI2 (3) into the EcoRI and XhoI sites of the pFLAG-CMV4 vector (Sigma) by using primers GDI2-f: AGAATTCAAATGACTGAAAAAGCCCCAGAG, and GDI2-r: GCCTCGAGTCATTCTGTCCACTCCTTC. All RhoGDI2 mutant inserts were verified by sequencing. SrcY527F constitutively active (23) and Src-K− kinase-inactive (24) plasmids were as described previously. Transient transfections of FLAG-tagged RhoGDI2, its mutants, and its empty vector into 293T and UMUC3 cells were performed by FuGENE 6 (Roche) using the manufacturer's instructions. To establish the GFP-tagged RhoGDI2 stable cell lines, WT RhoGDI2 and RhoGDI2-Y153E mutants were amplified and inserted into EGFP-C1 vector (Clontech) at BamHI and EcoRI with primers GFPGDI2b: CGGATCCTCATTCTGTCCACTCCT, and GFPGDI2a: GGAATTCTACTGAAAAAGCCCCAGAG. GFP-tagged RhoGDI2 was cotransfected with p-Babe plasmid into UMUC3. Two days after the transfection, 2 μg/mL puromycin (Invitrogen) was added to cultures. Puromycin-resistant cells were harvested and subjected to FACS sorting to select GFP-positive cells using a Becton Dickinson FACS Vantage SE Turbo Sorter with DIVA Option (Becton Dickinson). By using FACS, cell populations were selected for similar GFP levels.
Western Blot, Immunoprecipitation, Cell Fractionation, and Mass Spectrometry Analyses.
Whole-cell extracts were prepared and immunoprecipitations carried out with RhoGDI1 (Santa Cruz Biotechnology), RhoGDI2 (Spring Bioscience), or HA antibody (Y-11; Santa Cruz Biotechnology) as described previously (25). ECL reagents were purchased from Pierce, phosphotyrosine antibody 4G10 (05-321) from Upstate Biotechnology, FLAG antibody from Sigma, and GFP antibody from Santa Cruz Biotechnology. Src antibody clone 327 was a gift from Sarah Parsons. Membrane and cytosol fractionation was carried out as described in ref. 26. Membrane and cytosol fractions were then assayed for total protein and analyzed by Western blot analysis. The membrane fractions were checked for the fractionation purification by immunoblotting for the cytosolic and membrane markers α-tubulin (Calbiochem) and Caveolin-1 or integrin β1 (BD Biosciences), respectively. UMUC3 cells were transiently transfected with pFLAG-CMV4 and pFLAG-CMV4-RhoGDI2, and mass spectrometry analysis of the immunoprecipitate was carried out as described previously (27, 28).
In Vitro Kinase Assay.
N-terminal-tagged GST-RhoGDI1 or GST-RhoGDI2 fusion proteins were expressed in Escherichia coli strain BL21 (DE3) and purified by GST-Bind Kits (Novagen) according to the manufacturer's instructions. A total of 1 μg of GST, GST-RhoGDI1, or GST-RhoGDI2 immobilized on glutathione agarose was incubated with 10 units of Src(p60c-src) kinase (Upstate Biotechnology) in Src Kinase Reaction Buffer (Upstate Biotechnology) and 100 μM ATP for 1 h at 30 °C. The phosphorylation was detected by Western blotting using 4G10 antibody. The protein level in the reaction was checked by anti-GST blotting.
Gene Expression, TMA Construction, and Src IHC.
Gene expression profiles of human bladder cancer (12, 25) were used to evaluate the relationship of RhoGDI2 to Src expression. Two human bladder cancer TMAs were constructed by using previously described techniques (5). The first TMA contained four 0.6-mm cores obtained from 151 primary human urothelial carcinomas of the bladder and was constructed from zinc formalin-fixed, paraffin-embedded blocks (Beecher Instruments). All slides from the resections were reviewed by one pathologist (H.F.F.) for histologic subtype, grade of urothelial carcinoma, and pathologic stage [according to the AJCC Cancer Staging Manual (29)]. The second TMA was constructed from 31 previously described human bladder cancer cell lines (12). For the cell line TMA, 106 cells were scraped with a rubber policeman and suspended in ice-cold phosphate-buffered zinc formalin. At 24 h following fixation, the cells were pelleted and embedded in paraffin. For Src IHC, deparaffinized zinc formalin-fixed, paraffin-embedded tissue sections were placed in citrate buffer, pH 6.0, and heated in a microwave oven for 20 min. After incubation with anti-c-src (clone GD11 monoclonal at 1:100 dilution) for 1 h at room temperature, followed by the biotinylated secondary antibody, avidin-biotin immunoperoxidase was applied. Diaminobenzidine was used as the chromogen, and sections were counterstained with hematoxylin. The staining was cytoplasmic and cytoplasmic membrane. The staining score included percent of stained cells (0; 1: 1–25% positive; 2: 26–75%; and 3: 76–100%) × staining intensity (1: weak; 2: strong), leading to a score range of 0 to 6.
In Vitro and in Vivo Tumor Cell Assays.
In vitro cell growth assays were carried out by using Alamar Blue fluorescence emission as described previously (30). Colony formation in agar was assessed by using 2 × 104 cells per well of 6-well plates as described previously (13). Six-week-old nude mice were obtained from the National Cancer Institute breeding facility and maintained according to the National Institutes of Health and institutional guidelines. The s.c. tumorigenicity of 106 UMUC3 cells per site was evaluated as described previously (13). For evaluation of lung metastasis, mice were injected via tail vein with 106 UMUC3 cells suspended in 0.1 mL of serum-free media and evaluated by using a highly quantitative and sensitive DNA PCR metastasis assay described previously by our laboratory (14). Bioluminescent in vivo imaging of lung metastases was carried out as we have described previously (13) on UMUC3 cells stably transfected with a vector expressing firefly luciferase (13).
Supplementary Material
Acknowledgments.
We thank Drs. Alex Toker (Harvard Medical School, Boston) and Sarah Parsons (University of Virginia, Charlottesville) for Src constructs. This work was supported by National Institutes of Health Grant CA075115 (to D.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810094106/DCSupplemental.
References
- 1.Jemal A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Gildea JJ, et al. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- 3.Seraj MJ, Harding MA, Gildea JJ, Welch DR, Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- 4.DerMardirossian C, Bokoch GM. GDIs: Central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Theodorescu D, et al. Reduced expression of metastasis suppressor RhoGDI2 is associated with decreased survival for patients with bladder cancer. Clin Cancer Res. 2004;10:3800–3806. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 6.Golovanov AP, et al. Structure-activity relationships in flexible protein domains: Regulation of rho GTPases by RhoGDI and D4 GDI. J Mol Biol. 2001;305:121–135. doi: 10.1006/jmbi.2000.4262. [DOI] [PubMed] [Google Scholar]
- 7.Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol. 2000;7:122–126. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- 8.Unwin RD, Pierce A, Watson RB, Sternberg DW, Whetton AD. Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol Cell Proteomics. 2005;4:924–935. doi: 10.1074/mcp.M400193-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Unwin RD, et al. Global effects of BCR/ABL and TEL/PDGFRbeta expression on the proteome and phosphoproteome: Identification of the Rho pathway as a target of BCR/ABL. J Biol Chem. 2005;280:6316–6326. doi: 10.1074/jbc.M410598200. [DOI] [PubMed] [Google Scholar]
- 10.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 11.DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell. 2006;17:4760–4768. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Titus B, et al. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7327. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, et al. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. 2007;26:765–773. doi: 10.1038/sj.onc.1209835. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson BE, et al. Profiling the evolution of human metastatic bladder cancer. Cancer Res. 2004;64:7813–7821. doi: 10.1158/0008-5472.CAN-04-0826. [DOI] [PubMed] [Google Scholar]
- 15.Hu LD, Zou HF, Zhan SX, Cao KM. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol Rep. 2007;17:1383–1389. [PubMed] [Google Scholar]
- 16.Nakata Y, et al. Mutated D4-guanine diphosphate-dissociation inhibitor is found in human leukemic cells and promotes leukemic cell invasion. Exp Hematol. 2008;36:37–50. doi: 10.1016/j.exphem.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Gorvel JP, Chang TC, Boretto J, Azuma T, Chavrier P. Differential properties of D4/LyGDI versus RhoGDI: Phosphorylation and rho GTPase selectivity. FEBS Lett. 1998;422:269–273. doi: 10.1016/s0014-5793(98)00020-9. [DOI] [PubMed] [Google Scholar]
- 18.Groysman M, Hornstein I, Alcover A, Katzav S. Vav1 and Ly-GDI two regulators of Rho GTPases, function cooperatively as signal transducers in T cell antigen receptor-induced pathways. J Biol Chem. 2002;277:50121–50130. doi: 10.1074/jbc.M204299200. [DOI] [PubMed] [Google Scholar]
- 19.Scherle P, Behrens T, Staudt LM. Ly-GDI, a GDP-dissociation inhibitor of the RhoA GTP-binding protein, is expressed preferentially in lymphocytes. Proc Natl Acad Sci USA. 1993;90:7568–7572. doi: 10.1073/pnas.90.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell. 2004;15:117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 21.Fanning P, et al. Elevated expression of pp60c-src in low grade human bladder carcinoma. Cancer Res. 1992;52:1457–1462. [PubMed] [Google Scholar]
- 22.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 23.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 2003;22:109–120. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizawar RC, Tice DA, Karaoli T, Parsons SJ. The C terminus of c-Src inhibits breast tumor cell growth by a kinase-independent mechanism. J Biol Chem. 2004;279:23773–23781. doi: 10.1074/jbc.M312368200. [DOI] [PubMed] [Google Scholar]
- 25.Smith SC, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 26.Moissoglu K, Slepchenko BM, Meller N, Horwitz AF, Schwartz MA. In vivo dynamics of Rac-membrane interactions. Mol Biol Cell. 2006;17:2770–2779. doi: 10.1091/mbc.E06-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herlevsen M, Oxford G, Owens CR, Conaway M, Theodorescu D. Depletion of major vault protein increases doxorubicin sensitivity and nuclear accumulation and disrupts its sequestration in lysosomes. Mol Cancer Ther. 2007;6:1804–1813. doi: 10.1158/1535-7163.MCT-06-0372. [DOI] [PubMed] [Google Scholar]
- 28.Herlevsen MC, Theodorescu D. Mass spectroscopic phosphoprotein mapping of Ral binding protein 1 (RalBP1/Rip1/RLIP76) Biochem Biophys Res Commun. 2007;362:56–62. doi: 10.1016/j.bbrc.2007.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene FL, et al., editors. AJCC Cancer Staging Manual. 6th Ed. New York: Springer; 2002. [Google Scholar]
- 30.Havaleshko DM, et al. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther. 2007;6:578–586. doi: 10.1158/1535-7163.MCT-06-0497. [DOI] [PubMed] [Google Scholar]
- 31.Blaveri E, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 33.Blom N, Gammeltoft S, Brunak S. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.