Abstract
Urea denaturation studies were carried out as a function of transthyretin (TTR) concentration to quantify the thermodynamically linked quaternary and tertiary structural stability and to better understand the relationship between mutant folding energetics and amyloid disease phenotype. Urea denaturation of TTR involves at least two equilibria—dissociation of tetramers into folded monomers, and monomer unfolding. To deal with the thermodynamic linkage of these equilibria, we analyzed concentration-dependent denaturation data by global fitting to an equation that simultaneously accounts for the two-step denaturation process. Using this method, the quaternary and tertiary structural stabilities of well-behaved TTR sequences, wild type (WT) TTR and the disease-associated variant V122I, were scrutinized. The V122I variant is linked to late onset familial amyloid cardiomyopathy, the most common familial TTR amyloid disease. V122I TTR exhibits a destabilized quaternary structure and a stable tertiary structure relative to WT TTR. Three other variants of TTR were also examined, L55P, V30M, and A25T TTR. The L55P mutation is associated with the most aggressive familial TTR amyloid disease. L55P TTR has a complicated denaturation pathway that includes dimers and trimers, and so globally fitting its concentration-dependent urea denaturation data yielded error-laden estimates of stability parameters. Nevertheless, it is clear that L55P TTR is substantially less stable than WT TTR, primarily because its tertiary structure is unstable, although its quaternary structure is destabilized as well. V30M is the most common mutation associated with neuropathic forms of TTR amyloid disease. V30M TTR is certainly destabilized relative to WT TTR, but like L55P TTR it has a complex denaturation pathway that cannot be fit to the aforementioned two-step denaturation model. Literature data suggest that V30M TTR has stable quaternary structure but unstable tertiary structure. The A25T mutant, associated with central nervous system amyloidosis, is highly aggregation-prone and exhibits drastically reduced quaternary and tertiary structural stability. The observed differences in stability amongst the disease-associated TTR variants highlight the complexity and the heterogeneity of TTR amyloid disease, an observation having important implications for the treatment of these diseases.
Transthyretin (TTR) is one of more than thirty amyloidogenic proteins that, when partially denatured, can misassemble into several aggregate structures, including cross-β sheet amyloid fibril quaternary structures (1–6). The process of amyloidogenesis is causatively linked to numerous human diseases, many of which are associated with neurodegeneration (1–3). To date, all of the TTR variants associated with familial amyloidosis that have had their folding energetics characterized—21 of them—are destabilized relative to WT TTR, based on the observation that their denaturation transitions occur at lower concentrations of urea than WT TTR (7–15). TTR is tetrameric under physiological conditions. The thermodynamic linkage of the TTR tetramer dissociation and monomer unfolding equilibria make it challenging to extract thermodynamic data from the urea denaturation curves (7). How disease-associated mutations influence quaternary and tertiary structural stability and how folding and tetramerization energetics correlate with disease phenotypes is some of the valuable information sought herein.
Generally, rate-limiting tetramer dissociation enables monomer misfolding that is required for TTR to form amyloid (5, 16–22), by a very efficient downhill polymerization process (23). Most of the disease-associated TTR mutations form normal tetrameric structures based on crystallographic analysis (24) and function normally, including transporting and binding thyroid hormone and holo retinol binding protein in the plasma and cerebrospinal fluid (CSF). Thus, it is the propensity of these variants to misfold, not their inability to fold and function, that results in disease through a gain of toxic function mechanism(s), the details of which remain elusive but appear to be associated with TTR aggregation (7, 17, 18, 25, 26).
Senile systemic amyloidosis (SSA) is a disease that typically presents after age 60, characterized by the deposition of WT TTR in the heart, ultimately leading to congestive heart failure (27). SSA appears to affect as much as 25% of the population over age 80 (27). A number of autosomal dominant TTR amyloidoses have also been described that are associated with the deposition of variant transthyretin. These disorders have been subcategorized as familial amyloid polyneuropathy (FAP), familial amyloid cardiomyopathy (FAC), or central nervous system selective amyloidosis (CNSA), depending on the tissue(s) affected by deposition of the mutant TTR proteins (8, 12, 13, 28, 29).
The liver secretes TTR into the blood, whereas the choroid plexus secretes TTR into the CSF—secreted TTR from the liver and the choroid appear to be the source of amyloidogenic transthyretin in the periphery and the brain, respectively. Gene therapy mediated by replacing a disease-associated variant TTR/WT TTR expressing liver by a WT TTR/WT TTR expressing liver through transplantation has proven useful as a strategy for treating familial amyloid polyneuropathy (30). We have discovered small molecule TTR tetramer binders that impose kinetic stability on the tetramer, preventing dissociation required for amyloidogenesis (20, 31–34). Two such small molecules are currently being tested as agents to ameliorate FAP in placebo-controlled clinical trials (see http://www.clinicaltrials.gov/). This approach is expected to be efficacious because kinetic stabilization of tetrameric transthyretin incorporating disease-associated subunits by interallelic trans suppression (the incorporation of T119M TTR subunits) ameliorates FAP symptoms (17, 18, 20, 35).
Since TTR tetramer dissociation, partial monomer denaturation and amyloidogenesis are associated with several familial gain-of-function proteotoxicity diseases, we set out to examine the correlation between the quaternary and tertiary structural stabilities of TTR and disease phenotype. Folding and association energetics can be quantified by globally fitting chaotrope denaturation data obtained at several protein concentrations (36–40). Systematic urea denaturation studies were therefore carried out as a function of the concentration of TTR. These data corroborated our previous report that the tetramer dissociation and monomer unfolding equilibria are thermodynamically linked (7). For well-behaved TTR sequences, we analyzed the concentration-dependent denaturation data by global fitting to an equation that simultaneously accounts for the two-step denaturation process (tetramer to folded monomer and folded monomer to unfolded monomer). The concentration-dependent denaturation data for less-well behaved variants were analyzed qualitatively. Quantitative and qualitative analysis of the denaturation data, combined with literature reports on TTR folding energetics, reveals that each TTR variant has a unique stability profile that can be used to rationalize their disease phenotypes by considering how their folding energetics influence factors like their in vivo aggregation propensities and their interaction with the endoplasmic reticulum-associated degradation machinery (13, 41).
EXPERIMENTAL PROCEDURES
Expression and Purification of TTR
All TTR variants were expressed in BL21(DE3) Epicurian Gold Escherichia coli cells (Stratagene, La Jolla, California), transformed with the appropriate pMMHa expression vector containing the TTR and ampicillin resistance genes (42). E. coli cultures were grown, harvested, and lysed in the presence of protease inhibitors, and the cell lysates were centrifuged, as previously described (23). All of the variants examined were expressed as soluble proteins and were therefore purified from cell supernatants.
a. Tetrameric TTR variants (WT TTR, V30M, L55P, V122I, and A25T)
The supernatant was fractionated by ammonium sulfate precipitation; the 50–100% ammonium sulfate pellet was resuspended in a minimal volume of 25 mM Tris, 1 mM EDTA (pH 8.0) and was dialyzed in 10,000 MWCO dialysis tubing (Snakeskin from Pierce Biotechnology, Rockford, Illinois) against 4 L of 25 mM Tris, 1 mM EDTA (pH 8.0) overnight at 4 °C. After dialysis, the sample was filtered through 0.22 μm filters and applied to a Source 15Q anion exchange column (Amersham Biosciences, Piscataway, New Jersey), which had been equilibrated with 25 mM Tris, 1 mM EDTA, 50 mM NaCl (pH 8.0) and was run at 4 °C. TTR was eluted with a linear gradient of 50–350 mM NaCl in 1.5 column volumes followed by a 350 mM NaCl wash for 1.5 column volumes. Fractions containing TTR (the major peak in all cases) were pooled, concentrated, and further purified on a Superdex 75 gel filtration column (Amersham Biosciences, Piscataway, New Jersey) in 50 mM sodium phosphate, 100 mM KCl, 1 mM EDTA (pH 7.4) at 4 °C, in order to remove any soluble TTR aggregates. The identity of each purified TTR variant was confirmed by its mass, determined by electrospray LC/MS on a HP Series 1100-MSD liquid chromatography/mass spectrometer (Agilent Technologies, Palo Alto, California), as previously described (8). The masses determined for each TTR protein (WT TTR, 13,890 Da; V122I, 13,904 Da; L55P, 13,874 Da; V30M, 13,922 Da; A25T, 13,920 Da; M-TTR, 13,892 Da) are consistently ~2 Da lower than the calculated values, which include the expected extra N-terminal Met present in all of our recombinant TTR variants. Concentrations of TTR solutions are expressed in micromolar (μM) units and were determined spectrophotometrically, using an ε280 of 1.88×104 M−1 cm −1. A typical purification yields 60–100 mg of tetrameric TTR from 2 L of cell culture. All TTR solutions were stored at 4 °C and were used within a week of purification, except for solutions of the aggregation-prone A25T variant, which was used immediately following purification.
b. Monomeric TTR Variant (M-TTR)
M-TTR was purified according to the above procedure, with slight modifications to the ammonium fractionation step: A 25–90% ammonium sulfate pellet was obtained, resuspended, and dialyzed in 3,500 MWCO Snakeskin dialysis tubing (Pierce Biotechnology, Rockford, Illinois). The typical yield is 40–60 mg of M-TTR from 2 L of cell culture; M-TTR thus obtained is >95% monomeric, as previously reported (19, 23).
Urea denaturation
Stock solutions of urea were prepared in buffer, with final concentrations of ~10 M urea, 50 mM sodium phosphate, 100 mM KCl, 1 mM EDTA, 1 mM DTT (pH 7.4); the exact concentration of urea was determined from refractive index measurements (43). Denaturation reactions (200 μL total volume) containing varying concentrations of TTR (0.72–144μM) and urea (0–8 M) in 50 mM sodium phosphate, 100 mM KCl, 1 mM EDTA, 1 mM DTT (pH 7.4) were incubated for 96 h at 4 °C, unless otherwise noted. This incubation time was found previously to be sufficient for the samples to reach equilibrium (44), with the exception of V30M (see Results and Discussion); incubation times longer than 96 h were avoided, because they resulted in some urea-mediated covalent modifications of TTR, as previously reported (44). After 96 h, the samples were removed from the cold room and equilibrated for 1 h at 25 °C prior to the acquisition of fluorescence data. For each TTR variant, denaturation curves were obtained at 7 different TTR concentrations. Each denaturation curve, at a given TTR concentration, was derived from 20 separate denaturation reactions at different urea concentrations, in order to accurately define the pre- and post-transition baselines and the transition region of each curve.
Renaturation experiments
For some experiments, TTR was first completely denatured at a high urea concentration and then allowed to undergo renaturation triggered by dilution of the chaotrope. TTR was concentrated in centrifugal filters (Centriprep or Microcon from Millipore Corporation, Billerica, Massachusetts; 10,000 MWCO for tetrameric TTR variants, and 3,000 MWCO for monomeric TTR (M-TTR) to the desired concentration, and filtered or spun to remove any aggregates. Denaturation reactions containing 8 M urea and TTR at high concentration (360–720μM) were incubated for 96 h at 4 °C in 50 mM sodium phosphate buffer (pH 7.4), containing 100 mM KCl, 1 mM EDTA, 1 mM DTT. After 96 h, complete denaturation was verified using intrinsic tryptophan fluorescence, and renaturation assays were initiated by dilution of the denatured TTR sample into buffer containing varying concentrations of urea (from > 0 M up to 8 M). The lowest concentration of urea achievable in the renaturation reactions depended on the exact experiment, but was always > 0 M, due to the presence of urea in the denatured TTR sample. Reactions were incubated for an additional 24 h at 25 °C to allow complete refolding of TTR monomers and reassembly of TTR tetramers (renaturation) prior to fluorescence measurements.
Tertiary structure assessed by tryptophan fluorescence
The extent of unfolding of TTR was evaluated by intrinsic protein fluorescence spectroscopy (44). Samples were excited at 295 nm (2 nm bandwidth), and fluorescence emission spectra were recorded from 310–410 nm (6 nm bandwidth) at 1 nm intervals with an averaging time of 0.3 s on an AVIV ATF-105 fluorometer (AVIV Instruments, Lakewood, New Jersey). This excitation wavelength was chosen to allow selective excitation of the tryptophan residues in TTR (Trp41 and Trp79). Upon denaturation, these tryptophan residues become more solvent-exposed, and the λmax,emission shifts from ~335 nm in the native state to ~355 nm in the unfolded state; the tryptophan emission spectrum is therefore a sensitive probe of the tertiary structure of TTR. Spectra were corrected by subtraction of a buffer blank, and the fluorescence intensities at 335 and 355 nm were recorded for each sample. The ratio of the fluorescence intensities at these two wavelengths (F355/F335) was used as a measure of the extent of tertiary structure unfolding, to allow direct comparisons of the various denaturation curves. For all of the tetrameric TTR variants, the F355/F335 ratio for the native state in the absence of urea is ~0.85, and that of the denatured state in the presence of 8 M urea is 1.3–1.35. The F355/F335 ratios for M-TTR are similar to those of the tetrameric variants, with the ratio for the native state in the absence of urea consistently being a little lower, ~0.82.
Quaternary structure assessed by resveratrol binding
The binding of the small molecule resveratrol to the TTR tetramer was used to assess the quaternary structure of TTR as a function of urea concentration and TTR concentration, as previously described (7, 12). Resveratrol binding to tetrameric TTR (occupying at least one of the two thyroxine-binding sites) results in a blue shift in the fluorescence spectrum of resveratrol and a concomitant large increase in its fluorescence quantum yield. In contrast, studies with M-TTR have shown that resveratrol does not bind to monomeric TTR (X. Jiang and J.W. Kelly, unpublished results). An aliquot (1.44 μL) of a stock solution of resveratrol (0.5–12.5 mM in DMSO) was added to denatured TTR samples (200 μL containing 1.44, 7.2, or 36μM TTR) immediately prior to fluorescence measurements, in order to minimally perturb the slowly attained tetramer monomer equilibrium. The final concentration of resveratrol (3.6, 18, or 90 μM) varied according to the concentration of TTR and was present in each assay at a 10-fold stoichiometric excess over the maximum concentration of TTR tetramer (1.44 μM monomer = 0.36 μM tetramer; 36 μM monomer = 9.0 μM tetramer). Fluorescence emission spectra were recorded from 350–550 nm (5 nm bandwidth, 1 nm data interval, 0.3 s averaging time) on an AVIV ATF-105 fluorometer (AVIV Instruments, Lakewood, New Jersey), with an excitation wavelength of 320 nm (2 nm bandwidth). Spectra were corrected by subtraction of a buffer blank, and the fluorescence intensities at 390 nm (F390) were used directly as a measure of the amount of tetramer present in each sample.
Quaternary structure assessed by glutaraldehyde cross-linking and SDS–PAGE
To independently assess the TTR quaternary structural changes quantified by resveratrol fluorescence, glutaraldehyde crosslinking was also employed. Glutaraldehyde (5 μL of a 25% w/w solution in water, from Sigma-Aldrich, St. Louis, Missouri) was added to 50 μL aliquots of TTR samples in variable concentrations of urea (containing 7.2 μM, 36 μM, or 144 μM TTR), and the cross-linking reaction was allowed to proceed for 4 min at room temperature prior to quenching with NaBH4 (5 μL of a 7% w/w solution made in 0.1 M NaOH). Samples were immediately mixed with SDS reducing sample loading buffer, boiled for 5 min, and analyzed by discontinuous SDS–PAGE (3.9% acrylamide stacking gel, 12% acrylamide separating gel). A25T samples were run on gradient gels (Novex 10–20% Tris–glycine precast gels from Invitrogen, Carlsbad, California) to allow visualization of high molecular weight aggregates present in those samples. The total protein loaded in each lane was ~1.8 μg for the A25T samples, and ~1.0 μg for all other samples. Gels were stained with colloidal Coomassie Blue G-250 (GelCode Blue Stain Reagent from Pierce Biotechnology, Rockford, IL), and protein bands were identified by comparison to molecular weight standards (BenchMark Pre-stained Protein Ladder from Invitrogen). After staining, the gels were scanned, and the bands were quantified by densitometry, using NIH Image analysis software (National Institutes of Health; http://rsb.info.nih.gov/nih-image).
Data analysis
Denaturation curves were plotted, and stabilities of TTR were determined by fitting the data to a two-state model for either tetramer dissociation or for monomer unfolding in the presence of urea.
A. Determination of apparent Cm values for monomer unfolding (Cm,unfold)
The midpoint urea concentration values for monomer unfolding (Cm,unfold) were determined by fitting individual tryptophan fluorescence denaturation curves to the following equation (45):
| (1) |
where y is the observed signal (F355/F335 in this case); αfolded is the y-intercept and βfolded is the slope of the pre-transition baseline, which describes the F355/F335 value of the folded monomer at a given concentration of urea; αunfolded is the y-intercept and βunfolded is the slope of the post-transition baseline, describing the F355/F335 value of the unfolded monomer at a given concentration of urea; Cm,unfold and munfold describe the transition region of the denaturation curve, with Cm,unfold corresponding to the urea concentration where the protein is 50% unfolded (M urea), and munfold the constant of proportionality relating the free energy of unfolding to the urea concentration (kcal/mol M urea); R is the universal gas constant (kcal/K mol), and T is the absolute temperature (K). Cm,unfold values thus determined were used either for comparisons of apparent stability of single variants, as a function of TTR concentration, or for comparisons of apparent stability among all TTR variants examined at the same protein concentration. Apparent unfolding free energies, or ΔGunfold,appH2O were calculated from these fits by multiplying Cm,unfold by munfold.
B. Determination of apparent Cm values for tetramer dissociation (Cm,diss)
Equation 1 was slightly modified to analyze the native tetramer–folded monomer equilibrium, measured either by resveratrol binding or by glutaraldehyde cross-linking, as follows:
| (2) |
where αtetramer and βtetramer describe the pre-transition baseline, αmonomer and βmonomer describe the post-transition baseline, and Cm,diss (M urea) and mdiss (kcal/mol M urea) describe the transition region of each denaturation curve. For resveratrol binding data, the blank-subtracted fluorescence intensities at 390 nm were used as the observed signal, y. For analysis of cross-linking data, the % tetramer and % monomer values derived from densitometry of the various samples were each plotted as a function of urea concentration, and both curves were fit separately to eq 2. Cm,diss values derived from both measurements (% tetramer and % monomer) were generally in good agreement, except in cases where intermediates were detected (dimmers and trimers for L55P and aggregates for A25T; see Results and Discussion).
C. Global fitting of concentration-dependent data to extract thermodynamic parameters for TTR variants
Fraction unfolded (Funfold) values for all TTR variants were calculated as follows: the blank-corrected fluorescence intensity at 355 nm was subtracted from that at 335 nm for each sample, and these values (F335–F355) were normalized for each denaturation curve to a value of 1.00 for the spectrum of native WT TTR in the absence of urea. This transformation retains the linear dependence of the fluorescence signal of the folded and unfolded states of the protein on denaturant concentration and yielded F335–F355 values ranging from 1.00 at 0 M urea to approximately −1.8 at 8 M urea (−2.0 for M-TTR). Pre- and post-transition baselines (corresponding to the signal for 100% folded monomer and 100% unfolded monomer, respectively) were determined from this data by plotting the F335–F355 values vs. urea concentration, then fitting the linear portions of the plots at low and high urea concentrations to lines. The fraction of unfolded TTR (Funfold) was calculated at each urea concentration in the transition region, where Funfold = (ysample − y100%folded)/(y100%unfolded − y100%folded), where y100%folded is the pre-transition baseline and y100%unfolded is the post-transition baseline.
For each variant, the Funfold values determined over a range of TTR and urea concentrations were fit globally to a three-state model comprising unfolded monomer, folded monomer, and tetramer:
where Kunfold is the equilibrium constant for monomer folding and Kdiss is the equilibrium constant for tetramer dissociation: Kdiss = [monomerfolded]4/[tetramer] (quantities in brackets are molar concentrations). ΔGunfold is related to Kunfold through the equation ΔGunfold = −RT lnKunfold. Similarly, ΔGdiss is related to Kdiss through the equation ΔGdiss = −RT lnKdiss. ΔGunfold and ΔGdiss are expected to depend on the urea concentration as follows: ΔGunfold = ΔGunfoldH2O − munfold[urea] and ΔGdiss = ΔGdissH2O − mdiss[urea], where ΔGunfoldH2O and ΔGdissH2O are the standard state free energies of monomer unfolding and tetramer dissociation in 0 M urea, respectively. These equations can be combined to yield
| (3) |
The fraction of unfolded protein at a given urea and protein concentration can be calculated for given values of ΔGunfoldH2O, munfold, ΔGdissH2O, and mdiss using the relation
where the numerator is the real solution of the following polynomial equation such that 0 < [monomerunfolded] < [total protein]
| (4) |
The Funfolded data were fit by choosing initial values for ΔGunfoldH2O, munfold, ΔGdissH2O, and mdiss, computing the sum of squared differences between the calculated and experimental Funfolded data at all protein and urea concentrations, and then varying ΔGunfoldH2O, munfold, ΔGdissH2O, and mdiss until this sum of squared differences was a minimum. Note that using the relation ΔGdiss = −RT lnKdiss implies that ΔGdiss values are for a standard state in which the protein concentration is 1 M.
RESULTS AND DISCUSSION
Characterizing the quaternary and tertiary structural stability of the various TTR sequences requires that the energetics of both tetramer dissociation and monomer unfolding be examined. Methodology for examining these two equilibria has already been published: resveratrol binding for monitoring TTR tetramer dissociation (7, 12), and, since tryptophan fluorescence is unaffected by tetramerization (19), intrinsic tryptophan fluorescence spectroscopy for following TTR monomer unfolding (44) in the presence of chaotropic agents such as urea (see Scheme 1). These methods are useful for determining thermodynamic parameters (Keq and ΔG) for dissociation and unfolding, provided that the two equilibria can be examined independently. This is only possible if conditions exist where only the native tetramer and folded monomer are in equilibrium (and no unfolding occurs), and where only the folded monomer and unfolded monomer exist together in the absence of any tetramer. When this is the case, the expressions for the equilibrium constants have the following form: Kdiss = 256 × [TTRtotal]3 × ((1 − α)4/α), where α represents the fraction of tetrameric TTR, and the equilibrium position depends on the total concentration of TTR; and Kunfold = [monomerunfolded]/[monomerfolded] = Funfold/(1 − Funfold), where Funfold is the fraction of unfolded TTR, and the equilibrium is independent of TTR concentration.
Scheme 1.
Thermodynamic stability of tetrameric TTRa
a Denaturation of tetrameric TTR by urea involves at least two equilibria, as shown–dissociation of the tetramer into its component monomers, which are still natively folded, and subsequent unfolding of the monomers. These equilibria are defined by the equilibrium constants, Kdiss and Kunfold, which can in principle be determined independently from resveratrol binding and intrinsic Trp fluorescence, respectively. The data presented herein, however, show that the equilibria for dissociation and unfolding are thermodynamically linked, meaning that there is a negligible concentration of folded monomer under all the conditions examined. When the equilibria are linked, the apparent value of Kunfold is dependent on TTR concentration, and the concentration-dependence of Kunfold is a measure of the magnitude of Kdiss. Multiple urea denaturation curves at varying TTR concentration can hence be analyzed simultaneously to determine thermodynamic properties of both the tetramers (Kdiss) and the monomers (Kunfold) of wild-type and variant TTRs. Attempts to unlink the equilibria by decreasing the concentration of TTR were unsuccessful, due to limitations in the sensitivity of the measurement techniques.
In contrast to this ideal scenario, results from earlier denaturation studies suggest that TTR dissociation and unfolding are thermodynamically linked under the conditions typically used in these experiments (TTR concentrations of 7.2–36 μM). In particular, it was found that the urea denaturation curves for tetramer dissociation, measured by resveratrol binding, are nearly coincident with those monitoring monomer unfolding, as assessed by intrinsic protein fluorescence, for all the TTR variants examined (WT TTR, L55P, V122I, V30M, T119M) (7). Furthermore, the urea concentration required for monomer unfolding appeared to depend on the concentration of TTR (7), even though the equilibrium for this process should be independent of TTR concentration. These data strongly suggest that TTR tetramer stability influences the apparent stability of TTR monomers, and the true thermodynamic parameters for either of the two equilibria cannot be determined under these experimental conditions because they are linked.
Our initial studies in this investigation focused on identifying conditions under which the equilibria could be unlinked, i.e., where the dissociation of TTR tetramers proceeds to completion (100% of TTR present as the folded monomer) at denaturant concentrations that are too low to initiate subsequent unfolding of the monomers. To achieve this goal, we tried to exploit differences in the TTR concentration-dependence of the two equilibria. Whereas the equilibrium for monomer unfolding should be independent of TTR concentration, the tetramer monomer equilibrium is highly dependent on the total concentration of TTR, as seen in the expression for Kdiss (vide supra). A decrease in the total TTR concentration should shift this equilibrium toward the monomer, favoring dissociation and increasing the fraction of monomeric TTR present (as [TTRtotal] decreases, the (1 − α)4/α term concomitantly increases, which means a lower fraction of tetrameric TTR) without affecting the unfolding equilibrium. This approach gave encouraging results, but was only partially successful in unlinking the equilibria. For example, decreasing the concentration of WT TTR to 0.72 μM resulted in some separation of the Cm values for tetramer dissociation and monomer unfolding, determined to be 2.7 M and 3.0 M urea, respectively; nevertheless, the transition regions of the two denaturation curves still overlapped significantly (data not shown). Although complete unlinking of the equilibria could in principle be achieved by decreasing the TTR concentration further, in practice this is problematic because the sensitivity of the fluorometric methods precludes the use of TTR concentrations lower than ~0.72 μM.
An alternative approach, described herein, was used instead to determine the stabilities of TTR tetramers and monomers. Since the equilibria are thermodynamically linked, the observed dependence of the monomer unfolding equilibrium on the concentration of TTR is a measure of the stability of the TTR tetramer. In other words, not only are the denaturation curves measured by tryptophan fluorescence expected to shift to higher denaturant concentrations as the concentration of TTR is increased, as observed, but the extent to which the curves shift reflects the free energy of tetramer dissociation, with greater tetrameric stability effecting larger shifts. Therefore, global analysis of a series of tryptophan denaturation curves at varying TTR concentrations should allow the determination of the true thermodynamic parameters for both tetramer and monomer stability, provided that tetramer and monomer are the only species populated during denaturation. We describe this analysis method and its application for determination of the thermodynamic stability of wild-type TTR tetramers and monomers. We then use this method to assess the quaternary and tertiary structural stabilities of known disease-associated variants of TTR, enabling comparisons with those parameters determined for WT TTR.
Concentration-dependence of WT TTR denaturation
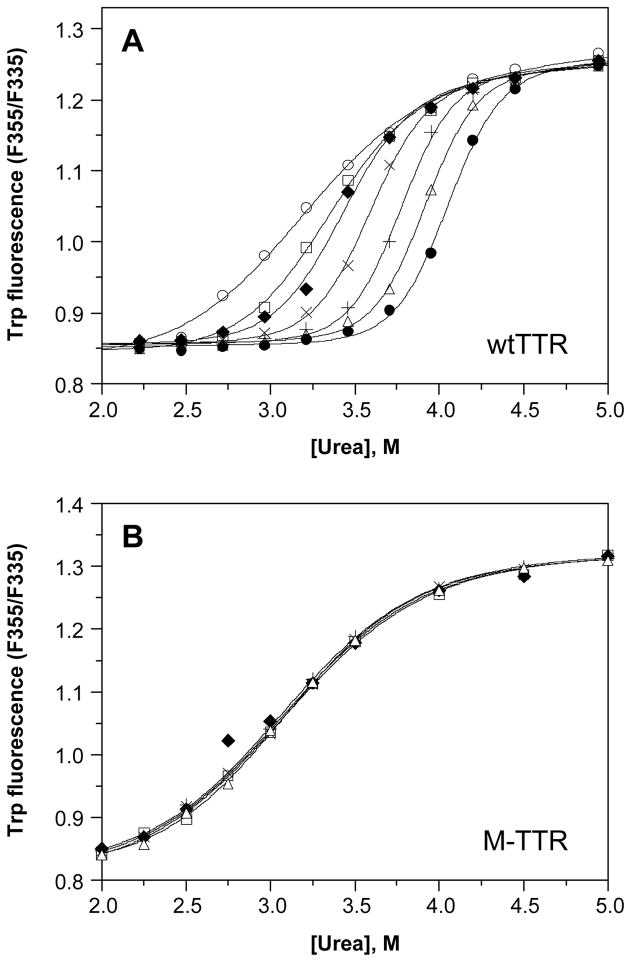
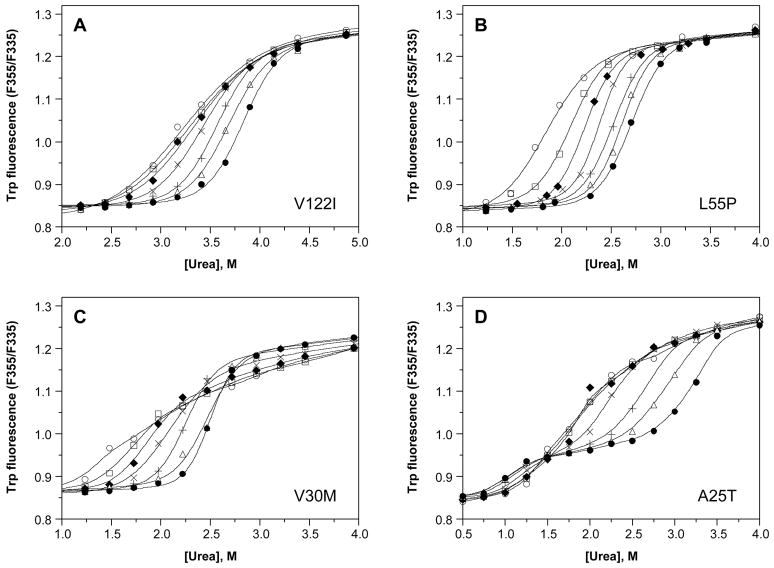
Figure 1A shows the effect of TTR concentration on the stability of WT TTR to urea denaturation, monitored by tryptophan fluorescence, for a representative experiment. Each denaturation curve was fit independently by eq 1 to a two-state denaturation model; the resulting fits are shown as curves overlaid with the corresponding data (symbols). At the lowest WT TTR concentration shown (1.44 μM), the midpoint for unfolding (or the urea concentration at which WT TTR is 50% unfolded, Cm,unfold) occurs at ~3.1 M urea, with a value of −munfold (the dependence of the free energy of unfolding on urea concentration) of ~1.6 kcal/mol/M urea. As the concentration of WT TTR is increased, two obvious changes in the denaturation curves are revealed: 1) The curves shift to the right, with the apparent Cm,unfold gradually increasing up to ~4.0 M urea at 144μM WT TTR, and 2) the steepness of the curve in the transition region increases, which is reflected in larger apparent values of −munfold, up to ~3.9 kcal/mol/M urea at WT TTR concentrations of 36 μM and above. The magnitude of −munfold reflects the amount of surface area that gets exposed upon unfolding (46). The values of −munfold are large for TTR at high concentration because tetramer dissociation and monomer unfolding are so tightly linked that the surface area exposed by these two processes is essentially combined. Tetramer dissociation and monomer unfolding are less tightly linked at lower concentrations and the apparent −munfold values are correspondingly lower. In contrast to these results, denaturation curves for M-TTR, a monomeric variant of WT TTR, exhibit no dependence on TTR concentration (Figure 1B; discussed further below).
Figure 1.
Dependence of urea denaturation of WT TTR (panel A) and M-TTR (panel B) on TTR concentration. Denaturation reactions contained TTR (1.44–144 μM) and were incubated for 96 h at 4 °C in the presence of varying concentrations of urea (0–8 M) prior to fluorescence measurements (see Experimental Procedures for details). The intrinsic tryptophan fluorescence of TTR shifts from a maximum emission wavelength around 335 nm in the native state to 355 nm upon denaturation; hence the ratio of the fluorescence intensity at these two wavelengths (F355/F335) is a measure of the extent of denaturation. TTR concentrations in this representative experiment were 1.44 (❍), 3.6 (□), 7.2 (◆), 14.4 (×), 36 (+), 72 (Δ), and 144μM (λ), and the fits of each denaturation curve to eq 1 are shown as solid lines. The apparent stability of WT TTR monomers to urea denaturation increases with increasing TTR concentration (from Cm,unfold = 3.1 to 4.0 M urea); in contrast the concentration of M-TTR has no effect on its apparent stability (Cm,unfold = 3.1 M urea at all M-TTR concentrations examined).
The apparent Gibbs free energy of monomer unfolding in the absence of denaturant, ΔGunfold,appH2O, can be calculated by multiplying the values of Cm,unfold and −munfold obtained as described above. Calculated values of ΔGunfold,appH2O range from 5.0 kcal/mol at 1.44 μM WT TTR to over 15 kcal/mol at 72 and 144μM WT TTR. We call the parameter obtained from fitting the individual denaturation curves to a two-state model an “apparent” ΔGunfoldH2O, because TTR denaturation is at least a two-step process. Since in reality we are simultaneously evaluating the tetramer dissociation and monomer unfolding linked equilibria, ΔGunfold,appH2O is a composite stability parameter that “combines” the free energy of monomer unfolding and the free energy of tetramer dissociation. As the TTR concentration is increased, the two equilibria become more linked, and the free energy of tetramer dissociation, ΔGdissH2O, influences ΔGunfold,appH2O to a greater extent, resulting in greater values of ΔGunfold,appH2O. Although we are unable to measure the folded monomer unfolded monomer concentrations independently in these experiments, the equilibria become less tightly linked at low TTR concentration, and hence the ΔGunfold,appH2O of 5.0 kcal/mol determined at 1.44 μM begins to approximate the true stability of the WT TTR monomer. The thermodynamic parameters—Cm, unfold, −munfold, and ΔGunfoldH2O —calculated from these analyses of the individual tryptophan denaturation curves are summarized in Table 1.
Table 1.
Apparent stability of WT TTR at varying TTR concentrationa
| TTR variant, concentration | Cm,unfoldb (M urea) | − munfoldb (kcal/mol M urea) | ΔGunfoldH2Ob (kcal/mol) |
|---|---|---|---|
| WT TTR, μM | |||
| 1.44 | 3.14 | 1.58 | 4.96 |
| 3.6 | 3.32 | 2.30 | 7.64 |
| 7.2 | 3.42 | 2.92 | 9.99 |
| 14.4 | 3.59 | 3.49 | 12.53 |
| 36 | 3.77 | 3.86 | 14.55 |
| 72 | 3.99 | 3.90 | 15.56 |
| 144 | 4.05 | 3.78 | 15.31 |
|
| |||
| M-TTRc, 1.44–72 μM | 3.07 ± 0.02 (n = 7) | 1.50 ± 0.07 (n = 7) | 4.61 ± 0.22 (n = 7) |
Apparent stabilities of WT TTR and M-TTR monomers were calculated from fitting of individual data sets at the indicated TTR concentration and are shown for a representative experiment (see Experimental Procedures for details).
Apparent values of Cm,unfold and −munfold were derived from fitting of the data shown in Figure 1A and 1B to eq 1, using the ratio of fluorescence intensities (F355/F335) as a measure of the extent of denaturation; Cm,unfold is the urea concentration where TTR is 50% unfolded and −munfold is the urea dependence of the free energy of unfolding. Apparent values of ΔGunfoldH2O were determined by multiplying Cm,unfold by −munfold.
Denaturation curves of M-TTR were identical at all concentrations examined (see Figure 1B for data and fits), and the values shown represent the mean and standard deviation of 7 different data sets at varying M-TTR concentration (1.44–72 μM).
Thermodynamic stability of monomeric TTR (M-TTR)
Further evidence that the concentration-dependence of monomer unfolding observed for WT TTR is due to the stability of the wild-type TTR tetramer is provided by control experiments carried out with M-TTR, an engineered monomeric variant of TTR that is >95% monomeric as purified and does not tetramerize under the conditions employed in these experiments, as determined by gel filtration and cross-linking studies (19, 23). Hence, urea denaturation of this variant involves only two species—the folded and unfolded monomers—and, as expected, the denaturation curves show no dependence on the M-TTR concentration (Figure 1B). Over a concentration range that varies more than 50-fold (1.44–72μM), the M-TTR data are superimposable, allowing the direct determination of the tertiary structural stability of the M-TTR monomer from any one of the curves obtained. The Cm, unfold for M-TTR occurs at 3.07 ± 0.02 M urea, with an −munfold of 1.50 ± 0.07 kcal/mol/M urea (n = 7). Calculated values of ΔGunfoldH2O for denaturation curves at varying M-TTR concentrations are also indistinguishable, with an average of 4.61 ± 0.22 kcal/mol (n = 7). These values of ΔGunfoldH2O and −munfold for M-TTR are close to those obtained for WT TTR at the lowest concentration examined (1.44 μM) and are in reasonable agreement with the previously reported stability parameters of M-TTR: ΔGunfoldH2O of 5.5 ± 0.8 kcal/mol and −munfold of 1.7 ± 0.2 kcal/mol/M urea (19). The small differences in measured stability may be due to slightly different experimental conditions used in the two studies. The thermodynamic parameters determined herein for M-TTR monomers are shown in Table 1, where they can be readily compared to the apparent stability of WT TTR monomers at varying TTR concentrations. For each of these parameters—Cm, unfold, −munfold, and ΔGunfoldH2O—the apparent values determined from WT TTR experiments at 1.44 μM approach those calculated for M-TTR, suggesting that the tertiary structural stability of M-TTR is similar to that of WT TTR monomers.
Evaluating quaternary structural changes in WT TTR
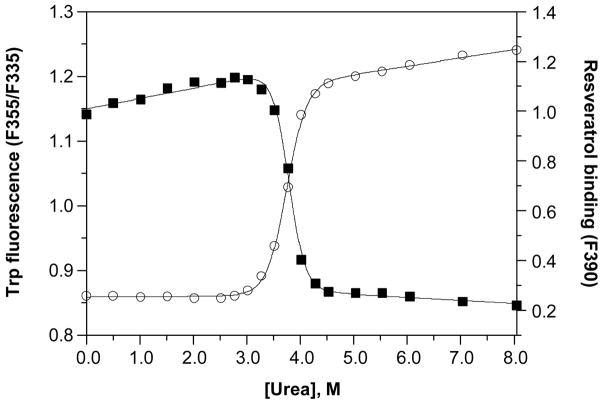
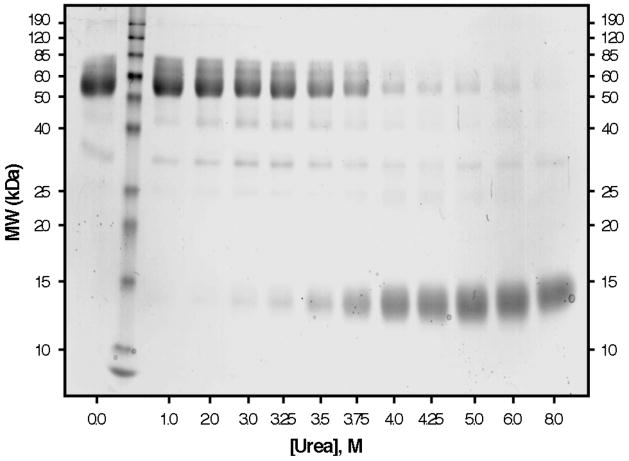
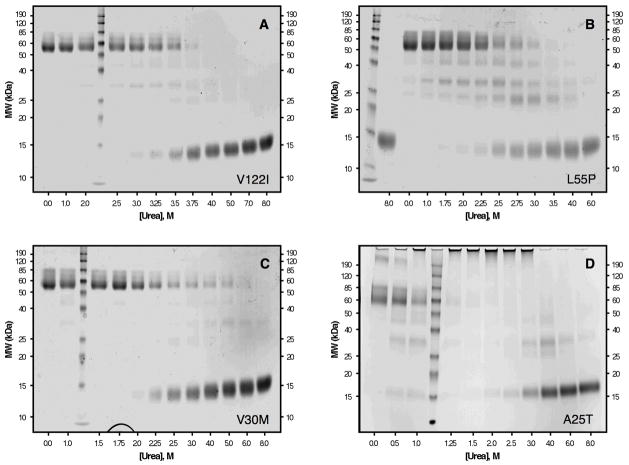
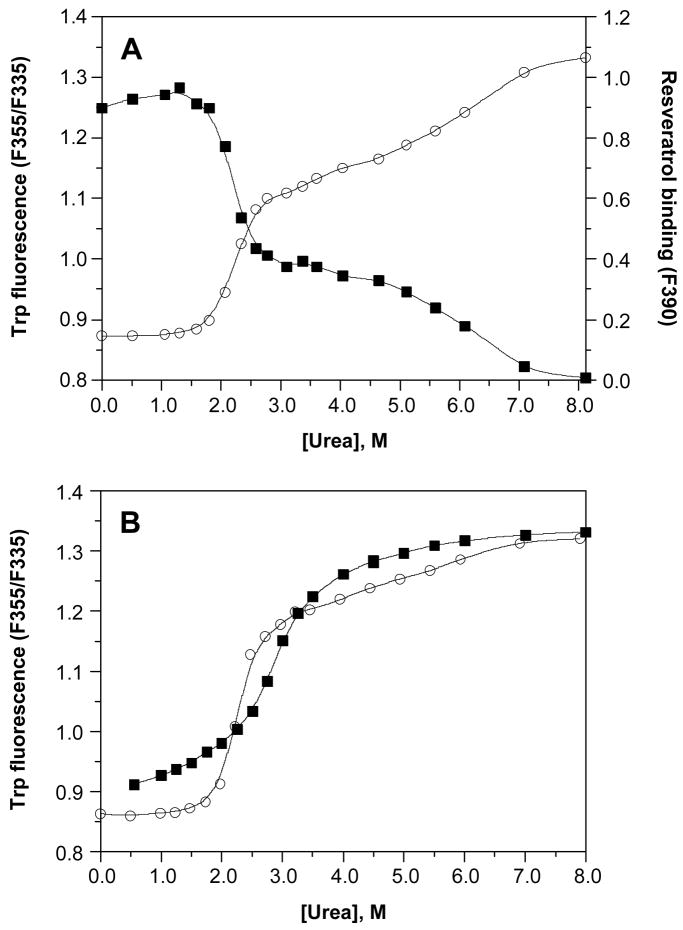
Although the described behavior for WT TTR monomer unfolding is that expected for a protein having linked quaternary and tertiary structural transitions at higher WT TTR concentrations, we also examined the tetramer monomer equilibrium directly by two different methods, to verify that the two equilibria are indeed linked under these experimental conditions. Figure 2 shows the superposition of resveratrol binding data, monitoring the dissociation of WT TTR tetramers, with tryptophan fluorescence data, following tertiary structural changes, in a representative experiment utilizing 36 μM WT TTR. Independent fitting of each data set to a two-state denaturation model, corresponding either to the tetramer–monomer equilibrium in the case of the resveratrol binding data (fit to eq 2), or the folded monomer–unfolded monomer equilibrium for the tryptophan fluorescence data (fit to eq 1), gave Cm values of 3.8 M and 3.7 M urea for tetramer dissociation and monomer unfolding, respectively. The tetramer–monomer equilibrium as a function of urea concentration (36 μM WT TTR) was also assessed by chemical cross-linking quantified by densitometric analysis of SDS–PAGE gels (Figure 3). As the concentration of urea is increased in these samples, the fraction of WT TTR present as tetramers (MW ~55 kDa) decreases, with the concomitant appearance of a diffuse band corresponding to monomeric TTR (MW ~14 kDa, the band becomes broad because several Lys residues are covalently modified by glutaraldehyde). Densitometry of the bands in each lane shows that dissociation proceeds from tetramer to monomer without any substantial accumulation of intermediates. The observed bands at ~42 and 28 kDa, which correspond to the molecular weights of trimeric and dimeric TTR, respectively, are present in all lanes and account for ~10% of the total TTR; a small increase in the intensity of these two bands (up to a maximum ~18%) is apparent in lanes 4–7 (corresponding to 2.0 to 3.5 M urea, concentrations just below the Cm,diss determined in this experiment), which indicates that these may be intermediate species in the dissociation of tetramers into monomers. Surprisingly, a small amount of tetrameric WT TTR (~5%) remains, even in the presence of 8 M urea, suggesting that a fraction of WT TTR tetramers is highly resistant to urea denaturation. Although we cannot rule out the possibility that this result may be due to a cross-linking artifact, it was only observed for WT TTR, with none of the other TTR variants displaying this behavior (vide infra). Fitting the densitometry data to eq 2 yielded a Cm for tetramer dissociation (Cm,diss) of 3.7 or 3.8 M urea, depending on whether the % tetramer or % monomer data were used, in excellent agreement with the value obtained above from resveratrol binding (3.8 M urea). As expected, Cm,diss increases with increasing TTR concentration and is nearly coincident with the Cm,unfold obtained at all WT TTR concentrations examined (0.72–36 μM; data not shown). When considered together, these WT TTR data strongly support the conclusion that urea-induced tetramer dissociation and unfolding of TTR occur simultaneously at the higher concentrations used in these denaturation experiments, demonstrating that the equilibria are inextricably linked under these conditions, which represent the physiological concentration range of TTR (7.2–28.8 μM in plasma and 0.72–7.2 μM in CSF). It is therefore reasonable to assume that tetramer dissociation and monomer unfolding equilibria are also thermodynamically linked in vivo.
Figure 2.
Urea denaturation of WT TTR. The denaturation of WT TTR (36 μM) was monitored both by intrinsic tryptophan fluorescence (❍, left y-axis) to assess changes in the tertiary structure, and by resveratrol binding (■, right y-axis) to assess tetramer dissociation. Samples were incubated for 96 h at 25 °C in the presence of varying concentrations of urea (0–8 M) prior to fluorescence measurements. Each data set was fit to a two-state denaturation model (eq 1 for tryptophan fluorescence data; eq 2 for resveratrol binding data) to give apparent Cm values of 3.8 and 3.7 M urea for tetramer dissociation and monomer unfolding, respectively.
Figure 3.
Chemical cross-linking of urea-denatured WT TTR. The same WT TTR samples (36μM) analyzed by fluorescence in Figure 2 were also cross-linked with glutaraldehyde and analyzed by SDS–PAGE with Coomassie Blue staining (see Experimental Procedures for details). Molecular weights of protein standards (Lane 2) are shown on the y-axis, and the urea concentration for each of the TTR samples is shown on the x-axis. The bands ~55 kDa correspond to TTR tetramers; these disappear as the concentration of urea is increased, with concomitant appearance of a diffuse monomer band ~14 kDa. The bands observed ~42 and 28 kDa probably correspond to trimeric and dimeric TTR, respectively. The intensity of each band was quantified by densitometry (not shown), and the Cm,diss determined by this method was 3.7 or 3.8 M urea, depending on whether the %tetramer or %monomer data were used, in good agreement with the value determined by resveratrol binding (3.8 M; see Figure 2).
Determining quaternary and tertiary structural stability simultaneously from concentration-dependent WT TTR denaturation curves by global fitting
Due to the thermodynamic linkage of the WT TTR quaternary (tetramer dissociation) and tertiary (monomer unfolding) structural changes, we were unable to find conditions where we could evaluate either equilibrium independently of the other. Therefore, we globally fit the concentration-dependent TTR urea denaturation data to a single set of parameters, allowing simultaneous quantification of quaternary and tertiary structural thermodynamic stability parameters. While a detailed explanation of the equations and methods used to determine WT TTR tetramer and monomer stabilities can be found in the Experimental Procedures, the methodology is briefly discussed here. Because the two steps under investigation are thermodynamically linked, we assume there are three species—native tetramer, folded monomer, and unfolded monomer—that coexist in equilibrium, and the concentration of each species under a given set of conditions depends on the corresponding equilibrium constants, Kdiss and Kunfold (Scheme 1; a model that accounted for only two species—native tetramer and unfolded monomer—gave a poor fit to the data, as shown in the Supporting Information). Starting with an equation for the total concentration of TTR (as molar concentration of TTR monomers) that accounts for all three species, [TTRtotal] = 4[tetramer] + [monomerfolded] + [monomerunfolded], and substituting in expressions for [tetramer] and [monomerfolded] in terms of [monomerunfolded] and the two equilibrium constants, we obtain [TTRtotal] = 4([monomerunfolded]4/KdissKunfold4) + ([monomerunfolded]/Kunfold) + [monomerunfolded] (see eq 4 in the Experimental section). This equation now relates the total TTR concentration, a known quantity, to the extent of monomer unfolding, which can be determined experimentally from the tryptophan fluorescence measurements, and the two unknowns of interest, Kdiss and Kunfold. The values of Kdiss and Kunfold are related to the free energies of tetramer dissociation and unfolding (ΔGdiss and ΔGunfold), which are in turn linearly related to the urea concentration with slopes −mdiss and −munfold; this relationship is summarized by eq 3 in the Experimental Procedures section. The values of ΔGdiss, ΔGunfold, −mdiss and −munfold can now be estimated if the extent of monomer unfolding is determined over a range of TTR concentrations, and the resulting data are analyzed globally by fitting to eq 4.
The power of this method is that we can, for the first time, assess the true stability of TTR tetramers and monomers, so long as other intermediates (e.g., dimers or trimers) are not extensively populated. Thus, the free energies determined for tetramer dissociation and monomer unfolding by this analysis are actual, rather than apparent, stability values determined at specified conditions and useful only for comparisons among variants under the same conditions. Hence, the stability of WT TTR and various disease-associated variants can be compared and integrated with known dissociation and unfolding rates (7, 8, 12, 13, 18, 47), in order to gain insight into factors that may lead to amyloid disease. Another advantage of the method is that stability parameters can be derived using only tryptophan fluorescence data. This advantage is significant not only because of the ready availability and simplicity of the fluorescence spectroscopy, but also because intrinsic protein fluorescence does not perturb the equilibrium when assessing the extent of denaturation. In contrast, both methods used previously to measure tetramer quaternary structural stability (resveratrol binding and chemical cross-linking) have the potential to shift the tetramer monomer equilibrium. Globally fitting the concentration-dependent urea denaturation data obviates our dependence on these techniques to measure tetramer concentrations as a function of TTR and urea concentrations.
As described in full detail in the Experimental Procedures, global analysis of concentration-dependent TTR denaturation data yields information about the thermodynamics of dissociation, expressed as ΔGdissH2O and −mdiss, and the thermodynamics of unfolding, expressed as ΔGunfoldH2O and −munfold. The results of this fluorescence analysis for the tetrameric TTR variants that conform to the two-step model denaturation model (WT TTR, V122I, and to a lesser extent L55P) are discussed in turn in the following paragraphs, and the corresponding data are summarized in Table 2. Other variants (V30M and A25T) that do not conform to this model are then discussed in more qualitative terms.
Table 2.
Comparison of Thermodynamic Stability of TTR Tetramers and Monomersa
| Tetramers | Monomers | |||
|---|---|---|---|---|
| TTR variant | ΔGdissH2O (kcal/mol) | −mdiss(kcal/mol M urea) | ΔGunfoldH2O (kcal/mol) | −munfold(kcal/mol M urea) |
| WT | 32.8±2.2 | 2.7±0.1 | 4.0±0.5 | 1.4±0.1 |
| V122I | 25.6±1.0 | 1.5±0.4 | 5.1±0.2 | 1.6±0.1 |
| L55P | 33±7 | 2.8±0.4 | 2.0±1.5 | 1.6±0.5 |
| V30M | n.d.b | n.d.b | n.d.b | n.d.b |
| A25T | n.d.c | n.d.c | n.d.c | n.d.c |
Thermodynamic stabilities were calculated by global analysis of concentration-dependent denaturation data, as described in Experimental Procedures. The stability of TTR tetramers is described by ΔGdissH2O, the free energy of tetramer dissociation in the absence of denaturant, and −mdiss, the dependence of ΔGdiss on urea concentration. TTR monomer stability is described by ΔGunfoldH2O, the free energy of monomer unfolding in the absence of denaturant, and −munfold, the urea dependence of ΔGunfold.
Thermodynamic stabilities could not be calculated by global fitting because urea denaturation curves did not fit a two-state model (See Figures 4C, 5C, and 6).
Thermodynamic stabilities could not be calculated by global fitting due to aggregation of A25T at low urea concentrations (See Figures 4D and 5D).
WT TTR tetramer and monomer thermodynamic stability determined by global fitting analysis
Global analysis of the concentration-dependent fluorescence-based denaturation data affords the following WT TTR stability parameters: ΔGdissH2O = 32.8 ± 2.2 kcal/mol and −mdiss = 2.7 ± 0.1 kcal/mol/M urea; ΔGunfoldH2O = 4.0 ± 0.5 kcal/mol and −munfold = 1.4 ± 0.1 kcal/mol/M urea. The global fit to the denaturation data is shown in the Supporting Information. The thermodynamic stability of the WT TTR monomer determined by global analysis is lower than that of M-TTR, by ~0.6 kcal/mol (19), probably owing to the two Met mutations in the latter sequence (F87M, L110M) and the different conditions used. Although the nearly superimposable denaturation curves obtained for M-TTR and 1.44 μM WT TTR seem to indicate that the stability of the monomers of WT TTR and M-TTR are very similar, it is clear from the results presented above that WT TTR monomer unfolding is still thermodynamically linked to tetramer dissociation, even at the lowest WT TTR concentration examined here. Hence, the true tertiary structural stability of WT TTR must be lower than the ΔGunfold,appH2O for denaturation of 1.44 μM WT TTR (5.0 kcal/mol), and the value of ΔGunfoldH2O of 4.0 kcal/mol obtained for WT TTR by global fitting is reasonable.
We had previously noted that the WT TTR quaternary and tertiary structural equilibria appear to become partially unlinked when the WT TTR concentration is decreased to ≤0.72 μM, and it is useful to reexamine how low a concentration of WT TTR would be needed to completely unlink these equilibria. The ΔGdiss calculated here, 33 kcal/mol, corresponds to a dissocation constant, Kdiss, of 9 × 10−25 M3. The magnitude of this tetramer dissociation constant means that even at a concentration of 0.72 μM, WT TTR is >95% tetrameric in the absence of urea. Furthermore, if we assume that >90% dissociation of tetramer to monomers (fraction tetramer, α = 0.10) would be required to completely unlink the two equilibria, then we can estimate that unlinking would only be accomplished at WT TTR concentrations below 3.6 nM, much lower than attainable in these experiments.
Having calculated the thermodynamic parameters for WT TTR by globally fitting the data, we can now do the same for the TTR disease-associated variants and comment on the extent of thermodynamic linkage exhibited by their quaternary and tertiary structural transitions. We can also determine whether the mechanism of denaturation and unfolding of WT TTR applies to the mutant disease-associated TTR sequences.
V122I TTR forms a destabilized tetramer and a stable monomer
The V122I mutation is examined first because it affects more people than any other TTR mutation; only WT TTR deposition in senile systemic amyloidosis is clinically more significant. Figure 4A shows representative concentration-dependent V122I denaturation curves monitored by tryptophan fluorescence. The individual denaturation curves fit well to eq 1, revealing Cm values ranging from ~3.3 M up to ~3.8 M urea, and −mufold values ranging from ~1.5 up to ~3.3 kcal/mol/M urea. Notably, the apparent value of each parameter increases gradually as the V122I concentration is increased from 1.44 to 144 μM; however, the denaturation curves appear to be much closer together, especially for the first three curves (corresponding to concentrations of 1.44, 3.6, and 7.2 μM), than observed with WT TTR, indicating the defect with V122I may be in tetramer stability.
Figure 4.
Dependence of urea denaturation of V122I (panel A), L55P (panel B), V30M (panel C), and A25T TTR (panel D) on protein concentration. Denaturation reactions contained TTR (1.44–144μM) and were incubated for 96 h at 4 °C in the presence of varying concentrations of urea (0–8 M) prior to fluorescence measurements (see Experimental Procedures for details). TTR concentrations for each variant were 1.44 (❍), 3.6 (□), 7.2 (◆), 14.4 (×), 36 (+), 72 (Δ), and 144μM (λ). The denaturation curves of V122I (panel A) and L55P (panel B) resemble those observed for WT TTR (see Figure 1A), with the steepness of the transition slope increasing and the curves shifting progressively further to the right with increasing TTR concentration. In contrast, for the remaining variants, V30M (panel C) and A25T (panel D), two transitions are observed in each curve, and the data cannot be fit to a two-state denaturation model (solid lines shown in these panels represent smoothing curves, rather than fits of the data to eq 1). Nevertheless, for all of the variants, the apparent stability of TTR to urea denaturation increases with increasing TTR concentration (Cm increases from 3.3 to 3.8 M urea for V122I, from 1.9 to 2.8 M urea for L55P, from 1.5 to 2.5 M urea for V30M, and from 1.7 to 3.1 M urea for the 2nd transition of A25T; note that the panels have different x-axis scales.)
Global analysis of the concentration-dependent data (see Supporting Information) confirms that the quaternary structure of V122I is destabilized, as reflected in the thermodynamic parameters determined for V122I tetramers: ΔGdissH2O, 25.6 ± 1.0 kcal/mol and −mdiss, 1.5 ± 0.4 kcal/mol M urea. This destabilization of the tetramer, ~7 kcal/mol relative to WT TTR, translates into a dissociation constant for tetramerization that is about 6 orders of magnitude larger for V122I than for WT TTR (10−18 M3 vs. 10−24 M3, respectively).
From the global fitting results, it appears that the tertiary structural stability of V122I is ΔGunfoldH2O = 5.1 ± 0.2 kcal/mol and −munfold, 1.6 ± 0.1 kcal/mol M urea. These results indicate that V122I monomers are more stable than those of WT TTR, by 1.1 ± 0.5 kcal/mol; however the previously reported stability of M-V122I, is ~0.8 kcal/mol lower than that of M-TTR, with ΔGunfoldH2O of 4.7 ± 0.2 kcal/mol and −munfold of 1.4 ± 0.1 kcal/mol/M urea (9); compare to ΔGunfoldH2O for M-TTR of 5.5 ± 0.8 kcal/mol under the same conditions (19). Again, this discrepancy is likely attributable to the F87M and L110M mutations in M-TTR constructs and the differences in the denaturation conditions used in this and previous work.
The effects of the V122I mutation on tetramer stability (less stable than WT TTR) and monomer stability (possibly slightly more stable than WT TTR) offset each other to some extent, accounting for our previous conclusion that V122I and WT TTR have similar overall thermodynamic stability (7). However, this offsetting is not perfect and there are significant differences between the two proteins at equilibrium. In particular, at physiological concentrations ([TTRtotal] = 14.4 μM), the amount of unfolded monomer for V122I TTR (0.15 nM) is 3-fold higher than for WT TTR (0.05 nM). Although these concentrations of unfolded monomer are low, this thermodynamic difference, combined with the 2-fold faster rate of dissociation than WT (t½= 19 h vs. 41 h for WT TTR) (7), likely contributes to the relatively early development of V122I cardiomyopathy.
Chemical cross-linking of V122I TTR (36 μM) in increasing urea concentrations shows that dissociation of tetramers of this variant proceeds completely to monomers without a significant accumulation of intermediates (Figure 5A). Fitting the densitometry data with eq 2 gives Cm,diss of 3.5 M (from % tetramer data) or 3.6 M (from % monomer data) urea. This value is in good agreement with the Cm,diss determined from resveratrol binding (3.4 M urea), and with the Cm determined from tryptophan fluorescence (3.6 M urea) for these same samples (data not shown).
Figure 5.
Chemical cross-linking of urea-denatured V122I (panel A), L55P (panel B), V30M (panel C), and A25T (panel D). TTR samples (36 μM for panels A–C and 144 μM for panel D) were cross-linked with glutaraldehyde and analyzed by SDS–PAGE with Coomassie Blue staining (see Experimental Procedures for details). Molecular weights of protein standards are shown on the y-axis, and the urea concentration for each of the TTR samples is shown on the x-axis. The bands ~55 kDa correspond to TTR tetramers; these disappear as the concentration of urea is increased, with concomitant appearance of a diffuse monomer band ~14 kDa. The denaturation of V122I (panel A) and V30M (panel C) resemble the data obtained for WT TTR (see Figure 3), in that for these 3 variants, the main species observed are the native tetramer and the denatured monomer (although some trimer and dimer are observed for V30M TTR). In contrast, when L55P is denatured (panel B), a significant fraction of dimeric TTR species (seen as bands ~28 and ~23 kDa) accumulate at intermediate urea concentrations. For the A25T variant (panel D), aggregation is observed rather than denaturation at low concentrations of urea. The aggregates formed are large, do not enter the gel, and accumulate maximally around 2.0 M urea; higher concentrations of urea denature the aggregates into monomers.
L55P exhibits a complex denaturation pathway
L55P is a rare but extremely pathogenic mutation in TTR, associated with amyloidosis in the second to third decade of life (the earliest onset amyloid disease known) (48). Urea denaturation curves for L55P obtained by intrinsic tryptophan fluorescence over a range of L55P concentrations are shown in Figure 4B. Although the apparent values of Cm,unfold are lower than those obtained for WT TTR at each TTR concentration examined, the pattern of behavior is similar to that observed for WT TTR, namely that each denaturation curve fits nicely to a two-state denaturation model, and that L55P TTR appears to become more stable (curves shifting to higher urea concentration and to steeper transition regions) as the L55P concentration increases. Fitting the individual denaturation curves to eq 1 gives apparent values of Cm,unfold increasing from ~1.9 up to ~2.8 M urea and of −munfold increasing from ~2.5 to ~3.5 kcal/mol M urea as the concentration of L55P is increased from 1.44 to 144 μM.
Global analysis of the concentration-dependent data affords the following thermodynamic parameters for L55P (see Supporting Information): ΔGdissH2O, 33 ± 7 kcal/mol and −mdiss, 2.8 ± 0.4 kcal/mol/M urea; ΔGunfoldH2O, 2.0 ± 1.5 kcal/mol and −munfold, 1.6 ± 0.5 kcal/mol/M urea. The value of ΔGdissH2O for L55P TTR derived from the fit is similar to that of WT TTR, even though a plethora of other data suggests that the L55P tetramer is less stable (e.g., the dissociation rate of L55P tetramer is ~9-fold faster than that of WT TTR tetramer (7) and L55P TTR tetramers dissociate at higher pH values than WT TTR tetramers (10, 42)). This observation and the large error in the parameter estimates from the global fit raise the suspicion that the two-step model used in the global analysis does not describe the denaturation pathway of L55P TTR well. Consistent with this supposition, chemical cross-linking of the same samples reveals several intermediate species (Figure 5B). Three different bands, with apparent molecular weights of 42, 28, and 23 kDa, are observed in turn as the tetramer dissociates. These bands are likely attributable to trimeric and dimeric TTR, respectively, and appear to be intermediates in the denaturation of L55P TTR.
The effect of the complicated denaturation mechanism of L55P TTR on the parameter estimates derived from our global fit is difficult to predict in detail, but should affect tetramer stability more than monomer stability. Supporting this notion, the difference between the values of ΔGunfoldH2O for L55P (2.0 kcal/mol) and WT TTR (4.0 kcal/mol) is 2.0 kcal/mol, consistent with previous experiments with the engineered monomeric version of L55P (M-L55P, with L55P/F87M/L110M mutations), which also exhibits a destabilization of ~2 kcal/mol relative to M-TTR (F87M/L110M mutations only). M-L55P affords a ΔGunfoldH2O of 3.3 ± 0.1 kcal/mol and −munfold of 1.9 ± 0.1 kcal/mol/M urea (X. Jiang, P. Hammerström, and J.W. Kelly, unpublished results) compared to the ΔGunfoldH2O for M-TTR of 5.5 ± 0.8 kcal/mol under the same conditions (19).
V30M TTR denaturation cannot be fit to a two-state model
The V30M mutation in TTR, which leads to the vast majority of FAP cases, is second in clinical importance among the disease-associated TTR mutations (3). Urea denaturation curves for V30M TTR are shown in Figure 4C, carried out over a V30M TTR concentration range from 1.44–144μM. The curves shift to the right with increasing V30M concentration, giving apparent Cm values for denaturation (estimated visually from the graph) that increase from ~1.5 M urea at 1.44 μM V30M up to ~2.5 M urea at 144 μM V30M. However, examination of any of the individual data sets obtained for this mutant reveals an unusual shape (this is best seen in Figure 6A for data obtained in a separate experiment at a V30M concentration of 36 μM). Fitting the concentration-dependent denaturation curves to eq 1 gives poor fits in all cases–yielding a small amplitude for the transition region and an apparently very steep post-transition slope, with the data points in this region deviating significantly and in a non-random manner from the fit. In fact, each individual curve appears to display two transition regions, both of which vary with the concentration of V30M. When denaturation curves derived from Trp fluorescence measurements are plotted with resveratrol binding data obtained in parallel (36 μM V30M, incubation for 96 h at 25 °C instead of 4 °C; Figure 6A), the two transition regions observed in each curve are symmetrical and exactly coincident, with apparent Cm values estimated to be ~2.2 and 6.5 M urea.
Figure 6.
Urea denaturation of V30M. (A) The denaturation of V30M (36 μM) was monitored both by intrinsic tryptophan fluorescence (❍, left y-axis) to assess changes in the tertiary structure, and by resveratrol binding (■, right y-axis) to assess tetramer dissociation. Samples were incubated for 96 h at 25 °C in the presence of varying concentrations of urea (0–8 M) prior to fluorescence measurements. Using either method, two transitions can be clearly seen in the denaturation curves, and the data cannot be fit to a two-state denaturation model. (B) Curves for the denaturation (❍) and renaturation (■) of V30M (36 μM), assessed by intrinsic tryptophan fluorescence, are compared. For the denaturation experiment, samples were incubated for 96 h at 4 °C in the presence of varying concentrations of urea (0–8 M); note that the lower temperature used here minimizes but does not eliminate the 2nd transition (compare with panel A). For the renaturation experiment, V30M (520 μM) was completely denatured by incubating the sample for 96 h at 4 °C in 8 M urea. The denatured V30M was subsequently diluted into renaturation reactions containing varying concentrations of urea (0.55–8 M), and the samples were incubated for 24 h at 25 °C prior to fluorescence measurements. The 2nd transition observed in the denaturation curve is absent in the renaturation curve; however, V30M cannot be completely refolded under these conditions, and furthermore the two curves do not overlap in the transition region. Solid lines in both panels represent smoothing curves of the data.
The same samples were also examined by glutaraldehyde cross-linking using SDS–PAGE quantification by densitometry (Figure 5C). Similar to WT TTR, the cross-linking data reveal an small accumulation of trimeric and dimeric V30M quaternary structures between 3.0–6.0 M urea (Figure 5C, Lanes 8–11), although with V30M TTR this accumulation occurs between the two transition regions observed by Trp fluorescence, whereas with WT TTR it occurs before the single transition. Resveratrol binding is weakly dependent on urea concentration in this range, suggesting that V30M TTR dissociation intermediates may be able to bind resveratrol. Such a dissociation pathway would be in stark contrast to that of WT TTR, which dissociates into dimers that do not contain a small molecule binding site (16, 49).
V30M is kinetically stable relative to WT TTR (7), consistent with a more robust small-molecule-binding dimer interface. The possibility that a slow rate of tetramer dissociation (t½= 68 h, compared with 41 h for WT TTR) might be responsible for the biphasic denaturation curves was examined. The samples were incubated for varying lengths of time (24–334 h rather than the usual 96 h), and the extent of denaturation at each time point was measured by Trp fluorescence. Although longer incubation times do increase the amplitude of the 1st transition and decrease the amplitude of the 2nd transition, both transitions are still apparent, even after 2 weeks of incubation time (t = 334 h; data not shown). Fitting the time-dependent fluorescence changes at each urea concentration to single exponentials indicates that, while the rates of approach to equilibrium increase non-linearly with urea concentration, as previously reported (7), equilibrium should be reached in all cases at t ≤ 334 h (data not shown). Since the two transitions are still apparent at this time point, where no further changes in the fluorescence signal are observed, there may be a thermodynamic as well as a kinetic explanation for the observed phenomenon, i.e., the stabilization of a TTR dimer that is not populated during WT TTR denaturation. The amplitude of the second transition is smaller at lower incubation temperatures (compare the tryptophan fluorescence denaturation data in panels A (25 °C) and B (4 °C) of Figure 6), presumably because TTR dissociation is faster at lower temperature (47), but is still present. While the 2nd transition could be minimized, it could not be eliminated when equilibrium is approached by denaturing the tetramer.
To gain further insight into the complex behavior exhibited by V30M TTR, renaturation was studied by diluting a V30M solution completely denatured in the presence of 8 M urea (complete within 96 h). Samples diluted to a final urea concentration of 0.55–8.0 M were incubated for an additional 24 h (25 °C) and studied by Trp fluorescence. Although the 2nd transition was absent in this renaturation experiment, and the post-transition region (4.0–8.0 M urea) appeared more typical, V30M did not completely refold at low urea concentrations (Figure 6B), suggesting significant kinetic barriers in both directions, especially at intermediate urea concentrations. Furthermore, the poor overlap of the two data sets in the transition region is evidence that denaturation and renaturation are not fully reversible for this variant. Increasing the incubation time in the renaturation experiment had no effect on the data (not shown). In contrast to these observations, coincident denaturation and renaturation curves are observed for WT TTR and several other TTR variants (L55P, V122I, M-TTR) demonstrating that equilibrium is reached regardless of whether it is approached in a denaturation or renaturation direction (data not shown).
Although V30M is clearly less stable than WT TTR, as evidenced by its much lower Cm for denaturation (Cm ~1.5 M for V30M vs. ~3.1 M urea for WT TTR at 1.44 μM), its peculiar behavior under both denaturation and renaturation conditions precludes quantification of its thermodynamic parameters. Previous denaturation studies reveal that M-V30M is destabilized by ~2.5 kcal/mol relative to M-TTR, with ΔGunfoldH2O of 3.0 ± 0.2 kcal/mol and −munfold of 2.0 ± 0.1 kcal/mol M urea (X. Jiang, P. Hammerström, and J.W. Kelly, unpublished results); compare to ΔGunfoldH2O for M-TTR of 5.5 ± 0.8 kcal/mol under the same conditions (19). Furthermore, the magnitude of the shift in Cm values (to ~1 M higher urea) observed when V30M concentration is increased 100-fold, from 1.44 to 144 μM, is similar to the shift observed for WT TTR (to ~0.9 M higher urea) over the same concentration range, perhaps reflecting a similar quaternary structural stability, albeit one with a distinct mechanism of dissociation. Thus, it appears likely that V30M TTR tetramers are at least as stable as those of WT TTR but they have an unusual dissociation mechanism and a destabilized tertiary structure, with the latter two effects conspiring to predispose individuals with this mutation to TTR amyloidogenesis. The kinetic stability of the V30M tetramer probably protects carriers of V30M against a more severe FAP pathology (7).
A25T is highly destabilized and prone to aggregation
Urea denaturation of A25T shows that this TTR variant behaves unlike any of the others examined. The concentration-dependent tryptophan fluorescence curves for A25T TTR are shown in Figure 4D. Denaturation curves obtained at low A25T TTR concentrations (0.72–7.2 μM) appear to fit well to eq 1, but unlike what is observed with the other variants, there is virtually no concentration dependence on the fitting parameters, with values of Cm ~1.7 M urea and −munfold ~1.3 kcal/mol M urea obtained for each curve in this concentration range (0.72 μM curve not shown). As the A25T TTR concentration is increased further (from 7.2 to 144 μM), the characteristic shifting of the curves to the right and an increased steepness of the transition region is observed. The most striking feature of these denaturation curves, however, is the appearance of a unique 2nd transition region at A25T TTR concentrations greater than 7.2 μM. The Cm for this 2nd transition, visually estimated from Figure 4D, increases from ~1.7 M urea at 7.2 μM up to ~3.1 M urea at 144 μM A25T TTR. The shapes of the curves superficially resemble those of V30M TTR, but the nature and origin of the additional transition is clearly different.
The first indication that these data were unusual came from direct observation of the fluorescence spectra collected at low urea concentrations, where dramatic increases in the fluorescence intensity at each of the wavelengths examined (335 and 355 nm) were apparent, even when the fluorescence ratio (F355/F335) was not changing appreciably (data not shown). The effect was greater for samples containing higher concentrations of A25T TTR. The samples were slightly cloudy to the eye, and turbidity measurements showed a corresponding increase in turbidity in this same region of the denaturation curves (data not shown), suggesting that aggregation of A25T TTR occurs under these conditions. Additional direct evidence for aggregation was obtained by cross-linking followed by SDS–PAGE, which clearly shows the appearance of very large aggregates of A25T at low urea concentration (Figure 5D). In the 144 μM samples shown here (where aggregation is most favored), the aggregates accumulate maximally at 2.0 M urea (Lane 7), where they represent nearly 85% of the total A25T TTR in the sample. These large aggregates are apparently disrupted at higher concentrations of urea (> 4.0 M; Lanes 10–12), yielding monomeric A25T TTR as the predominant species. Trimeric (MW ~42 kDa) and dimeric (MW ~28 kDa) species are observed as intermediates in both the aggregation and the disaggregation of A25T TTR, accumulating to a maximum of ~30% at 1.0 M urea (aggregation phase) and ~32% at 4.0 M urea (disaggregation phase). A25T TTR apparently aggregates in the absence of any denaturation stress, as evidenced by the presence of a species with a molecular weight >190 kDa (~15% of total A25T) and the very large aggregates that do not enter the gel (~5% of A25T), in the 0.0 M urea sample (Lane 1).
Fitting the densitometry data for the disappearance of A25T TTR tetramers or the appearance of A25T TTR monomers to eq 2 gives Cm values of 1.2 M urea and 3.1 M urea, respectively. Each of the two transitions is coincident with either the appearance (Cm 1.2 M urea) or disappearance (Cm 3.1 M urea) of the aggregates (data not shown). These observations suggest that the 1st transition in the fluorescence data corresponds to the aggregation of A25T TTR (presumably preceded by tetramer dissociation and partial denaturation of the monomers), and the 2nd transition is the disaggregation of the large aggregates, resulting in unfolded monomers. At low A25T TTR concentrations, the methods employed do not detect aggregates, which presumably would form to a lesser extent at low protein concentration, and the denaturation curves monitored by tryptophan fluorescence show a single transition.
Because A25T clearly does not undergo a tetramer-folded monomer-unfolded monomer denaturation process, the data cannot be fit by global analysis to yield thermodynamic constants. However, fitting the individual denaturation curves at low A25T TTR concentration (where aggregation is not observed) to eq 1 gives us an upper limit on the stability of the A25T TTR monomer. The apparent tertiary structural stability, given by ΔGunfoldH2O of 1.8 ± 0.2 kcal/mol and −munfold of 1.1 ± 0.0 kcal/mol/M urea (n = 3), is lower than the previously reported ΔGunfoldH2O of 2.8 kcal/mol (12). Although the calculated ΔGunfoldH2O represents an apparent stability value, it may in fact closely approximate the true monomeric stability of this variant, since the overlapping curves over a 10-fold A25T concentration range (0.72–7.2 μM) suggest that tetramer dissociation and monomer unfolding are not appreciably linked at low concentration. The observation that the equilibria are at least partially unlinked at A25T concentrations as high as 7.2 μM indicates that the tetrameric structure is probably highly destabilized as well, consistent with tetramer disappearance at lower urea concentrations than any of the other TTR variants studied herein (see Figures 3 and 5).
In summary, the thermodynamic stability profile of A25T shows that both tetramers and monomers of this variant are highly destabilized. In addition, the rate of A25T TTR tetramer dissociation is extremely fast, with a t½~2.1 min (~1200-fold faster than the dissociation of WT TTR) (12), reflecting a high degree of kinetic destabilization of the quaternary structure. These factors together probably contribute to the high propensity of A25T to aggregate in vitro (12, 13). Paradoxically, the extreme instability of A25T appears to be protective against aggressive systemic amyloid disease, in that the serum concentrations of variant protein are very low due to extensive endoplasmic reticulum-associated degradation (ERAD) by cellular quality control during secretion. ERAD occurs to a greater extent in the liver than in the choroid plexus, partially explaining the CNS phenotype of this amyloid disease (12, 13). That this is one of the few disease-associated TTR variants that is substantially degraded by the cellular secretory pathway supports the fact the quaternary and tertiary structural stability of A25T are significantly compromised.
CONCLUDING REMARKS
Our results are broadly consistent with previous findings on the stabilities of TTR variants (7–15, 17, 18, 22, 42). However, the concentration-dependent denaturation data obtained herein and the global fits to these data (where possible) add substantial detail. Combining our data with that available in the literature yields the following picture of TTR stability and the effect of mutations thereon. WT and V122I TTR both have simple denaturation pathways (tetramer-folded monomer-unfolded monomer) and stable monomers, but the WT forms more stable tetramers than V122I. V30M and L55P TTR both have complicated denaturation pathways and unstable monomers, but V30M tetramers are quite stable (possibly even more so than WT tetramers). L55P tetramers are less stable than V30M but are still stable enough to be the dominant species at physiological TTR concentrations. Finally, A25T TTR has a complicated denaturation pathway that includes substantial amounts of aggregate formation, unstable monomers, and dramatically destabilized tetramers.
Categorizing the TTR variants studied herein according to the phenotypes of their associated amyloid diseases reveals groupings that are strikingly similar to those above. WT TTR and V122I TTR are associated with late onset cardiac amyloid diseases, with the latter being more aggressive than the former (29). V30M TTR and L55P TTR are associated with early onset polyneuropathy, again with the latter being more aggressive than the former (48). Finally, A25T TTR is not associated with systemic amyloidosis, because it is extensively degraded in liver cells by ERAD (12, 13). Rather, the high concentration of thyroxine in the choroid plexus stabilizes A25T TTR enough for it to be secreted into the central nervous system where it appears to cause central nervous system selective amyloidosis after dissociation of thyroxine in the CSF (12, 13).
The correspondence between the thermodynamic and amyloidosis phenotype groupings of TTR variants suggests a clear relationship between TTR folding and tetramerization energetics and disease pathology. TTR variants with unstable monomers and complex tetramer dissociation pathways (V30M and L55P TTR; A25T is discussed separately below) appear to cause early onset TTR amyloid diseases. In contrast, TTR variants with stable monomers and straightforward tetramer dissociation pathways (WT and V122I TTR) appear to cause TTR amyloid diseases with substantially later onset. Thus, monomer instability and/or complicated tetramer dissociation pathways, both of which should increase the partitioning of TTR into aggregation pathways (50), seem to be a primary source of TTR pathogenicity. Tetramer stability also affects TTR pathogenicity but in a more complicated way. Moderate decreases in tetramer stability appear to shift the age of onset of TTR amyloid disease. For example, V122I TTR amyloidosis is earlier onset than WT TTR amyloidosis and L55P TTR amyloidosis is earlier onset than V30M TTR amyloidosis. This effect, however, is secondary to the effect of monomer instability and complicated dissociation pathways, as V122I TTR amyloidosis is substantially later onset than V30M TTR amyloidosis. As tetramer instability becomes extreme, TTR variants with unstable monomers like A25T TTR become susceptible to ERAD and do not cause systemic amyloid disease (8, 12, 13, 41). Notably, the stabilities of V30M and L55P TTR monomers are about the same as those of several monomeric TTR variants that have been shown to undergo extensive ERAD (13). It seems that for V30M and L55P TTR, tetramerization stabilizes the proteins enough to “chaperone” them out of the ER and through the export pathway.
Finally, the relationship between TTR stability and pathology presented above provides insight into therapeutic strategies for TTR amyloidosis. Based on the above discussion, the small molecule binding strategy to treat TTR amyloidosis described in the Introduction would be only marginally beneficial if small molecule binding did nothing more than stabilize TTR tetramers; as noted above, tetramer stabilization appears to have a secondary role in TTR amyloidosis. Tetramer dissociation kinetics, however, trump the effects of tetramer stability, monomer stability, and complicated dissociation pathways. This assertion is supported by the observation mentioned in the Introduction that T119M TTR subunits, which slow the dissociation of TTR tetramers into which they have been incorporated, can ameliorate TTR amyloidosis in T119M/V30M TTR compound heterozygotes (17, 18, 35). Fortunately, small molecule binding greatly slows tetramer dissociation by imparting kinetic stabilization (18, 20, 21), increasing the likelihood that this therapeutic strategy will be effective. The clinical trials for small molecule kinetic stabilizer treatment of TTR amyloidosis are now underway and will soon inform us if the relationship between TTR energetics and TTR amyloid disease phenotype proposed above is correct.
Supplementary Material
Acknowledgments
We thank Maria Dendle and Michael Saure for their help with protein expression, and Nora Green for providing A25T TTR. We also thank Per Hammarstrom and R. Luke Wiseman for critically reading the manuscript, and Natalia Reixach and members of the Kelly laboratory for helpful discussions.
Footnotes
Abbreviations. TTR, transthyretin; WT, wild-type; M-TTR, a monomeric variant of transthyretin harboring F87M and L110M mutations; SSA, senile systemic amyloidosis; FAP, familial amyloid polyneuropathy; FAC, familial amyloid cardiomyopathy; Tris, Tris(hydroxymethyl)aminomethane; EDTA, ethylenediamine-N,N,N′,N′-tetraacetic acid; DTT, dithiothreitol; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; CSF, cerebrospinal fluid.
This research was supported by NIH Grant DK46335, the Skaggs Institute for Chemical Biology, and the Lita Annenberg Hazen Foundation. A.R.H.B. was supported by NIH Grants T32-AG00080 and F32-GM067348.
SUPPORTING INFORMATION AVAILABLE
Plots showing the global fit of our three state model (native tetramer, folded monomer, unfolded monomer) to the concentration-dependent urea denaturation data for WT, V122I, and L55P TTR, and a plot showing the relatively poor global fit to a two state model (native tetramer, unfolded monomer) for WT TTR. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Kelly JW. Alternative conformations of amyloidogenic proteins govern their behavior. Curr Opin Struct Biol. 1996;6:11–17. doi: 10.1016/s0959-440x(96)80089-3. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB. Amyloidosis. Annu Rev Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 3.Buxbaum JN, Tagoe CE. The genetics of the amyloidoses. Annu Rev Med. 2000;51:543–569. doi: 10.1146/annurev.med.51.1.543. [DOI] [PubMed] [Google Scholar]
- 4.Colon W, Kelly JW. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 5.Lai Z, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 6.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat Struct Biol. 2000;7:754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 7.Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16427–16432. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammarstrom P, Sekijima Y, White JT, Wiseman RL, Lim A, Costello CE, Altland K, Garzuly F, Budka H, Kelly JW. D18G transthyretin is monomeric, aggregation prone, and not detectable in plasma and cerebrospinal fluid: a prescription for central nervous system amyloidosis? Biochemistry. 2003;42:6656–6663. doi: 10.1021/bi027319b. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Buxbaum JN, Kelly JW. The V122I cardiomyopathy variant of transthyretin increases the velocity of rate-limiting tetramer dissociation, resulting in accelerated amyloidosis. Proc Natl Acad Sci U S A. 2001;98:14943–14948. doi: 10.1073/pnas.261419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCutchen SL, Colon W, Kelly JW. Transthyretin mutation Leu-55-Pro significantly alters tetramer stability and increases amyloidogenicity. Biochemistry. 1993;32:12119–12127. doi: 10.1021/bi00096a024. [DOI] [PubMed] [Google Scholar]
- 11.McCutchen SL, Lai Z, Miroy GJ, Kelly JW, Colon W. Comparison of lethal and nonlethal transthyretin variants and their relationship to amyloid disease. Biochemistry. 1995;34:13527–13536. doi: 10.1021/bi00041a032. [DOI] [PubMed] [Google Scholar]
- 12.Sekijima Y, Hammarstrom P, Matsumura M, Shimizu Y, Iwata M, Tokuda T, Ikeda S, Kelly JW. Energetic characteristics of the new transthyretin variant A25T may explain its atypical central nervous system pathology. Lab Invest. 2003;83:409–417. doi: 10.1097/01.lab.0000059937.11023.1f. [DOI] [PubMed] [Google Scholar]
- 13.Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Ferrao-Gonzales AD, Palmieri L, Valory M, Silva JL, Lashuel H, Kelly JW, Foguel D. Hydration and packing are crucial to amyloidogenesis as revealed by pressure studies on transthyretin variants that either protect or worsen amyloid disease. J Mol Biol. 2003;328:963–974. doi: 10.1016/s0022-2836(03)00368-1. [DOI] [PubMed] [Google Scholar]
- 15.Shnyrov VL, Villar E, Zhadan GG, Sanchez-Ruiz JM, Quintas A, Saraiva MJ, Brito RM. Comparative calorimetric study of non-amyloidogenic and amyloidogenic variants of the homotetrameric protein transthyretin. Biophys Chem. 2000;88:61–67. doi: 10.1016/s0301-4622(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 16.Foss TR, Kelker MS, Wiseman RL, Wilson IA, Kelly JW. Kinetic stabilization of the native state by protein engineering: implications for inhibition of transthyretin amyloidogenesis. J Mol Biol. 2005;347:841–854. doi: 10.1016/j.jmb.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 17.Hammarstrom P, Schneider F, Kelly JW. Trans-suppression of misfolding in an amyloid disease. Science. 2001;293:2459–2462. doi: 10.1126/science.1062245. [DOI] [PubMed] [Google Scholar]
- 18.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X, Smith CS, Petrassi HM, Hammarstrom P, White JT, Sacchettini JC, Kelly JW. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40:11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 20.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc Chem Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 21.Wiseman RL, Johnson SM, Kelker MS, Foss T, Wilson IA, Kelly JW. Kinetic stabilization of an oligomeric protein by a single ligand binding event. J Am Chem Soc. 2005;127:5540–5551. doi: 10.1021/ja042929f. [DOI] [PubMed] [Google Scholar]
- 22.Quintas A, Vaz DC, Cardoso I, Saraiva MJ, Brito RM. Tetramer dissociation and monomer partial unfolding precedes protofibril formation in amyloidogenic transthyretin variants. J Biol Chem. 2001;276:27207–27213. doi: 10.1074/jbc.M101024200. [DOI] [PubMed] [Google Scholar]
- 23.Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 24.Hornberg A, Eneqvist T, Olofsson A, Lundgren E, Sauer-Eriksson AE. A comparative analysis of 23 structures of the amyloidogenic protein transthyretin. J Mol Biol. 2000;302:649–669. doi: 10.1006/jmbi.2000.4078. [DOI] [PubMed] [Google Scholar]
- 25.Reixach N, Deechongkit S, Jiang X, Kelly JW, Buxbaum JN. Tissue damage in the amyloidoses: Transthyretin monomers and nonnative oligomers are the major cytotoxic species in tissue culture. Proc Natl Acad Sci U S A. 2004;101:2817–2822. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa MM, Cardoso I, Fernandes R, Guimaraes A, Saraiva MJ. Deposition of transthyretin in early stages of familial amyloidotic polyneuropathy: evidence for toxicity of nonfibrillar aggregates. Am J Pathol. 2001;159:1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westermark P, Sletten K, Johansson B, Cornwell GG., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci U S A. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson MD. Familial amyloidotic polyneuropathy. Trends Neurosci. 1989;12:88–92. doi: 10.1016/0166-2236(89)90162-8. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson DR, Pastore RD, Yaghoubian R, Kane I, Gallo G, Buck FS, Buxbaum JN. Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black Americans. N Engl J Med. 1997;336:466–473. doi: 10.1056/NEJM199702133360703. [DOI] [PubMed] [Google Scholar]
- 30.Stangou AJ, Hawkins PN. Liver transplantation in transthyretin-related familial amyloid polyneuropathy. Curr Opin Neurol. 2004;17:615–620. doi: 10.1097/00019052-200410000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Miroy GJ, Lai Z, Lashuel HA, Peterson SA, Strang C, Kelly JW. Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc Natl Acad Sci U S A. 1996;93:15051–15056. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razavi H, Palaninathan SK, Powers ET, Wiseman RL, Purkey HE, Mohamedmohaideen NN, Deechongkit S, Chiang KP, Dendle MT, Sacchettini JC, Kelly JW. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action. Angew Chem Int Ed Engl. 2003;42:2758–2761. doi: 10.1002/anie.200351179. [DOI] [PubMed] [Google Scholar]
- 33.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. Diflunisal analogues stabilize the native state of transthyretin. Potent inhibition of amyloidogenesis. J Med Chem. 2004;47:355–374. doi: 10.1021/jm030347n. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SM, Connelly S, Wilson IA, Kelly JW. Biochemical and structural evaluation of highly selective 2-arylbenzoxazole-based transthyretin amyloidogenesis inhibitors. J Med Chem. 2008;51:260–270. doi: 10.1021/jm0708735. [DOI] [PubMed] [Google Scholar]
- 35.Coelho T, Carvalho M, Saraiva MJ, Alves I, Almeida MR, Costa PP. A strikingly benign evolution of FAP in an individual found to be a compound heterozygote for two TTR mutations: TTR Met 30 and TTR Met 119. J Rheumatol. 1993;20:179. [Google Scholar]
- 36.Deu E, Kirsch JF. The unfolding pathway for Apo Escherichia coli aspartate aminotransferase is dependent on the choice of denaturant. Biochemistry. 2007;46:5810–5818. doi: 10.1021/bi602621t. [DOI] [PubMed] [Google Scholar]
- 37.Dev S, K ND, Sinha S, Surolia A. Thermodynamic analysis of three state denaturation of Peanut Agglutinin. IUBMB Life. 2006;58:549–555. doi: 10.1080/15216540600902228. [DOI] [PubMed] [Google Scholar]
- 38.Grimsley JK, Scholtz JM, Pace CN, Wild JR. Organophosphorus hydrolase is a remarkably stable enzyme that unfolds through a homodimeric intermediate. Biochemistry. 1997;36:14366–14374. doi: 10.1021/bi971596e. [DOI] [PubMed] [Google Scholar]
- 39.Hobart SA, Ilin S, Moriarty DF, Osuna R, Colon W. Equilibrium denaturation studies of the Escherichia coli factor for inversion stimulation: implications for in vivo function. Protein Sci. 2002;11:1671–1680. doi: 10.1110/ps.5050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornby JA, Luo JK, Stevens JM, Wallace LA, Kaplan W, Armstrong RN, Dirr HW. Equilibrium folding of dimeric class μ glutathione transferases involves a stable monomeric intermediate. Biochemistry. 2000;39:12336–12344. doi: 10.1021/bi000176d. [DOI] [PubMed] [Google Scholar]
- 41.Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 42.Lashuel HA, Wurth C, Woo L, Kelly JW. The most pathogenic transthyretin variant, L55P, forms amyloid fibrils under acidic conditions and protofilaments under physiological conditions. Biochemistry. 1999;38:13560–13573. doi: 10.1021/bi991021c. [DOI] [PubMed] [Google Scholar]
- 43.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 44.Hammarstrom P, Jiang X, Deechongkit S, Kelly JW. Anion shielding of electrostatic repulsions in transthyretin modulates stability and amyloidosis: insight into the chaotrope unfolding dichotomy. Biochemistry. 2001;40:11453–11459. doi: 10.1021/bi010673+. [DOI] [PubMed] [Google Scholar]
- 45.Fersht AR. Structure and Mechanism in Protein Science. W. H. Freeman and Company; New York: 1999. [Google Scholar]
- 46.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider F, Hammarstrom P, Kelly JW. Transthyretin slowly exchanges subunits under physiological conditions: A convenient chromatographic method to study subunit exchange in oligomeric proteins. Protein Sci. 2001;10:1606–1613. doi: 10.1110/ps.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobson DR, McFarlin DE, Kane I, Buxbaum JN. Transthyretin Pro55, a variant associated with early-onset, aggressive, diffuse amyloidosis with cardiac and neurologic involvement. Hum Genet. 1992;89:353–356. doi: 10.1007/BF00220559. [DOI] [PubMed] [Google Scholar]
- 49.Foss TR, Wiseman RL, Kelly JW. The pathway by which thetetrameric protein transthyretin dissociates. Biochemistry. 2005;44:15525–15533. doi: 10.1021/bi051608t. [DOI] [PubMed] [Google Scholar]
- 50.Wiseman RL, Powers ET, Kelly JW. Partitioning conformational intermediates between competing refolding and aggregation pathways: insights into transthyretin amyloid disease. Biochemistry. 2005;44:16612–16623. doi: 10.1021/bi0511484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.