Abstract
The neonatal Fc receptor for IgG (FcRn) functions to transport maternal IgG to a fetus or newborn and to protect IgG from degradation. Although FcRn is expressed in a variety of tissues and cell types, the extent to which FcRn expression is regulated by immunological and inflammatory events remains unknown. Stimulation of intestinal epithelial cell lines, macrophage-like THP-1, and freshly isolated human monocytes with the cytokine TNF-α rapidly up-regulated FcRn gene expression. In addition, the TLR ligands LPS and CpG oligodeoxynucleotide enhanced the level of FcRn expression in THP-1 and monocytes. Treatment of TNF-stimulated THP-1 cells with the NF-κB-specific inhibitor or overexpression of a dominant negative mutant inhibitory NF-κB (IκBα; S32A/S36A) resulted in down-regulation of FcRn expression. By using chromatin immunoprecipitation we identified three NF-κB binding sequences within introns 2 and 4 of the human FcRn gene. An EMSA confirmed the p50/p50 and/or p65/p50 complex (s) bound to intron 2- or 4-derived oligonucleotides containing putative NF-κB binding sequences, respectively. The intronic NF-κB sequences in combination with the promoter or alone regulated the expression of a luciferase reporter gene in response to TNF-α stimulation or overexpression of NF-κB p65 and p50. DNA looping interactions potentially occurred after the stimulation between intronic NF-κB sequences and the FcRn promoter as shown by a chromosome conformation capture assay. Finally, TNF-α stimulations enhanced IgG transport across an intestinal Caco-2 epithelial monolayer. Together, these data provide the first evidence that NF-κB signaling via intronic sequences regulates FcRn expression and function.
Immunoglobulin G is a most important Ig isotype for neutralizing agents of infectious diseases and modifies inflammatory and autoimmune diseases. The functional consequences of these actions are achieved by interaction of the IgG molecule with several conventional IgG Fc receptors (FcγRs) at cell surfaces, including FcγRI (CD64), FcγRII (CD32), FcγRIII (CD16) (1), and recently described FcγRIV (2). The so-called neonatal Fc receptor for IgG (FcRn)3 differs from these FcγRs because it is structurally related to the MHC class I family with a membrane-bound H chain in noncovalent association with β2-microglobulin (β2m) (3, 4). The overall exonintron organization of the FcRn gene is similar to that of MHC class I molecules, with the exception of a very large 10-kb intron between exons 4 and 5. In addition, FcRn displays pH-dependent binding of IgG; specifically, FcRn preferentially binds IgG at acidic pH (6 – 6.5) and releases it at neutral pH (7–7.4) (5, 6). FcRn is a transport receptor involved in controlling the movement of IgG from the maternal to the fetal blood of rodents and humans in placental and/or intestinal tissues. FcRn, therefore, plays a major role in the passive acquisition of maternal immunity by newborn mammals. FcRn also functions in the maintenance of IgG homeostasis in mammals of all ages by salvaging IgG from degradation. In the proposed model, IgG is taken up into cells by pinocytosis or endocytosis from the surrounding tissue fluid or blood. FcRn in acidic compartments, such as the endosome, binds and recycles IgG out of the cell to avoid IgG degradation in the lysosome (6, 7). The IgG transport and protective functions of FcRn are evidenced by several studies in which mice deficient in either β2m or the FcRn H chain fail to transport maternal IgG and show a significant reduction in the serum half-life of IgG (6, 8).
Although first observed in the intestinal epithelial cells of the neonatal rodent, FcRn has more recently been shown to express in a variety of cell types and tissues, including epithelial cells, endothelial cells, macrophages, and dendritic cells in adult life (6, 9). The level of FcRn expression appears to be critical for controlling IgG levels in tissues and blood (10). In autoimmune situations, FcRn expression may act as a rheostat that enables sufficient levels of pathogenic IgG to permit downstream conventional FcγR-mediated immune responses, immune complexes, and inflammatory cascades. Indeed, FcRn has been shown to be associated with the development of pathogenic IgG-mediated autoimmune diseases (11, 12). Although an understanding of the operative mechanism of FcRn function is emerging, evidence of how FcRn is regulated, especially under immune responses or inflammatory reactions, is not understood. The central roles that FcRn plays in the protection and transportation of IgG under normal or inflammatory situations have led to an increased interest in the mechanism that controls FcRn expression with regard to both constitutive and stimuli-mediated receptor expression.
NF-κB is a family of transcription factors that coordinate the expression of numerous genes in the innate and adaptive immune responses and the development of inflammatory and autoimmune diseases (13). NF-κB is composed of five members of the Rel family, including NF-κB 1 (p50), NF-κB 2 (p52), RelA (p65), RelB, and c-Rel (Rel). These proteins form homodimers and heterodimers, which are usually sequestered in the cytosol of un-stimulated cells via noncovalent interactions with specific inhibition proteins known as IκBs (14). Upon stimulation, signals that induce NF-κB activity subsequently cause the phosphorylation of IκBs by an IκB kinase complex (15, 16). As a result, the ubiquitin ligase complex interacts with the phosphorylated IκB, mediates polyubiquitination of IκB, and leads to its subsequent proteasomal degradation (17). The degradation of IκB proteins, thereby, allows NF-κB dimers to translocate to the nucleus and bind to the cognate NF-κB sequences of target genes.
In the present study, we investigate the involvement of NF-κB activation in the regulation of human FcRn expression and function. Our study showed that an NF-κB-specific inhibitor significantly down-regulated expression of the human FcRn gene. Using several complementary strategies, we have further identified the direct involvement of NF-κB-specific binding sites located in human FcRn introns 2 and 4 (18) that may function as modulators in response to immunologic and inflammatory stimuli, such as agonists of TLR and the proinflammatory cytokines TNF-α and IL-1β.
Materials and Methods
Cell lines, Abs, reagents
The human intestinal epithelial cell lines HT-29, Caco-2, and THP-1 cells (gifts from Dr. R. S. Blumberg, Harvard Medical School, Boston, MA) were maintained in DMEM or RPMI 1640 medium (Invitrogen Life Technologies) supplemented with 10 mM HEPES, 10% FCS, 1% L-glutamine, nonessential amino acids, and 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2 in an incubator at 37°C.
The anti-human FcRn mAbs DVN24 and ADM31 were made by immunizing C57BL/6 murine FcRn−/− mice first with 2 × 107 spleen cells from C57BL/6 human FcRn transgenic mice in complete Freund’s adjuvant (Sigma-Aldrich), followed 2 wk later with the same cells in IFA (Sigma-Aldrich). Spleen cells from seropositive responder mice were fused with SP2-0 cells using established procedures to make hybridomas. HRP-conjugated donkey anti-rabbit Ab was purchased from Pierce and purified human IgG from Jackson ImmunoResearch Laboratories. Abs against NF-κB p65, p50, Rel B, and p52 were obtained from Santa Cruz Biotechnology (Santa Cruz Biotechnology) and IL-1β and TNF-α from R&D Systems. Affinity-purified rabbit anti-FcRn Ab has been described (9). The phosphorothioate CpG oligodeoxynucleotide (5′-TCGTCGTTTTGTCGTTTTGTCGTT-3′) and the mutant GpC oligodeoxynucleotide (5′-TCTGGCTTTTCTCATTTTCTGGTT-3′) were from Operon.
Semiquantitative RT-PCR and quantitative real-time RT-PCR
Total RNA was isolated from cells (2 × 106/ml) in TRIzol (Invitrogen Life Technologies). cDNA was generated by the amplification of total RNA using FcRn-specific primers (5′-CCGGAATTGGAGCCCCCCTCCAT-3′ and 5′-TGCTCTAGAGGAGGACTTGGCTGGAGATT-3′) with a onestep RT-PCR kit (Qiagen). GAPDH was amplified by primers (5′-GAGA AGGCTGGGGCTCAT-3′ and 5′-TGCTGATGATCTTGAGGCTG-3′). Thirty cycles of PCR amplification were performed under optimized conditions.
The total RNA samples were extracted from freshly isolated human CD14+CD11+ monocytes (Cambrex). The 106 cells/ml monocytes were stimulated with TNF-α (50 ng/ml) or LPS (1 μg/ml). Total RNA (150 ng/reaction) was reversely transcribed to yield first-strand cDNA using SuperScript III (Invitrogen Life Technologies). Real-time RT-PCR was performed using FcRn and GAPDH primers and the SYBR Green Super-mix kit (Bio-Rad) in Chromo 4 (MJ Research). FcRn expression was calculated following normalization to GAPDH levels by the comparative delta delta threshold cycle (ΔΔCT) method. All reactions were performed for 40 cycles: 15 s at 94°C, 15 s at 58°C, and 20 s at 72°C. The specificity of the amplification reactions was confirmed by melt curve analysis. Opticon Monitor version 3.1 software was used for real-time RT-PCR.
Gel electrophoresis, Western blotting, and IgG binding assay
Gel electrophoresis and Western blotting were performed as previously described (19, 20). In brief, cell lysates were prepared in PBS with 0.5% CHAPS by adding a protease inhibitor mixture (Sigma-Aldrich). A post-nuclear supernatant was analyzed for total protein concentrations by the Bradford method with BSA as a standard (Bio-Rad). The proteins were separated by electrophoresis on 12% SDS-polyacrylamide gels under reducing conditions and transferred onto nitrocellulose (Schleicher & Schuell). The membranes were blocked with 5% nonfat milk and probed with affinity-purified, anti-human FcRn Ab and then with HRP-conjugated goat anti-rabbit or anti-mouse Ab. All washing steps were performed in 5% milk containing 0.05% Tween 20. The final product was visualized by ECL (Pierce).
IgG binding assay was performed as previously described (20). Cells were lysed in PBS (pH 6.0 or 7.5) with 0.5% CHAPS (Sigma-Aldrich) and a protease inhibitor mixture. Postnuclear supernatants containing 0.5–1 mg of soluble proteins were incubated with human IgG-Sepharose (Amersham Biosciences). The unbound proteins were washed away with PBS (pH 6.0 or 7.5) containing 0.1% CHAPS. The adsorbed proteins were boiled with reducing electrophoresis sample buffer. The eluted proteins were subjected to electrophoresis with 12% SDS-PAGE gels. Proteins were visualized by Western blotting.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed according to the manufacturer’s recommendations (Upstate Biotechnology). In brief, THP-1 cells (5 × 106 cells) were incubated with or without TNF-α (50 ng/ml) for 20 – 60 min. The cells were fixed with formaldehyde. The nuclei were isolated and sonicated. Chromatin was immunoprecipitated overnight at 4°C by mild agitation with 5 μg of Ab specific for p65 or p50 or with 5 μg of normal IgG as negative control. Immune complexes were collected by incubation with protein A-agarose. The DNA samples were amplified by PCR primers (Table I) in an optimized condition.
Table I.
ChIP PCR primers used in this study
| Genes | Forward Primer | Reverse Primer | ||
|---|---|---|---|---|
| FcRn | −1178 | 5′-GGCCTGGTGGCAAGTGTGTG-3′ | −1058 | 5′-GACAGGGTCTGGCTCTGTCA-3′ |
| −386 | 5′-TTGTATTATTAGTAGAGACG-3′ | −161 | 5′-CCTAGCGCTCGCTGTTAATC-3′ | |
| 1050 | 5′-GCTGCCTTCTCCGCTCAATTAAC-3′ | 1330 | 5′-CTGCAGACAGAAACAGAGCT-3′ | |
| 1259 | 5′-ATAATAGATTCTTCTCCCTC-3′ | 1451 | 5′-GCAGGCTATTGTAGCTCAG-3′ | |
| 3430 | 5′-GCTCTGTAGGGCAGAGTTCA-3′ | 3548 | 5′-GGGTGAATGTGTGTCACTTC-3′ | |
| 5429 | 5′-GATTACAGGCATAAGCCACC-3′ | 5688 | 5′-ACTGGACATCTGATGCCATT-3′ | |
| 7651 | 5′-GCAGTAGCACGATCTCGGCT-3′ | 7951 | 5′-ATGGCAGCGCATGCCTGTAG-3′ | |
| 9582 | 5′-AGGCTGGAGTGCAGTGGCTA-3′ | 9815 | 5′-GCAGTGGCTCACTCCTACAAT-3′ | |
| 10370 | 5′-CAAGTGGCTGGGACTACAGG-3′ | 10576 | 5′-ATGAGGTCAGCATCATCTTG-3′ | |
| 12542 | 5′-CGGGTCTTCCTGGAATCTG-3 | 12857 | 5′-CAGTCAGGACACAGAACTG-3′ | |
| IkBα | 1618 | 5′-GACGACCCCAATTCAAATCG-3′ | 1918 | 5′-TCAGGCTCGGGGAATTTCC-3′ |
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared by a nuclear and cytoplasmic extraction kit (Pierce). The double-stranded oligonucleotides containing the tested NF-κB sequences from the FcRn introns were used as follows: intron +1104 (5′-GAGGCTGGGAACCACCTGTCGC-3′); intron +5561 (5′-AAGTGCGGGAGTCACCGTGCCC-3′); and intron +9651 (5′-GCTCAAGGATTCCTCCGTGCCT-3′). The NF-κB binding sequences are underlined. The DNA was labeled by a biotin 3′ end DNA labeling kit (Pierce). For competition assays, a 100-fold excess of nonlabeled oligonucleotide was incubated during the preincubation time. For the supershift assays, 0.8 μg of each Ab specifically directed against NF-κB p65, p50, RelB, or p52 was preincubated with the nuclear extracts. The samples were run on a 5% native polyacrylamide gel. The gels were blotted onto a nylon membrane, blocked, incubated with HRP-avidin, and developed using the LightShift chemiluminescent EMSA kit (Pierce, IL). Signal visualization was achieved by exposing the membrane to x-ray film.
Construction of expression or reporter plasmids and mutagenesis
The pRc/CMV-IκB (S32A/S36A) plasmid (16) was a gift from Dr. G. Cheng (University of California Los Angeles, CA). The plasmids p50, p65, and p50 plus p65 were constructed by cloning RT-PCR-amplified fragments for p50, p65, or both p50 and p65 into the pBUDCE4.1 (Invitrogen Life Technologies) vector. The pBudCE4.1 vector is designed for the independent expression of two genes from a single plasmid in mammalian cells. The resultant vector placed p65 or p50 under the promoter of CMV or EF-1α. The DNA fragments were amplified with primer pairs for human p65 (5′-CGGGGTACCCGATATGGACGAACTGTTCCCCCTCATCTTC-3′ and 5′-CGCTCGAGCGTTAGGAGCTGATCTGACTCAGCAGGGCTG-3′), and human p50 (5′-CCCAAGCTTGGGATGGCAGAAGATGATCCATATTTGGGAAG-3′ and 5′-GCTCTAGAGCTTAGTCCTTTTTAGATTCAGTGTCCATGGTTC-3′). The KpnI and XhoI or the HindIII and XbaI sites (underlined) were used for cloning.
Luciferase reporter plasmid P1, containing sequences from −1801 to +863 of the FcRn gene promoter, was constructed by cloning the PCR-amplified products (2664 bp) into the pGL3 vector (Promega) through XhoI and NcoI digestion. The PCR primer pairs (5′-CCGCTCGAGATGGCGCCGCATCATGACTCTGAACCAG and 5′-AGCCATGGTGAGAGGACGACCTGGGGCG) were used for amplification. Plasmid P2 was synthesized by PCR amplification using the forward primers described above and a reverse primer (5′-AGCCATGGAGAGGTGGCTTTCTGCAGACAGAAACAG-3′) followed by subcloning into a XhoI/NcoI-digested pGL3 vector. In the P2 plasmid the start codon methionine was mutated by overlapping PCR mutagenesis. Plasmid P3 was constructed by overlapping PCR mutagenesis to disable the NF-κB binding site (Fig. 6A) with pGL3ATG* (P2) used as a template.
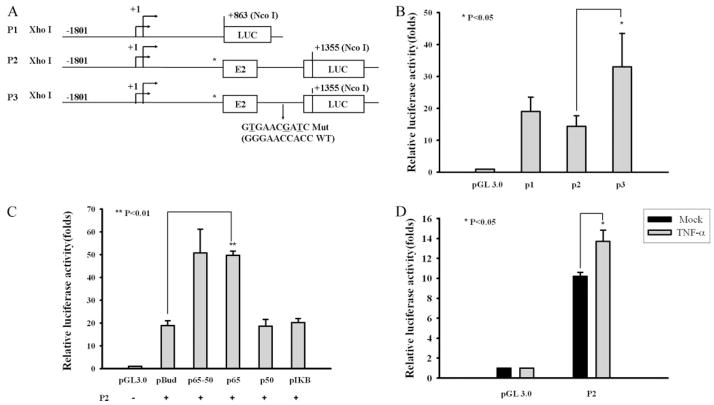
FIGURE 6.
NF-κB binding sequence in intron 2 modulates the promoter activity of the FcRn gene. A, Designations and diagrams of the luciferase reporter constructs. The reporter construct (P1) contains the FcRn promoter sequence from −1801 to +863 kb. The P2 construct represents the FcRn promoter sequence, exon 1, exon 2, and partial exon 3. Asterisks represent the start codon ATG mutation (Mut) in FcRn. Arrows denote the two putative initiation sites for transcription (17). The mutated nucleotides in the P3 construct are underlined. Exons (E) are drawn as boxes and introns are shown as roman numerals. LUC, Luciferase. B, Effects of NF-κB binding sequence (+1104) in intron 2 in regulating the FcRn promoter in untreated THP-1 cells. THP-1 cells were transiently transfected with the indicated vector alone or the luciferase reporter constructs P1, P2, and P3, respectively. After 24 h the cells were harvested and protein extracts were prepared for the luciferase assays. Results are expressed as relative luciferase activity and represent the mean value from at least three experiments. C, Effects of NF-κB site (+1104) in intron 2 in moderating FcRn promoter activity in the presence of NF-κB or the IκBα dominant negative (S32A/S36A). THP-1 cells were transiently transfected with the P2 construct together with the plasmids as indicated. Luciferase activity was measured 24 h posttransfection. Transcriptional activity was measured as firefly luciferase activity and normalized to Renilla luciferase activity as described above. The results show the mean of three independent experiments. pBUD, pBudCE4 vector. D, Effects of NF-κB site (+1104) in intron 2 in moderating FcRn promoter activity in TNF-α -treated cells. THP-1 cells were transiently transfected with the P2 luciferase reporter constructs. Twenty-four hours after transfection, cells were either mock treated (solid bar) or treated with TNF-α (gray bar). After 12 h of stimulation the cells were harvested and protein extracts were prepared for the luciferase assay as described above. The results show the mean value from three independent experiments.
The pLuc-MCS-FcRn vectors were constructed by cloning the pairs of complementary oligonucleotides encompassing the three tandem NF-κB binding sequences from the FcRn introns (Fig. 7A; +1104, +5561, and +9651). An FcRn DNA sequence (+3463) was also used as negative control. A double-stranded oligonucleotide was cloned into the pLuc-MCS plasmid (Stratagene) digested with HindIII and XhoI. All plasmids were verified by DNA sequencing analysis.
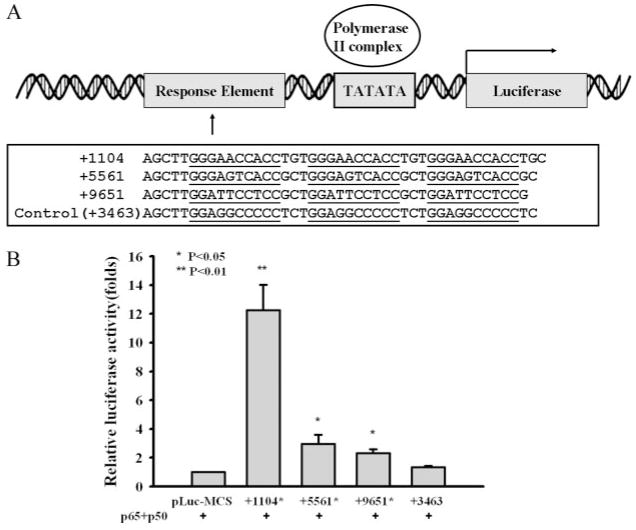
FIGURE 7.
NF-κB binding sequences from FcRn introns can enhance the transcription of the luciferase gene. A, Schematic representation of constructs pLuc-MCS containing NF-κB binding sequences from human FcRn introns 2 and 4. The pLuc-MCS plasmid has the minimal promoter with a TATA box. The plasmids pLuc-MCS +1104, pLuc-MCS +5561, pLuc-MCS +9651, and pLuc-MCS +3463, were constructed as described in Materials and Methods. The NF-κB sequences from FcRn introns are underlined. Numbers represent the locations of NF-κB sites in the human FcRn gene. A DNA sequence corresponding to +3463 in Fig. 4B was used as a negative control. B, HT-29 monolayers were cotransfected with the pLuc-MCS reporter plasmids as indicated and the NF-κB transactivator plasmid p50/65 in addition to the Renilla luciferase pRL-TK control plasmid. The pLuc-MCS backbone serves as a negative control. Luciferase activity was measured 24 h posttransfection. The results represent the mean of three independent experiments.
Transient transfection and luciferase assay
THP-1 and HT-29 cells were transiently transfected with Effectene (Qiagen). In each cotransfection, 2 × 106 cells were transfected with a DNA mix containing 0.95 μg of a firefly luciferase reporter plasmid and 0.05 μg of a Renilla luciferase pRL-TK control plasmid. On the following day the cells were cultured with or without TNF-α (10 –50 ng/ml). The cells were harvested 24 h after stimulation and assayed for the expression of Renilla and firefly luciferase using the dual luciferase kit (Promega). The values for firefly luciferase were normalized to the Renilla luciferase activity and expressed as fold activation over the vector background.
Chromosome conformation capture (3C) assay
The 3C experiment was modified according to previously described procedures (21–24). Briefly, THP-1 cells (1 × 107) were fixed with 2% formaldehyde. The nuclei were harvested and suspended in the HindIII digestion buffer containing 0.3% SDS at 37°C for 1 h. Triton X-100 was added to a 1.8% final concentration to sequester the SDS. Samples were digested with HindIII overnight at 37°C. Samples were then diluted with a ligase buffer. Triton X-100 was added to a final concentration of 1%. T4 DNA ligase was then added for incubation at 16°C for 4.5 h. The intramolecular ligation of cross-linked fragments was optimized and the ligation product was monitored by PCR. Proteinase K was added at a 200 μg/ml final concentration. The samples were incubated at 65°C overnight to reverse cross-linking and the DNA was isolated.
The PCR amplifications were determined by primer pairs in Table II. Primer pairs were designed to span each of six HindIII sites positioned along the 15-kb region of the human FcRn gene. Primers were used for cross-linked and control templates in all pairwise combinations. In general, DNA amplifications were done in 20-μl reaction mixtures with an initial denaturing step for 5 min at 94°C and then 35 cycles of PCR. PCR products were cloned and sequenced to confirm the presence of the FcRn DNA fragments ligated in a HindIII site.
Table II.
3C primers used in this study
| Name | Sequence |
|---|---|
| H1F | 5′-GAACTCGGATAGAGGTGACAGTTGCAC-3′ |
| H1R | 5′-CCGAGATTGCACCACTGCACTCCAGAC-3′ |
| H2F | 5′-CGCAGCAGTACCTGAGCTACAATAGC-3′ |
| H2R | 5′-GAGGTGTTGTCAGGGCCCAGTTCACAG-3′ |
| H3F | 5′-AGGAAGCGAGCATCCCATCACTGAGAC-3′ |
| H3R | 5′-GCAGTGAGCCGAGACTGAGCCACTACAC-3′ |
| H4F | 5′-GCTTTGGTAAATCTCAGACATCACAGTG-3′ |
| H4R | 5′-GTCAGGAGTTCAAGACCAGCCTGGCC-3′ |
| H5F | 5′-GCCCTTTGCATCCTGTGATGTTGCTG-3′ |
| H5R | 5′-GTCCACAGAACAGCACAGAAGCAAGC-3′ |
IgG transcytosis
IgG transport was performed as previously described with modification (25). Caco-2 cells growing onto Transwell inserts (Corning Costar) with a monolayer exhibiting resistances (250 –300 ohm/cm2) were equilibrated in HBSS. Monolayers were stimulated with TNF-α (10 ng/ml) for 1 h. Thereafter, human IgG or chicken IgY (1 mg/ml) was added to the apical medium. Monolayers were incubated for 1 h with IgG at either 37°C or 4°C. An aliquot of the buffer was collected in which apically and basolaterally directed IgG transports were conducted. Transported IgG was analyzed by SDS-PAGE gel and Western blot-ECL methods. NIH Image software (National Institutes of Health, Bethesda, MD) was used to determine relative band intensities.
Statistical analysis
Data from three independent studies were analyzed using ANOVA to identify significant changes between stimulated and mock-stimulated cells. All results are expressed as mean ± SEM from three independent experiments. A value of p < 0.05 is considered significant.
Results
Regulation of FcRn expression by TNF-α and IL-1β stimulations
NF-κB is well known to be activated by the exposure of cells to proinflammatory mediators such as TNF-α and IL-1β (13, 26). To show the possibility that TNF-α and IL-1β regulate the gene expression of human FcRn, we treated human THP-1 cells, a human macrophage-like cell line, with TNF-α (50 ng/ml) or IL-β (20 ng/ml). Our data showed that FcRn was constitutively expressed at a basal level as expected (9). However, FcRn expression was rapidly induced in response to TNF-α treatment as well as IL-β, as shown by semiquantitative RT-PCR, Northern blotting (data not shown), and real-time RT-PCR (Fig. 1A). TNF-α increased the mRNA levels 3.3-fold over those of the unstimulated cells after 1 h and ~6-fold after 2 h (Fig. 1A). We also found that FcRn expression induced by TNF-α was exhibited in a dose-dependent manner. IL-1β increased the mRNA level 1.5-fold after 20 min and up to 1.7-fold after 1 h as measured by semiquantitative RT-PCR (data not shown). To determine whether newly synthesized proteins, including transcription factors, are required for TNF-dependent induction of FcRn mRNA, we treated THP-1 cells with cycloheximide (CHX; 25 μg/ml) and subsequently with cytokines for 20 min and 1 h. CHX failed to inhibit the TNF-α-dependent induction of FcRn expression from both semiquantitative or real-time RT-PCR (data not shown). These data indicate that de novo protein biosynthesis was not required for the induction of FcRn transcription by TNF-α. Enhanced expression of the FcRn protein in THP-1 cells was shown by Western blotting in TNF-α -stimulated cells (Fig. 1B, lanes 4 and 5) in comparison with mock-stimulated cells (lane 3). Lysates from HeLa and HeLa-FcRn were used as negative (Fig. 1B, lane 1) and positive (lane 2) controls in Western blotting. Furthermore, induction of the FcRn protein could be significantly detected by staining with FcRn-specific Abs in THP-1 cells using flow cytometry or immunofluorescent microscopy (data not shown).
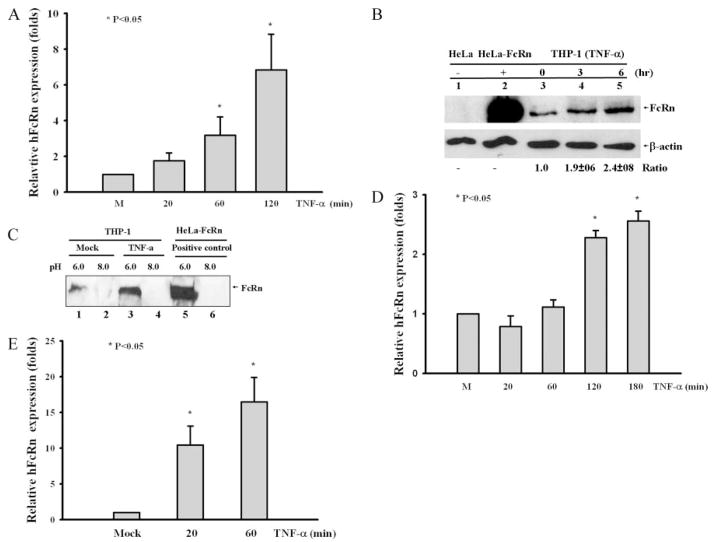
FIGURE 1.
FcRn expression in response to cytokine stimulation. Data are mean ± SD of three independent experiments. A, Quantitative real-time RT-PCR analysis of FcRn mRNA in THP-1 cells treated with TNF-α (50 ng/ml) for the indicated times (20, 60, and 120 min). B, Western blot. The cell lysates (10 μg) from HeLa (lane 1), HeLa-FcRn (lane 2), and TNF-stimulated THP-1 (lanes 3–5) were separated by electrophoresis in a 12% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and blotted with an FcRn-specific (top panel) or a β-actin-specific Ab (bottom panel). Blots were then incubated with a HRP-conjugated secondary Ab and visualized with the ECL method. The ratio of the mock group is assigned a value of 1.0, and the values from other groups are normalized to this value. The ratios of FcRn to β-actin are shown as indicated. C, The pH-dependent FcRn binding of IgG. The cells were lysed in sodium phosphate buffer (pH 6.0 or 8.0) with 0.5% CHAPS. Approximately 1 mg of soluble protein was incubated with human IgG-Sepharose at 4°C. The eluted proteins were subjected to electrophoresis with 12% SDS-polyacrylamide gels and subjected to Western blot analysis. Proteins were probed with an affinity-purified rabbit anti-FcRn Ab (9) and a HRP-conjugated donkey ant-rabbit Ab. Immunoblots were developed with ECL. D, Quantitative real-time RT-PCR analysis of FcRn mRNA by analyzing total RNA extracted from HT-29 cells treated with TNF-α (10 ng/ml) at the indicated times. E, Quantitative real-time RT-PCR analysis of FcRn mRNA in freshly isolated human monocytes treated with TNF-α (50 ng/ml) for the indicated time ors mock treated.
FcRn binds IgG at acidic pH 6.0 and releases IgG at neutral pH (5). We tested whether the enhanced expression of FcRn after TNF-α stimulation affects its ability to bind to its natural ligand, IgG. We incubated cell lysates from cells at either pH 6.0 or pH 8.0 with human IgG-Sepharose. Cell lysates from HeLa cells transfected with FcRn (9) were used as positive control. As expected, FcRn from HeLa-FcRn cells bound IgG at pH 6.0 but not at pH 8.0 (Fig. 1C, lanes 5 and 6). Our result showed that TNF-α stimulation enhanced cellular FcRn binding to IgG at pH 6.0 (Fig. 1C, lane 3) in comparison with mock-stimulated cells (Fig. 1C, lane 1), suggesting that the enhanced level of FcRn led to increased FcRn-IgG complexes. To evaluate other cell types in response to TNF-α induction, human intestinal HT-29 cells were treated with TNF-α (10 ng/ml). The mRNA level of FcRn was increased ~2.5-fold over the unstimulated level (Fig. 1D). Furthermore, FcRn mRNA from freshly isolated human monocytes treated with TNF-α (50 ng/ml) was increased 12-fold over the mock-stimulated cell mRNA after 20 min and ~17-fold after 1 h as assessed by real-time RT-PCR (Fig. 1E). Taken together, we conclude that TNF-α and IL-1β up-regulated the FcRn expression in human macrophage-like THP-1, the intestinal epithelial cell line, and freshly isolated human monocytes.
Regulation of FcRn expression in THP-1 cells by a TLR-mediated signaling pathway
Similar to TNF-α receptor engagement, the activation of TLRs by their cognate ligands can effectively result in NF-κB activation (27). We investigated the activation of TLR on cells in the induction of human FcRn expression. TLR9 binds nonmethylated CpG-containing DNA, and TLR4 in association with CD14 recognizes the ligand LPS. THP-1 cells express both TLR4 and TLR9 (27). FcRn mRNA appeared to be rapidly augmented in response to CpG in comparison with the mutant GpC treatment (Fig. 2A). CpG increased the FcRn mRNA level 1.7-fold and 2-fold over the mock-stimulated cell after 60 and 120 min, respectively. Furthermore, the FcRn protein was increased 2.1-fold after 3 h and up to ~2.5-fold after 6 h of stimulation (Fig. 2B, lane 5) in comparison with mock-stimulated cells (lane 3) in Western blotting. THP-1 cells expressed nearly undetectable levels of CD14 expression on the surface. PMA and 1,25-dihydroxy vitamin D3 are known to increase the expression of CD14 (28). Thus, THP-1 cells were first stimulated with 100 nM PMA and 100 nM 1,25-dihydroxy vitamin D3. THP-1 cells were then mock treated (Fig. 2C, left lane) or treated by LPS (1 μg/ml) (Fig. 2C, lanes 2–5 from the left). Semi-quantitative RT-PCR showed that the FcRn mRNA level was enhanced 1.8-fold after 1 h (Fig. 2C, lane 3 from the left) and up to 2-fold after 3 h (right lane) following LPS stimulation. Furthermore, human freshly isolated monocytes treated with LPS (1 μg/ml) increased FcRn mRNA level ~6-fold over the mock-stimulated cells after 1 h and ~12-fold after 2 h as measured by real-time RT-PCR (Fig. 2D). As such, the studies in Fig. 2 suggest that activation of TLR signal pathways, similar to TNF-α and IL-β, can also enhance FcRn expression.
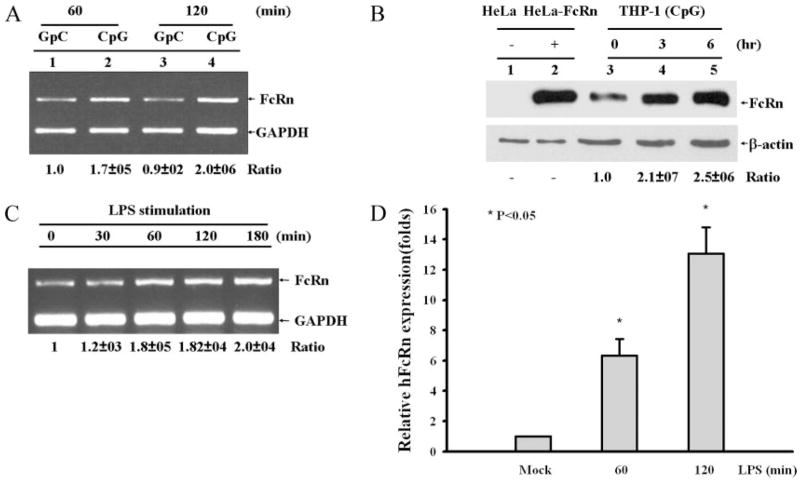
FIGURE 2.
FcRn expression in response to CpG or LPS stimulation. Densitometric intensity analyses of the RT-PCR experiments were conducted. The ratios of FcRn to GAPDH are shown. Similar results were seen in three independent experiments for each condition. A, THP-1 cells were stimulated with 4 μg/ml CpG or mutant GpC for 60 and 120 min. A representative sample for RT-PCR analysis using total RNA extracted from the cells was shown for FcRn and GAPDH. B, Western blot analysis of FcRn expression. The cell lysates (10 μg) from HeLa (lane 1), HeLa-FcRn (lane 2), and CpG-stimulated THP-1 (lanes 3–5) were subjected to electrophoresis with 12% SDS-polyacrylamide gels under a reducing condition and transferred to nitrocellulose membrane. Immunoblotting was performed with affinity-purified rabbit anti-FcRn Ab (top) or β-actin (bottom) and a HRP-conjugated secondary Ab with development by ECL. The ratio of the mock group is assigned a value of 1.0, and the values from other groups are normalized to this value. The ratios of FcRn to β-actin are shown as indicated. C, THP-1 cells were pretreated with PMA and 1,25-dihydroxy vitamin D3 for 6 days and then stimulated with 1 μg/ml LPS for the indicated time. A representative sample for RT-PCR analysis using total RNA extracted from the cells is shown for FcRn and GAPDH. D, Quantitative real-time RT-PCR analysis of FcRn mRNA from freshly isolated human monocytes treated with LPS (1 μg/ml) for the indicated times or left untreated. Data are mean ± SD of three independent experiments. *, p < 0.05.
Effect of NF-κB inhibition on FcRn expression
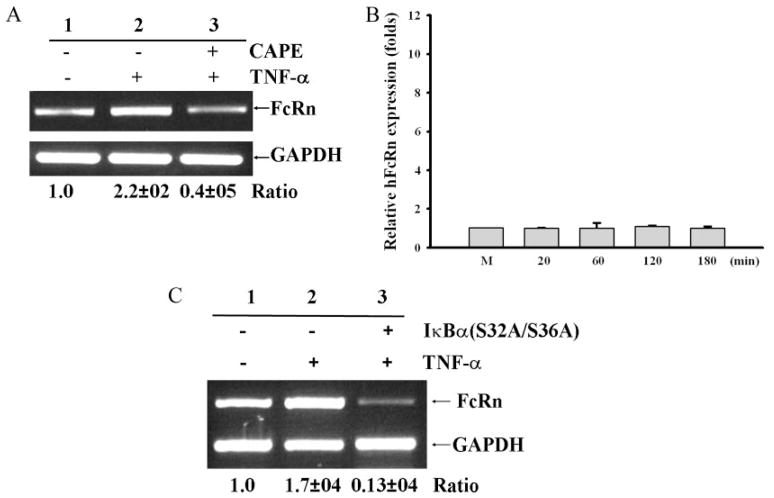
Because downstream activations of TNF-α, IL-β, and TLRs undergo similar NF-κB -mediated signaling pathways, we studied further the function of NF-κB signaling in FcRn transcription. Several steps of the NF-κB activation pathway, such as IκB kinase activation, IκB phosphorylation and degradation, and NF-κB nuclear translocation, can be targeted by a variety of inhibitors (29). Caffeic acid phenethyl ester (CAPE) has an inhibitory effect on the translocation of NF-κB p65 to the nucleus and on the binding of NF-κB to DNA (29). THP-1 cells were pretreated with CAPE (25 μg/ml) for 2 h and subsequently stimulated by TNF-α. Treatment with CAPE significantly reduced TNF-α -stimulated FcRn levels to that of the mock-stimulated THP-1 as assessed by semiquantitative RT-PCR (Fig. 3A) and real-time RT-PCR in a time course experiment (Fig. κB). We further tested the effects of CAPE on CpG-stimulated THP-1 cells, and similar inhibitions were observed. Treatment with CAPE significantly reduced the CpG-stimulated FcRn yield to the level of the mutant GpC-stimulated THP-1 (data not shown). In addition, CAPE did not exert the obvious toxicity to cells in our assay.
FIGURE 3.
Effect of NF-κB inhibitors on the expression of FcRn. Results are representative of three independent experiments. Ratios of FcRn to GAPDH mRNA are indicated below. A and B, Effects of CAPE on expression of FcRn mRNA. THP-1 cells were incubated with or without the NF-κB -specific inhibitor CAPE (25 μg/ml) for 2 h. THP-1 cells were subsequently stimulated with or without TNF-α. At the end of the incubation periods for the indicated times, RNA was isolated and analyzed by semiquantitative RT-PCR (A) or quantitative real-time RT-PCR (B). C, Effect of NF-κB blockage by IκBα on expression of FcRn. Human THP-1 cells were transfected with the dominant negative construct pCMV-IκBα (S32A/S36A). RNA was isolated and analyzed by semiquantitative RT-PCR.
NF-κB can be activated by the phosphorylation of its inhibitory subunit, IκB-α, on serine residues 32 and 36 by IκB kinases (16). Signaling through the TNF-α or IL-1β receptor can cause rapid phosphorylation of IκBα. The substitution of alanine residues for serines 32 and 36 within the N-terminal signal response domain can abolish the signal-induced IκBα phosphorylation and ubiquitination for degradation, resulting in a blockage of NF-κB activation (15). Overexpression of this IκBα (S32A/S36A) decreased FcRn expression in response to TNF-α by at least 80% (Fig. 3C, lane 3), compared with cells transfected with a control plasmid (Fig. 3C, lanes 1 and 2). Overall, this selective NF-κB inhibition by CAPE in conjunction with the overexpression of dominant IκB mutant indicates that the blockade of NF-κB activation inhibited FcRn transcription in response to TNF-α.
Screening for NF-κB binding sites adjacent to the FcRn gene
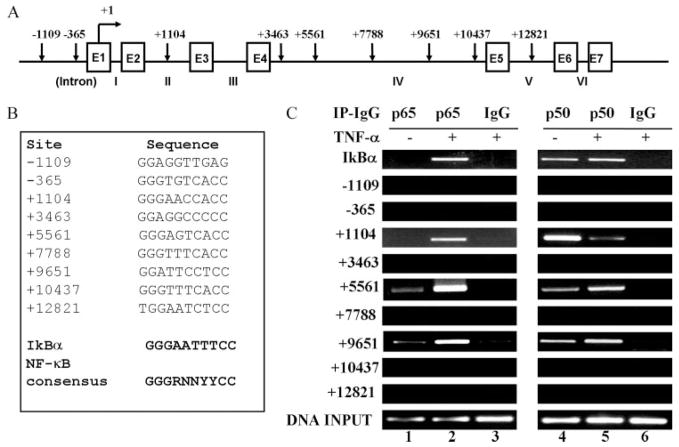
The canonic NF-κB DNA binding sequence is a common 10-bp consensus DNA element that has been identified as 5′-GGGRNNYYCC-3′ or 5′-HGGARNYYCC-3′ (where H is A, C, or T; R is an A or G purine; and Y is a C or T pyrimidine) (13). Because FcRn expression does not require newly synthesized proteins (Fig. 1A), we hypothesized that NF-κB regulates FcRn expression through a mechanism that involves direct binding to a putative regulatory NF-κB binding sequences located in the FcRn gene. To test this hypothesis, we searched for putative NF-κB-binding sequence(s) along the entire human FcRn genomic sequence (Fig. 4A) (GenBank accession no. AC010619). Computational inspection revealed that the promoter and introns of the FcRn gene contained sequences with a similarity to the NF-κB consensus sequence (Fig. 4B). To verify that these putative NF-κB binding sequences do have the capability to directly bind NF-κB proteins in living cells, we used a ChIP assay to precipitate the NF-κB-DNA complexes with Ab specific for p65 or p50 after cross-linking the DNA with bound NF-κB proteins in situ in TNF-stimulated verus mock-stimulated THP-1 cells. The DNA fragments containing the putative NF-κB binding sequences in FcRn gene were measured by PCR. As shown in Fig. 4C, PCR with primers flanking the putative NF-κB binding sequences (Fig. 4C, +1104, +5561, and +9651) produced a band from DNA coprecipitated with p65 or p50. In a negative control, immunoprecipitation with normal rabbit IgG did not generate any corresponding PCR products (Fig. 4C, lane 6). The NF-κB binding sequence in the IκB gene promoter was used as a positive control. These data suggest that NF-κB p65 and p50 interacted with the three NF-κB binding sequences of FcRn gene in THP-1 cells.
FIGURE 4.
Mapping of NF-κB binding sequence(s) in the human FcRn gene by ChIP. A, Organization of the human FcRn gene. The exon/intron organization of the 15-kb FcRn gene is schematically shown. The positions of the exons/introns and the base counts are shown (GenBank accession no. AC010619). Positions of the putative NF-κB binding sequences relevant to the transcription start site of the FcRn gene are shown at the top. Transcriptional start sites are shown (17). Arrows indicate the position of candidate NF-κB sequences. Exons (E) are drawn as boxes and intron numbers are shown as roman numerals. B, The putative NF-κB binding sequences in the FcRn gene are listed. Numbers represent the putative NF-κB binding sequences relevant to the transcription start site of the FcRn gene. The consensus NF-κB sequence is bolded. R is an A or G purine and Y is a C or T pyrimidine. The NF-κB binding sequence in the promoter of IκB is used as a positive control. C, NF-κB p65 and p50 components are present at FcRn introns in vivo in response to TNF-α. THP-1 cells were treated with TNF-α (50 ng/ml) for 30 min. ChIP assays were performed using p65-specific (lanes 1–3) and p50-specific (lanes 4–6) Abs. IgG was used as a negative control (lanes 3 and 6). Immunoprecipitated chromatin was prepared and subjected to PCR analysis using primer pairs (Table I). ChIP was performed at least three times. IP, Immunoprecipitation.
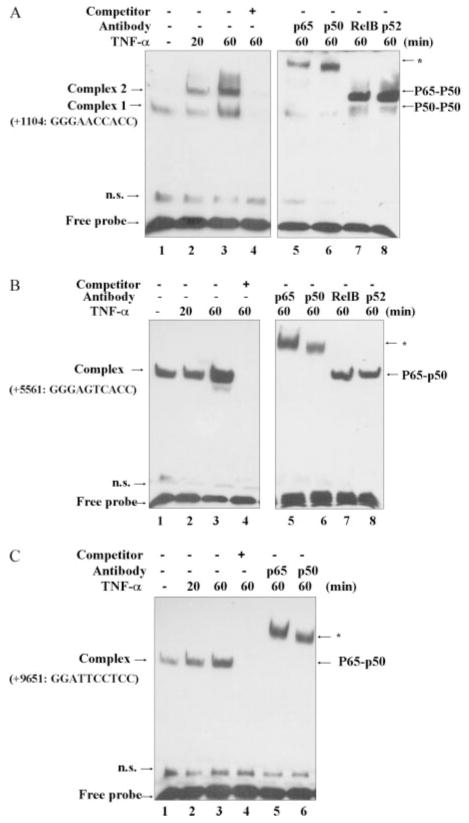
To further test the proposition that NF-κB factors have the capability to directly bind the FcRn NF-κB sequences identified from the ChIP assay, EMSAs were conducted using oligonucleotides containing NF-κB binding sequences (+1104, +5561, and +9651, Fig. 5). As shown in Fig. 5, a higher amount of complexes formed when bound to nuclear extracts from TNF-α-stimulated cells than when bound to nuclear extracts from mock-stimulated cells. To verify the binding specificity, a competition assay was performed. The inducible band could be competed completely by unlabeled oligonucleotides in all three intronic NF-κB binding sequences (Fig. 5, lanes 4). Supershift experiments indicated that the complexes contained p50 and p65 (Fig. 5, lanes 5 and 6), but not RelB and p52 components of the NF-κB subfamily (Fig. 5, A and B, lanes 7 and 8; Fig. 5C, data not shown). These results suggest that TNF-stimulation of THP-1 cells led to NF-κB interaction with FcRn mainly through the p50 and p65 components. Additionally, it should be noted that unlike the NF-κB binding sequences (+5561 and +9651) in intron 4 (Fig. 5, B and C), the sequence (+1104) in intron 2 bound the NF-κB p50/50 dimer in untreated cells; however, it appeared to bind p65/p50 in the presence of TNF-α stimulation (Fig. 5A). The p50/50 dimer was also verified by migrating to the same position as that of recombinant p50 proteins (data not shown). Taken together, the results indicate that the three novel NF-κB binding sequences are in far downstream intronic regions of human FcRn.
FIGURE 5.
Analysis of NF-κB binding. DNA binding was performed with nuclear extracts from THP-1 cells treated with or without TNF-α (50 ng/ml) for the indicated times (20 or 60 min). A 24-bp fragment, spanning the NF-κB binding sequences corresponding to the FcRn intron 2 (+1104) (A), intron 4 (+5561) (B), or intron 4 (+9651) (C), was used as a biotin-labeled probe. A 100-fold molar excess of unlabeled oligonucleotide was added (lanes 4 and 8) for competition. Supershift experiments (lanes 5 and 6) were performed in the presence of the indicated Abs. Free-labeled probes are also indicated by an arrow. n.s., Nonspecific bands.
NF-κB binding sequences in FcRn introns can regulate the expression of the luciferase gene
The activation of a signal transduction pathway can be monitored by the level of the luciferase controlled by the promoter and the regulatory elements. To test the function of the NF-κB sites, we first constructed the luciferase reporter gene driven by the FcRn promoter in the presence or absence of a NF-κB binding sequence (+1104) from the FcRn intron 2 (Fig. 6A). In addition, we generated a construct containing mutations on the NF-κB binding sequence of intron 2 (Fig. 6A, P3). Transactivation of the luciferase gene was tested. Transient transfection revealed that both constructs caused high expressions of luciferase. However, transient transfection of the P3 construct in THP-1 cells revealed that mutation of this NF-κB binding site significantly increased the basal promoter activity ~2-fold above that of the wild-type NF-κB binding sequence in mock-stimulated cells (Fig. 6B). Furthermore, co-transfection of P3 with P65, P50, or P65/P50 failed to affect the luciferase activity (data not shown).
To directly assess the involvement of NF-κB in FcRn expression, we investigated whether overexpression of NF-κB p65 and p50 affects the promoter activity of FcRn. We transiently transfected cells with the P2 plasmid and plasmids encoding p65 and p50, alone or in combination. We found that overexpression of NF-κB p65/p50 or p65 caused a significant increase in the transcriptional activity of the FcRn promoter to drive the luciferase expression when compared with cells transfected with p50 or inhibitory IκB (S32A/S36A) plasmids (Fig. 6C). It is not clear why p65 transfection alone had an enhancing effect (Fig. 6C). These results indicate that the overexpression of NF-κB p65 and p50 is sufficient to drive the up-regulation of FcRn expression. Furthermore, we tested whether FcRn promoter activity could be up-regulated by TNF-α stimulation. The results showed that promoter activity of human FcRn in our P2 construct was induced in response to TNF-α in comparison with mock-treated THP-1 cells (Fig. 6D). Together, these data strongly suggest that the presence of the NF-κB binding site in intron 2 was important for TNF-α regulation of FcRn transcription. We conclude that NF-κB regulates human FcRn expression in THP-1 cells by binding to a specific sequence in the intron 2 of the FcRn gene.
Because of the relatively long distances (≈10 kb) of the NF-κB binding sequences in intron 4 from the promoter, it is difficult to differentiate the functions of each κB binding sequence in FcRn regulation. To further determine the nature of the involvement of these NF-κB binding sequences, we constructed a luciferase reporter plasmid in which three tandem copies of NF-κB binding sequences (+1104, +5561, and +9651) were respectively linked to a minimal promoter containing only a TATA box (Fig. 7A). The pLuc-MCS plasmids (Fig. 7A) were transiently transfected into intestinal HT-29 cells with or without cotransfection of the NF-κB p50/p65 plasmid to validate the results obtained from THP-1 cells. The data showed that each NF-κB binding sequence significantly amplified the luciferase signal from 2- to 12-fold in cooperation with p65/p50 proteins (Fig. 7B). A relevant DNA sequence (Fig. 4B) that did not bind p50 or p65 in vivo from our ChIP assay was used a negative control, and it failed to enhance the luciferase activity over the vector alone (Fig. 7B). However, the NF-κB binding sequence (+1104) from intron 2 supported the strongest induction (12-fold) in comparison with the other intronic NF-κB binding sequences (2.1-fold). Overall, the results support the notion that these NF-κB binding sequences are functional.
Mutual interactions between promoter and intronic NF-κB of the human FcRn gene
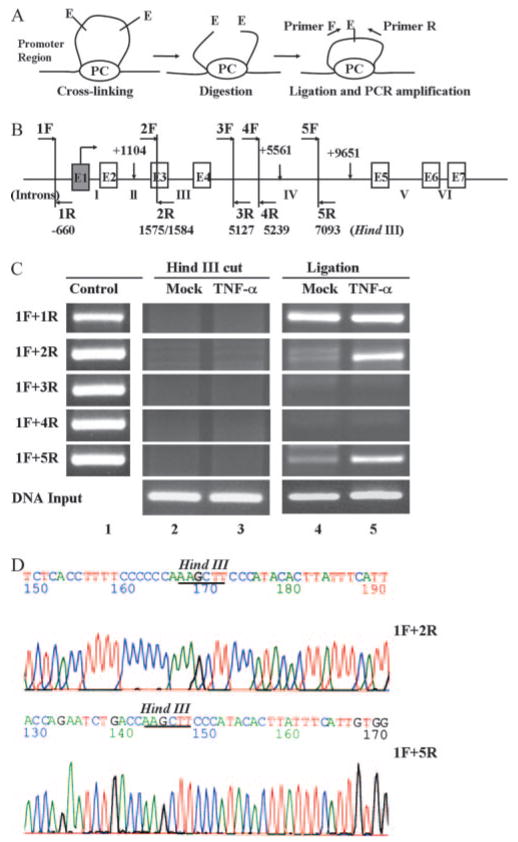
Considering that NF-κB-dependent induction of genes involves the cooperative interactions between NF-κB and other transcription factors (30, 31), we speculated that the intronic NF-κB binding sequences might also act upon the upstream FcRn promoters through a DNA looping mechanism. The 3C assay has proven to be an effective method to analyze the interactions between genomic regions, such as promoter-enhancer or enhancer-enhancer interactions, in gene regulation (21, 32). We used the 3C assay to investigate the mechanism through which the distant NF-κB binding sequences regulate the FcRn gene. Intact nuclei isolated from live THP-1 cells were cross-linked with formaldehyde to fix segments of genomic DNA that are in close physical proximity (Fig. 8A). The cross-linked DNA was then digested with HindIII (Fig. 8B) and ligated at a low concentration of DNA, and the ligated DNA was then analyzed by PCR using primers in all possible combinations (1F plus 2R, 1F plus 3R, 1F plus 4R, and 1F plus 5R, where F is forward and R is reverse) (Fig. 8B). The appearance of a positive PCR product signals successful ligation and DNA looping.
FIGURE 8.
3C analysis of interaction between the promoter and the downstream human FcRn gene. A, Schematic representation of the 3C technology in analyzing human FcRn gene regulation. Chromatin from formaldehyde-fixed cells was digested with a restriction enzyme (E) and diluted for “intramolecular” ligation. Purified DNA fragments were assayed by PCR. PC, Protein complex ; F and R, forward and reverse primer, respectively. B, Locations of HindIII sites and PCR primers in the 15-kb FcRn gene are shown. Positions of the six HindIII restriction sites (perpendicular bars) from the transcriptional start site of the FcRn gene are indicated. HindIII sites (1575/1584) are spaced by only 9 bp and are considered as one site in our assay. Arrows in the HindIII fragments represent the primers used. Exons are shown as boxes and intron numbers are shown as roman numerals. C, The 3C assay. THP-1 cells were mock treated (lanes 2 and 4) or stimulated by TNF-α (50 ng/ml) for 20 min (lanes 3 and 5). Chromatin material was cross-linked and digested with excessive amounts of HindIII, ligated, and amplified by PCR. Primer 1F was the anchor primer, and the others amplified from 5′ to 3′. In lane 1 random ligation control templates were generated by amplifying the genomic DNA fragment with primers that amplify across the HindIII sites. Equimolar amounts of five different PCR products were mixed and digested with HindIII overnight at 37°C. Ligated DNA was used to generate control PCR products by using a combination of the primer pairs listed in Table II to monitor the efficiency of ligation. HindIII-digested chromosomal DNA was used as template for PCR to examine the efficiency of HindIII digestion (lanes 2 and 3). Ligated templates after dilution were used to PCR amplify after HindIII digestion and ligation (lanes 4 and 5). Input DNA was used as an internal control for PCR. D, Sequence analysis of PCR products from 3C analysis. PCR products were sequenced to confirm the fidelity of HindIII digestion and ligation. The HindIII sites were underlined in ligated products.
As shown in Fig. 8B, the primer H1F (Table II), which anneals to the promoter region sequence of the FcRn gene, was used as an anchor primer to pair with other test primers (Table II) in the HindIII-digested and ligated fragments. In Fig. 8C, PCR from three primer pairs (1F plus 1R, 1F plus 2R, and 1F plus 5R) yielded positive products (Fig. 8C, lanes 4 and 5). The PCR products from the primer pairs 1F plus 2R and 1F plus 5R were sequenced to confirm the fidelity of ligations and PCR amplifications (Fig. 8D). The primer 1F plus 1R produced positive products from the ligated DNA template in both stimulated and unstimulated cells (Fig. 8C, lanes 4 and 5). To exclude the possibility that the observed PCR products were a result of random collisions as the result of the inherent flexibility of chromatin (21, 22) and were independent of HindIII digestion, we also analyzed unligated samples after HindIII treatment overnight (Fig. 8C, lanes 2 and 3). We failed to detect corresponding bands amplified by PCR from the nonligated DNA template. To monitor the differences in PCR amplification and ligation efficiencies, an additional control was used by mixing all restriction fragments from HindIII-digested PCR fragments (1F plus 1R, 2F plus 2R, 3F plus 3R, 4F plus 4R, and 5F plus 5R) in equimolar amounts. After ligation at a high DNA concentration, all possible ligation products were present in the sample and amplified by PCR (Fig. 8C, lanes 2 and 3). Taken together, we conclude that there were specific looping interactions between NF-κB binding sequences and FcRn promoters.
TNF-α enhanced bidirectional transport of IgG in polarized intestinal epithelial monolayers
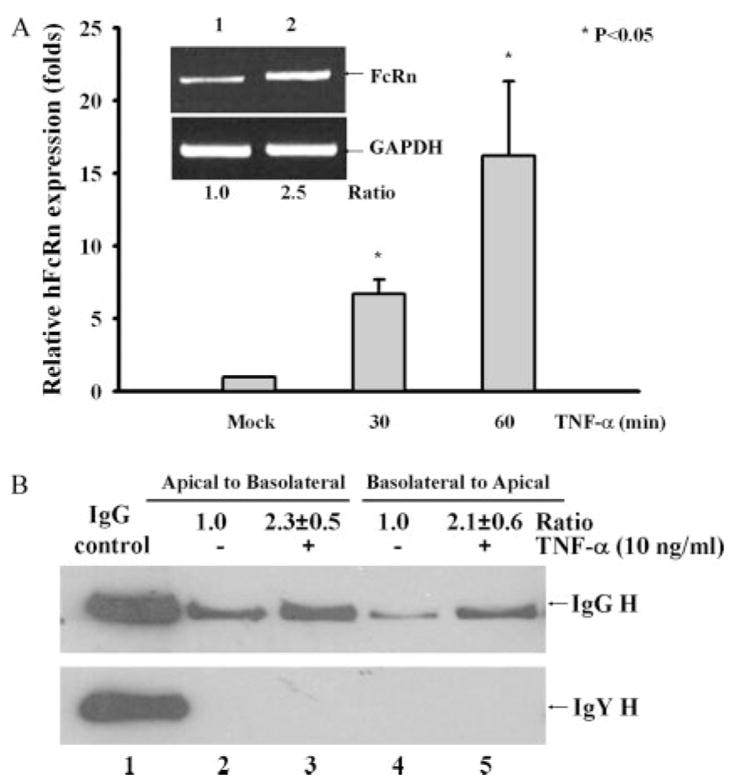
The FcRn protein has been shown to transport IgG bidirectionally, namely from the apical to the basolateral side or vice versa (25). In addition, NF-κB is a crucial modulator of innate immunity in intestinal epithelial cells induced by bacterial infection (33). We therefore address the possibility that NF-κB signaling for the regulation of human FcRn expression in intestinal epithelial cells would influence the regulation of IgG transcytosis. First, we established the enhancement of FcRn expression in Caco-2 intestinal epithelial cells by TNF-α stimulation as assessed by real-time RT-PCR (Fig. 9A) and verified by semiquantitative RT-PCR (Fig. 9A, inset). In our transport experiment, after adding human IgG to the apical or basolateral surface of a Caco-2 cell monolayer we assessed the IgG transported to the opposite basolateral or apical chamber, respectively. As expected, after 1 h at 37°C intact human IgG applied to the apical or basolateral side was transported across this monolayer. The IgG H chain was detected in medium incubated at 37°C (Fig. 9B, upper panel). Importantly, IgG transport was enhanced ~2.3-fold in the apical to basolateral (Fig. 9B, lane 3) direction or 2.1-fold in the basolateral to apical (Fig. 9B, lane 5) direction by TNF-α stimulation in comparison to the mock-treated monolayer (Fig. 9B, lanes 2 and 4). Chicken IgY is structurally similar to human IgG but does not bind to human FcRn (25). However, chicken IgY influx in both directions was absent when monolayers were incubated at 37°C (Fig. 9B, bottom panel), suggesting that transepithelial flux of IgG did not occur by passive diffusion through intercellular tight junctions or monolayer leaks. Therefore, we conclude that NF-κB signaling, at least by TNF-α, can augment the IgG transport across the polarized epithelial cells.
FIGURE 9.
Effects of TNF-α stimulation on the IgG transcytosis. A, Quantitative real-time RT-PCR analysis of FcRn mRNA in Caco-2 treated with TNF-α (10 ng/ml) for the indicated times or left untreated (mock). Data, normalized to GAPDH, are the means of three independent experiments. *, p < 0.05. Inset is a representative result for RT-PCR analysis of FcRn expression in Caco-2 left untreated (lane 1) or treated with TNF-α for 20 min (lane 2). Ratios of FcRn to GAPDH are shown as indicated. B, Caco-2 cells (5 × 104/well) were grown in a 12-well transwell plate. When the resistance of the monolayer reached 250 –300 ohm/cm2, cells were stimulated with or without TNF-α for 30 min. Cells were loaded with human IgG (top panel) or chicken IgY (bottom panel) (1 mg/ml) at 4°C in either the apical (lanes 2 and 3) or basolateral (lanes 4 and 5) chamber. Lane 1 represents an IgG (top panel) or IgY (bottom panel) H chain. Cells were warmed to 37°C to stimulate transcytosis and the medium was collected from the nonloading compartment 1 h later and subjected to Western blotting/ECL analysis. The results are representative of at least three independent experiments. Band intensities of IgG H were compared by densitometry against IgG transported from mock-stimulated cells.
Discussion
Activation or inhibition of the NF-κB signaling pathway can regulate human FcRn expression. We used several experimental approaches and provided data to strongly implicate NF-κB as a regulator for FcRn expression. First, proinflammatory cytokines, TNF-α as well as TLR ligands, are well known to activate the NF-κB signaling pathway. Our results showed that stimulation by TNF-α, IL-β, CpG, and LPS augmented FcRn expression in the THP-1 and HT-29 cell lines or in freshly isolated human monocytes at both the mRNA and the protein levels (Figs. 1 and 2). These results suggest that the FcRn gene might be coordinately induced through a common signaling pathway, perhaps NF-κB -mediated, by these stimuli. This result is in concordance with studies showing that TNF-α induces MHC class I (34) and polymeric IgA receptor (26, 35) expression. Interestingly, the activation dynamics of FcRn expression exhibited an oscillatory behavior when cells were stimulated by TNF-α but a lengthened period when stimulated by LPS treatment (data not shown). This observation is in agreement with the NF-κB expression patterns induced by TNF-α and LPS (36). Furthermore, FcRn expression induced by TNF-α was strongly counteracted by the NF-κB -specific inhibitor CAPE in the THP-1 (Figs. 3, A and B) and HT-29 (data not shown) cell lines. This was corroborated by the fact that overexpression of the dominant negative IκBα (S32A/S36A) almost completely abrogated FcRn transcriptional activation induced by TNF-α (Fig. 3C). This complementary experiment lessens the concern of specificity or toxicity of the chemical inhibitor. Interestingly, overexpression of either NF-κB or IκBα did not affect FcRn basal expression in the absence of TNF-α stimulation (data not shown). It is possible that TNF-α treatment may modify the chromatin structure, allowing NF-κB or IκBα to become more accessible to its binding site. It has been shown that TNF-α stimulation can remodel chromatin by histone acetylation (30, 37). More importantly, TNF-α also induces the binding of NF-κB to κB-like sequences in the promoter of the human β2m gene (38). Therefore, the activation of NF-κB by TNF-α and possibly other NF-κB activators could coordinately up-regulate the expression of both the FcRn and the β2m genes. This balanced regulation of both genes through an NF-κB -mediated signaling pathway may have functional significance because the noncovalent association of the FcRn H chain and β2m is critical for FcRn to exit the endoplasmic reticulum (19).
NF-κB binding sequences were identified in the intronic regions of the human FcRn gene. It is unclear whether NF-κB regulates FcRn expression directly or through an indirect mechanism by inducing other transcription factors that, in turn, could bind to the FcRn gene. FcRn mRNA exhibited rapid kinetics, usually within 20 –30 min, in response to stimuli in our experiments. Our inhibition experiment with CHX further showed that the induction of FcRn mRNA was mediated without newly synthesized protein factors. In fact, three NF-κB binding sequences were mapped in our ChIP experiment to the second and fourth introns of the human FcRn gene (Fig. 4). This result was further confirmed by binding of the NF-κB p65/p50 heterodimer or the p50/p50 homodimer to a synthesized NF-κB DNA oligonucleotide covering the NF-κB binding sequences in the EMSA method (Fig. 5). This is in concert with the observed TNF-α -induced expression of other genes through intronic NF-κB -dependent mechanisms (30, 31, 35, 38). Notably, a large number of studies identify NF-κB binding sequences within noncoding introns (26, 31, 39–41). The data presented here extend this concept and support an inducer-specific role for intronic regulatory elements in the transcription of human FcRn. Several interesting aspects should be perceived. First, the NF-κB binding sequences identified in the introns of human FcRn were not completely conserved, based on the NF-κB consensus binding sequence (Fig. 4B). However, the apparent binding sequence for NF-κB is very broad and displays a certain degree of degeneration (42). Second, multiple NF-κB isoforms can be detected in nuclear extracts of activated cells. These isoforms may compensate for each other and/or induce gene expression in a sequence-dependent manner. In our experiments, we failed to detect either p52 or RelB in the complex by supershift (Fig. 5). Therefore, the predominant NF-κB isoform in the regulation of human FcRn is the p50/p65 heterodimer or the p50/p50 homodimer.
Human FcRn is constitutively expressed in cells that do not have NF-κB activated. The inducible binding of NF-κB p50/p65 (Figs. 4 and 5) suggested a role for these NF-κB binding sequences in regulating FcRn expression. However, the relative importance of the three NF-κB binding sequences is not clear. Their function may be, to some extent, different. Our EMSAs demonstrated that p50/p50 homodimers and p65/p50 heterodimers constituted NF-κB complexes that bound to the intronic sequence (+1140; Fig. 5A). We failed to identify a p50/p50 homodimer bound to either NF-κB sequence in the fourth intron of the human FcRn gene in THP-1 cells (Fig. 5, B and C). Most interestingly, the NF-κB binding pattern was changed in Fig. 5A, at least in some population of stimulated cells, from p50/p50 to p65/p50 binding following TNF-α stimulation (Fig. 5A). This shift implies the NF-κB binding sequence in intron 2 (+1140) may play a dual role in regulating FcRn expression, namely repressing FcRn expression in un-stimulated cells but activating the expression of human FcRn in stimulated cells. It has been well documented that the p50/p50 homodimer is implicated in the inhibition of NF-κB-regulated gene expression (13). This is due to the fact that p50 lacks a classic activation domain. However, p50/p50 can also lead to a transcriptional activation through other mechanisms (43). Our results supported the repression model because mutation of the NF-κB binding sequence in FcRn intron 2 (+1140) increased the activity of the human FcRn promoter to drive the luciferase expression in unstimulated cells (Fig. 6B). Overall, the NF-κB binding sequence in FcRn intron 2 may demonstrate a transcriptional balance between the p65/p50 and p50/p50 dimers that ultimately would determine the nature and level of FcRn gene expression. Therefore, a mechanism may be determined involving the competitive binding of inhibitory (p50/50) and activatory (p50/65) NF-κB isoforms to the κB sequence in the FcRn. However, the functional significance of these NF-κB binding sequences in regulating FcRn expression in vivo merits further investigation.
The intronic location of the NF-κB binding sequences leads to an intriguing question about how these regulatory elements communicate over the ~12-kb distance to influence the FcRn promoter. In general, the packaging of eukaryotic DNA into the nucleosome and higher order structures of chromatin allow for the possibility that the distant regulatory regions are physically close enough for direct interaction or communication through bridging molecules. For example, the promoter region of Ig κ alleles in plasmacytoma cells exhibits a variety of interactions with downstream chromatin segments over 22 kb (24). In addition, Th2-type cytokine genes IL-4, IL-5, and IL-13 can be coordinately regulated by the recently described Th2 locus control region and promoters of each cytokine gene, which are separated by >120 kb (32). It is tempting to speculate that the transcriptional factors binding to the proximal region of the FcRn promoter interact functionally with NF-κB dimer bound to the intronic sequences after induction by stimuli such as TNF-α or TLR ligands. In support of this, intronic NF-κB binding sequences appeared to interact with FcRn promoter-specific DNA elements in our 3C experiments (Fig. 8), and these NF-κB binding sequences maximally activated luciferase gene expression in an FcRn promoter-dependent (Fig. 6) or -independent manner (Fig. 7B). Thus, our 3C experiment suggests that a protein complex might be formed between an FcRn intron and an FcRn promoter region via a chromatin looping mechanism.
NF-κB per se is clearly involved in the transcriptional regulation of many genes. Its activity can be modulated significantly by factors that bind to motifs adjacent to, overlapping with, or distant from that of NF-κB binding sequences (13, 14, 17). In this regard, Sp1 elements may be strong candidates because they are often found in the enhancers or promoters of NF-κB -regulated genes (30, 44, 45). Additional studies have suggested that NF-κB dimers can act synergistically with NF-IL-6, AP-1, and Ets transcriptional factors to influence gene regulation (46–48). Most importantly, the 5′-proximal promoter region of human and rodent FcRn shares numerous putative consensus sequences that are recognized by Sp1, AP-1, Ets, and NF-IL6 (18, 49–51). These protein-protein interactions may be involved in mediating the transcriptional regulation of the FcRn gene in response to stimuli and can functionally cooperate to elicit maximal activation of the promoter. Further studies are needed to determine whether and how NF-κB and other transcriptional factors in the introns and promoter cooperatively regulate FcRn expression.
What might be the biological significance of the regulation of FcRn by NF-κB signaling via intronic binding sequences? NF-κB is activated by the exposure of cells to many physiological and nonphysiological stimuli (13). Thus, the regulation of FcRn expression in vivo likely involves the coordinated action of numerous modulatory factors. Tight control of FcRn may be especially important because FcRn plays a critical role in maintaining IgG homeostasis (6, 8). This function results in the maintenance of much higher serum concentrations of immunoprotective IgG. However, FcRn also extends the life span of pathogenic or autoimmune IgG, potentially promoting the progression of autoimmune diseases (11, 12). Humans with lupus have been shown to have elevated TNF serum levels (52) that could exacerbate their condition based on these findings. Therefore, the level of FcRn expression may be directly coupled to the pathogenesis and treatment of autoimmune diseases. NF-κB-based regulation of FcRn expression may have certain advantages. Because the promoter of the IκBα gene contains NF-κB binding sites, NF-κB is able to autoregulate the transcription of its own inhibitor (53). As a result, the NF-κB activation of gene expression is transient in nature. Therefore, this autoregulatory control of NF-κB and IκBα expression may, in turn, maintain FcRn expression and be the basis of IgG homeostasis. Additionally, FcRn transports normal or neutralizing IgG across polarized epithelial cells, potentially “seeding” neonatal and mucosal immunity (25). FcRn expression elevated from the basal level by cytokines or TLR ligands, or during mucosal infections, could facilitate a local immune response and/or promote the transport of IgG to mucosal surfaces and thereby allow the rapid eradication of infectious agents. From our findings, one might speculate whether cytokines or TLR ligands inducing the expression of the FcRn receptor might lead to the enhancement of IgG transport. Our results showed that TNF-α functionally enhanced the IgG transport in the intestinal Caco-2 cell line (Fig. 9). This result is further supported by the fact that the activated macrophages are a major source of TNF-α in intestinal inflammation (54). By examining the molecular mechanisms by which NF-κB regulates FcRn expression using human cell lines or freshly isolated cells, our studies may contribute toward the general understanding of FcRn-mediated mucosal immunity and IgG-mediated autoimmune diseases. The in vitro results described in the present study are likely paralleled by in vivo events when NF-κB activation causes enhanced FcRn-mediated transport and protection of IgG. These questions are being investigated further.
In summary, the unusually long intron of the FcRn gene contains sequences that bind either p65/p50 heterodimers or p50/p50 homodimers of the NF-κB transcription factors. The presence of NF-κB binding sequences located in distant intronic regions suggests that NF-κB complexes may play an important role in the regulated expression of FcRn, possibly in cooperation with other transcriptional elements in the FcRn promoter. Because the FcRn protein may exert both beneficial and detrimental effects in a variety of infectious and autoimmune diseases, FcRn biosynthesis may be under the control of multiple complex regulatory mechanisms in response to an extracellular stimulus. Understanding the complex regulation of this critically important receptor will require additional studies both in vivo and in vitro.
Acknowledgments
We thank Drs. David M. Mosser, Shanjin Cao, Wenxia Song, and Xia Zhang for critically reading the manuscript. We acknowledge receipt of the HT-29, Caco-2, and THP-1 cell lines from Dr. Richard S. Blumberg, the pGL3-luciferase plasmid from Dr. Gerald Crabtree, and the IκB (S32A/S36A) plasmid from Dr. Genhong Cheng. We thank Dr. Haichen Song, Kumar Kadavil, and Yun Yun for technical help. We also acknowledge the helpful editing of the manuscript by Ireen Dryburgh-Barry.
Footnotes
This work was in part supported by the faculty start-up package and Maryland Agricultural Experiment Station competitive grants from the University of Maryland (to X.Z.), National Institutes of Health Grants AI67965 and AI65892 (to X.Z.) and DK56597, and a grant from The Alliance for Lupus Research (to D.C.R.).
Abbreviations used in this paper: FcRn, neonatal Fc receptor; β2m, β2-microglobulin; CAPE, caffeic acid phenethyl ester; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; 3C, chromosome conformation capture.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 2.Nimmerjahn F, Bruhns P, Horiuchi K, Ravetch JV. FcγRIV: a novel FcR with distinct IgG subclass specificity. Immunity. 2005;23:41–51. doi: 10.1016/j.immuni.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184– 187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 4.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 5.Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I- related receptor FcRn. Annu Rev Immunol. 2000;18:739–766. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. J Exp Med. 2003;197:315–322. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brambell FW, Hemmings WA, Morris IG. A theoretical model of γ-globulin catabolism. Nature. 1964;203:1352–1354. doi: 10.1038/2031352a0. [DOI] [PubMed] [Google Scholar]
- 8.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, Petkova S, Avanessian L, Choi EY, Shaffer DJ, et al. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J Immunol. 2003;170:3528–3533. doi: 10.4049/jimmunol.170.7.3528. [DOI] [PubMed] [Google Scholar]
- 9.Zhu X, Meng G, Dickinson BL, Li X, Mizoguchi E, Miao L, Wang Y, Robert C, Wu B, Smith PD, et al. MHC class I-related Fc receptor for IgG is functionally expressed in monocytes, intestinal macrophages, and dendritic cells. J Immunol. 2001;166:3266–3276. doi: 10.4049/jimmunol.166.5.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petkova SB, Akilesh S, Sproule TJ, Christianson GJ, Al Khabbaz H, Brown AC, Presta LG, Meng YG, Roopenian DC. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18:1759–1769. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 11.Akilesh S, Petkova S, Sproule TJ, Shaffer DJ, Christianson GJ, Roopenian D. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J Clin Invest. 2004;113:1328–1333. doi: 10.1172/JCI18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li N, Zhao M, Hilario-Vargas J, Prisayanh P, Warren S, Diaz LA, Roopenian DC, Liu Z. Complete FcRn dependence for intravenous Ig therapy in autoimmune skin blistering diseases. J Clin Invest. 2005;115:3440–3450. doi: 10.1172/JCI24394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, I, Verma M. NF-κB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 14.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 15.Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621– 663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 18.Mikulska JE, Simister NE. Analysis of the promoter region of the human FcRn gene. Biochim Biophys Acta. 2000;1492:180– 184. doi: 10.1016/s0167-4781(00)00068-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Peng J, Raychowdhury R, Nakajima A, Lencer WI, Blumberg RS. The heavy chain of neonatal Fc receptor for IgG is sequestered in the endoplasmic reticulum by forming oligomers in the absence of β2m association. Biochem J. 2002;367:703–714. doi: 10.1042/BJ20020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X, Peng J, Chen D, Liu X, Ye L, Iijima H, Kadavil K, Lencer WI, Blumberg RS. Calnexin and ERp57 facilitate the assembly of the neonatal Fc receptor for IgG with β2-microglobulin in the endoplasmic reticulum. J Immunol. 2005;175:967– 876. doi: 10.4049/jimmunol.175.2.967. [DOI] [PubMed] [Google Scholar]
- 21.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 22.Eivazova ER, Aune TM. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc Natl Acad Sci USA. 2004;101:251–256. doi: 10.1073/pnas.0303919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickinson BL, Badizadegan K, Wu Z, Ahouse JC, Zhu X, Simister NE, Blumberg RS, Lencer WI. Bidirectional FcRn-dependent IgG transport in a polarized human intestinal epithelial cell line. J Clin Invest. 1999;104:903–911. doi: 10.1172/JCI6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schjerven H, Brandtzaeg P, Johansen FE. A novel NF-κB/Rel site in intron 1 cooperates with proximal promoter elements to mediate TNF-α -induced transcription of the human polymeric Ig receptor. J Immunol. 2001;167:6412– 6420. doi: 10.4049/jimmunol.167.11.6412. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 28.Hmama Z, Nandan D, Sly L, Knutson KL, Herrera-Velit P, Reiner NE. 1α, 25-dihydroxyvitamin D3-induced myeloid cell differentiation is regulated by a vitamin D receptor-phosphatidylinositol 3-kinase signaling complex. J Exp Med. 1999;190:1583–1594. doi: 10.1084/jem.190.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natarajan K, Singh S, Burke TR, Grunberger D, Aggarwal BB. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-κB. Proc Natl Acad Sci USA. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z, Boekhoudt GH, Boss JM. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J Biol Chem. 2003;278:23570–23578. doi: 10.1074/jbc.M303431200. [DOI] [PubMed] [Google Scholar]
- 31.Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, Costas MA, Zwirner NW. NF-κB regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J Immunol. 2004;173:5583–5590. doi: 10.4049/jimmunol.173.9.5583. [DOI] [PubMed] [Google Scholar]
- 32.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 33.Elewaut D, DiDonato JA, Kim JM, Truong F, Eckmann L, Kagnoff MF. NF-κB is a central regulator of the intestinal epithelial cell innate immune response induced by infection with enteroinvasive bacteria. J Immunol. 1999;163:1457–1466. [PubMed] [Google Scholar]
- 34.Israel A, Le Bail O, Hatat D, Piette J, Kieran M, Logeat F, Wallach D, Fellous M, Kourilsky P. TNF stimulates expression of mouse MHC class I genes by inducing an NF-κB-like enhancer binding activity which displaces constitutive factors. EMBO J. 1989;8:3793–3800. doi: 10.1002/j.1460-2075.1989.tb08556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kvale D, Lovhaug D, Sollid LM, Brandtzaeg P. Tumor necrosis factor-α up-regulates expression of secretory component, the epithelial receptor for polymeric Ig. J Immunol. 1988;140:3086–3089. [PubMed] [Google Scholar]
- 36.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-κB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 37.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF-α promoter in monocytes and macrophages. J Leukocyte Biol. 2003;73:862– 871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 38.Gobin SJ, Biesta P, van den Essen PJ. Regulation of human β 2-microglobulin transactivation in hematopoietic cells. Blood. 2003;101:3058–3064. doi: 10.1182/blood-2002-09-2924. [DOI] [PubMed] [Google Scholar]
- 39.Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Distribution of NF-κB -binding sites across human chromosome 22. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-κB with chromatin: the art of being at the right place at the right time. Nat Immunol. 2005;6:439– 445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 41.Ge B, Li O, Wilder P, Rizzino A, McKeithan TW. NF-κB regulates BCL3 transcription in T lymphocytes through an intronic enhancer. J Immunol. 2003;171:4210– 4218. doi: 10.4049/jimmunol.171.8.4210. [DOI] [PubMed] [Google Scholar]
- 42.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453– 464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Cha-Molstad H, Agrawal A, Zhang D, Samols D, Kushner I. The Rel family member P50 mediates cytokine-induced C-reactive protein expression by a novel mechanism. J Immunol. 2000;165:4592– 4597. doi: 10.4049/jimmunol.165.8.4592. [DOI] [PubMed] [Google Scholar]
- 44.Ping D, Boekhoudt G, Zhang F, Morris A, Philipsen S, Warren ST, Boss JM. Sp1 binding is critical for promoter assembly and activation of the MCP-1 gene by tumor necrosis factor. J Biol Chem. 2000;275:1708–1714. doi: 10.1074/jbc.275.3.1708. [DOI] [PubMed] [Google Scholar]
- 45.Boekhoudt GH, Guo Z, Beresford GW, Boss JM. Communication between NF-κB and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J Immunol. 2003;170:4139– 4147. doi: 10.4049/jimmunol.170.8.4139. [DOI] [PubMed] [Google Scholar]
- 46.Stein B, Baldwin AS, Ballard DW, Greene WC, Angel P, Herrlich P. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-κB p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- 48.Tomaras GD, Foster DA, Burrer CM, Taffet SM. ETS transcription factors regulate an enhancer activity in the third intron of TNF-α. J Leukocyte Biol. 1999;66:183–193. doi: 10.1002/jlb.66.1.183. [DOI] [PubMed] [Google Scholar]
- 49.Kandil E, Noguchi M, Ishibashi T, Kasahara M. Structural and phylogenetic analysis of the MHC class I-like Fc receptor gene. J Immunol. 1995;154:5907–5918. [PubMed] [Google Scholar]
- 50.Jiang L, Wang J, Solorzano-Vargas RS, Tsai HV, Gutierrez EM, Ontiveros LO, Kiela PR, Wu SV, Martin MG. Characterization of the rat intestinal Fc receptor (FcRn) promoter: transcriptional regulation of FcRn gene by the Sp family of transcription factors. Am J Physiol. 2004;286:G922–G931. doi: 10.1152/ajpgi.00131.2003. [DOI] [PubMed] [Google Scholar]
- 51.Tiwari B, Junghans RP. Functional analysis of the mouse Fcgrt 5′ proximal promoter. Biochim Biophys Acta. 2005;1681:88–98. doi: 10.1016/j.bbaexp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Svenungsson E, Fei GZ, Jensen-Urstad K, de Faire U, Hamsten A, Frostegard J. TNF-α: a link between hypertriglyceridaemia and inflammation in SLE patients with cardiovascular disease. Lupus. 2003;12:454– 461. doi: 10.1191/0961203303lu412oa. [DOI] [PubMed] [Google Scholar]
- 53.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 54.Rugtveit J, Nilsen EM, Bakka A, Carlsen H, Brandtzaeg P, Scott H. Cytokine profiles differ in newly recruited and resident subsets of mucosal macrophages from inflammatory bowel disease. Gastroenterology. 1997;112:1493–1505. doi: 10.1016/s0016-5085(97)70030-1. [DOI] [PubMed] [Google Scholar]