Summary
Regulatory inactivation of DnaA is dependent on Hda, a protein homologous to the AAA+ ATPase region of the replication initiator DnaA. When bound to the sliding clamp loaded onto duplex DNA, Hda can stimulate the transformation of active DnaA-ATP into inactive DnaA-ADP. The crystal structure of Hda from Shewanella amazonensis SB2B at 1.75 Å resolution reveals that Hda resembles typical AAA+ ATPases. The arrangement of the two subdomains in Hda (residues 1-174, 175-241) differs dramatically from that of DnaA. A CDP molecule anchors the Hda domains in a conformation which promotes dimer formation. The Hda dimer adopts a novel oligomeric assembly for AAA+ proteins in which the arginine finger, crucial for ATP hydrolysis, is fully exposed and available to hydrolyze DnaA-ATP through a typical AAA+ type mechanism. The sliding clamp binding motifs at the N-terminus of each Hda monomer are partially buried and combine to form an antiparallel β-sheet at the dimer interface. The inaccessibility of the clamp binding motifs in the CDP bound structure of Hda suggests that conformational changes are required for Hda to form a functional complex with the clamp. Thus, the CDP-bound Hda dimer likely represents an inactive form of Hda.
Keywords: Hda, DnaA, RIDA, AAA+ ATPase
Introduction
In Escherichia coli, the initiation protein DnaA, when complexed with ATP (DnaA-ATP), binds to the 9-base pair DnaA boxes within the chromosomal replication origin (oriC) to initiate DNA replication 1; 2. At the beginning of initiation, 20–30 DnaA-ATP proteins bind to the origin and oligomerize into a large nucleoprotein complex, which facilitates melting of the adjacent, AT-rich, 13-base pair, DNA unwinding element (DUE) 3. The initiation of chromosomal DNA replication is tightly regulated to ensure only one replication per cell cycle 4; 5. Three regulation mechanisms have been identified. Firstly, an oriC sequestration mechanism inactivates the newly replicated oriC through binding of the SeqA protein. SeqA has a higher affinity for hemimethylated GATC sites within the oriC. These hemimethylated GATC sites exist in newly synthesized oriC for about one-third of a cell cycle after initiation until the oriC is fully methylated by the DNA-adenine methyltransferase. Secondly, a DnaA titration mechanism limits the availability of the DnaA protein at the origin by diverting DnaA to other binding sites outside the oriC region. For example, a single copy of the datA locus, located 470kb following oriC, could bind usually large number of DnaA molecules. DnaA boxes are also present in the promoter regions of many genes. Thirdly, a mechanism known as RIDA (regulatory inactivation of DnaA) negatively regulates DnaA by converting active DnaA-ATP into inactive DnaA-ADP following the initiation 6; 7; 8. DnaA-ADP cannot induce melting of the origin and, thus, is ineffective for replication initiation 1.
The two essential components of the RIDA system are the sliding clamp of DNA polymerase III (DnaN or β subunit) and Hda (homologous to DnaA, also referred to as IdaB) 9; 10. When the sliding clamp is loaded onto duplex DNA (dsDNA), Hda can promote hydrolysis of DnaA-ATP to DnaA-ADP 11; 12; 13. As a result of RIDA, the level of DnaA-ATP in the cell, which peaks in vivo around the initiation of replication, decreases rapidly after the start of replication 8. The Hda-mediated RIDA process is an important mechanism for preventing over-initiation 14; 15, and hda-deficient cells do, in fact, over-initiate 9; 16. Hda is a dimer in solution and binds to the sliding clamp via its N-terminal sliding clamp binding motif 13; 17. Besides being involved in RIDA, Hda may also have other cellular functions, since it also interacts with plasmid RK2 replication initiation protein TrfA 10; 18. Details of how and where RIDA occurs in the cell are not currently clear. In vitro, the RIDA interaction of DnaA-Hda is dependent on the sliding clamp loaded onto dsDNA 12. In vivo, RIDA may occur on the post-replication sliding clamps that remain on the lagging strands after synthesis of Okazaki fragments 13. It has also been suggested that Hda binds to the sliding clamp at the replication fork and RIDA occurs at the fork 5. Another possibility is that Hda could interact with the DnaA-ATP origin complex after opening of the DUE 4; 6; 19.
A typical Hda is about 250 residues in length. It is classified as an AAA+ (ATPases associated with diverse cellular activities) ATPase due to its sequence homology to the AAA+ ATPase region of DnaA (domains III and IIIb). Recent in vitro reconstitution of E. coli RIDA activity revealed that a conserved arginine in the box VII motif of Hda is required to stimulate ATP hydrolysis in DnaA and suggests that a conserved AAA+ type interaction takes place between Hda and DnaA during RIDA 13.
The AAA+ superfamily of ATPases are found in all kingdoms of living organisms. They act as motors or switches and control a wide range of biological processes 20; 21; 22; 23; 24. Structural characterization of AAA+ modules has revealed that they consist of two domains: an N-terminal P-loop NTPase homologous domain (the base domain) and a smaller C-terminal helical bundle domain (the lid domain). The nucleotide binding site is located at the domain interface (see section “Nucleotide binding site”). AAA+ proteins generally form ring-shaped oligomeric assemblies, where one AAA+ subunit inserts residues from a conserved motif (box VII) into the ATP-interaction site of its adjacent subunit. ATP hydrolysis is enabled once the bipartite nucleotide-binding pocket is formed and contact is made between a conserved arginine (‘arginine finger’) from the box VII motif in one subunit with the γ-phosphate of the bound nucleotide in the adjacent monomer. ATP hydrolysis often results in intra- and inter-subunit conformational changes, which can be employed in many cellular events 24. The AAA+ ATPases are key components of DNA replication and repair complexes, such as the replication initiator DnaA 25. The opening of DUE by DnaA does not involve ATP hydrolysis, since a non-hydrolysable ATP analog can functionally replace DnaA-ATP 1. Thus, it appears that ATP hydrolysis in DnaA is employed in its inactivation.
Recent structures of inactive DnaA with ADP 26; 27, active DnaA with an ATP analog 19, the DNA binding domain of DnaA complexed with DNA 28, archaeal Orc1-DNA complex 29 and the heterodimeric Orc1-DNA complex 30, have greatly enhanced our understanding of the role of AAA+ proteins in the initiation of replication 4. Here, we present the crystal structure of Hda from Shewanella amazonensis, the essential component of RIDA. S. amazonensis is likely to share a similar RIDA system with E. coli based on the similarity in the replication origins 31 and the presence of highly homologous DnaA and Hda proteins (sequence identities of 81% and 48%, Figure 1a). Thus, the crystal structure of Hda presented here provides an excellent framework for understanding the corresponding experimental results in E. coli and offers valuable insights into the regulation of DnaA, as well as interactions between Hda, the sliding clamp and DnaA.
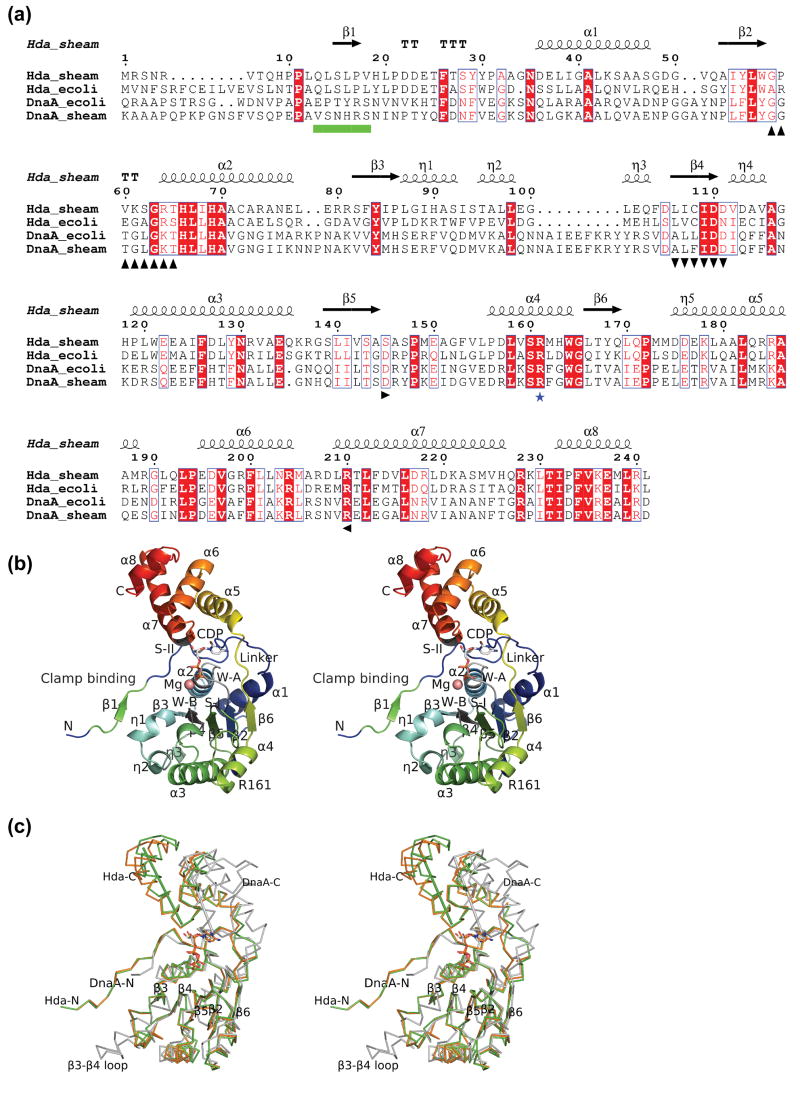
Figure 1.
Structure of Hda from Shewanella amazonensis. (a) Sequence alignment between Hda and DnaA (ATPase region) of E. coli and S. amazonensis. The Hda of S. amazonensis (Hda_sheam) is highly homologous to Hda of E. coli (Hda_ecoli) with 48% sequence identity and to the DnaA ATPase regions of E. coli (residues 107-365) and S. amazonensis (residues 95-355) with sequence identity of 24% and 21% respectively. The secondary structure elements and sequence numbering of the S. amazonensis Hda structure are shown in the top row. The conserved Walker A, Walker B, sensor I, sensor II motifs are marked in black with up, down, right and left triangles in the bottom row. The ‘arginine finger’ of box VII is shown with a blue star. The sliding clamp binding motif is denoted as green boxes. (b) Structure of an Hda monomer with bound CDP and magnesium. The Walker A (W-A, or P-loop), Walker B (W-B), sensor I (S-I), sensor II (S-II), the sliding clamp binding and box VII (‘arginine finger’) motifs are labeled corresponding to a). (c) Comparison of Hda monomers (orange and green) with the DnaA III and IIIb (PDB id 2hcb, gray). The Hda and DnaA are superimposed based on their respective NTPase domains (i.e. I and III). The sensor II helices of Hda (210–225) and DnaA (276–289) are shown as rods to highlight the significant difference in the domain Ib orientation.
Results and Discussion
Overall structure
The structure of Hda from Shewanella amazonensis, complexed with CDP and magnesium (Fig1a–b), was determined to 1.75Å resolution by multi-wavelength anomalous dispersion (MAD) with selenomethionine-labeled protein (Table I). The structure was refined to Rcryst and Rfree of 16.6 and 20.1% respectively. The model displays good geometry with 99.3% favorable main chain torsion angles (no Ramachandran outliers) and 98.4% favorable side chain rotamers according to MOLPROBITY 32. The final model contains a dimer (A12-241, B11-241) in which each monomer is complexed with a CDP and a magnesium ion. In addition, 571 solvent molecules (1 thiocyanate, 14 ethylene glycols, 1 sodium, 1 chloride, and 554 waters) were modeled. The main chain for residues A0-11, B0-10, A32-33 and the side chains for residues A12, A36, A134, A150, B11-12, B81, B102-103, B136, B192, B221 and B229 were disordered and were not included in the final model.
Table I.
Data collection and refinement statistics
| Data collection | λ1 MADSe | λ2 MADSe | |
|---|---|---|---|
| Wavelength (Å) | 0.9793 | 0.9184 | |
| Resolution range (Å) | 29.6-1.75 | 29.5-1.75 | |
| Number of observations | 204,590 | 204,972 | |
| Number of unique reflections | 55,436 | 55,361 | |
| Completeness (%) | 99.7 (99.7)a | 99.7 (100)a | |
| Mean I/σ(I) | 9.1 (1.8)a | 8.3 (1.6)a | |
| Rsym on I (%) | 0.10 (0.64)a | 0.11 (0.81)a | |
| Model and refinement statistics | |||
| Resolution range (Å) | 29.6-1.75 | Data set used in refinement | λ1 MADSe |
| No. of reflections (total) | 55,402b | Cutoff criteria | |F|>0 |
| No. of reflections (test) | 2,813 | Rcryst | 0.166 |
| Completeness (% total) | 99.5b | Rfree | 0.201 |
| Stereochemical parameters | |||
| Restraints (RMS observed) | |||
| Bond angle (°) | 1.47 | ||
| Bond length (Å) | 0.014 | ||
| Average isotropic B-value (Å2) | 24.1 | ||
| ESU based on Rfree (Å) | 0.10 | ||
| Protein residues / atoms | 461 / 3,682 | ||
| Ligand and water molecules | 575 | ||
Highest resolution shell (1.80-1.75) in parentheses.
Typically, the number of unique reflections used in refinement is less than the total number that were integrated and scaled. Reflections are excluded due to systematic absences, negative intensities, and rounding errors in the resolution limits and cell parameters.
ESU = Estimated overall coordinate error.
Rsym = Σ|Ii−<Ii>| / Σ|Ii|, where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Rcryst = Σ| |Fobs|−|Fcalc| | / Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.1% of the total reflections chosen at random and omitted from refinement.
The Hda monomer (Figure 1b) displays a molecular architecture typical of AAA+ ATPases 22; 23. The N-terminal domain I (residues 1-174, the base domain) has a RecA-like fold containing the Walker A (the P-loop), Walker B and sensor I motifs. An extended N-terminal loop contains a short strand (β1), which has previously been shown to bind to the sliding clamp 13. The C-terminal domain Ib (residues 175-241, the lid domain) is a short four-helix bundle (α5–α8), which contains the sensor II motif. The two domains of Hda are similar to domain III (rmsd 1.57 Å for 138 Cα) and domain IIIb (rmsd 1.56 Å for 48 Cα) of DnaA (PDB ids 2hcb and l8q) when compared separately. The most significant difference between domain I of Hda and domain III of DnaA is the nature of the insertion between β3 and β4 (the ‘steric wedge’ in DnaA 19) (Figure 1c). This region often varies among different AAA+ ATPases and is used to classify them 21; 22. In Hda, this insertion is shorter than most others, and contains three 310 helices (η1, η2 and η3). Helices η2-η3 are equivalent to the second helix of a prototypical AAA+ base domain 22. The η1 of Hda is shorter than the equivalent helix of DnaA. The Hda insertion η1-η3 is structurally similar to the clamp loaders, such as the clamp loader delta subunit 33. However, Hda does not possess the corresponding long η3-β4 loop found in the clamp loader, which is critical in binding the sliding clamp. Since the β3-β4 insertion resides next to the clamp binding motif (β1), it could play a role in binding the sliding clamp.
The orientation between the base and lid domains varies significantly between DnaA and Hda. A superposition of the base domains of DnaA (PDB id 2hcb) and Hda, reveals that the lid domain of Hda is rotated a further 47° towards its base domain around the principal axis of the bound nucleotide (Figure 1c).
Hda dimer
The crystallographic asymmetric unit contains a homodimer (Figure 2). The two monomers are similar to each other with an rmsd of 0.99 Å for all Cα atoms, with the most significant differences in the orientation of the two C-terminal helices, α7 and α8 (Figure 1c). The Hda dimer resembles a flat rhombic prism, with dimensions of 79 Å × 62 Å × 43 Å, in which the two N-terminal base domains are aligned along the short axis, while the two C-terminal lid domains are aligned along the long axis of the rhombus. The dimer buries a surface area of 2045 Å2 from each monomer (Figure 2). Molecular weights of 52,480 Da and 54,030 Da were determined by two independent runs of analytical size exclusion chromatography in combination with static light scattering (SEC/SLS). Because an Hda monomer has a calculated molecular weight of 27,059.1 Da (mass determined by LC/MS was 27,060.8 Da), SEC/SLS suggested that it forms a dimer in solution, consistent with its packing in the crystal. The bulk of the dimer interface is formed from interactions between the two base domains. These interactions are clustered around β1 and η1. The positions of the two N-terminal extended loops (11–21) are swapped, with the N-terminal loop from one subunit making extensive contacts with the β3, η1 and η2 from the second subunit. Together, these N-terminal loops form a short anti-parallel β-sheet, which contributes 25% of the buried surface area to the dimer interface. Additional dimer interactions are between the lid domain of one monomer and the base domain of the second monomer. Specifically, the N-terminal portion of α3 of the base domain is docked between the α7 and α8 of the lid domain from the other subunit.
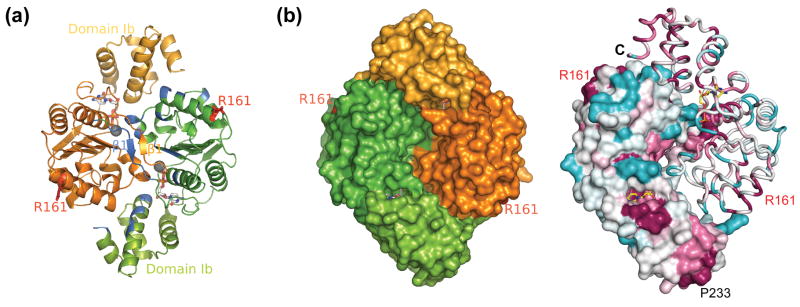
Figure 2.
The Hda dimer. (a) The two monomers are colored orange and green. The residues which are involved in dimer formation are colored blue in one monomer. The two ‘arginine fingers’ (Arg161) in the dimer are shown as red sticks. The bound nucleotides and magnesium ions are shown in sticks and spheres respectively. (b) The surface representation shows the opposite side of the dimer in (a) (left). The right panel shows the Hda dimer colored by sequence conservation. The most conserved residues are shown in magenta, while the least conserved residues are in cyan.
The Hda homodimer represents a novel mode of AAA+ oligomerization. AAA+ proteins frequently form hexameric rings in a ‘head-to-tail’ manner such that the arginine finger from one subunit can interact with the nucleotide of the adjacent one. However, the two monomers in the Hda dimer are arranged in a ‘head-to-head’ manner. As a result, the dimerization of Hda places the two nucleotide binding sites close to each other, while the two corresponding arginine fingers are located far apart on the perimeter (Figure 2).
The transcription activator NtrC1 is an example of an AAA+ protein that forms a dimer in its inactive form through a swapped N-terminal linker, when its receiver domain is unphosphorylated 34. However, the NtrC1 dimer differs significantly from the Hda dimer. The non-crystallographic two-fold axis in the NtrC1 dimer is parallel to the linker region consisting of two long, anti-parallel helices. The NtrC1 dimer does not involve the base domains of the AAA+ regions. In Hda, the non-crystallographic two-fold axis is perpendicular to the anti-parallel β-sheet in the N-terminal region. In contrast to the critical role of the NtrC1 linker in its dimerization, the N-terminus of Hda does not appear to be essential for dimerization, since HdaΔN26 can still dimerize 13.
Nucleotide binding site
Well-defined electron density at the nucleotide binding site identified a bound cytidine-5′-diphosphate (CDP), that presumably was obtained during protein expression in E. coli (Figure 3a). The possibility of ADP (or GDP) was excluded due to poor fit to the electron density, lack of favorable hydrogen bonds to stabilize the adenosine base, and steric clashes with the protein. Bound CDP is surprising since ADP or ATP was expected based on similarity to DnaA and other AAA+ proteins. However, CTP has previously been shown to bind to ATPases. For example, E. coli DnaA-CTP complex was effective in opening the DUE (but not effective in subsequent steps of the initiation process) 35. The conformation of the CDP is very similar to the nucleotides ADP and AMP-PCP bound to DnaA 19; 26. The cytidine base is stabilized through hydrophobic stacking interactions and three hydrogen bonds with the protein, along with two water-mediated hydrogen contacts (Figure 3b). The binding site clearly favors CDP, and replacement of cytidine by uracil would result in the loss of two hydrogen bonds. These hydrogen bond interactions involve the main chain of Tyr30 and the side chain of Arg185. Since Arg185 is the only side chain involved in hydrogen bonding interaction with the base, it could play a role in specifying the substrate. Arg185 and the cytidine binding pocket are conserved in E. coli and highly conserved in other putative Hda proteins. Thus, it seems likely that other Hda proteins can also bind CDP.
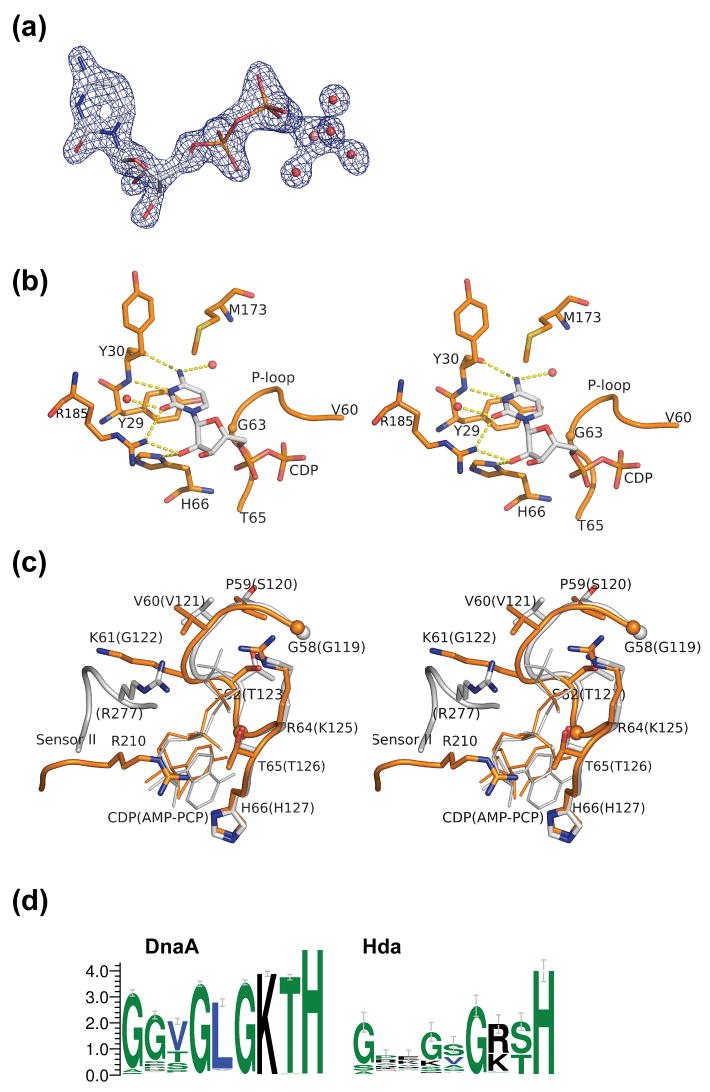
Figure 3.
Nucleotide binding site of Hda. (a) The CDP (shown in sticks) with the experimental density contoured at 2.5σ. The map was calculated after density modification improvement of the initial MAD phases. The conserved magnesium ion (pink), coordinated by waters (red), and Thr65 (not shown) stabilize the β-phosphate of the CDP. (b). Interaction between the protein and the cytidine base and sugar of CDP. (c) Comparison of motifs Walker A and sensor II of Hda (orange) and DnaA (gray). The corresponding residues for DnaA (PDB id 2hcb) are shown in parenthesis. (d). Sequence conservation pattern of the P-loop region of putative DnaA and Hda proteins. The DnaA group of sequences (785 sequences) are selected based on the presence of both a C-terminal DNA binding motif, as well as an ATPase domain. The putative Hda sequences (94 sequences) are selected by full-length homology to E. coli Hda and of length between 210 and 270 residues.
The conserved motifs Walker A, Walker B, sensor I and sensor II found in AAA+ proteins are all present in S. amazonensis Hda. The position of the sensor II motif of Hda is significantly different from DnaA due to the change in orientation of the lid domain Ib (Figure 1c, 3c). The conserved arginine residue in the sensor II motif (e.g. Arg277 in Aquifex aeolicus DnaA) typically interacts with the γ-phosphate of ATP 23. However, the corresponding residue in Hda (Arg210) interacts with Asp24 of the base domain I, as well as theα-phosphate of CDP (Figure 3c). Hda has a variant (58-GPVKSGRT-65) of the Walker A consensus sequence (GXXGXGK[ST]). Lys61 physically occupies the same space as the DnaA box VIII (sensor II) Arg277. The Arg64 side chain interacts with the β-phosphate of CDP (Figure 3c). The conformation of Arg64 is identical to the Lys64Arg mutant of RuvB from Thermotoga maritima 36.
NTP hydrolysis and possible roles of nucleotides
Hda shares many of the structural features found in DnaA and other members of the AAA+ ATPase superfamily, including a conserved nucleotide binding site. However, no experimental evidence is available to suggest that it has NTPase activity. It has been proposed that Hda could form a typical AAA+ ‘head-to-tail’ dimer 13. Such a model is not consistent with the crystal structure determined here. The homodimer of Hda precludes a mechanism through a canonical ‘arginine finger’ type of interaction between two Hda monomers. The CDP binding site is buried within the dimer interface (Figure 2) and, despite a few solvent channels nearby, access to CDP is limited. No space is available to form a bipartite complex similar to other AAA+ ATPases. Significant structural changes, such as dimer dissociation, would be required to achieve NTP hydrolysis.
It seems likely that Hda has lost its ability to bind tightly or hydrolyze efficiently NTP. The presence of Arg64 in the Walker A motif appears to be detrimental to NTP binding and hydrolysis. All structurally known P-loops have lysines at this position. Mutational studies in other AAA+ ATPases have indicated that the conserved lysine in the Walker A motif is essential. For example, Lys to Arg mutants of E. coli RuvB and DnaC are defective in both ATP binding and hydrolysis 37; 38. The requirement of a conserved lysine becomes more apparent when the Walker A motif is analyzed in all pertinent bacterial sequences currently in Pfam PF00308 39. The sequences fall into two groups: one with a C-terminal DNA binding domain (likely DnaA proteins) and one which only has the prototypical DnaA AAA+ module (likely Hda proteins). In the first group, for which ATP is required, the conserved lysine is invariant (Figure 3d). In the second group, both the Walker A motif and the lysine position show greater sequence variability, as do the Walker B and sensor I motifs. The loss of the strictly conserved lysine suggests that the γ-phosphate of NTP is less important for Hda function. Structurally, an overlay of AMP-PCP of DnaA onto the CDP site of Hda indicates that the guanidinium group of Arg64 would clash with the γ-phosphate (Figure 3c) and would, therefore, disrupt NTP binding.
Hda has retained its ability to bind NDP as the residues involved in binding ribose (His65) and α- or β-phosphate (Gly63, Arg210, Arg64) are invariant or highly conserved. A possible role of the CDP is to promote dimer formation and maintain its conformation. The smaller cytidine base may facilitate greater plasticity between the two domains compared to adenosine.
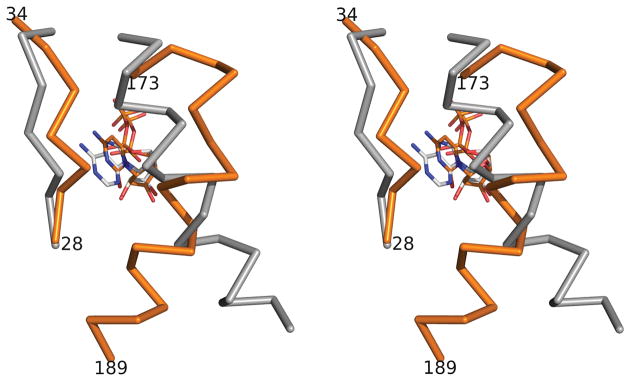
As ATP/ADP is found in all other AAA+ structures and the nucleotide binding pockets are highly conserved in AAA+ proteins22, the possibility that Hda can also bind ADP can not be excluded from this study. If Hda were to bind ADP like DnaA26; 27, it is likely that ADP may affect the conformation of Hda locally or globally. Due to the larger size of the adenosine base, the region between 28 and 34 of Hda must move outward to accommodate it (Figure 4). Changes in this region could propagate to nearby regions, such as the linker region between domains I and Ib, the first helix of domain Ib (α5), as well as the clamp binding motif region. Thus, any association of ADP with Hda could disrupt interactions between domains I and Ib in Hda, and potentially alter the dimerization state or the conformation of the clamp binding region.
Figure 4.
Structural comparisons of the regions around the cytidine/adenosine bases in Hda-CDP (PDB id 3bos, orange) and DnaA-ADP (PDB id 1l8q, gray). The superimposition is obtained by overlapping the base domains of Hda and DnaA.
The sliding clamp binding motif and the clamp interaction
Hda can form a stable functional complex with the sliding clamp in vitro with a stoichiometry of Hda2-clamp or, less likely, Hda4-clamp 12; 13. Previous studies have shown that a conserved hexapeptide motif, QL[SP]LPL, at the N-terminus of E. coli Hda is essential for binding the sliding clamp 13; 17. This motif is also widely conserved in other clamp binding proteins 40.
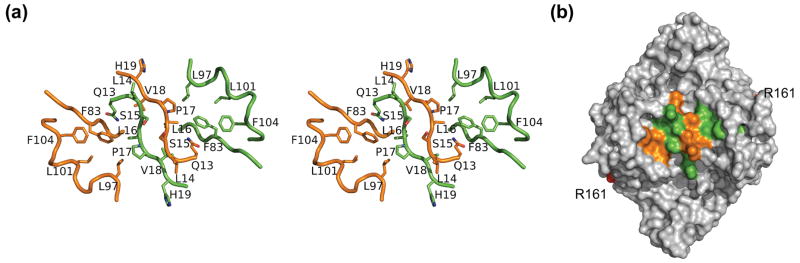
Hda from S. amazonensis possesses a similar conserved sequence motif (13-QLSLPV-18) at its N-terminus, (Figure 1a, b). In the Hda dimer, the clamp binding motifs of each subunit form a short, antiparallel β-sheet at the non-crystallographic twofold axis of the dimer. Mutational studies on this motif 13; 17 have suggested that Gln13, Leu16 and Val18 (equivalent to Gln21, Leu24 and Leu26 of E. coli) are critical for binding the sliding clamp. Interestingly, the side chains of these residues are buried in the dimer interface. The Gln13 from one subunit makes contact with the backbone of the adjacent subunit through two hydrogen bonds (Figure 5a). Leu14, Ser15 and Pro17 in this motif are solvent exposed and, along with conserved residues Leu97, Leu101 and Phe104, located at the β3-β4 insertion (η2 and η3), and Phe83 from β3, form a hydrophobic patch of dimensions 25 Å × 22 Å, with a central ridge and a shallow hole on either side (Figure 5b).
Figure 5.
The clamp binding motifs in the Hda dimer. (a). Close up view of the conserved residues including the clamp binding motif. The orientation of the dimer is the same as the Hda dimer shown in ribbons in Figure 2. (b). Surface representation of the hydrophobic pocket.
Several characterized clamp binding motifs adopt extended conformations and bind to a conserved location between the second and third domain of the β-subunit (Figure 6a) 33; 40; 41; 42. The clamp binding motif of Hda is also likely equivalent to other clamp binding motifs and interacts with the same binding site on the clamp 17; 43. The clamp binding motif of Hda (QL[SP]LPL) is similar in sequence to the Pol IV clamp binding motif (QLVLGL) 41; thus, it is expected that the Hda clamp binding motif will adopt a similar conformation when bound to the clamp. However, the two-fold symmetric clamp binding motifs of the Hda dimer are partially buried and have an unusual conformation (Figure 5). Clearly, they cannot bind the clamp in a canonical manner in their current conformation without significant steric clashes. As a result, the CDP-bound dimer of Hda observed in the crystal structure is likely unable to bind the clamp and, thus, is likely inactive in RIDA, unless the hydrophobic patch of Hda binds the sliding clamp in an unconventional manner.
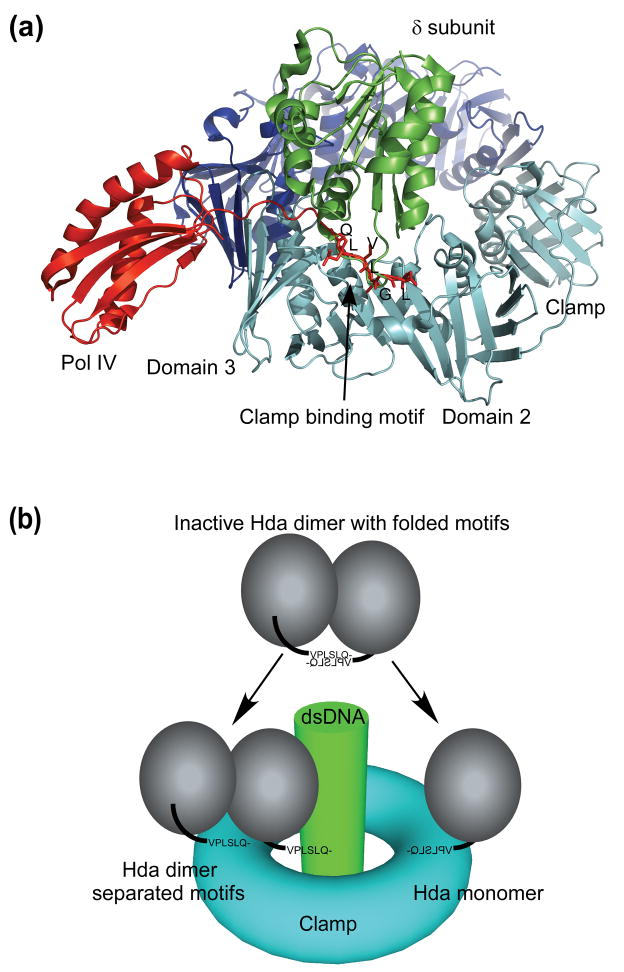
Figure 6.
Proposed model for interaction between Hda and the sliding clamp. (a) The clamp binding motifs of Pol IV (little finger domain, red) and the clamp loader (δ subunit, green) interact with the clamp (cyan and blue) in a conserved fashion. The clamp binding motif of Pol IV (residues 346-351) is also shown in stick model. (b) The clamp binding motifs of Hda (or the whole Hda dimer) dissociate and one of the motifs binds the clamp in a conserved fashion.
Hda must then adopt a different conformation in order to bind to the clamp in a canonical manner. This alternative structure could involve dissociation of the clamp binding motifs from the rest of the dimer to an extended conformation, or the Hda dimer itself may dissociate and interact with the clamp in a monomeric form (Figure 6b). Dissociation of Hda clamp binding motifs (or the dimer) may occur spontaneously in solution. However, deuterium exchange mass spectroscopy (DXMS) experiments have indicated that the clamp binding motifs of the Hda-CDP dimer are not flexible in solution (data not shown), consistent with the crystal structure. Thus, it appears that additional factors are needed to regulate the conformation of the clamp binding motifs of Hda, such as a conformational switch associated with binding of different nucleotides (e.g. ADP or CDP, see above) or a weak intrinsic NTPase activity. The physiological role of the inactive Hda dimer is currently unclear, it may offer additional means to regulate Hda activity so that it only become active when associated with the sliding clamp.
Since the clamp binding motif of Hda is located at its N-terminus rather than at the C-terminus as in Pol IV 41, the Hda molecule will likely be located close to the middle domain of the β-subunit (Figure 6a). It is not clear whether the functional Hda-clamp complex for RIDA involves an Hda monomer or an Hda dimer at each site of the β-subunit (Figure 6b). Although the conserved site on the clamp generally interacts with a monomer 33; 41, Hda was previously shown to interact with the clamp as a dimer 11; 13. However, a recent study suggested a possible monomeric interaction between Hda and the clamp 12. Further biochemical experiments and the complex structure of Hda and the sliding clamp will help resolve such questions.
Arginine finger and Hda-DnaA interaction
In order to utilize the conserved mode of ATP hydrolysis in AAA+ proteins, it is expected that DnaA and Hda form a heterodimeric complex during the RIDA reaction. The ‘arginine finger’, Arg168, of E. coli Hda is critical for the deactivation of ATP-DnaA 13, whereas the corresponding Arg161 of S. amazonensis Hda is located near the middle of a short helix α4 (box VII). The ‘arginine fingers’ are fully exposed to solvent.
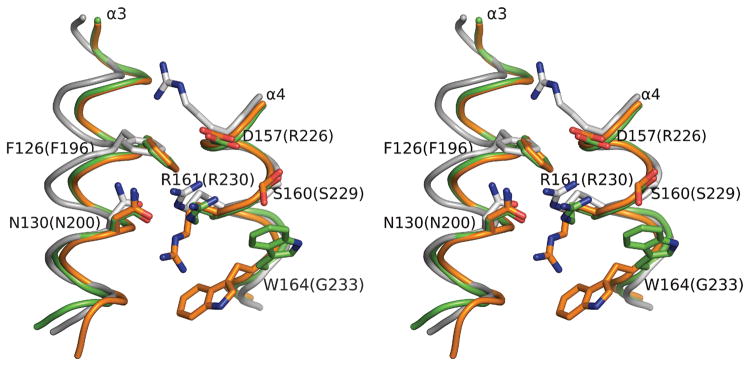
Residues in the immediate surrounding of Arg161 are also highly conserved among Hda, as well as DnaA (Figure 1a, Figure 7). Arg161 is preceded by an acidic (D/E) residue (Asp157) and a smaller polar (S/T) residue (Ser160), and followed by a hydrophobic residue (Trp164). Additionally, two other residues, Phe126 and Asn130, from the neighboring α3 (the central helix) are also highly conserved. Asn130 hydrogen bonds with Arg161 in one of the Hda subunits and both are located on the same helical surface and are fully exposed to solvent. Arg161 and Trp164 are in different conformations in the two Hda monomers, which indicate that these residues are flexible.
Figure 7.
Arginine fingers of Hda. The superimposed conformations of the two arginine fingers (Arg161) in the dimer and surrounding conserved residues of Hda (orange and green). The positional equivalent residues of DnaA (PDB id 2hcb, gray) are also shown. The corresponding residue numbers for DnaA are shown in parenthesis.
In contrast to DnaA, which is present in all bacteria, Hda is predominantly found in proteobacteria, which suggests that Hda originally evolved from DnaA to acquire its regulatory function. Due to the high similarity in both sequence and structure in the arginine finger regions (i.e. Phe126, Asn130, Glu134, Ser160, Arg161, Trp164 and Gly165) of both DnaA and Hda (Figure 1a, Figure 7), it has been proposed that Hda may interact with DnaA in a similar fashion to the DnaA self-assembly 4; 19. Structural studies of A. aeolicus DnaA with ADP and an ATP analog have illustrated a rigid body movement in the two AAA+ sub-domains between the ATP and ADP state 19; 26. Although there is no Hda homolog and likely no Hda-dependent RIDA in A. aeolicus, the structural changes of DnaA associated with different nucleotide states and the mode of DnaA self-assembly are likely to be generally applicable to DnaA in other bacteria due to the highly conserved AAA+ region (37% sequence identity between A. aeolicus and S. amazonensis)27. We, therefore, made use of these DnaA models in order to gain further insights in how the base domain and the arginine finger of Hda interacts with DnaA.
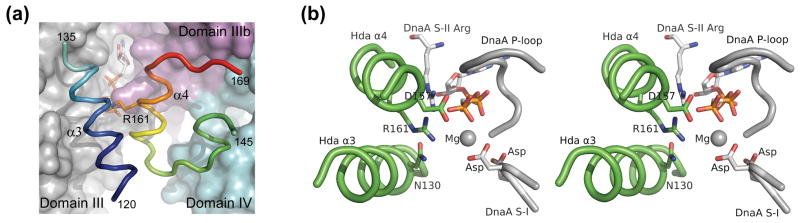
The Hda dimer seems to be compatible with the mechanism suggested for the DnaA self-assembly structure 4; 19. A geometrically plausible model can be built by replacing a DnaA molecule with an Hda dimer (or monomer) at the ‘ATP end’ (the side where the γ-phosphate of the ATP bound to DnaA is accessible 19) of the DnaA self-assembly through superimposition of the bases domains of DnaA and Hda. Extensive contacts are made through the domain III of DnaA in the DnaA filament self-assembly 19. The corresponding Hda secondary structure elements that make contact with the DnaA ATP site are α3 (120–135) and α4 (156–166). Helix α3 is close to a groove on the surface of domain III and α4 makes contacts with domain IIIb (Figure 8a).
Figure 8.
Hypothetical models for DnaA-Hda interaction. (a) Putative interaction between the base domain of Hda (shown in ribbon representation) and DnaA. The domain III, IIIb and IV of DnaA are shown as surface and colored as gray, magenta and cyan respectively. (b) The bipartite active site for ATP hydrolysis is formed by the conserved residues in the DnaA-Hda (white/green) heterodimeric complex shown in (a).
This model is consistent with known AAA+ interactions for ATP hydrolysis. The γ-phosphate of ATP sits next to the strictly conserved Arg161 and Asp157 (Figure 8b) and therefore, both these residues likely play a catalytic role in ATP hydrolysis. An acidic residue, corresponding to Asp157, is conserved in many AAA+ modules, such as glutamate in Pol III delta subunit or aspartate in NSF-D1 20. Asp157 is close to the Walker B motif of DnaA and located at the gate of the binding pocket where the γ-phosphate of the ATP would contact the solvent. Thus, a possible role for Asp157 is to help the Walker B motif stabilize a nucleophilic water, which would interact with the γ-phosphate. Asp157 is not, however, conserved in DnaA (Figure 1a, 4) and may explain why DnaA-ATP in the origin of replication assembly does not self-hydrolyze efficiently.
In the transition from DnaA-ATP to DnaA-ADP, the wedge opening, where Hda binds, will narrow through interdomain movement between the III and IIIb domains of DnaA 19; 26. It is plausible that the conformational changes induced by ATP hydrolysis in DnaA could drive the dissociation of Hda and DnaA-ADP. The released Hda could then be recycled for the next reaction. The cellular concentration of Hda (~50 dimers/cell) is much less than that DnaA (500–2000 molecules/cell) 13; thus, re-use of the Hda is important in RIDA.
Possible association of Hda with the membrane
The replicatively active DnaA-ATP is regenerated from DnaA-ADP by acidic phospholipids in the presence of oriC and ATP 44. A region near the N-terminus of the bridging helix (residues 349-383) of E. coli DnaA, between the lid domain (IIIb) and the DNA binding domain (IV), is critical in membrane-mediated nucleotide release 4; 44. Hda may also interact with the membrane since the T7-epitope-tagged form of Hda was associated with the inner membrane by immunoblot analysis 10. Interestingly, the lipid binding region of DnaA corresponds to a highly conserved region of Hda (residues 225-241, Figure 1a). The α8 helix of Hda is equivalent to the bridging helix of DnaA, although shorter in length. Additionally, Arg328 of E. coli DnaA, whose mutation affects membrane binding in DnaA 45, is also highly conserved in Hda and located in the same region (adjacent to the side chain of Lys236 of Hda). Thus, it is possible that Hda could interact with the membrane in a similar fashion to DnaA.
Materials and Methods
Protein production
The gene encoding a putative Hda from Shewanella amazonensis SB2B (GenBank: YP_927791, gi|119775051) was amplified by polymerase chain reaction (PCR) from genomic DNA using PfuTurbo DNA polymerase (Stratagene) and primers corresponding to the predicted 5′ and 3′ ends. The PCR product was cloned into plasmid pSpeedET, which encodes an expression and purification tag followed by a tobacco etch virus (TEV) protease cleavage site (MGSDKIHHHHHHENLYFQG) at the amino terminus of the protein. The cloning junctions were confirmed by DNA sequencing. Protein expression was performed in a selenomethionine-containing medium using the Escherichia coli strain GeneHogs (Invitrogen). At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 μg/mL, and the cells were harvested. After one freeze/thaw cycle, the cells were homogenized in Lysis Buffer [50 mM HEPES pH 8.0, 50 mM NaCl, 10 mM imidazole, 1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP)] and passed through a Microfluidizer (Microfluidics). The lysate was clarified by centrifugation at 32,500 × g for 30 minutes and loaded onto nickel-chelating resin (GE Healthcare) pre-equilibrated with Lysis Buffer. The resin was washed with Wash Buffer [50 mM HEPES pH 8.0, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP], and the protein was eluted with Elution Buffer [20 mM HEPES pH 8.0, 300 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP]. The eluate was buffer exchanged with HEPES Crystallization Buffer [20 mM HEPES pH 8.0, 200 mM NaCl, 40 mM imidazole, 1 mM TCEP] and treated with 1 mg of TEV protease per 10 mg of eluted protein. The digested protein was passed over nickel-chelating resin (GE Healthcare) pre-equilibrated with HEPES Crystallization Buffer, and the resin was washed with the same buffer. The flow-through and wash fractions were combined and concentrated for crystallization assays to 14 mg/mL by centrifugal ultrafiltration (Millipore). Hda was crystallized using the nanodroplet vapor diffusion method 46 with standard JCSG crystallization protocols 47. Initial screening for diffraction was carried out using the Stanford Automated Mounting system (SAM) 48 at the Stanford Synchrotron Radiation Laboratory (SSRL, Menlo Park, CA). To determine its oligomeric state, S. amazonensis Hda was analyzed using a 0.8 cm × 30 cm Shodex PROTEIN KW-803 column coupled with miniDAWN static light scattering and Optilab differential refractive index detectors (Wyatt Technology). The mobile phase consisted of 20 mM Tris pH 7.4 and 150 mM NaCl. The molecular weight was calculated using ASTRA 5.1.5 software (Wyatt Technology).
Crystallization and data collection
The crystallization reagent contained 0.2 M sodium thiocyanate and 20% (w/v) polyethylene glycol (PEG) 3350 at pH 6.9. Ethylene glycol was added as a cryoprotectant to a final concentration of 10% (v/v). Multi-wavelength anomalous diffraction (MAD) data were collected at SSRL on beamline 11-1 at wavelengths corresponding to the peak (λ1) and high energy remote (λ2) of a selenium MAD experiment. The data sets were collected to 1.75 Å at 100K using a Mar CCD 325 detector (Mar USA).
Structure determination and refinement
The crystals were indexed in orthorhombic space group P212121 with unit cell dimensions a=55.04 Å, b=64.03 Å and c=153.64 Å. Data processing and structure solution were carried using an automatic structure solution pipeline XSOLVE developed at Joint Center for Structure Genomics. The MAD data were integrated and reduced using Mosflm49 and then scaled with the program SCALA50 of the CCP4 suite51. Phasing was performed with SHELXD 52 and autoSHARP 53. The automated model building was performed with ARP/wARP 54. Refinement was carried out using REFMAC5 55 interspersed with manual building using COOT 56. The model geometry was analyzed with MOLPROBITY 32 to assess Ramachandran plot, side-chain rotamers, hydrogen bonds, steric clashes and van der Waals contacts. Data and refinement statistics are summarized in Table I.
Acknowledgments
We greatly appreciate the very helpful comments of Dr. James Berger and his group. The project is sponsored by National Institutes of General Medical Sciences Protein Structure Initiative (P50 GM62411, U54 GM074898). Portions of this research were carried out at the SSRL. SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Accession code. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB, http://www.wwpdb.org/) under accession code 3bos.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 2.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 3.Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 4.Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 5.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 6.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- 7.Katayama T. Feedback controls restrain the initiation of Escherichia coli chromosomal replication. Mol Microbiol. 2001;41:9–17. doi: 10.1046/j.1365-2958.2001.02483.x. [DOI] [PubMed] [Google Scholar]
- 8.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. Embo J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. Embo J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim PD, Banack T, Lerman DM, Tracy JC, Camara JE, Crooke E, Oliver D, Firshein W. Identification of a novel membrane-associated gene product that suppresses toxicity of a TrfA peptide from plasmid RK2 and its relationship to the DnaA host initiation protein. J Bacteriol. 2003;185:1817–1824. doi: 10.1128/JB.185.6.1817-1824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su’etsugu M, Takata M, Kubota T, Matsuda Y, Katayama T. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells. 2004;9:509–522. doi: 10.1111/j.1356-9597.2004.00741.x. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami H, Su’etsugu M, Katayama T. An isolated Hda-clamp complex is functional in the regulatory inactivation of DnaA and DNA replication. J Struct Biol. 2006;156:220–229. doi: 10.1016/j.jsb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Su’etsugu M, Shimuta TR, Ishida T, Kawakami H, Katayama T. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J Biol Chem. 2005;280:6528–6536. doi: 10.1074/jbc.M412060200. [DOI] [PubMed] [Google Scholar]
- 14.Camara JE, Breier AM, Brendler T, Austin S, Cozzarelli NR, Crooke E. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 2005;6:736–741. doi: 10.1038/sj.embor.7400467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riber L, Olsson JA, Jensen RB, Skovgaard O, Dasgupta S, Marinus MG, Lobner-Olesen A. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006;20:2121–2134. doi: 10.1101/gad.379506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camara JE, Skarstad K, Crooke E. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J Bacteriol. 2003;185:3244–3248. doi: 10.1128/JB.185.10.3244-3248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurz M, Dalrymple B, Wijffels G, Kongsuwan K. Interaction of the sliding clamp beta-subunit and Hda, a DnaA-related protein. J Bacteriol. 2004;186:3508–3515. doi: 10.1128/JB.186.11.3508-3515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kongsuwan K, Josh P, Picault MJ, Wijffels G, Dalrymple B. The plasmid RK2 replication initiator protein (TrfA) binds to the sliding clamp beta subunit of DNA polymerase III: implication for the toxicity of a peptide derived from the amino-terminal portion of 33-kilodalton TrfA. J Bacteriol. 2006;188:5501–5509. doi: 10.1128/JB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 20.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 21.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 23.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 24.Tucker PA, Sallai L. The AAA+ superfamily-a myriad of motions. Curr Opin Struct Biol. 2007;17:641–652. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- 26.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. Embo J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 28.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 30.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 31.Gao F, Zhang CT. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007;23:1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O’Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 34.Lee SY, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 36.Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- 37.Hishida T, Iwasaki H, Yagi T, Shinagawa H. Role of Walker motif A of RuvB protein in promoting branch migration of Holliday junctions. Walker motif A mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of Ruvb. J Biol Chem. 1999;274:25335–25342. doi: 10.1074/jbc.274.36.25335. [DOI] [PubMed] [Google Scholar]
- 38.Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. Embo J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijffels G, Dalrymple B, Kongsuwan K, Dixon NE. Conservation of eubacterial replicases. IUBMB Life. 2005;57:413–419. doi: 10.1080/15216540500138246. [DOI] [PubMed] [Google Scholar]
- 41.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. Embo J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wijffels G, Dalrymple BP, Prosselkov P, Kongsuwan K, Epa VC, Lilley PE, Jergic S, Buchardt J, Brown SE, Alewood PF, Jennings PA, Dixon NE. Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 44.Boeneman K, Crooke E. Chromosomal replication and the cell membrane. Curr Opin Microbiol. 2005;8:143–148. doi: 10.1016/j.mib.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Makise M, Mima S, Koterasawa M, Tsuchiya T, Mizushima T. Biochemical analysis of DnaA protein with mutations in both Arg328 and Lys372. Biochem J. 2002;362:453–458. doi: 10.1042/0264-6021:3620453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. J Appl Crystallogr. 2002;35:278–281. [Google Scholar]
- 47.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact samples cassettes and a small-scale robot. J Appl Cryst. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 50.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 51.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 53.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 54.Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, Lamzin VS, Kleywegt GJ, Perrakis A. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr D. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]